Abstract
Purpose
To evaluate the response of cells over-expressing dominant negative (DN) Ku70 to single and multiple small radiation doses.
Methods and Materials
Clones of fibroblasts over-expressing DNKu70, DNKu70-7, DNKu70-11, and parental Rat-1 cells were irradiated under oxic or hypoxic conditions with single or multiple doses. Cells were trypsinized 0 or 6 h after irradiation to determine surviving fraction (SF).
Results
Oxic DNKu70-7 or -11 cells trypsinized 6 h after irradiation were 1.52 or 1.25 and 1.28 or 1.15 times more sensitive than oxic Rat-1 at SF of 0.5 and 0.1, respectively. Hypoxic DNKu70-7 or -11 cells trypsinized 6 h after irradiation were 1.44 or 1.70 and 1.33 or 1.51 times more sensitive than hypoxic Rat-1 at SF of 0.5 and 0.1, respectively. To the multiple doses, oxic and hypoxic DNKu70-7 or -11 cells were 1.35 or 1.37 and 2.23 or 4.61 times more sensitive than oxic and hypoxic Rat-1, respectively, resulting in very small oxygen enhancement ratios. Namely, enhancement caused by DNKu70 under hypoxia after multiple doses was greater than that under oxic conditions and greater than that after single dose.
Conclusions
Over-expression of DNKu70 enhances cells’ response to radiation given as a single dose and as multiple small doses. The enhancement after multiple doses was stronger under hypoxic than under oxic conditions. These results encourage the use of DNKu70 fragment in a gene-radiotherapy.
Keywords: Gene-radiotherapy, Ku70, dominant-negative (DN) Ku70, fractionation, DNA repair, PLD repair
INTRODUCTION
Ionizing radiation induces several types of DNA lesions, such as base damage, DNA single-strand breaks (SSBs), and DNA double-strand breaks (DSBs). Deoxyribonucleic acid nonhomologous end joining represents the major pathway for the repair of DNA-DSBs in mammalian cells and is essential for the survival of irradiated cells (1–8). One of the major participants in this pathway is the DNA dependent protein kinase (DNA-PK) complex, which consists of two components: a 450-kd catalytic subunit, DNA-PKcs, and a heterodimeric protein named Ku. The Ku protein, consisting of two tightly associated but different polypeptides of 70 kd and 80 kd (Ku70 and Ku80, respectively), has double-strand DNA end-binding activity, thereby targeting the complex to DNA ends (9, 10). In the past decades, studies using various mutant cell lines, knock-out mice, and their respective mouse embryo fibroblast cell lines have elucidated the important roles of DNA-PK in many biological processes in cell survival. Of significance for radiation oncology is the evidence that defect in or absence of Ku70, Ku80, or DNA-PKcs subunit results in deficiencies in DNA-DSB repair, leading to hypersensitivity to ionizing radiation (11–13). This crucial role of the DNA-PK complex in repairing radiation-induced DNA damage has suggested that targeting the compartment(s) of this complex could enhance the radiation response of mammalian cells (14, 15).
Our research group has been actively studying the potential of inhibiting the function of the DNA-PK complex to enhance radiation treatment and recently extended our investigation to gene-radiotherapy. Our previous study has identified a dominant negative construct of Ku70, DNKu70, and demonstrated the feasibility of using adenovirus-mediated expression of the DNKu70 fragment in a gene-radiotherapy paradigm to sensitize cells to ionizing radiation (22). Data obtained from structure–function analyses of Ku70 and Ku80 (16–21) have led to a hypothesis that a construct with a deletion of the N-terminal region of Ku70 might be a potential candidate. An N-terminal deleted mutant of Ku70 was constructed, and Rat-1 cells stably over-expressing DNKu70 were generated. We have demonstrated increased radiation sensitivity of these Rat-1 cells and U-87 human glioma cells infected with recombinant adenovirus containing cytomegalovirus (CMV) promoter-driven DNKu70. The increased response to radiation was also observed in hypoxic cells (22).
The purpose of this study was to evaluate the radiation response of cells over-expressing DNKu70 when radiation doses are given in multiple fractions. We hypothesized that, if hypersensitivity is due to the deficiencies in DNA-DSB repair, the enhancement ratio after multiple radiation doses would be greater than that after a single radiation dose. This hypothesis was tested in vitro by giving multiple doses under both oxic and hypoxic conditions.
METHODS AND MATERIALS
Cell culture
DNKu70 cells (rat embryo fibroblasts over-expressing DNKu70) and parental Rat-1 cells were used. Construction of DNKu70 cells has been described previously (22). Briefly, we constructed the expression vector containing the DNKu70 fragment (amino acid residues 62–609 of human Ku70) under the control of CMV promoter and a hygromycin-resistant gene. Rat-1 cells were stably transfected with this plasmid. Drug-resistant cells were selected by culturing these cells in medium containing hygromycin (300 μg/mL) for 2 to 3 weeks, and individual colonies were isolated (designated DNKu70) and grown in monolayer. Among five clones previously tested (22), strongly over-expressing clone 7 (DNKu70-7) and moderately over-expressing clone 11 (DNKu70-11) were studied.
