Abstract
Interactions of the antimicrobial peptide protegrin-1 (PG-1) with phospholipid monolayers have been investigated by using grazing incidence X-ray diffraction (GIXD) and specular X-ray reflectivity (XR). The structure of a PG-1 film at the air-aqueous interface was also investigated by XR for the first time. Lipid A, dipalmitoyl-phosphatidylglycerol (DPPG) and dipalmitoyl-phosphatidylcholine (DPPC) monolayers were formed at the air-aqueous interface to mimic the surface of the bacterial cell wall and the outer leaflet of the erythrocyte cell membrane, respectively. Experiments were carried out under constant area conditions where the pressure changes upon insertion of peptide into the monolayer. GIXD data suggest that the greatest monolayer disruption produced by PG-1 is seen with the DPPG system at 20 mN/m since the Bragg peaks completely disappear after introduction of PG-1 to the system. PG-1 shows greater insertion into the lipid A system compared to the DPPC system when both films are held at the same initial surface pressure of 20 mN/m. The degree of insertion lessens at 30 mN/m with both DPPC and DPPG monolayer systems. XR data further reveal that PG-1 inserts primarily in the head group region of lipid monolayers. However, only the XR data of the anionic lipids suggest the existence of an additional adsorbed peptide layer below the head group of the monolayer. Overall the data show that the extent of peptide/lipid interaction and lipid monolayer disruption depends not only on the lipid composition of the monolayer, but the packing density of the lipids in the monolayer prior to the introduction of peptide to the subphase.
Introduction
The last decade has seen a considerable increase in the study of antimicrobial peptides due to the interest for their potential as future pharmaceutical drug compounds.1 Antimicrobial peptides have a broad spectrum of antimicrobial properties and serve as inducers and modulators of various chemical components of the innate immune system.2
Recent work has shown that antimicrobial peptides act by permeating cell membranes.3–4 Although it is evident that the cell membrane is a barrier which must be overcome by antimicrobial peptides for cell disruption, the exact mechanism of action of antimicrobial peptides remains unclear.5 It has been established that antimicrobial peptides have the ability to differentiate between foreign and native cells4, 6–8 and one of the main reasons for this is thought to be due to the different lipid components and physical properties of the cell membranes involved.
Eukaryotic and bacterial membranes have very different lipid compositions. The mammalian cell membrane consists mainly of phosphatidylcholine (PC), phosphatidylethanolamine (PE), sphingomyelin, and cholesterol,6, 9 with the outer leaflet being enriched in PC.5, 10 This is vastly different from bacterial membranes which contain a high proportion of negatively charged lipids such as phosphatidylglycerol (PG) and PG derivatives such as diphosphatidylglycerol or cardiolipin.5–6 The lipid components of bacterial membranes also depend on the classification of the bacterium into Gram-positive or Gram-negative classes.11 Gram-positive bacteria such as Staphylococcus aureus have a single membrane which contains a high level of PG lipids, whereas Gram-negative bacteria have, in addition to the plasma membrane, an outer cell wall coated with lipopolysaccharide (LPS), of which lipid A acts as the lipophilic anchor.12 Since lipid A is an important component of the outer membrane of Gram-negative bacteria, it is a prospective target for the design of novel antibacterial drug compounds.
In this paper the interactions of an antimicrobial peptide, protegrin-1 (PG-1§), with DPPC§, DPPG§ and lipid A monolayers are examined under constant area conditions. These lipids are present in the outer leaflet of human erythrocytes, Gram-positive bacterial cell membranes and Gram-negative bacterial outer membranes, respectively.
Protegrin-1 is an arginine- and cysteine-rich antimicrobial peptide, originally isolated from porcine leukocytes. 13, 14 It contains 18 amino acid residues, and NMR studies show that it has a one-turn β-hairpin structure that includes two disulfide bonds when in solution. 15 Members of the protegrin family have been shown to exhibit anti-human immunodeficiency virus type 1 activity in vitro 16 and activity against Chlamydiae, 17 among other antimicrobial properties.
Previous work 18 using oriented circular dichroism has studied the alignment of PG-1 in lipid bilayers. It has been proposed that the PG-1 peptide exists in membranes in two different states which correspond to the surface adsorbed state and the inserted state, similar to that of the α-helical antimicrobial peptide, alamethicin. 18 In the surface state, PG-1 is adsorbed to the head group region of a lipid bilayer with the peptide being oriented parallel to bilayer, whereas in the inserted state, the peptide is orientated perpendicular to the lipid bilayer plane. Ma et al. 19 observed a PG-1 induced thickness change and a kinetic profile that corroborates with this model by using surface plasmon resonance.
More recently, Mani et al. 20, 21 used solid-state NMR to investigate the oligomeric structure and insertion of PG-1 into lipid bilayers. Their studies on the dimeric structure of PG-1 molecules in palmitoyloleoylphosphatidylcholine (POPC) bilayers suggest that two PG-1 molecules align in a parallel fashion.20 Further work was carried out with lipids which mimic the bacterial inner membrane or the red blood cell membrane. The results of Mani et al. suggest that PG-1 adopts dramatically different oligomeric structures depending on the lipid composition of the membrane with much greater contact of the peptide with anionic lipids than with those which mimic the red blood cell membrane (POPC/cholesterol). 21
The interactions of PG-1 with various lipid monolayers have also been recently studied to model the initial interaction of PG-1 with the outer leaflet of the cell membrane. 22–23 Different lipid compositions were used to represent the membrane of various cell types, and these systems were studied by insertion assay, epifluorescence microscopy and X-ray scattering techniques under constant pressure and constant area conditions. These studies showed that PG-1 readily inserts into lipid monolayers of phosphatidylglycerol lipids and lipid A, but significantly less so into lipid monolayers of phosphatidylcholine lipids. 22–23
In this paper, the interaction of PG-1 with lipid monolayers was carried out by monitoring insertion under constant area conditions where changes in surface pressure indicate the degree of interaction of peptide with the lipid layer. This paper not only consolidates and reinforces earlier work carried out using epifluorescence microscopy and X-ray scattering methods 22–23 but it also presents a thorough investigation of the interaction of PG-1 with different lipids which has not been published to date. Moreover, the importance of the effect of the initial surface pressure, and hence lipid packing density, of the different monolayers on subsequent peptide induced disorder is discussed.
