Abstract
The signaling pathways by which the phytochrome (phy) family of photoreceptors transmits sensory information to light-regulated genes remain to be fully defined. Evidence for a relatively direct pathway has been provided by the binding of one member of the family, phyB, to a promoter-element-bound, basic helix–loop–helix protein, PIF3, specifically upon light-induced conversion of the photoreceptor molecule to its biologically active conformer (Pfr). Here, we show that phyA also binds selectively and reversibly to PIF3 upon photoconversion to Pfr, but that the apparent affinity of PIF3 for phyA is 10-fold lower than for phyB. This result is consistent with previous in vivo data from PIF3-deficient Arabidopsis, indicating that PIF3 has a major role in phyB signaling, but a more minor role in phyA signaling. We also show that phyB binds stoichiometrically to PIF3 at an equimolar ratio, suggesting that the resultant complex is the unit active in transcriptional regulation at target promoters. Deletion mapping suggests that a 37-aa segment present at the N terminus of phyB, but absent from phyA, contributes strongly to the high binding affinity of phyB for PIF3. Conversely, deletion mapping and point mutation analysis of PIF3 for determinants involved in recognition of phyB indicates that the PAS domain of PIF3 is a major contributor to this interaction, but that a second determinant in the C-terminal domain is also necessary.
Light controls the growth and development of higher plants. Efforts to understand the molecular basis for this phenomenon have included investigations of how the phytochrome (phy) family of sensory photoreceptors (phyA to phyE in Arabidopsis) sense and transduce light stimuli into molecular signals that control these plant responses (1, 2). These responses certainly involve changes in gene expression, although other phenomena, such as rapid changes in membrane properties, postulated to involve other pathways, have also been reported (1). Phytochromes exert their regulatory activity by switching between their biologically inactive, Pr, and active, Pfr, conformers upon absorption of red (R) and far-red (FR) light. The photoreceptor molecule consists of a dimer of two ≈125-kDa polypeptide subunits, each with a covalently linked tetrapyrrole chromophore that is autocatalytically attached by the protein (3, 4).
In recent years, efforts to identify components that function as early signaling intermediates relaying information from photoactivated phytochromes to photoresponsive genes have included genetic screens for signaling-defective mutants and yeast two-hybrid screens for phytochrome-interacting factors (4–6). A considerable number of Arabidopsis mutants have been identified that appear to selectively affect signaling through phyA or phyB, consistent with the existence of separate transduction pathway segments dedicated to one or the other photoreceptor (2, 5, 7–12). On the other hand, mutants apparently affecting both phyA and phyB signaling have also been identified, indicating convergence of these pathways (5).
Yeast two-hybrid library screens for phytochrome-interacting factors as potential primary signaling partners have thus far led to reports of three apparently unrelated proteins that bind to one or more phytochromes (6): phytochrome kinase substrate 1 (PKS1) (13), nucleoside diphosphate kinase 2 (14), and phytochrome-interacting factor 3 (PIF3) (15). In addition, targeted interaction studies have provided evidence that the blue-light receptors cryptochromes 1 (CRY1) and 2 (CRY2) can also interact with phyA consistent with crosstalk between the photoreceptor systems (16). Both PKS1 and the cryptochromes have been reported to be phosphorylated by preparations of recombinant oat phyA isolated from yeast, leading to the proposal that the phytochromes are light-regulated protein kinases that signal in vivo by transphosphorylation of protein substrates (13, 16).
PIF3 is a nuclear-localized, basic helix–loop–helix (bHLH) factor initially isolated as interacting with the nonphotoactive, C-terminal domain of Arabidopsis phyB, but found also to interact with the equivalent domain of phyA (15). Subsequently, full-length photoactive phyB was shown to bind to PIF3 only upon R-induced conversion to the biologically active Pfr form, and to dissociate upon FR-induced reconversion to the Pr form (17). Most recently, it has been shown that PIF3 binds specifically to a G-box DNA-sequence motif present in a variety of light-regulated promoters, and that phyB binds reversibly to G-box-bound PIF3 only upon photoconversion to the Pfr form (18). Together with evidence that phyA and phyB are induced to translocate from cytoplasm to nucleus upon Pfr formation (19–22), these data suggest a direct signaling pathway from phyB to target gene promoters in which the photoreceptor molecule becomes an integral, light-switchable component of a transcription-regulator complex (6, 18).