Rat-1 and DNKu70 cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) (Mediatech, Herndon, VA) supplemented with 10% fetal calf serum (Gemini Bio-Product, West Sacramento, CA) and antibiotics (1% penicillin–streptomycin [Mediatech] for Rat-1 and hygromycin [Sigma, St. Louis, MO] for DNKu70 cells). Cell doubling times of exponentially growing Rat-1, DNKu70-7, and DNKu70-11 cells were 17.8, 24.5, and 38.7 h, respectively.
Irradiation under oxic and hypoxic conditions
Cells were irradiated with a 137Cs unit (Mark 1 model 68; JL Shephard and Associates, San Fernando, CA) at a dose rate of approximately 2.3 Gy/min. For oxic irradiations, cells were plated in 35-mm or 60-mm Petri dishes and irradiated in air. For hypoxic irradiation, cells were plated in glass flasks; on the day of irradiation, medium was replaced with those containing N-2-hydroxyethylpi-perazine-N′-2-ethanesulfonic acid (25 mmol/L). Flasks were tightly shielded with rubber stoppers, two needles were inserted through a stopper into each glass flask, and 100% nitrogen gas was flushed through needles (one for inflow and another for outflow) for 10 min at a flow rate of 1.5 L/min. Subsequently needles were removed to secure hypoxia in the flasks, and irradiation was initiated approximately 10 min later. Either immediately or 6 h after irradiation, stoppers were removed and the cells prepared for colony formation assays.
Single-dose irradiation and colony formation assay
To determine cell survival after irradiations, either single cells or 3-day colonies (3 days after cell plating) were irradiated.
Single cells
Exponentially growing cells were trypsinized and counted, and cells were plated in 60-mm Petri dishes. The number of cells plated was estimated to form approximately 50 colonies per dish. Cells were irradiated in air with graded doses approximately 24 h after plating and incubated in a humidified incubator with 5% CO2 gas flow for 10–14 days. Colonies were stained with crystal violet and counted under a dissecting microscope. This method was only applied for single-dose irradiation.
Three-day colonies
Exponentially growing cells were trypsinized and counted. Thirty thousand Rat-1 cells, forty thousand DNKu70-7, or fifty thousand DNKu70-11 cells were plated in each 35-mm diameter Petri dish or glass flask and incubated for 3 days. During this incubation cells formed colonies, and the majority of colonies contained >30 cells. Because a large number of cells were plated, each colony was likely derived from two or more cells. Cells were irradiated under oxic or hypoxic conditions as described above. Two dishes or glass flasks were used for each radiation dose and analyzed separately. For survival assay after a single-dose irradiation, cells were trypsinized either immediately or 6 h after irradiation, then counted and plated in 60-mm diameter Petri dishes for colony formation assay as mentioned for single-cell experiments. In all the colony formations, 105 heavily irradiated cells (with 50 Gy) were added as feeder cells in each 60-mm diameter Petri dish at the time of cell plating.
Surviving fractions (SF) were calculated by dividing the number of colonies counted by the number of cells plated [PE (test)] and then corrected by plating efficiency of nonirradiated control cells [PE (control)]. For survival after irradiation of single cells, cell multiplicity correction was applied.
Multiple radiation doses
For the multiple irradiations, 3 days after cells were plated in either 35-mm diameter Petri dishes or glass flasks, irradiation was initiated. Cells were given 0 to 5 doses of 3 Gy each (total of 0–15 Gy) with an interval of 6 h. Six hours after the last fraction, cells were trypsinized, counted, and plated for colony formation assays as described above. Between the irradiations, cells were kept in the 37°C incubator. For the hypoxic irradiation, cells were kept hypoxic until trypsinization.
During the time of multiple doses (i.e., from 0 to 30 h after the first dose), either an increase or a decrease in the cell number was noticed. These changes in the population size are likely due to cell proliferation or loss, suggesting that a conventional SF estimate would not fully express the effects of fractionated doses, because it takes no account of these changes in the population size. Accordingly, in addition to the SF obtained, it was further corrected by the change in cell number; that is, the SF [PE (test)/PE (control)] was multiplied by the cell ratio [number of cells (test)/number of cells (control)], whereby the latter indicates the change in cell number.
Notably, this ratio equals the “clonogen ratio” that is derived by dividing the number of clonogens in the test group by the number of clonogens in the control group, whereby the number of clonogens is simply the number of counted cells multiplied by the PE. Namely,
Survival curve analysis
The linear-quadratic model was fitted to the measured cell survival curve after single-dose irradiation, and α and β were obtained. The exponential regression analysis was used to obtain the cell survival curve after multiple doses. These analyses were made using commercially available softwares.