Results
GIXD systems at 20 mN m−1
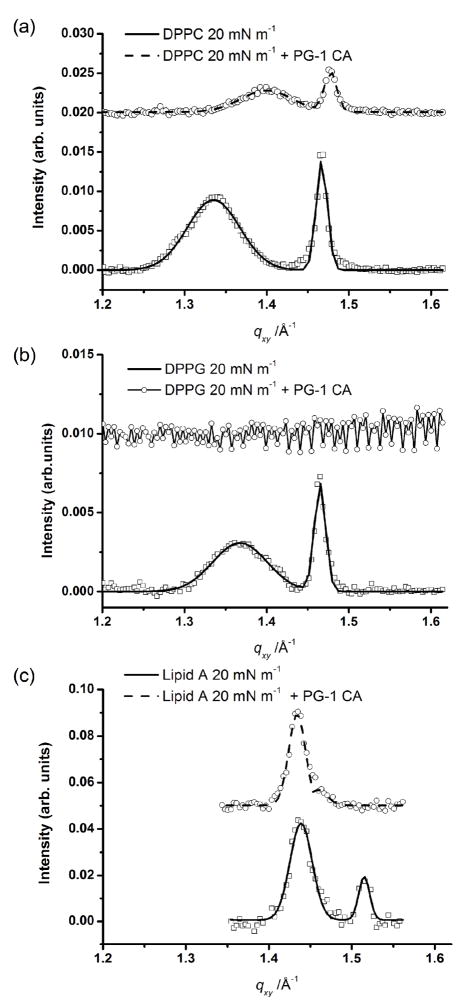
GIXD measurements of lipid monolayers before and after injection of PG-1 were carried out for DPPC, DPPG and lipid A monolayers under constant area conditions after compression to 20 mN m−1. GIXD data of these systems are shown in Fig. 1. The top and the bottom scans correspond to the GIXD scan after and before the injection of PG-1, respectively. The corresponding values of unit cell dimensions and d-spacings can be found in Table 1. In all cases, two Bragg peaks indicate the presence of an ordered structure with centered rectangular packing for pure lipid monolayers.
Fig. 1.
Bragg peak plot of scattering vector qxy as a function of intensity monolayers at 20 mN m−1. (a) Data (□) and fit (—) of DPPC monolayer at 20 mN m−1; data (○) and fit (---) of DPPC monolayer after 0.025 mg ml−1 PG-1 injection into the subphase. (b) Data (□) and fit (—) of DPPG monolayer at 20 mN m−1; data (-○-) of DPPG monolayer at 20 mN m−1 after 0.025 mg ml−1 PG-1 injection into the subphase. C) Data (□) and fit (—) of lipid A monolayer at 20 mN m−1 (—); data (○) and fit (---) of lipid A monolayer after 0.025 mg ml−1 PG-1 injection into the subphase. For clarity the data have been offset vertically.
Table 1.
Grazing Incidence X-ray diffraction data lipid systems before and after injection of PG-1 under constant area conditions.
| Experiment | d11a/Å | d02a/Å | a/Å | b/Å | Area per molecule/Å2 |
|---|---|---|---|---|---|
| DPPC 20 mN m−1 | 4.71 | 4.28 | 5.63 | 8.57 | 48.3 |
| DPPC 20 mN m−1 + PG-1 | 4.49 | 4.25 | 5.29 | 8.5 | 44.9 |
| DPPG 20 mN m−1 | 4.59 | 4.29 | 5.44 | 8.58 | 46.7 |
| DPPG 20 mN m−1 + PG-1 | No peaks | ||||
| Lipid A 20 mN m−1 | 4.34 | 4.15 | 5.09 | 8.3 | 126.9 |
| Lipid A 20 mN m−1 + PG-1 | 4.38 | 4.3 | 5.09 | 8.61 | 131.4 |
| DPPC 30 mN m−1 | 4.60 | 4.26 | 5.47 | 8.53 | 46.6 |
| DPPC 30 mN m−1 + PG-1 | 4.61 | 4.28 | 5.47 | 8.56 | 46.8 |
| DPPG 30 mN m−1 | 4.51 | 4.25 | 5.32 | 8.51 | 45.2 |
| DPPG 30 mN m−1 + PG-1 | 4.60 | 4.28 | 5.46 | 8.56 | 46.7 |
“11” and “02” are used to denote (hk) for a set of Bragg rods with equal in-plane components which cannot be resolved from the GIXD data. In this case data are calculated using the rectangular unit cell, thus {11} lattice means {(11), (11̄), (1̄1), (1̄1̄)} and {02} means {(02), (02̄)}.
The data for the DPPC systems after PG-1 injection under constant area conditions (Fig. 1a) show a decrease in peak intensity and a shift in the peak positions. The insertion of PG-1 into this system gave an increase in surface pressure of 12 mN m−1 over the time scale of 20 minutes, corroborating the shift in the positions of the Bragg peaks observed in Fig. 1a, signifying that the area per molecule decreases on peptide injection due to lipid compression.
The effect of PG-1 insertion on DPPG monolayers at 20 mN m−1 (Fig. 1b) is very different from that of the DPPC system after PG-1 insertion. Here the previously ordered structure, shown by the two Bragg peaks, completely disappears (Fig. 1b). The pressure increased on insertion of PG-1 under the DPPG monolayer and was 28.7 mN m−1, 25 minutes after peptide injection.
GIXD data for the pure lipid A monolayer at an initial surface pressure of 20 mN m−1 are compared with data for lipid A system after the injection of PG-1 (Fig. 1c). The d-spacings and unit cell dimensions calculated for lipid A before and after PG-1 injection have been performed based on the assumption that the molecule contains six acyl chains on average, as determined previously. 31 Unlike DPPG, the lipid A monolayer remains crystalline after introduction of PG-1, although the area per lipid A molecule has apparently grown. This apparent increase in area per lipid A molecule seems to be due to the effect of an increase in intermolecular lipid A spacing (seen from the negative shift of the {02} peak, most likely due to PG-1 penetration) rather than the intramolecular spacing (spacing between lipid A tails, {11} peak) which is very similar before and after peptide injection. There was also an increase in pressure upon insertion of peptide of 19 mN m−1, which strongly suggests that peptide penetration occurred and forced the lipid A molecules apart, whilst the conformation of the individual lipid A molecules remained the same.
GIXD – 30 mN m−1 systems
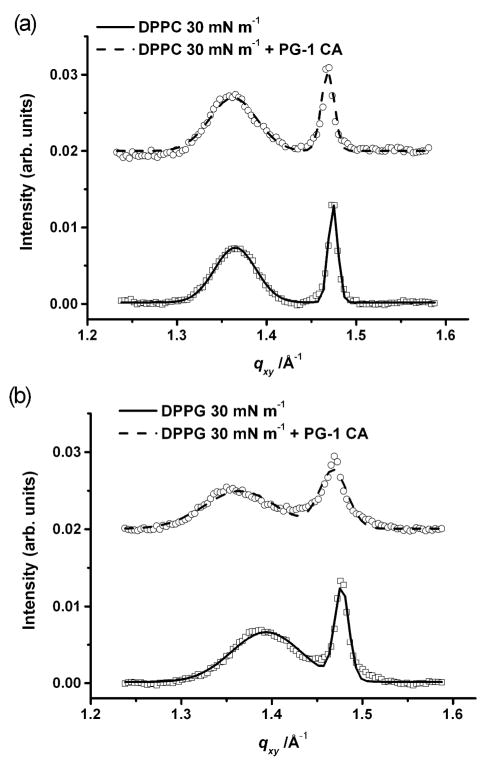
The GIXD data for the DPPC system at 30 mN m−1 before and after injection of PG-1 are shown in Fig. 2a. The data after injection of PG-1 show that the area per molecule remains roughly the same as the pure DPPC monolayer at 30 mN m−1 (Table 1). This suggests that the higher pressure of 30 mN m−1 and constant area conditions hinder peptide incorporation into the monolayer.
Fig. 2.
Bragg peak plot of scattering vector qxy as a function of intensity monolayers at 30 mN m−1. A) Data (□) and fit (—) of DPPC monolayer at 30 mN m−1; data (○) and fit (---) of DPPC monolayer after 0.025 mg ml−1 PG-1 injection into the subphase; B) Data (□) and fit (—) of DPPG monolayer at 30 mN m−1 (—); data (○) and fit (---) of DPPG monolayer at 30 mN m−1 after 0.025 mg ml−1 PG-1 injection into the subphase. For clarity the data have been offset vertically.