Although the nonphotoactive C-terminal domain of phyA has been shown to interact with PIF3 in a yeast two-hybrid assay (15), the question has remained open as to whether full-length, photoactive phyA, like phyB, would recognize this protein, and whether differential binding of the Pr and Pfr forms would occur. Moreover, although PIF3-depleted seedlings exhibit reduced responsiveness to both phyB-active, continuous R, and phyA-active, continuous FR, striking quantitative differences between the two treatments suggest that PIF3 may have a major role in phyB signaling, but a more minor role in phyA signaling (15). This observation raises the question of whether phyA and phyB have differential binding activities toward PIF3. We have examined these questions here by using a quantitative, in vitro pull-down assay that employs labeled recombinant protein synthesized by coupled in vitro transcription-translation. In addition, to begin to identify regions or sequence motifs within the interacting partners involved in specific, mutual recognition, we have investigated the binding activities of a series of deletion derivatives of phyB and PIF3. Both molecules contain domains of potential importance to the specificity of partner recognition, including PER-ARNT-SIM (PAS)-related domains (3, 15) implicated in mediating protein–protein interactions in other systems (23).
Materials and Methods
Expression Vector Constructs.
The various T7-promoter driven templates for in vitro transcription and translation were constructed as follows. Gal4 activation domain (GAD), GAD-PIF3, FL-phyB, phyB N-terminal domain (amino acids 1–644), and phyB C-terminal domain (residues 645–1,211) were from Ni et al. (17). The template for the phyB-GAD bait protein with GAD fused to the C terminus of phyB was constructed by PCR amplification of both the full-length PHYB coding sequence (18) and the GAD sequence from the vector pGAD424 (CLONTECH) and cloning into the pET-17b vector (Novagen) by three-way ligation. Full-length PIF3 (15) and its deletion mutants were also amplified by PCR and cloned into the pRSETB vector (Invitrogen). PhyB deletion derivatives were amplified from a pET-3c vector (17) and cloned into pET-17b. The full-length phyA coding sequence with a T7 promoter at its 5′ terminus was cloned into pBluescript KS+. Gly165 of the PIF3 N-120 deletion was converted to Val by using a Quick Change Site-Directed Mutagenesis Kit from Stratagene.
In Vitro Synthesis of Bait and Prey Proteins.
All bait and prey polypeptides were synthesized by using the T'n'T in vitro transcription-translation system from Promega with 35S-Met as a radioactive label according to the manufacturer's instructions. Each 50-μl T'n'T reaction mix contained 2 μg template DNA and 20 pmol (20 μCi) of labeled 35S-Met. All synthesis reactions were carried out for 1.5 h at 30°C. Phycocyanobilin chromophore, prepared as in Scheer (24), was added to all phytochrome T'n'T products to a final concentration of 10 μM, and the mixture was incubated at 4°C in darkness for 1 h.
Processing of Bait and Prey Proteins.
Bait proteins, GAD, GAD-PIF3, and phyB-GAD synthesized in separate T'n'T reactions were attached to agarose beads by addition of 1 μg monoclonal antibody against GAD and 20 μl protein-A agarose beads (Santa Cruz Biotechnology). The mixture was incubated at 4°C for 1 h in PBS binding buffer that contained 137 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4, 1.4 mM KH2PO4, 0.1% BSA, 0.1% Nonidet P-40, pH 7.3, and complete protease inhibitor mixture (Roche Molecular Biochemicals). Bead bound proteins were washed three times with 1.0 ml of PBS binding buffer without BSA, recovered by centrifugation at 5,000 × g for 5 min at 4°C, and resuspended in 100 μl of PBS buffer. These preparations were generally used for five separate binding reactions in equal aliquots. Upon completion of prey protein synthesis, the reactions were precleared by adding 25 μl of protein A/G plus agarose beads to each 50-μl T'n'T reaction. The mixture was incubated in darkness at 4°C for 1 h after addition of 75-μl 2× PBS binding buffer containing complete protease inhibitor mixture. Supernatants containing the labeled prey protein were recovered by centrifugation at 6,000 × g for 5 min at 4°C and used for binding reactions.
In Vitro Interaction Assay.
Twenty microliters of agarose beads bearing approximately 2.5 fmol of bait protein were mixed with 30 μl of various precleared prey-protein preparations (at roughly equimolar ratio) for binding studies that were carried out in a 50-μl reaction, unless otherwise indicated. For all R lanes, the reaction mixture received 10-min R illumination on ice by using an LED light source (QBEAM 2200) with 343 μmol m−2 s−1 at 664 nm (Quantum Devices, Madison, WI). For all R/FR lanes, the mixture received 1-min FR immediately after the R treatment by using the same LED light source with 353 μmol m−2 s−1 at 748 nm. All binding reactions were then incubated in darkness for 2 h at 4°C. The beads containing bait and bound prey molecules were then pelleted at 5,000 × g for 5 min, and washed with 2× 500 μl of PBS binding buffer without BSA before being solubilized in SDS sample buffer at 95°C for 5 min and analyzed by 10% SDS/PAGE. A 15% gel was used for the PIF3 PAS domain fragment. The gel was then fixed, dried, and exposed overnight in a phosphorimager cassette.
Data Processing.