RESULTS
Single dose survival curves
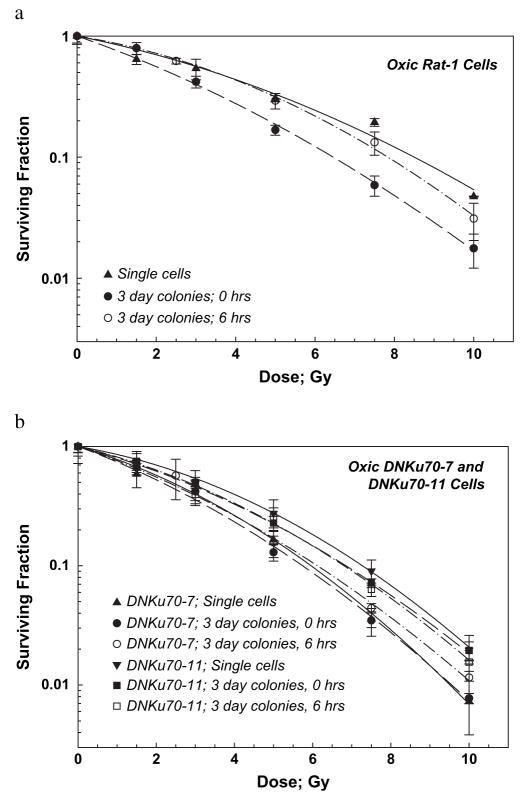
Cell survival curves for oxic Rat-1 and DNKu70 cells irradiated with graded single doses are shown in Fig. 1a and 1b, respectively. Survival of single cells (irradiated 24 h after plating single cells) showed typical linear-quadratic curves for both cell lines (solid triangles or reverse triangles in Fig. 1). The β values of the DNKu70-7 and -11 cell survival curves were slightly larger, and the α/β ratio slightly smaller than those of Rat-1 cells. The DNKu70-7 or -11 single cells were 1.46 (3.5/2.4) or 1.06 (3.5/3.3) and 1.42 (8.5/6.0) or 1.16 (8.5/7.3) times more sensitive at SF of 0.5 and 0.1, respectively, than the Rat-1 single cells (all numbers, including shown below, are tabulated in Table 1).
Fig. 1.
Radiation dose–cell survival curves for Rat-1 cells (a) and DNKu70-7 and -11 cells (b) irradiated under oxic conditions. Solid triangles and reverse triangles indicate that they were irradiated 24 h after plating and incubated without trypsinization; circles and squares indicate that cells were irradiated 3 days after plating (3-day colonies) and trypsinized immediately (solid symbols) or 6 h (open symbols) thereafter.
Table 1.
Alpha and beta values, α/β ratios, OERs, and DNKu70 ERs
| Dose (Gy) and OER |
DNKu70 ER |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Cell line | α ± SE | β ± SE | α/β ± SE | SF 0.5 | SF 0.1 | SF 0.01 | SF 0.5 | SF 0.1 | SF 0.01 | |
| Oxic cells | ||||||||||
| Rat-1 | ||||||||||
| Single cell | −0.150 ± 0.052 | −0.0142 ± 0.006 | 10.55 ± 5.80 | Dose | 3.5 | 8.5 | 13.5 | |||
| 3-day colony, 0 h | −0.263 ± 0.033 | −0.0145 ± 0.004 | 18.13 ± 5.33 | Dose | 2.3 | 6.5 | 10.9 | |||
| 3-day colony, 6 h | −0.121 ± 0.028 | −0.0221 ± 0.003 | 5.47 ± 1.52 | Dose | 3.5 | 7.8 | 12.0 | |||
| DNKu70-7 | ||||||||||
| Single cell | −0.223 ± 0.037 | −0.0275 ± 0.004 | 8.11 ± 1.85 | Dose | 2.4 | 6.0 | 9.5 | 1.46 | 1.42 | 1.42 |
| 3-day colony, 0 h | −0.279 ± 0.031 | −0.0212 ± 0.004 | 13.16 ± 2.72 | Dose | 2.1 | 5.8 | 9.6 | 1.90 | 1.12 | 1.14 |
| 3-day colony, 6 h | −0.256 ± 0.043 | −0.0196 ± 0.005 | 13.08 ± 3.99 | Dose | 2.3 | 6.1 | 10.1 | 1.52 | 1.28 | 1.19 |
| DNKu70-11 | ||||||||||
| Single cell | −0.127 ± 0.032 | −0.0261 ± 0.003 | 4.87 ± 1.34 | Dose | 3.3 | 7.3 | 11.1 | 1.06 | 1.16 | 1.22 |
| 3-day colony, 0 h | −0.198 ± 0.030 | −0.0201 ± 0.003 | 9.85 ± 2.03 | Dose | 2.7 | 6.8 | 11.0 | 0.85 | 0.96 | 0.99 |
| 3-day colony, 6 h | −0.181 ± 0.061 | −0.0232 ± 0.006 | 7.82 ± 3.30 | Dose | 2.8 | 6.8 | 10.7 | 1.25 | 1.15 | 1.12 |
| Hypoxic cells | ||||||||||
| Rat-1 | ||||||||||
| 3-day colony, 0 h | −0.0611 ± 0. 0071 | −0.0018 ± 0.0003 | 33.94 ± 6.55 | Dose | 9.0 | 22.6 | 36.4 | |||
| OER | 3.9 | 3.5 | 3.3 | |||||||
| 3-day colony, 6 h | −0.0627 ± 0.0008 | −0.0014 ± 0.0000 | 44.79 ± 1.10 | Dose | 9.2 | 23.9 | 39.1 | |||
| OER | 2.6 | 3.1 | 3.3 | |||||||
| DNKu70-7 | ||||||||||
| 3-day colony, 0 h | −0.0718 ± 0.0057 | −0.0026 ± 0.0002 | 27.62 ± 2.19 | Dose | 7.6 | 19.1 | 30.6 | 1.38 | 1.30 | 1.26 |
| OER | 3.6 | 3.3 | 3.2 | |||||||
| 3-day colony, 6 h | −0.0970 ± 0.0343 | −0.0017 ± 0.0001 | 57.06 ± 15.58 | Dose | 6.4 | 18.0 | 30.7 | 1.44 | 1.33 | 1.27 |