The PG-1 peptide imparts a smaller degree of disordering on the DPPG monolayer at 30 mN m−1 (Fig. 2b) compared to that at 20 mN m−1 (Fig. 1b). The peak intensities decrease upon PG-1 injection, suggesting that the area fraction of the ordered phase has diminished due to the presence of the peptide. However, unlike at the lower surface pressure, two Bragg peaks can still be observed even in the presence of the peptide.
XR PG-1 at the air-aqueous interface
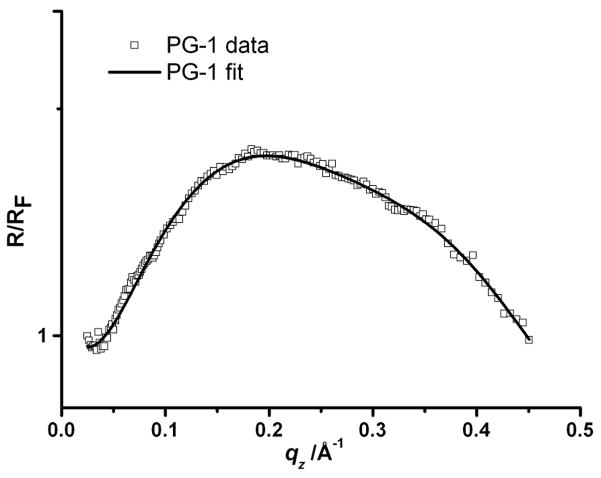
To assess the surface adsorption of the peptide, a study of PG-1 itself at the air-aqueous interface was carried out. The peptide was injected into the subphase of the trough under constant area conditions in the absence of a lipid monolayer (initial surface pressure was 0 mN m−1). Upon injection of PG-1 into the subphase, the surface pressure increased from 0 to 17 mN m−1, suggesting the formation of a PG-1 film at the air-aqueous interface. XR measurements were then carried out on this PG-1 film after the surface pressure reached equilibrium. Fig. 3 shows the XR data and the corresponding fit obtained from the analysis of the data. The data were fitted and a two-layer model was needed to adequately fit the data. The fitting returned values of layer thickness of 12.2 Å and 12.7 Å for L1 and L2 respectively, with the layer nearest the air, termed L1, and the layer nearest to the subphase, termed L2. The respective electron densities for these layers normalized to the electron density of the subphase (0.337 e− Å −3) were 1.45 and 1.05. This electron density profile suggests the formation of peptide bilayer at the air-aqueous interface.
Fig. 3.
X-ray reflectivity data and corresponding fits normalized by Fresnel reflectivity plotted against scattering vector (qz) of PG-1 at air-aqueous interface at 17 mN m−1. PG-1 reflectivity data (□) and fit (—). For clarity the data have been offset vertically.
XR - Interactions of PG-1 with lipid monolayers at 20 mN m−1
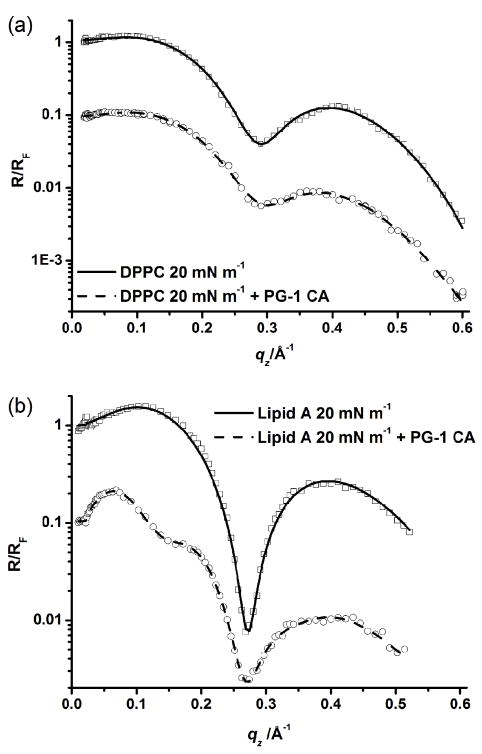
The reflectivity curves (normalized to the Fresnel reflectivity of a planar interface) for the DPPC and lipid A monolayers at 20 mN m−1 before and after injection of PG-1 into the subphase are shown in Fig. 4. Corresponding fit parameters are shown in Table 2.
Fig. 4.
X-ray reflectivity data and corresponding fits normalized by Fresnel reflectivity plotted against scattering vector (qz) of lipid monolayers at 20 mN m−1. (a) DPPC at 20 mN m−1 reflectivity data (□) and fit (—); DPPC + 0.025 mg ml−1 PG-1 (at 20 mN m−1 constant area conditions), data (○) and fit (---). (b) lipid A at 20 mN m−1 reflectivity data (□) and fit (—); lipid A + 0.025 mg ml−1 PG-1 (at 20 mN m−1 constant area conditions), data (○) and fit (---). For clarity the data have been offset vertically.
Table 2.
Grazing Incidence X-ray diffraction data lipid systems before and after injection of PG-1 under constant area conditions
| Experiment | L1/Åb | ρ 1b | L2/Åb | ρ2b | L3/Å | ρ3b | Roughness σ/Å |
|---|---|---|---|---|---|---|---|
| DPPC at 20 mN m−1 | 12.6 | 0.9 | 7.1 | 1.25 | - | - | 4.0 |
| DPPC 20 mN m−1 + PG-1 | 12.5 | 0.97 | 8.5 | 1.19 | - | - | 4.2 |
| Lipid A 20 mN m−1 | 13.3 | 0.99 | 5.1 | 1.65 | - | - | 4.0 |
| Lipid A 20 mN m−1 + PG-1 | 12.3 | 1.04 | 8.8 | 1.37 | 26.6 | 1.22 | 4.6 |
| DPPG at 20 mN m−1 [taken from 22] | 12.3 | 0.92 | 8.4 | 1.30 | - | - | - |
| DPPG 20 mN m−1 + PG-1[taken from 22] | 13.9 | 1.22 | 7.0 | 1.27 | 27.2 | 1.07 | - |
| DPPC at mN m−1 | 15.2 | 0.91 | 8.8 | 1.33 | - | - | 3.3 |
| DPPC 30 mN m−1 + PG-1 | 16.1 | 0.96 | 11.6 | 1.12 | - | - | 5.5 |
| DPPG at 30 mN m−1 | 18.1 | 0.98 | 5.9 | 1.55 | - | - | 3.7 |
| DPPG 30 mN m−1 + PG-1 | 14.9 | 0.98 | 6.9 | 1.39 | 6.1 | 1.21 | 5.2 |
L1 refers to the slab closest to the air and the subsequent slabs (L2–4) proceed in order from the slab next nearest to the air to the slab nearest the aqueous subphase. ρn is the electron densities of these layers, where n is the corresponding layer number. The reported electron densities are normalized by the electron density value of the DPBS subphase (0.337 e- Å−3). Data shows comparison of systems before and after addition of PG-1 to the different lipid systems.