The radioactivity in each protein band was quantified by using a PhosphorImager (Molecular Dynamics). To determine the absolute amounts of the labeled proteins in the different bands, we established a standard curve for 35S-Met using the PhosphorImager. To minimize the variation between R and R/FR treatments because of differences in bait protein recovery, we divided the value for the bait from the R lane by that of the R/FR lane, and used this factor to correct the radioactivity of the bound prey fraction in the corresponding R/FR lane. On average, 70% or more of the bait protein was recovered in the pellet fraction.
Results
PIF3 Binds PhyB and PhyA with Different Apparent Affinities.
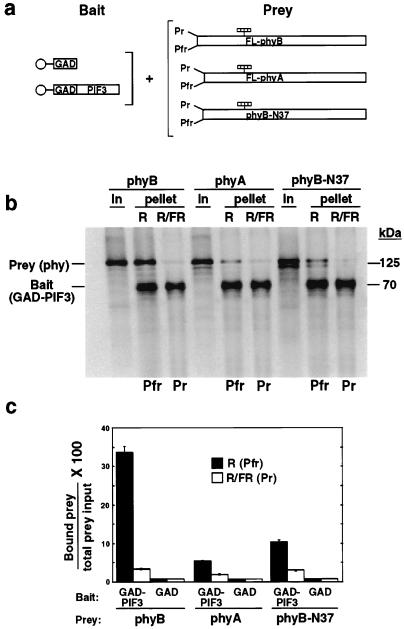
In previous studies, we used unlabeled recombinant proteins produced in E. coli and immobilized on beads as bait in in vitro binding assays involving full-length phyB as the prey (17). Here, to monitor the binding reaction quantitatively, we have implemented a pull-down assay procedure in which both bait and prey proteins were synthesized and radioactively labeled by in vitro transcription and translation, and the absolute molar quantities of individual proteins in different fractions were determined by PhosphorImager analysis. In initial studies using this procedure, we found that full-length phyB bound strongly and specifically to PIF3 in the Pfr form as expected (17), with up to 35% of the input phyB being bound (Fig. 1). PhyA also bound selectively to PIF3 in the Pfr form, but the level of phyA binding was substantially lower than for phyB (Fig. 1).
Figure 1.
PIF3 binds selectively to the Pfr forms of both phyA and phyB but with differential efficiency. (a) Experimental design. In vitro synthesized 35S-labeled bait proteins (GAD or GAD-PIF3) immobilized on beads (○) were each mixed with 35S-labeled, chromophore-ligated (striped bars), full-length (FL) phyB or phyA, or phyB-N37 deletion-mutant (lacking N-terminal residues 1–37) prey proteins. The phytochromes were established either in the Pfr form by a pulse of R only (R), or in the Pr form following sequential pulses of R and FR (R/FR), before incubation in darkness for 2 h. Bead-bound proteins were then pelleted, and analyzed by phosphorimager. (b) Autoradiogram showing phy proteins (phyB, phyA, and phyB-N37) initially in the prey input samples (In), and bead-associated (pellet) bait (GAD-PIF3) and prey (phy) proteins after the binding reaction. “In” lanes contain 1/15 of the total input protein used in each binding assay. R and R/FR lanes contain 1/6 of the total bound fraction from each assay. (c) Quantitative analysis of data from three separate experiments. The amount of bound prey protein in each treatment is expressed as a percentage of the relevant total prey input. GAD control lanes not shown in b, but presented quantitatively here.
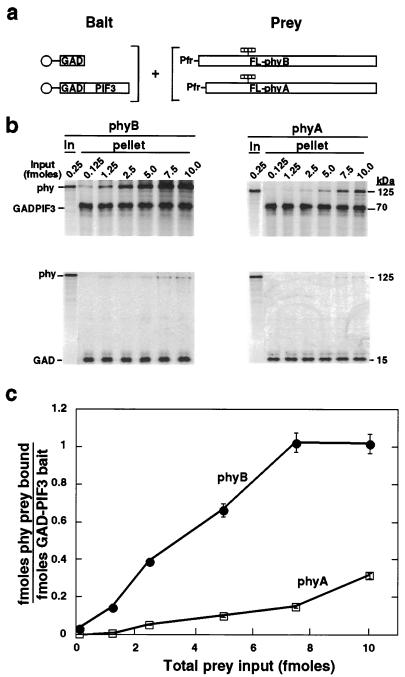
To compare the relative binding efficiencies of phyA and phyB for PIF3 more systematically, increasing amounts of each photoreceptor were added in the Pfr form as prey to a constant amount of bead-immobilized PIF3 bait. The molar ratio of bound phyA or phyB to immobilized PIF3 was then plotted as a function of input photoreceptor level (Fig. 2). The data indicate that phyB binds with up to 10-fold higher apparent affinity to PIF3 than does phyA.