| OER | 2.8 | 3.0 | 3.0 | |||||||
| DNKu70-11 | ||||||||||
| 3-day colony, 0 h | −0.0923 ± 0.0130 | −0.0023 ± 0.0004 | 40.77 ± 9.12 | Dose | 6.5 | 17.5 | 29.1 | 1.32 | 1.29 | 1.25 |
| OER | 2.4 | 2.6 | 2.6 | |||||||
| 3-day colony, 6 h | −0.1190 ± 0.0358 | −0.0017 ± 0.0011 | 69.96 ± 21.56 | Dose | 5.4 | 15.8 | 27.7 | 1.70 | 1.51 | 1.41 |
| OER | 1.9 | 2.3 | 2.6 | |||||||
Abbreviations: OER = oxygen enhancement ratio; ER = enhancement ratio; SF = surviving fraction.
Data obtained from single dose–cell survival curves for Rat-1 and DNKu70 cells irradiated under oxic and hypoxic conditions. 0 h and 6 h indicate that cells were trypsinized immediately or 6 hours after irradiation.
For Rat-1 cells irradiated 3 days after plating, those trypsinized immediately (0 h) after irradiation (3-day colonies, solid circles in Fig. 1a) were more sensitive than those trypsinized 6 h after irradiation (open circles in Fig. 1a). However, this difference was not observed for both DNKu70-7 and -11 cells irradiated 3 days after plating (compare open and solid symbols in Fig. 1b). The α/β ratios of these 0-h and 6-h survival curves for both DNKu70 cells were also identical (Table 1). The α/β ratio of the survival curve for the Rat-1 cells trypsinized immediately after irradiation was larger than that for cells trypsinized 6 h after irradiation. This small α/β ratio for cells trypsinized 6 h after irradiation seemed to be due to the low survival after a dose of 10 Gy.
The DNKu70-7 and -11 cells in 3-day colonies trypsinized immediately after irradiation were approximately 1.1 and approximately 0.9 times, respectively, more sensitive than the Rat-1 cells in 3-day colonies at all calculated survival levels of 0.5–0.01. DNKu70-7 or -11 cells trypsinized 6 h after irradiation were 1.52 or 1.25 and 1.28 or 1.15 times more sensitive than the Rat-1 cells in 3-day colonies at SF of 0.5 and 0.1, respectively.
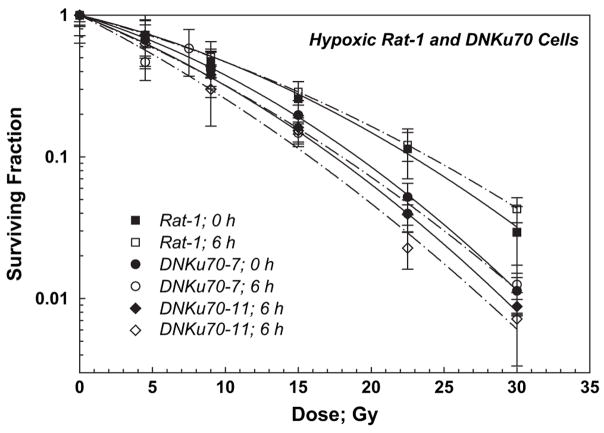
Cell survival curves for hypoxic Rat-1 and DNKu70-7 and -11 cells are shown together in Fig. 2. Three-day colonies were irradiated and trypsinized either immediately (solid symbols, solid lines) or 6 h after irradiation (open symbols, dash–dot lines). Survival curves for these cells trypsinized immediately and 6 h after irradiation were nearly identical, although α/β ratios for both cells trypsinized immediately were smaller than those for cells trypsinized 6 h later in all three lines (Table 1). The hypoxic DNKu70-7 or -11 cells in 3-day colonies trypsinized immediately after irradiation were 1.38 or 1.32 and 1.30 or 1.29 times more sensitive than the hypoxic Rat-1 cells at SF of 0.5 and 0.1, respectively. The hypoxic DNKu70- and -11 cells in 3-day colonies trypsinized 6 h after irradiation were 1.44 or 1.70 and 1.33 or 1.51 times more sensitive than the hypoxic Rat-1 cells at SF of 0.5 and 0.1, respectively. The PEs for Rat-1 and DNKu70-7 cells kept under no oxygen for 6 h were identical to those of oxic cells, but the PE for DNKu70-11 cells kept under no oxygen for 6 h decreased to 55% of that for oxic cells; thus, the aforementioned survival curve for this cell line was normalized.