For pure lipid monolayers, the L1 layer corresponds to the tail group region of the phospholipid, and the L2 layer the head group region. After injection of PG-1 under the DPPC monolayer at 20 mN m−1 (Fig. 4a), the layer thicknesses remain similar, but the normalized electron densities increase and decrease for the L1 and L2 layers, respectively (Table 2). The corresponding GIXD data suggest that there is insertion of some peptide into the film as a decrease in intensity of the Bragg peaks was observed. Put together, the XR and GIXD data indicate that the insertion is mainly into the head group region, perhaps with some minor partitioning of PG-1 into the base of the tail group region.
XR data of the pure lipid A monolayer at 20 mN m−1 were compared with those of the same lipid film at 20 mN m−1 but after PG-1 injection into the subphase (Fig. 4b). The lipid A with PG-1 system was modeled with three slabs and the results are listed in Table 2. An additional slab had to be added in order to model the system adequately. The slab nearest the air was named L1, the next slab L2, and the slab closest to the subphase, L3. The increase in electron density of L1 along with the decrease of electron density of L2 indicates that the PG-1 molecules have fully inserted into the lipid A monolayer. This is expected since the electron densities of the pure peptide layers are known to be higher and lower than the lipid tail and head group layers respectively. The third layer, L3, is 26.6 Å thick with a normalized electron density of 1.22. This suggests that the peptide may insert and hang down perpendicular to the monolayer or that multilayers may adsorb underneath the lipid monolayer.
For reasons of comparison and completeness, the fitting results for the DPPG at 20 mN m−1 system before and after injection of PG-1 as previously published 22 are presented in Table 2. It is interesting to note that the results from the data fitting for the DPPG and lipid A systems after PG-1 injection are quite similar with regards to layer thicknesses. Especially noteworthy is the fact that both anionic lipid systems require a third layer to be modeled which is of a similar thickness (~27 Å). It turns out that this value is approximately the known length of the PG-1 molecule, 22, 25 suggesting that at 20 mN m−1 the PG-1 peptide most likely orients with its long axis almost perpendicular to the interface or forms multi-adlayers in both the lipid A and DPPG systems.
XR - Interactions of PG-1 with lipid monolayers at 30 mN m−1
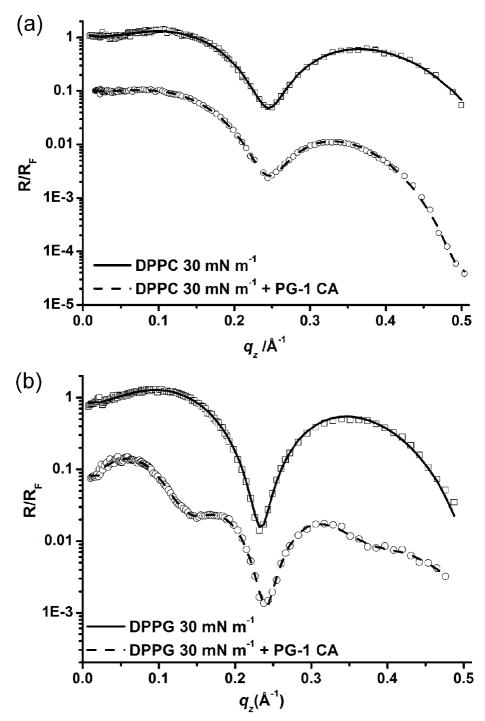
Fig. 5a shows the normalized reflectivity curves for the DPPC system at 30 mN m−1 before and after injection of PG-1 under constant area conditions. The fitted parameters for the DPPC monolayer at 30 mN m−1 can be seen in Table 2. After injection of PG-1, the data could again be fitted with two layers. The data suggest that the PG-1 peptides penetrate into the head group layer (as shown by the decrease in electron density and the increase in thickness of the head group layer).
Fig. 5.
X-ray reflectivity data and corresponding fits normalized by Fresnel reflectivity plotted against scattering vector (qz) of DPPC and DPPG monolayers at 30 mN m−1. (a) DPPC at 30 mN m−1 reflectivity data (□) and fit (—); DPPC + 0.025 mg ml−1 PG-1, data (○) and fit (---); (b) DPPG at 30 mN m−1 reflectivity data (□) and fit (—); DPPG + 0.025 mg ml−1 PG-1, data (○) and fit (---). For clarity the data have been offset vertically.
Fig. 5b shows the comparison of the X-ray reflectivity data for the DPPG system at 30 mN m−1 before and after injection of PG-1 under constant area conditions. The data suggest that the head group region was again targeted, as was the case with DPPC under the same experimental conditions. The data for the DPPG with PG-1 system had to be modeled using three layers (Table 2). However, the third layer did not have the same thickness as the third layer for the lipid A and DPPG systems with PG-1 at 20 mN m−1, suggesting that an adsorbed layer of peptide oriented perpendicular to the interface is not present at 30 mN m−1, and that it is more likely that the third layer is made up of peptide which has not fully inserted into the monolayer but rather with portions of it sticking out into the subphase.
Discussion
GIXD – 20 mN m−1 systems
Data from the DPPC with PG-1 system show that the calculated area per molecule value decreases slightly from the value of the pure DPPC monolayer at 20 mN m−1 (Fig. 1a). This is thought to be due to the monolayer being forced to pack more tightly upon peptide insertion under the constant area conditions since the surface area of the trough being used is kept constant. Comparing this case with that at 30 mN m−1, lipids at 20 mN m−1 have a lower packing density, and should have greater lipid mobility and hence allow greater ease for peptide insertion compared to the 30 mN m−1 DPPC system. Indeed, this is confirmed by our GIXD data where the DPPC system at 20 mN m−1 changes much more noticeably after PG-1 insertion (Fig. 1a) than the DPPC system at 30 mN m−1 after PG-1 injection (Fig. 2a). This is further confirmed by the fact that no discernible pressure change was observed for the 30 mN m−1 DPPC system after peptide injection.
GIXD data for the DPPG with PG-1 system at 20 mN m−1 indicate that the DPPG monolayer becomes completely disordered upon insertion of PG-1 even on the Angstrom-scale. This and the fact that there is a 44 % increase in surface pressure corroborates our earlier findings of micron-scale domains originally observed via epifluorescence microscopy in a DPPG monolayer disappearing upon the introduction of PG-1 into the subphase. 22–23
The lipid A data suggest that under constant area conditions the individual “tails” of each lipid A molecule remain with a similar conformation before and after injection of PG-1, and that the inferred peptide insertion based on the observation of an increased pressure is due to peptide penetration into the lipid A monolayer, mostly in the head group regions. Since the structure of the tail region of the lipid A molecule seems to be rigid, 31 it is reasonable to assume that the insertion occurs mostly in the head group regions. The data also show that the {11} peak is largely unaffected, but that the {02} peak decreases in size and is significantly shifted to a lower qxy value by around 0.05 Å−1. Since the lipid A molecule has six hydrocarbon chains on average, 31 the {11} peak corresponds to intramolecular tail - tail distances, while the {02} peak corresponds to intermolecular lipid A – lipid A molecule distances. Upon PG-1 insertion into the lipid A head group region, the area per lipid molecule increases, and the intermolecular distance increases correspondingly, as signified by a shift of the {02} peak to a lower qxy region. In other words, the data suggest that the apparent area per lipid A molecule is increasing as the PG-1 is inserting in between lipid A molecules.