Figure 2.
PIF3 binds with higher apparent affinity to phyB than phyA and exhibits 1:1 molar binding stoichiometry with phyB at saturation. (a) Experimental design. 35S-labeled bait proteins (GAD and GAD-PIF3) immobilized on beads (○) were each incubated with increasing levels of the prey proteins, 35S-labeled, chromophore-ligated (striped bars) full-length (FL) phyB or phyA in the Pfr form for 2 h in darkness before pelleting and PhosphorImager analysis. (b) Autoradiograms showing labeled proteins, phyB and phyA (0.25 fmol per lane), initially in the prey input samples (In), and bait (GAD or GAD-PIF3) and prey (phy) proteins associated with the beads (pellet) after the binding reaction. The level of bait protein was constant in all reactions (approximately 2.5 fmol), whereas increasing amounts (0.125–10.0 fmol) of prey protein were added. (c) Binding of phyB and phyA to PIF3 as a function of phy input level. Binding curves show the average of two parallel experiments as in b. The molar amounts of immobilized bait (GAD-PIF3) and its bound prey (phyB and phyA) in the same lane were obtained by using a standard curve constructed using known amounts of 35S-Met. Radioactivity bound to GAD alone at each input level was subtracted before calculation.
The Interaction Between PIF3 and PhyB Saturates at Equimolar Ratio.
The phyB binding curve plateaued at 7.5 fmol of input phyB (Fig. 2C), suggesting that the binding reaction was saturated. After correction for methionine content, the molar ratio of bound phyB to immobilized PIF3 was found to be 1:1 at saturation (Fig. 2C). The weaker interaction of phyA with PIF3 precluded testing for saturation binding with the present protocol.
Mapping PIF3-Interacting Domains Within PhyB.
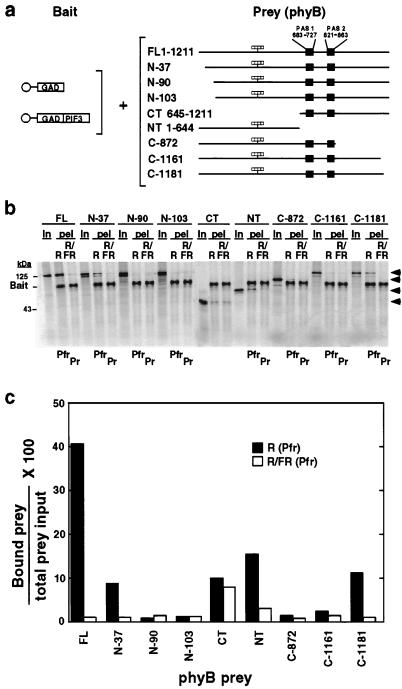
The Arabidopsis phyB molecule has a 37-aa residue extension at its N terminus compared with phyA (25). To determine whether this protein segment might account for the higher apparent binding affinity of phyB than of phyA for PIF3, we compared the binding activity of a deletion derivative of phyB lacking these 37 residues (phyB-N37) with that of full-length phyA (Fig. 1A). The data show that phyB-N37 binds selectively in its Pfr form to PIF3, but that the binding activity is strongly reduced relative to the full-length phyB, to a level about 2-fold greater than phyA (Fig. 1 B and C).
To more systematically investigate molecular determinants within phyB necessary for interaction with PIF3, we tested a series of deletion mutant derivatives of the photoreceptor for binding activity (Fig. 3A). The results show that removal of only 30 amino acids from the C terminus of phyB strongly reduces binding activity, similar to that caused by removal of the N-terminal 37 residues. As in the case of phyB-N37, the C-1181 mutant retains selective recognition of PIF3 in the Pfr form (Fig. 3 B and C). Further removal of another 53 and 30 amino acids from either N or C termini, respectively, essentially eliminated the binding activity (Fig. 3 B and C). Interestingly, as previously reported (17), the separate C-terminal (CT, 645-1,211) and N-terminal (NT, 1–644) domains of phyB, each retained moderate binding activity, with the N-terminal domain showing selectiveness for PIF3 in the Pfr form (Fig. 3 B and C).
Figure 3.
Deletion mapping of PIF3-interacting domains within the phyB polypeptide. (a) Experimental design. 35S-labeled bait proteins (GAD or GAD-PIF3) immobilized on beads (○) were each incubated with 35S-labeled, chromophore-ligated (striped bars) phyB prey proteins, as either Pfr [established by an R pulse (R)] or Pr [established by sequential R and FR pulses (R/FR)], for 2 h in darkness before pelleting and PhosphorImager analysis. phyB prey designations: FL, full-length; NT, N-terminal domain, residues 1–644; CT, C-terminal domain, residues 645-1,211; N, N-terminal deletion; C, C-terminal deletion; the residue at which each N- or C-terminal deletion was made is indicated. (b) Autoradiogram showing binding activity of phyB deletion mutants toward GAD-PIF3. Shown are each of the labeled phyB products (designations as in a) initially in the prey input samples (In), and the bead-associated (pel) bait (GAD-PIF3) and prey (phyB and derivatives) proteins after the binding reaction. (c) Quantitative analysis of the data in b. Amount of bound prey protein in each treatment is expressed as a percentage of the relevant total prey input. GAD bait alone was included as a negative control with FL phyB only and the amount of bead-bound phyB prey was always <1% of the total prey input.