Fig. 2.
Radiation dose–cell survival curves for Rat-1 (squares), DNKu70-7 (circles), and DNKu70-11 (diamonds) cells irradiated under hypoxic conditions. Cells were irradiated 3 days after plating and trypsinized immediately (0 h, solid symbols) or 6 h (open symbols) thereafter. Hypoxia had been maintained until cell trypsinization.
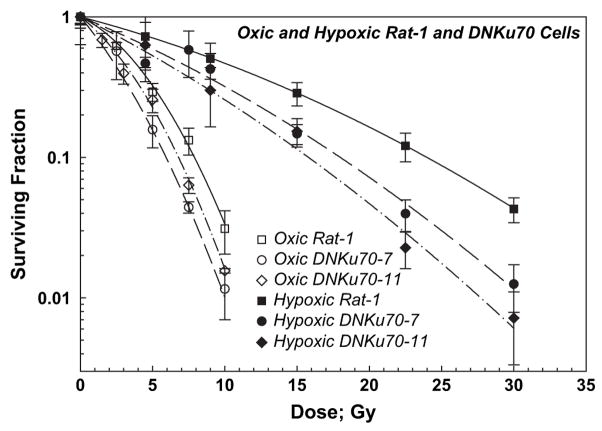
The survival curves for oxic and hypoxic Rat-1 and DNKu70 cells (3-day colonies trypsinized 6 h after irradiation) are summarized in Fig. 3 for comparison. The OERs (oxygen enhancement ratios) at SF 0.5–0.01 for DNKu70 cells trypsinized 6 h after irradiation were slightly smaller than those of corresponding Rat-1 cells and were between 1.9 and 3.0, depending on the survival level. All OER values are tabulated in Table 1.
Fig. 3.
Radiation survival curves for Rat-1 and DNKu70-7 and -11 cells irradiated under oxic and hypoxic conditions and trypsinized 6 h later, shown together for comparison. Squares, circles, and diamonds indicate Rat-1, DNKu70-7, and DNKu70-11 cells, respectively, and open and solid symbols indicate radiation given under oxic and hypoxic conditions, respectively.
Survival curves after multiple doses
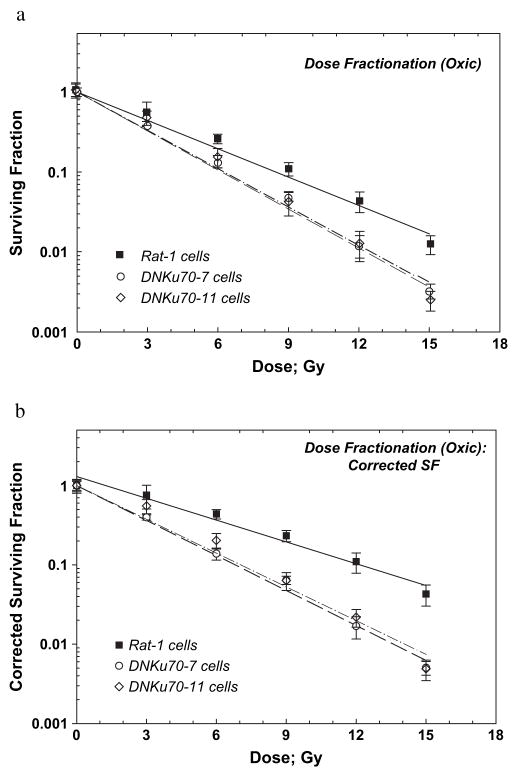
Oxic Rat-1 and DNKu70-7 and -11 cells in 3-day colonies received 0–5 doses of 3 Gy each and were trypsinized 6 h after the final dose. The survivals of these cells are plotted in Fig. 4a. The survival curves for both cells were exponential, and the D0 values for Rat-1 and DNKu70-7 and -11 cells were 3.67, 2.72, and 2.67 Gy, respectively (Table 2); that is, the DNKu70-7 and -11 cells were 1.35 and 1.37 times more sensitive than the Rat-1 cells. The enhancement ratio for DNKu70-7 was slightly smaller at SF 0.5 but greater at SF ≤0.1 than those found for cells irradiated with graded single doses (i.e., 1.28 and 1.19 at SF 0.1 and 0.01, respectively) (Tables 1 and 2). However, that for DNKu70-11 after multiple doses was greater at all survival levels tested.
Fig. 4.