GIXD – 30 mN m−1 systems
GIXD data for the DPPC systems at 30 mN m−1 (Fig. 2a) show some differences from those for DPPC at 20 mN m−1 (Fig. 1a). The DPPC monolayer at 30 mN m−1 (Fig. 2a) again shows an ordered structure, giving rise to Bragg peaks just as in the case at 20 mN m−1 (Fig. 1a). However, unlike at 20 mN m−1, upon injection of PG-1 at 30 mN m−1, there is very little change in both the peak values found and the unit cell dimensions (Table 1). This suggests that the higher initial pressure of 30 mN m−1 hinders the ability of the peptide to incorporate into the monolayer.
When the DPPG with PG-1 system was studied at 30 mN m−1 under constant area conditions, the calculated area per molecule of the condensed phase increased slightly after PG-1 insertion (Table 1, Fig. 2b). The data suggest that PG-1 inserts mostly into the disordered lipid phase, although some peptide molecules may adsorb underneath the solid domains due to charge interaction, giving rise to a dilated unit cell as observed with GIXD.
XR – PG-1 at the air-aqueous interface
The surface activity of the PG-1 peptide was observed as there was an increase in pressure upon peptide injection while the barriers were open, without any lipid film present. The jump of the surface pressure from 0 to 17 mN m−1 upon peptide introduction into the subphase clearly indicates that PG-1 is surface-active, even though the peptide is highly soluble in the aqueous subphase. The peptide surface layer is thus in equilibrium with the subphase, and the observed change in surface pressure is related to the amount of peptide adsorbed at the surface as given by the Gibbs adsorption equation. 24
Analysis of the XR data (Fig. 3) suggests that when PG-1 was injected into the subphase, it adsorbed to and aligned at the air-aqueous interface, with its long axis oriented along the interface. Data of the pure PG-1 peptide film at the air-aqueous interface seem to further suggest the formation of two peptide layers at the air-aqueous interface, with the lower layer (L2) being a partial layer due to the reduction in electron density in comparison to the upper layer, (L1). These layers are most likely two layers of PG-1 due to their similar thicknesses that match closely with the cross-sectional diameter of the peptide from previous NMR studies. 15
XR - Interactions of PG-1 with lipid monolayers at 20 mN m−1
Data for the DPPC system at 20 mN m−1 before and after injection of PG-1 into the subphase (Fig. 4a) suggest that there was some PG-1 insertion into the DPPC monolayer at 20 mN m−1 since the changes in the electron density are in accordance with a certain degree of PG-1 insertion, mainly into the head group region of the monolayer.
When analyzing the XR data of the lipid A with PG-1 system (Fig. 4b), the increase in electron density of L1, along with the decrease in electron density of L2, indicates that PG-1 molecules have fully inserted into the lipid A monolayer in some regions, with the majority of insertion occurring into the head group regions of the monolayer. However, one cannot discern from the data whether the peptide molecules are uniformly distributed between the lipid A molecules or they group together in between the lipid A molecules. It has been suggested that this grouping together of peptide molecules, which then align at the interface, may be more likely than the individual peptide molecules inserting randomly between lipid A molecules, as the lipid A molecules have bulky rigid tail group structures. 31
XR - Interactions of PG-1 with lipid monolayers at 30 mN m−1
XR data of the DPPC with PG-1 system at 30 mN m−1 show that there is some degree of insertion due to an increase in the electron density of L1 and a decrease in that of L2. The DPPC at 30 mN m−1 with PG-1 data (Fig. 5a) suggest that there may be some compaction of lipid molecules due to peptide insertion under constant area conditions. This is in contrast to the GIXD data which suggest little insertion. However, since GIXD is only sensitive to the ordered domains in the film, while XR takes into account both the ordered and the disordered regions, if the peptide were to insert only into the disordered region without disturbing the structure in the ordered region, we would expect to obtain XR results that indicate the incorporation of the peptide in the film and GIXD results which show little change from before peptide injection, both of which are observed here.
The XR data for the DPPG system at 30 mN m−1 (Fig. 5b) are best described by a two-layer model with thicknesses of 18.1 and 5.9 Å (Table 2). This is indicative of a DPPG phospholipid monolayer, with a tail group region and a head group region, respectively, as previously published. 8 After injection of PG-1, the data could no longer be modeled adequately with a two-layer model and a third layer was required. The data show that the tail layer reduces in thickness whilst the head group layer increases in thickness and decreases in electron density. The third layer has a thickness of 6.1 Å and a normalized electron density of 1.21. From the XR data for the DPPG system with PG-1 at 30 mN m−1, it can be suggested that the PG-1 inserts into the DPPG head groups with the remaining part of the peptide protruding into the subphase, possibly giving rise to a partial peptide adsorbed layer. The fact that the tail group layer electron density does not change implies that the peptide does not fully penetrate through the DPPG monolayer. As a third layer was required to model the data adequately, the data suggest that the peptide is protruding out from the head group layer. This third layer is around one quarter of the thickness of the adsorbed layers which occur at 20 mN m−1 (Table 2) suggesting that if peptide adsorption does occur at 30 mN m−1, the peptides are aligned with the long axis parallel to the interface rather than perpendicular to it, as was proposed to occur at 20 mN m−1.
The observation of an increase in area per molecule in the DPPG systems after PG-1 injection as compared to a decrease observed in equivalent DPPC systems suggests that the peptide interacts significantly differently with DPPG versus DPPC monolayers. These differences in interactions can only be due to differences in head group region, with DPPC being zwitterionic and DPPG anionic, since the tail groups of the lipid molecules are completely identical. 23
Schematic cartoons
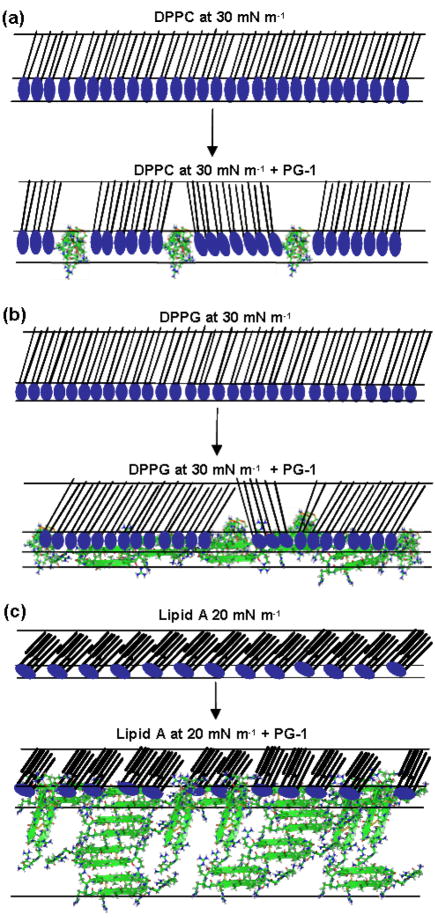
Complementary data from GIXD and XR allow us to speculate how the PG-1 peptide interacts with different lipid monolayers. The data analysis suggests that there is some low degree of interaction between PG-1 and DPPC monolayers, but its interaction is much more extensive with DPPG and lipid A monolayers. The data also show that PG-1 forms a surface active layer.