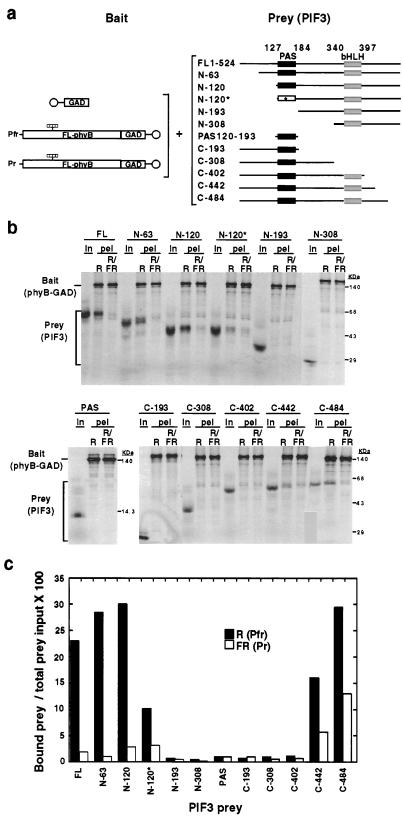
Mapping PhyB-Interacting Domains Within PIF3.
To identify regions within PIF3 necessary for interaction with phyB, we tested the binding activities of a series of deletion mutant derivatives using bead-immobilized, full-length phyB as bait (Fig. 4A). Both the N-63 and N-120 deletion mutants showed similar, or somewhat higher, binding activities compared with that of full-length PIF3, with retention of strong selectivity for the Pfr form of phyB (Fig. 4 B and C). However, the N-193 construct with the entire PAS domain deleted lost its ability to interact with phyB completely. To further assess the importance of this domain, we generated a clone containing a missense mutation by substituting a Val for the conserved Gly at position 165 in the N-120 construct. We also expressed the 74-aa PAS domain alone to test whether it is sufficient for binding. The data show that the single amino acid substitution caused a strong, albeit incomplete, loss of binding activity by the N-120* construct, confirming the importance of the PAS domain for phyB interaction. However, the PAS domain alone did not interact with phyB-GAD. Deletion from the C terminus of PIF3 resulted in a progressive affinity loss, coupled with a higher nonselective binding between the two conformers of phyB (C-484, C-442). The C-402 deletion mutant that ends immediately downstream of the bHLH motif, as well as all constructs that terminated downstream of the PAS domain, or upstream of the bHLH domain showed no binding activity (Fig. 3).
Figure 4.
The PAS domain of PIF3 is necessary but not sufficient for phyB binding. (a) Experimental design. 35S-labeled bait proteins (GAD or full-length, chromophore-ligated (striped bars) phyB-GAD fusion protein (FL-phyB-GAD), immobilized on beads (○), were each mixed separately with 35S-labeled PIF3 prey proteins [full-length (FL) PIF3, nine deletion mutants, one point mutant, and the PAS region alone]. The residues at which deletions were made from the N terminus (N) or C terminus (C) of PIF3 are indicated, as are the terminal residues of the PAS fragment. Asterisk indicates the point mutation (G165V) within the PAS domain of the N-120 deletion (N-120*). phyB was established either as Pfr by a pulse of R only (R) or as Pr by sequential R and FR pulses before incubation in darkness for 2 h, pelleting and PhosphorImager analysis. (b) Autoradiogram showing the binding activities of full-length PIF3 and its mutant derivatives with phyB-GAD. Shown are each of the labeled PIF3 products (designations as in a) initially in the prey input samples (In), and the bead-associated (pel) bait (phyB-GAD) and prey (PIF3 or derivatives) proteins after the binding reaction. (c) Quantitative analysis of the data in b. Bound prey protein in each treatment is expressed as a percentage of the relevant total prey input. GAD bait alone was included as a negative control with FL-PIF3 only and the amount of bead-bound PIF3 was always <1% of the total prey input.