Radiation dose–cell survival curves for Rat-1 (solid squares), DNKu70-7 (open circles), and DNKu70-11 (open diamonds) cells receiving 0–5 doses of 3 Gy each under oxic conditions. The first radiation dose was given 3 days after cell plating, and the interfraction interval was 6 h. Cells were trypsinized 6 h after each final dose for colony formation assay. Cell survivals are shown without correction for the change in cell numbers (a) and with correction (b).
Table 2.
D0 values, OERs, and DNKu70 ERs
| OER |
OER |
DNKu70 ER |
||||||
|---|---|---|---|---|---|---|---|---|
| Cells | D0 ± SE (Gy) | (D0 ratio) | SF 0.5 | SF 0.1 | SF 0.01 | SF 0.5 | SF 0.1 | SF 0.01 |
| Oxic Cells | ||||||||
| Rat-1 | 3.67 ± 0.12 | |||||||
| DNKu70-7 | 2.72 ± 0.06 | 1.35 | 1.35 | 1.35 | ||||
| DNKu70-11 | 2.67 ± 0.05 | 1.37 | 1.37 | 1.37 | ||||
| Rat-1, Corrected | 4.74 ± 0.92 | |||||||
| DNKu70-7, Corrected | 2.95 ± 0.09 | 2.25 | 1.79 | 1.70 | ||||
| DNKu70-11, Corrected | 2.79 ± 0.13 | 2.14 | 1.74 | 1.64 | ||||
| Hypoxic dells | ||||||||
| Rat-1 | 12.87 ± 0.88 | 3.51 | 3.51 | 3.51 | 3.51 | |||
| DNKu70-7 | 5.78 ± 0.17 | 2.13 | 2.13 | 2.13 | 2.13 | 2.23 | 2.23 | 2.23 |
| DNKu70-11 | 2.79 ± 0.09 | 1.04 | 1.04 | 1.04 | 1.04 | 4.61 | 4.61 | 4.61 |
| Rat-1, Corrected | 14.18 ± 2.24 | 3.00 | 4.31 | 3.46 | 3.24 | |||
| DNKu70-7, Corrected | 6.17 ± 0.18 | 2.09 | 2.09 | 2.09 | 2.09 | 4.51 | 2.97 | 2.64 |
| DNKu70-11, Corrected | 2.94 ± 0.10 | 1.05 | 1.05 | 1.05 | 1.05 | 9.70 | 6.21 | 5.55 |
Abbreviations as in Table 1.
Data obtained from survival curves after fractionated doses for Rat-1 and DNKu70 cells irradiated under oxic and hypoxic conditions.
Corrected survivals (survivals corrected for cell proliferation or loss) for Rat-1 cells showed a D0 of 4.74 Gy and an extrapolation number of 1.30 (Fig. 4b). This D0 was larger than that for uncorrected survival curve for the same cells, indicating that Rat-1 cells proliferated during the course of fractionated doses. The D0 values for DNKu70-7 and -11 cells were 2.95 and 2.79 Gy, respectively, only slightly larger than those of the uncorrected survival curve of the same cells, suggesting limited cell proliferation during the fractionation treatment. Corrected survival curve showed that the DNKu70-7 or -11 cells were 2.25 or 2.14 and 1.79 or 1.74 times more sensitive than the Rat-1 cells at SF of 0.5 and 0.1, respectively.
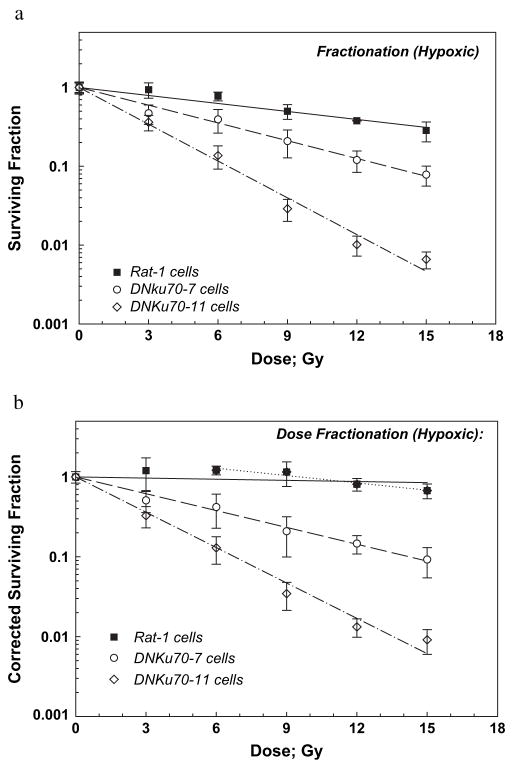
Survivals of hypoxic Rat-1 and DNKu70 cells are shown in Fig. 5a. Three-day colonies were made hypoxic, received 0–5 doses of 3 Gy each, and were trypsinized 6 h after the final dose. Hypoxia was kept until cell trypsinization. Similar to oxic cells, these survival curves were exponential, and D0 values for Rat-1 and DNKu70-7 and -11 cells were 12.87, 5.78, and 2.79 Gy, respectively; that is, DNKu70-7 and -11 cells were 2.23 and 4.61 times, respectively, more sensitive than the Rat-1 cells. Notably, enhancement ratios for DNKu70-7 or -11 cells after graded single doses were 1.44 or 1.70 and 1.33 or 1.51 at SF of 0.5 and 0.1, respectively (Table 1). The OERs for multiple doses calculated as the D0 ratio [D0 (oxic)/D0 (hypoxic)] were 3.51, 2.13 and 1.04 for Rat-1, DNKu70-7 and -11 cells, respectively (Table 2). It was noticed that the PE of DNKu70-11 cells decreased with time, but SFs were not normalized. This will be discussed later, together with the extremely low OER values for this cell line.