Our X-ray reflectivity results (Fig. 3) indicate that PG-1 aligns at the surface and that there is a partial second layer of PG-1 underneath the top layer. A schematic cartoon of the alignment of PG-1 molecules at the air-aqueous interface is shown in Fig. 6. It is possible to come up with this diagram as the approximate dimensions of the molecule are known 22, 25 and the thicknesses of the layers are taken from the X-ray reflectivity fitting results.
Fig. 6.
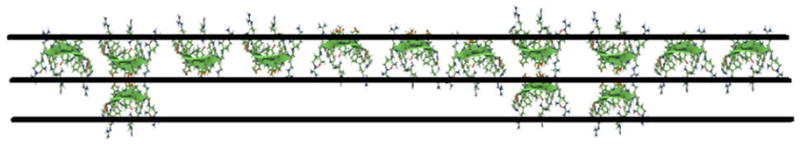
Schematic cartoon of PG-1 film at the air-aqueous interface.
When the GIXD and XR data for the DPPC with PG-1 system at 30 mN m−1 were further examined, it appeared that peptide penetration into the DPPC monolayer was more limited in comparison to the DPPG and lipid A systems, and that the majority of the peptide insertion occurred into the head groups of the DPPC monolayer. Although GIXD data show little difference after PG-1 injection, XR data indicate that the PG-1 peptide does interact with the DPPC monolayer, most likely in the disordered region of the film. The schematic model proposed here in Fig. 7a is based on the fact that the head group layer decreases in electron density and increases in thickness as a result of peptide insertion. The tail group also increases in thickness and electron density, which we interpret as being due to the compaction of lipid molecules induced by peptide insertion.
Fig. 7.
Cartoon schematic of possible interactions of PG-1 with A) DPPC monolayer at 30 mN/m; B) DPPG monolayer at 30 mN/m and C) lipid A monolayer at 20 mN/m.
Fig. 7b shows a schematic cartoon of the proposed mode of interaction of PG-1 with the DPPG monolayer at 30 mN m−1 based on results from the X-ray data. The DPPG monolayer has a tightly packed ordered structure at 30 mN m−1, which is disturbed after PG-1 injection, as the peptide penetrates the DPPG monolayer. Some regions of ordered lipid structure may remain intact, but the overall structure of the DPPG monolayer is altered, with regions of peptide inserted into it, regions of peptide protruding from the head group layer of the monolayer, as well as a partial peptide adsorption to the underside of the DPPG monolayer.
In contrast, Fig. 7c shows a schematic of how PG-1 molecules may penetrate lipid A monolayers at 20 mN m−1. The figure is based on insertion assay data where an increase in pressure to 39 mN m−1 was observed. The model also takes into account GIXD and X-ray reflectivity data, which show that PG-1 peptides penetrate the lipid A monolayer and adsorb to it on the subphase side as shown by the third slab in the model.
Experimental
Lipid monolayers
The cell membranes of different organisms have characteristic lipid compositions. In order to study the targeting selectivity o f P G-1, Langmuir monolayers composed of lipids representative of different membranes were examined. This approach has been utilized repeatedly over the years to elucidate peptide-membrane interactions. 26–27 In order to develop a full understanding of membrane interactions, each lipid component was studied separately in order to ascertain the contribution of each membrane component to the overall interaction of the peptide with the membrane. Dipalmitoyl-phosphatidylcholine (DPPC), dipalmitoyl-phosphatidylglycerol (DPPG) and lipid A were used to form Langmuir monolayers. In this work DPPC was used as it is a major component of the outer leaflet of human red blood cell membranes. 10, 28 DPPG is a representative lipid of the outer leaflet of the Gram-positive bacterial membrane 29–30 and lipid A is the lipid anchor of lipopolysaccharides from the outer membrane of Gram-negative bacteria. Lipids were prepared as described earlier. 8, 22, 31 DPPC and DPPG were purchased from Avanti Polar Lipids, and lipid A was purchased from Sigma-Aldrich. All lipids were used without further purification.
Peptides
Protegrin-1 (P G-1) (RGGRLCYCRRRFCVCVGR) is a porcine antimicrobial peptide. 15 The PG-1 peptide used in this study was synthesized in-house, and details of the synthesis have been published elsewhere. 15 The PG-1 solution was made up in 0.01 % w/v acetic acid to a working solution with a concentration of 1 mg ml−1. This, when injected into the subphase gave a final concentration of 0.025 mg ml−1. The concentration of PG-1 used here was used in previous studies on C. albicans, L. monocytogenes and E. coli where it was shown to have potent microbicidal activity. 13 The concentration used is also known to be around the value of the minimum inhibitory concentration of several bacteria including E. faecalis, P. aeruginosa 14 and L. interrogans. 32
Langmuir trough conditions
X-ray scattering measurements were taken on the ID10B (Troika II) synchrotron beamline at the European Synchrotron Radiation Facility 33 and at the 9-ID (CMC-CAT) beamline at the Advance Photon Source, Argonne National Laboratory. 34 Both setups used PTFE (Teflon)-lined Langmuir troughs equipped with a single moveable barrier as described previously. 8, 31, 33
Peptide insertion technique
Upon spreading, the lipid film was left undisturbed for 15 minutes to allow for solvent evaporation. Subsequently, barrier compression (at 2 cm2 min−1) was initiated to attain the target surface pressure, which corresponds to the condensed phase of lipids (20–40 mN m−1). These pressures were chosen so that the lipid packing density was equivalent to that of the cell membrane. 35 Moreover, the condensed phase condition also allowed us to monitor changes in lipid packing during PG-1 insertion. After reaching the target surface pressure, the insertion assay was then carried out in the constant area mode, where changes in the surface pressure indicate the degree of interaction of the peptide with the lipid monolayer. In this mode the barrier motors were simply switched off whilst the peptide solution was uniformly injected underneath the monolayer with an L-shaped needle (VDRL needle; Hamilton, Reno, NV, USA). The introduction of peptides under the compressed lipid monolayer mimics the approach by the peptide to the outer surface of the cell, as the hydrophilic head groups are closest to the subphase, with the lipid film simulating the outer leaflet of the membrane and the peptide in the subphase mimicking the peptide in the extracellular fluid. Changes in pressure were recorded as a function of time during and after peptide injection into the subphase. All experiments were carried out using the constant area mode, and the surface pressure condition mentioned refers to the target surface pressure to which the monolayer was compressed at the start of the experiment.
Grazing Incidence X-ray Diffraction (GIXD) and X-ray Reflectivity (XR)
Grazing incidence X-ray diffraction (GIXD) 36 is used to obtain in-plane information concerning the molecular structure of surfaces, 37 whereas specular XR measurements reveal information on the electron density distribution along the surface normal and may be used to determine the density and thickness of thin layers. 27, 36, 38
X-ray scattering measurements were carried out at the ID10B (Troïka II) beamline at the European Synchrotron Radiation Facility, France, as previously described, and data were analyzed as before. 8, 31, 33 Control measurements of pure lipid monolayers were followed by subsequent injection of the desired amount of peptide into the subphase under a monolayer compressed to the desired surface pressure.