Discussion
After initial identification of PIF3 as a factor that bound to the nonchromophoric C-terminal domain of phyB, it was found that this factor also bound to the equivalent domain of phyA in a yeast two-hybrid assay (15). Subsequently, in vitro pull-down experiments revealed that full-length, chromophore-conjugated phyB binds with substantially greater affinity to PIF3 than does the C-terminal domain alone, but only upon light-induced conversion of the photoreceptor to its biologically active Pfr conformer (17). Here, we have provided evidence that full-length, chromophore-conjugated phyA is also induced to bind PIF3 upon R-driven conversion to its Pfr form, and that reconversion of Pfr to Pr by subsequent FR irradiation blocks or reverses this interaction (Fig. 1). This selective binding of PIF3 to the biologically active conformer of phyA in a manner qualitatively similar to phyB suggests that this interaction may be functionally significant to phyA signaling.
These data are thus consistent with the proposition that PIF3 is a shared signaling partner for phyA and phyB, thereby implying immediate convergence of the two signaling pathways through this common factor (5, 15). On the other hand, our comparative binding studies demonstrate that the apparent affinity of phyA for PIF3 is of the order of 10-fold lower than that of phyB (Fig. 2). This observation is consistent with previous data from Arabidopsis seedlings expressing reduced PIF3 levels (15). These seedlings displayed a striking reduction in responsiveness to continuous R (perceived predominantly through phyB) but only a marginal reduction in responsiveness to continuous FR (perceived exclusively through phyA). Together, these data strongly suggest that PIF3 has a dominant role in phyB signaling, but may have only a minor role in phyA signaling.
This observation suggests the general possibility that, although different members of the phy family may share signaling partners, the strength of signal flow through each family member may be modulated at the level of the binding reaction as a result of intrinsic differences in their affinities for the shared partner. In the case of phyA and phyB, the potential for yet another layer of complexity exists because of the well-known photoregulation of the relative abundance of these molecules (26). Initially, in etiolated Arabidopsis seedlings, just transferred to light, the level of phyA is 50-fold greater than phyB (26, 27), thereby potentially permitting phyA to compete effectively for binding to PIF3 despite the lower apparent binding affinity than phyB. However, phyA is light-labile in the cell and its abundance declines rapidly to a level comparable to or less than that of phyB (26, 27). Under these conditions, the phyB-PIF3 interaction is likely to dominate. This postulated temporal, light-dependent shift in the balance of phyA and phyB signaling through PIF3 might account for some of the apparent complexities observed in photoperception and signaling in early deetiolation responses.
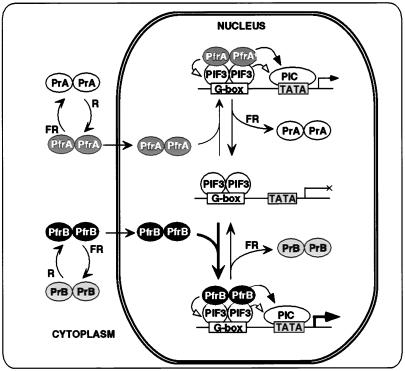
A schematic model depicting this postulated convergence of, and potential competition between, phyA and phyB signaling pathways through interaction with PIF3 is shown in Fig. 5. This model is based on the evidence that both phyA and phyB are induced to translocate into the nucleus as presumptive homodimers upon Pfr formation (20–22), where they can bind in a Pfr-dependent manner to PIF3 homodimers which are bound to G-box DNA sequence motifs in the promoters of phy-regulated genes (18). The evidence from our quantitative binding analysis with phyB (Fig. 2) indicates that the photoreceptor and bHLH proteins associate stoichiometrically into a complex containing one dimeric molecule of each binding partner. These results suggest that this equimolar phy-PIF3 complex is the active unit in transcriptional regulation at target promoters.
Figure 5.
Model depicting postulated convergence of phyA and phyB signaling pathways through shared interaction with the bHLH factor PIF3. R-induced conversion of phyA and phyB homodimers from their cytoplasmically localized, biologically inactive Pr conformers (PrA and PrB, respectively) to their active Pfr conformers (PfrA and PfrB, respectively) triggers translocation of the photoreceptors to the nucleus (19–21). Here, they can bind reversibly in the Pfr form to PIF3, which is constitutively nuclear (15) and bound as a presumptive dimer to G-box motifs in target promoters (18). The PfrB-PIF3 interaction (high affinity, heavy arrow) is postulated to dominate over the PfrA-PIF3 interaction (low affinity, light arrow) at equivalent levels of the two photoreceptors and limiting levels of PIF3. Bound Pfr molecules are postulated to regulate transcription either directly (solid arrowheads) by functioning as coregulators in recruiting or modifying components of the core transcriptional machinery (PIC), or indirectly (open arrowheads) by modifying the potential transcriptional regulatory activity of PIF3.