Fig. 5.
Radiation dose–cell survival curves for Rat-1 (solid squares), DNKu70-7 (open circles), and DNKu70-11 (open diamonds) cells receiving 0–5 doses of 3 Gy each under hypoxic conditions. Cells were made hypoxic 3 days after cell plating, and the first radiation dose was given approximately 10 min thereafter. The interfraction interval was 6 h. Cells were kept hypoxic until trypsinization 6 h after each final dose for colony formation assay. Cell survivals are shown without correction for the change in cell numbers (a) and with correction (b).
Corrected survivals of these cells are plotted in Fig. 5b. The corrected SF for Rat-1 cells slightly increased until a total dose of 9 Gy and then decreased slightly. An exponential regression line was almost flat, with a D0 of 90.9 Gy, and the correlation coefficient was 0.20; thus, alternative regression was fitted between 6 and 15 Gy, with a resultant D0 of 14.18 Gy, an extrapolation number of 1.96, and a correlation coefficient of 0.93. These regression analyses indicated that Rat-1 cells proliferated under no oxygen regardless of repeated 3-Gy doses, whereas each 3 Gy could reduce SF to 0.84 if the hypoxic cell survival curve shown in Fig. 2 is applied.
Corrected survival curve for DNKu70-7 and -11 showed D0 of 6.17 and 2.94 Gy, respectively, slightly larger than the D0 of 5.78 and 2.79 Gy of the uncorrected survival curves, suggesting that cell proliferation was negligible or compensated with cell death caused by hypoxia. On the basis of the corrected survival curves, DNKu70-7 or -11 cells were 4.51 or 9.70 and 2.97 or 6.21 times more sensitive than Rat-1 cells at SF of 0.5 and 0.1, respectively. The OERs based on corrected survivals for Rat-1 cells were 4.31 and 3.46 at SF of 0.5 and 0.1, respectively, whereas the OERs for DNKu70-7 and -11 cells were 2.09 and 1.05 (ratio of 2 D0s), respectively, and independent of the survival level.
DISCUSSION
The present study has shown that DNKu70 cells, rat fibroblasts over-expressing DNKu70, are more sensitive to radiation given under both oxic and hypoxic conditions than parental Rat-1 cells, and the radiation dose–cell survival curves for both cell lines were well fitted by the linear-quadratic model. This enhanced radiation sensitivity due to the over-expression of DNKu70 seemed to be greater at low dose levels than at high dose levels (Table 1, DNKu70 ER at SF 0.5–0.01). In the extreme situation whereby none of the damage is repaired, the radiation dose–cell survival curve could be exponential. However, this type of survival curve was not observed, suggesting that not all radiation-induced damage has been repaired.
Interestingly, oxic Rat-1 cells trypsinized immediately after irradiation were more sensitive than those trypsinized 6 h after irradiation, whereas oxic DNKu70 cells trypsinized either immediately and 6 h after irradiation showed identical survival curves. Three days after plating, both cells were still in exponential growth phase, but we observed that a majority of colonies contained more than 30 cells. This suggests that cells in large colonies could repair potentially lethal damage (PLD) (23, 24), and the difference in sensitivity of cells trypsinized immediately and 6 h after irradiation could be due to this repair. In other words, Rat-1 cells were able to repair PLD, but the DNKu70 cells were unable to repair PLD like cells irradiated with high linear energy transfer (LET) radiation (25, 26). These results suggest that Ku70 could be an essential protein for PLD repair. If the PLD is also the DNA-DSB (27), the involvement of Ku70 protein in PLD repair may be reasonable. Because of this difference, we have chosen to trypsinize cells 6 h after irradiation in fractionation experiments and, in this sense, single-dose survival curves of 3-day colonies trypsinized 6 h after irradiation are appeared to be the most meaningful for analysis.
We also studied survival curves of cells irradiated under hypoxic conditions and trypsinized either immediately and 6 h after irradiation. Both survival curves for each cell line were nearly identical (Fig. 2), suggesting hypoxic cells’ inability to repair PLD. However, the α/β ratios for both cells trypsinized 6 h after irradiation were larger than those for cells trypsinized immediately after irradiation (Table 1).