Grazing incidence X-ray diffraction measurements are made with variation of the X-ray momentum transfer component qxy that is parallel to the air aqueous interface. The reflections of the Bragg peaks observed with this geometry can be indexed by two Miller indices, hk. Their angular position 2θhk, corresponding to qhk = (4π/λ) sin θhk, yields the repeat distance dhk = 2π/qhk for the two-dimensional (2D) lattice structure. 39, 40 Bragg peak profiles (intensity against qxy) were fitted with Gaussians to obtain full-width half-maximum (FWHM) values and the peak position values were used to obtain unit cell dimensions of the lipid lattices. The observation of two Bragg peaks in the diffraction pattern of an amphiphilic monolayer is indicative of a distorted hexagonal unit cell (which may be better described 39 as a centered rectangular unit cell). Therefore, all unit cell dimensions in this paper have been calculated using the centered rectangular unit cell approximation.
XR is measured as a function of the scattering vector qz, where qz = 4π sin α/λ, α is the grazing angle of the incident beam and λ the wavelength of the X-ray beam. The reflectivity curve contains information regarding the gradient of the electron density profile in the direction normal to the surface. 8, 39 X-ray reflectivity measurements were carried out at a range of angles corresponding to qz values of approximately 0 to 0.65 Å−1. The reflected beam intensity was measured as a function of the incident angle using a position sensitive detector.
The XR data were then analyzed using data processing and fitting programs as carried out previously 8, 31, 33 in order to gain information on the electron density distribution in the direction normal to the surface. This provided an electron density profile averaged laterally over both ordered and disordered parts of the system. The whole monolayer and subphase system was modelled as slabs, or layers, where each slab has a constant electron density and thickness. 33
Conclusions
In conclusion, the work presented here consolidates and reinforces earlier work carried out using epifluorescence microscopy and X-ray scattering methods, 22–23 and thoroughly investigates the interaction of PG-1 with different lipids using X-ray scattering techniques. From the changes in ordering, area per molecule and unit cell dimensions, GIXD data show that PG-1 interacts with and inserts into DPPC monolayers. However, unlike the case of DPPG, under none of the conditions examined does PG-1 injection result in the complete disordering of DPPC packing, suggesting that electrostatic forces are playing an important role in the interaction. Our data further show that while PG-1 inserts into DPPC, DPPG and lipid A monolayers under constant area conditions and primarily into the head group region, the effect of the peptide on the DPPG and lipid A monolayers is much more pronounced than that on the DPPC monolayer, and an additional peptide adsorbed layer was only observed below the head group region for the two anionic lipid systems. Our data further suggest that there is a much greater extent of disordering in lipid systems with lower packing due to PG 1 insertion, in agreement with our earlier findings from epifluorescence microscopy. 22–23 The fact that PG-1 preferentially damages lipid monolayers composed of lipids prevalent in different types of bacteria suggests that PG-1 is a good candidate for antimicrobial drug targets to be designed in the future. This bodes well for the design and development of protegrins and other antimicrobial peptides as future therapeutic agents.
Acknowledgments
We acknowledge the European Synchrotron Radiation Facility for provision of synchrotron radiation facilities at the ID10B beamline. Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. W-31-109-ENG-38. YI and KYCL are grateful for the support of the Packard Foundation (99–1465). FN and DG would like to acknowledge the sponsorship of this work by the Engineering and Physical Sciences Research Council, UK
Footnotes
Abbreviations: DPPC, dipalmitoyl-phosphatidylcholine; DPPG, dipalmitoyl-phosphatidylglycerol; GIXD, grazing incidence X-ray diffraction; XR, X-ray reflectivity; PG 1, protegrin-1
Notes and references
- 1.Zasloff M. N Engl J Med. 2002;347:1199–1200. doi: 10.1056/NEJMe020106. [DOI] [PubMed] [Google Scholar]; Bals R, Wilson JM. Cell Mol Life Sci. 2003;60:711–720. doi: 10.1007/s00018-003-2186-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Devine DA, Hancock RE. Mammalian Host Defense Peptides. 1. Vol. 6 Cambridge University Press; Cambridge: 2004. [Google Scholar]; Yang D, Biragyn A, Hoover DM, Lubkowski J, Oppenheim JJ. Annu Rev Immunol. 2004;22:181–215. doi: 10.1146/annurev.immunol.22.012703.104603. [DOI] [PubMed] [Google Scholar]; Bowdish DME, Davidson DJ, Scott MG, Hancock REW. Antimicrob Agents Chemother. 2005;49:1727–1732. doi: 10.1128/AAC.49.5.1727-1732.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Oren Z, Shai Y. Biopolymers. 1998;47:451–463. doi: 10.1002/(SICI)1097-0282(1998)47:6<451::AID-BIP4>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]; Shai Y. Biochim Biophys Acta Biomembr. 1999;1462:55–70. doi: 10.1016/s0005-2736(99)00200-x. [DOI] [PubMed] [Google Scholar]
- 4.Matsuzaki K. Biochim Biophys Acta Biomembr. 1999;1462:1–10. doi: 10.1016/s0005-2736(99)00197-2. [DOI] [PubMed] [Google Scholar]
- 5.Pozo Navas B, Lohner K, Deutsch G, Sevcsik E, Riske KA, Dimova R, Garidel P, Pabst G. Biochim Biophys Acta Biomembr. 2005;1716:40–48. doi: 10.1016/j.bbamem.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 6.Lohner K, Prenner EJ. Biochim Biophys Acta Biomembr. 1999;1462:141–156. doi: 10.1016/s0005-2736(99)00204-7. [DOI] [PubMed] [Google Scholar]
- 7.Neville F, Cahuzac M, Nelson A, Gidalevitz D. J Phys: Condens Matter. 2004;16:S2413–S2420. [Google Scholar]
- 8.Neville F, Cahuzac M, Konovalov O, Ishitsuka Y, Lee KYC, Kuzmenko I, Kale GM, Gidalevitz D. Biophys J. 2006;90:1275–1287. doi: 10.1529/biophysj.105.067595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Graham JM. Membrane Analysis. Springer; New York: 1997. [Google Scholar]