Our deletion analysis aimed at identifying determinants within the phyB molecule involved in specific interactions with PIF3 indicate that a relatively short segment present at each extremity of the polypeptide is necessary, directly or indirectly, for effective binding (Fig. 3). Interestingly, both the C-terminal domain (CT, 645-1,211) and the N-terminal domain (NT, 1–645) “regained” binding activity relative to intermediate deletion derivatives in their respective series, albeit to a level less than that of full-length phyB (Fig. 3), as previously reported (17). This obervation suggests that the two halves of the bisected phyB molecule might act cooperatively in PIF3 binding. In addition, the inability of all intermediate-length deletion derivatives to bind may indicate the existence of previously unexposed inhibitory motifs within the molecule that either mask or disrupt the normal functions of the remaining determinants (Fig. 3). Unfortunately, our data do not indicate whether the deleted domains participate directly in the binding process, or have more indirect conformational effects on the photoreceptor. Nevertheless, the results are in rough general agreement with in vivo analysis of introduced phytochrome deletion derivatives in transgenic plants (3, 28). These studies indicate that deletion of relatively short regions from either extremity of the photoreceptor polypeptide results in a reduction in phytochrome signaling activity in the plants. Thus, regardless of the specific molecular basis for the reduced binding of phyB to PIF3 in these deletion derivatives, there is a correlation between the reduced apparent binding affinity and the functional activity of phyB in the cell.
Despite these inherent limitations to interpretation of data obtained from deletion derivatives, it is possible that the N-37 deletion mutant of phyB may provide some insight into the molecular basis for the difference in apparent binding affinities of phyA and phyB for PIF3. Arabidopsis phyB has an additional 37 amino acids at its N terminus, appearing as an extension beyond that of phyA. Deletion of these residues reduced the binding of phyB to PIF3 to a level approaching that of phyA, but at the same time retaining differential binding of Pr and Pfr conformers in a manner similar to phyA (Fig. 1). Because the residual phyB polypeptide in the N-37 derivative corresponds in length and sequence alignment to the native phyA molecule, it is possible that the 37-residue N-terminal extension does indeed contribute directly to the enhanced binding of PIF3 to phyB relative to that of phyA.
Previously, we showed that the bHLH domain of PIF3 is sufficient for sequence-specific binding to its cognate DNA G-box motif, but does not interact with phyB, thereby indicating that determinants outside this domain are necessary for phyB binding (18). The data presented here are consistent with this observation. Our deletion analysis indicates that the first N-terminal 120 amino acids of PIF3 are not involved in phyB recognition, but that the region between residues 120 and 193 containing the PAS-related domain, is necessary for phyB binding (Fig. 4). Substitution within this region of a residue generally conserved in PAS domains also caused a reduction in binding (Fig. 4). PAS domains are proposed to function in protein–protein interactions (23). It is possible, therefore, that the PIF3 PAS domain is directly involved in interacting with phyB. On the other hand, no binding was observed of phyB to the isolated PAS domain, indicating that this motif is not sufficient for phytochrome recognition. This observation is consistent with the finding that a region between residues 402 and 484 on the C-terminal side of the bHLH domain is also necessary for maximal binding (Fig. 4). In addition, the C-terminal domain from residues 442 to 524 may also be involved in discriminating between the Pr and Pfr conformers of phyB, as deletions within this region of PIF3 caused enhanced binding of PrB relative to PfrB (Fig. 4). Taken together, these data indicate that, whereas the bHLH region of PIF3 is involved in DNA binding and presumably dimerization, as expected of this class of factors (29), two domains on either side of this region may cooperate to provide high-affinity, conformer-specific recognition of the biologically active form of phyB.
The differential apparent binding affinities of phyA and phyB for PIF3 observed here provide a glimpse of the potential complexities involved in signal channeling among individual members of the photoreceptor family. Further dissection of the domains involved in mutually specific recognition by these binding partners may shed light on the molecular basis of this specificity, and on the biochemical nature of the proposed role of the photoreceptor molecules in the transcriptional process.
Acknowledgments
We thank S. Sato and E. Monte for contributions to generation of the FL-phyA and phyB-GAD constructs, respectively; and M. Hudson and E. Huq for constructive discussions and critical reading of the manuscript. This work was supported by grants from National Institutes of Health (GM47475), Department of Energy (DE-FG03-87ER 13742), and U.S. Department of Agriculture (5335-21000-010-00D). Y.Z. was the recipient of a Rockefeller Foundation Biotechnology Career Fellowship.
Abbreviations
- phy
phytochrome
- R
red light
- FR
far-red light
- Pr
R-absorbing form of phy
- Pfr
FR-absorbing form of phy
- GAD
Gal4 activation domain
- bHLH
basic helix–loop–helix
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.230433797.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.230433797
References
- 1.Kendrick R E, Kronenberg G H M. Photomorphogenesis in Plants. 2nd Ed. Dordrecht, The Netherlands: Kluwer; 1994. [Google Scholar]
- 2.Fankhauser C, Chory J. Annu Rev Cell Dev Biol. 1997;13:203–229. doi: 10.1146/annurev.cellbio.13.1.203. [DOI] [PubMed] [Google Scholar]
- 3.Quail P H. Plant Cell Environ. 1997;20:657–665. [Google Scholar]
- 4.Neff M M, Fankhauser C, Chory J. Genes Dev. 2000;14:257–271. [PubMed] [Google Scholar]
- 5.Deng X-W, Quail P H. Semin Cell Dev Biol. 1999;10:121–129. doi: 10.1006/scdb.1999.0287. [DOI] [PubMed] [Google Scholar]
- 6.Quail, P. H. (2000) Semin. Cell Dev. Biol., in press.
- 7.Quail P H. Philos Trans R Soc London B. 1998;353:1399–1403. doi: 10.1098/rstb.1998.0294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoecker U, Tepperman J M, Quail P H. Science. 1999;284:496–499. doi: 10.1126/science.284.5413.496. [DOI] [PubMed] [Google Scholar]
- 9.Hudson M, Ringli C, Boylan M T, Quail P H. Genes Dev. 1999;13:2017–2027. doi: 10.1101/gad.13.15.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huq E, Tepperman J M, Quail P H. Proc Natl Acad Sci USA. 2000;97:9789–9794. doi: 10.1073/pnas.170283997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fairchild C D, Schumaker M A, Quail P H. Genes Dev. 2000;14:2377–2391. [PMC free article] [PubMed] [Google Scholar]
- 12.Bolle C, Koncz C, Chua N-H. Genes Dev. 2000;14:1269–1278. [PMC free article] [PubMed] [Google Scholar]
- 13.Fankhauser C, Yeh K C, Lagarias J C, Zhang H, Elich T D, Chory J. Science. 1999;284:1539–1541. doi: 10.1126/science.284.5419.1539. [DOI] [PubMed] [Google Scholar]
- 14.Choi G, Yi H, Lee J, Kwon Y-K, Soh M S, Shin B, Luka Z, Hahn T-R, Song P-S. Nature (London) 1999;401:610–613. doi: 10.1038/44176. [DOI] [PubMed] [Google Scholar]
- 15.Ni M, Tepperman J M, Quail P H. Cell. 1998;95:657–667. doi: 10.1016/s0092-8674(00)81636-0. [DOI] [PubMed] [Google Scholar]
- 16.Ahmad M, Jarillo J A, Smirnova O, Cashmore A R. Mol Cell. 1998;1:939–948. doi: 10.1016/s1097-2765(00)80094-5. [DOI] [PubMed] [Google Scholar]
- 17.Ni M, Tepperman J M, Quail P H. Nature (London) 1999;400:781–784. doi: 10.1038/23500. [DOI] [PubMed] [Google Scholar]
- 18.Martínez-García J F, Huq E, Quail P H. Science. 2000;288:859–863. doi: 10.1126/science.288.5467.859. [DOI] [PubMed] [Google Scholar]
- 19.Sakamoto K, Nagatani A. Plant J. 1996;10:859–868. doi: 10.1046/j.1365-313x.1996.10050859.x. [DOI] [PubMed] [Google Scholar]
- 20.Yamaguchi R, Nakamura M, Mochizuki N, Kay S A, Nagatani A. J Cell Biol. 1999;145:437–445. doi: 10.1083/jcb.145.3.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kircher S, Kozma-Bognar L, Kim L, Adam E, Harter K, Schaefer E, Nagy F. Plant Cell. 1999;11:1445–1456. doi: 10.1105/tpc.11.8.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nagy F, Schäfer E. EMBO J. 2000;19:157–163. doi: 10.1093/emboj/19.2.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dunlap J. Science. 1998;280:1548–1549. doi: 10.1126/science.280.5369.1548. [DOI] [PubMed] [Google Scholar]
- 24.Scheer H. In: Techniques in Photomorphogenesis. Smith H, Holmes M G, editors. London: Academic; 1984. pp. 227–256. [Google Scholar]
- 25.Sharrock R A, Quail P H. Genes Dev. 1989;3:1745–1757. doi: 10.1101/gad.3.11.1745. [DOI] [PubMed] [Google Scholar]
- 26.Quail P H, Boylan M T, Parks B M, Short T W, Xu Y, Wagner D. Science. 1995;268:675–680. doi: 10.1126/science.7732376. [DOI] [PubMed] [Google Scholar]
- 27.Somers D E, Sharrock R A, Tepperman J M, Quail P H. Plant Cell. 1991;3:1263–1274. doi: 10.1105/tpc.3.12.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wagner D, Koloszvari M, Quail P H. Plant Cell. 1996;8:859–871. doi: 10.1105/tpc.8.5.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Littlewood T, Evan G I. Helix-Loop-Helix Transcription Factors. New York: Oxford Univ. Press; 1998. [Google Scholar]