- 10.Keller SL, Pitcher WH, Huestis WH, McConnell HM. Phys Rev Lett. 1998;81:5019–5022. [Google Scholar]
- 11.Ratledge C, Wilkinson SG. Microbial Lipids. Academic Press; London: 1988. [Google Scholar]
- 12.Burton AJ, Carter HE. Biochemistry. 1964;3:411–418. doi: 10.1021/bi00891a018. [DOI] [PubMed] [Google Scholar]; Takayama K, Qureshi N, Mascagni P, Nashed M, Anderson L, Raetz C. J Biol Chem. 1983;258:7379–7385. [PubMed] [Google Scholar]
- 13.Kokryakov VN, Harwig SSL, Panyutich EA, Shevchenko AA, Aleshina GM, Shamova OV, Korneva HA, Lehrer RI. FEBS Lett. 1993;327:231–236. doi: 10.1016/0014-5793(93)80175-t. [DOI] [PubMed] [Google Scholar]
- 14.Steinberg D, Hurst M, Fujii C, Kung A, Ho J, Cheng F, Loury D, Fiddes J. Antimicrob Agents Chemother. 1997;41:1738–1742. doi: 10.1128/aac.41.8.1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fahrner RL, Dieckmann T, Harwig SSL, Lehrer RI, Eisenberg D, Feigon J. Chem Biol. 1996;3:543–550. doi: 10.1016/s1074-5521(96)90145-3. [DOI] [PubMed] [Google Scholar]
- 16.Tamamura H, Murakami T, Horiuchi S, Sugihara K, Otaka A, Takada W, Ibuka T, Waki M, Yamamoto N, Fujii N. Chem Pharm Bull. 1995;43:853–858. doi: 10.1248/cpb.43.853. [DOI] [PubMed] [Google Scholar]
- 17.Yasin B, Harwig SS, Lehrer RI, Wagar EA. Infect Immun. 1996;64:709–713. doi: 10.1128/iai.64.3.709-713.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heller WT, Waring AJ, Lehrer RI, Huang HW. Biochemistry. 1998;37:17331–17338. doi: 10.1021/bi981314q. [DOI] [PubMed] [Google Scholar]
- 19.Ma C, Srinivasan MP, Waring AJ, Lehrer RI, Longo ML, Stroeve P. Colloids Surf B. 2003;28:319–329. [Google Scholar]
- 20.Mani R, Tang M, Wu X, Buffy JJ, Waring AJ, Sherman MA, Hong M. Biochemistry. 2006;45:8341–8349. doi: 10.1021/bi060305b. [DOI] [PubMed] [Google Scholar]
- 21.Mani R, Cady SD, Tang M, Waring AJ, Lehrer RI, Hong M. Proc Natl Acad Sci USA. 2006;103:16242–16247. doi: 10.1073/pnas.0605079103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gidalevitz D, Ishitsuka YJ, Muresan AS, Konovalov O, Waring AJ, Lehrer RI, Lee KYC. Proc Natl Acad Sci USA. 2003;100:6302–6307. doi: 10.1073/pnas.0934731100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ishitsuka Y, Pham DS, Waring AJ, Lehrer RI, Lee KYC. Biochim Biophys Acta Biomembr. 2006;1758:1450. doi: 10.1016/j.bbamem.2006.08.001. [DOI] [PubMed] [Google Scholar]
- 24.Lu JR, Thomas RK, Penfold J. Adv Colloid Interface Sci. 2000;84:143–304. doi: 10.1016/s0001-8686(99)00019-6. [DOI] [PubMed] [Google Scholar]; Yaseen M, Wang Y, Su TJ, Lu JR. J Coll Interface Sci. 2005;288:361–370. doi: 10.1016/j.jcis.2005.03.024. [DOI] [PubMed] [Google Scholar]
- 25.Zhang L, Vidu R, Waring AJ, Lehrer RI, Longo ML, Stroeve P. Langmuir. 2002;18:1318–1331. [Google Scholar]
- 26.Maget-Dana R. Biochim Biophys Acta Biomembr. 1999;1462:109–140. doi: 10.1016/s0005-2736(99)00203-5. [DOI] [PubMed] [Google Scholar]; Brockman H. Curr Opin Struct Biol. 1999;9:438–443. doi: 10.1016/S0959-440X(99)80061-X. [DOI] [PubMed] [Google Scholar]; Jensen TR, Balashev K, Bjornholm T, Kjaer K. Biochimie. 2001;83:399–408. doi: 10.1016/s0300-9084(01)01265-2. [DOI] [PubMed] [Google Scholar]; Losche M. Current Topics in Membranes. Vol. 2002. Academic Press; New York: pp. 117–161. [Google Scholar]; Sun F. Biophys J. 2002;82:2511–2519. doi: 10.1016/S0006-3495(02)75594-X. [DOI] [PMC free article] [PubMed] [Google Scholar]; Ambroggio EE, Separovic F, Bowie J, Fidelio GD. Biochim Biophys Acta Biomembr. 2004;1664:31–37. doi: 10.1016/j.bbamem.2004.03.013. [DOI] [PubMed] [Google Scholar]
- 27.Castano S, Desbat B, Dufourcq J. Biochim Biophys Acta Biomembr. 2000;1463:65–80. doi: 10.1016/s0005-2736(99)00175-3. [DOI] [PubMed] [Google Scholar]
- 28.Nouri-Sorkhabi MH, Wright LC, Sullivan DR, Kuchel PW. Lipids. 1996;31:765–770. doi: 10.1007/BF02522893. [DOI] [PubMed] [Google Scholar]
- 29.Goldfine H. Adv Microb Physiol. 1972;8:1–58. doi: 10.1016/s0065-2911(08)60187-3. [DOI] [PubMed] [Google Scholar]; Op den Kamp JAF. Annu Rev Biochem. 1979;48:47–71. doi: 10.1146/annurev.bi.48.070179.000403. [DOI] [PubMed] [Google Scholar]
- 30.Basañez G, Shinnar AE, Zimmerberg J. FEBS Lett. 2002;532:115–120. doi: 10.1016/s0014-5793(02)03651-7. [DOI] [PubMed] [Google Scholar]
- 31.Neville F, Hodges CS, Liu C, Konovalov O, Gidalevitz D. Biochim Biophys Acta Biomembr. 2006;1758:232–240. doi: 10.1016/j.bbamem.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 32.Sambri V, Marangoni A, Giacani L, Gennaro R, Murgia R, Cevenini R, Cinco M. J Antimicrob Chemother. 2002;50:895–902. doi: 10.1093/jac/dkf220. [DOI] [PubMed] [Google Scholar]
- 33.Konovalov O, Myagkov I, Struth B, Lohner K. Eur Biophys J Biophys Lett. 2002;31:428–437. doi: 10.1007/s00249-002-0233-3. [DOI] [PubMed] [Google Scholar]
- 34.Pao WJ, Zhang F, Heiney PA, Mitchell C, Cho W-D, Percec V. Phys Rev E: Stat, Nonlinear, Soft Matter Phys. 2003;67:021601. doi: 10.1103/PhysRevE.67.021601. [DOI] [PubMed] [Google Scholar]
- 35.Demel RA, Geurts van Kessel WSM, Zwaal RFA, Roelofsen B, van Deenen LLM. Biochim Biophys Acta Biomembr. 1975;406:97–107. doi: 10.1016/0005-2736(75)90045-0. [DOI] [PubMed] [Google Scholar]; Blume A. Biochim Biophys Acta Biomembr. 1979;557:32–44. doi: 10.1016/0005-2736(79)90087-7. [DOI] [PubMed] [Google Scholar]
- 36.Als-Nielsen J, McMorrow D. Elements of Modern X-ray Physics. 1. John Wiley & Sons; Chichester: 2001. [Google Scholar]
- 37.Jensen TR, Kjaer K. In: Novel Methods to Study Interfacial Layers. Mobius D, Miller R, editors. Elsevier; Amsterdam: 2001. pp. 205–254. [Google Scholar]
- 38.Schalke M, Losche M. Adv Colloid Interface Sci. 2000;88:243–274. doi: 10.1016/s0001-8686(00)00047-6. [DOI] [PubMed] [Google Scholar]
- 39.Als-Nielsen J, Jacquemain D, Kjaer K, Leveiller F, Lahav M, Leiserowitz L. Phys Rep. 1994;246:251–313. doi: 10.1126/science.252.5012.1532. [DOI] [PubMed] [Google Scholar]
- 40.Leveiller F, Jacquemain D, Leiserowitz L, Kjaer K, Als-Nielsen J. J Phys Chem. 1992;96:10380–10389. [Google Scholar]