Results of the α/β ratio were mixed. Both oxic and hypoxic DNKu70 cells in 3-day colonies trypsinized 6 h after irradiation showed greater α/β ratio than Rat-1 cells treated identically. However, oxic DNKu70 cells in 3-day colonies trypsinized immediately after irradiation showed rather smaller α/β ratio than identically treated Rat-1 cells, whereas hypoxic DNKu70 cells trypsinized immediately after irradiation showed either greater (DNKu70-11) or smaller (DNKu70-7) α/β ratio than Rat-1 cells treated identically.
Previous study has shown that DNKu70 expression inhibits DNA-DSB repair (22). However, to date we do not understand how DNKu70 expression affects the α/β ratio of the radiation survival curve. One could surmise that, if the β component (but not the α component) is related to the DNA-DSB repair, the α/β ratio of DNKu70 cells should be greater than that of parental Rat-1 cells. Our analysis of the radiation survival curves of DNKu70 and parental cells provide the first results on this topic. Notably, late-responding normal tissue with large repair capability is characterized by a small α/β ratio, whereas early-responding normal tissue with small repair capability is characterized by a large α/β ratio (28). As we discussed above, if we focus on the survival curve of cells trypsinized 6 h after irradiation, an increase in the α/β ratio of both oxic and hypoxic DNKu70 cells is due to an increase in the α values rather than a decrease in the β values (Table 1). These increased α values may suggest an increase in non-repairable damage in DNKu70 cells, or may reflect the absence or unavailability of the repair proteins in the vicinity of radiation-induced DNA damages, although more detailed experiments are needed for a definite conclusion. Increased α and α/β values are also reported for antisense Ku70 transfected human lung cancer cell lines (29).
Results of fractionated doses showed that DNKu70 cells were consistently more sensitive to radiation than Rat-1 cells, and the uncorrected survival curves for all cell lines irradiated under oxic and hypoxic conditions were exponential (Figs. 4a and 5a). DNKu70-7 or -11 cells were 1.35 or 1.37 and 2.23 or 4.61 times more sensitive than Rat-1 cells when irradiated under oxic and hypoxic conditions, respectively (Table 2). This indicates that DNKu70 expression enhanced response to multiple doses more substantially under hypoxic than oxic conditions. However, this difference may not be due to the differential enhancement of DNKu70 between oxic and hypoxic cells. Comparison between corrected and uncorrected survival curves suggests that this differential enhancement is likely due to the difference in cell proliferation and hypoxia-induced cell death between DNKu70 cells and Rat-1 cells.
The corrected survival curves for both oxic and hypoxic Rat-1 cells were less steep than uncorrected survival curves, indicating that Rat-1 cells proliferated throughout the treatment period. The interval between doses was set for 6 h in the present study for two reasons. First, PLD repair is completed within 6 h after irradiation (30, 31). Second, the doubling time of cells growing in vitro is usually shorter than that of cells growing in both animal and human tumors. During these short intervals, cells proliferated. The SF of oxic Rat-1 cells after a total dose of 15 Gy was 0.0168 in uncorrected survival curve and 0.0550 in corrected survival curve; that is, the number of cells increased by a factor of approximately 3.3 (0.0550/0.0168) during the total treatment period of 30 h. The SFs of oxic DNKu70-7 or -11 cells after a total dose of 15 Gy were 0.00403 or 0.00363 and 0.00624 or 0.00743 in uncorrected and corrected survival curves, respectively; that is, the number of DNKu70-7 or -11 cells increased by a factor of approximately 1.5 or 2.0, respectively.
The SFs of hypoxic Rat-1 cells after a total of 15 Gy were 0.312 and 0.681 in uncorrected and corrected survival curves, respectively, indicating an increase in the cell number by a factor of approximately 2.2 (0.681/0.312) under no oxygen. On the other hand, the SFs of hypoxic DNKu70-7 or -11 cells after the same total dose were 0.0746 or 0.00464 and 0.0878 or 0.00611 in uncorrected and corrected survival curves, respectively. This suggests that the number of DNKu70-7 or -11 cells increased only by a factor of approximately 1.2 or 1.3, much smaller compared with Rat-1 cells. These results indicate that cell proliferation under hypoxic conditions was less substantial than under oxic conditions; nevertheless, a 3-Gy dose given under hypoxia induced less cell lethality compared with the same dose given under oxic conditions.
It has been reported that cells kept under no oxygen for a prolonged period failed to survive (32, 33). Our preliminary experiments indicate that both Rat-1 and DNKu70 cells can proliferate at least for 24 h after placing them under no oxygen, but the survival of these cells decreases with time. Uncorrected survivals of Rat-1 and DNKu70-7 and -11 cells kept under no oxygen for 24 h were approximately 80%, 60%, and 15%, respectively. The present results together with these preliminary data suggest an involvement of Ku70/Ku80 heterodimers in cell proliferation and cell survival in a nonphysiologic environment. The role of Ku70 in cell proliferation has been discussed (12, 34), but further studies are undoubtedly needed. Furthermore, the repair of radiation damage is known to be less extensive in cells irradiated under hypoxia (35, 36). It is highly predictable that the repair inhibitory effect of DNKu70 might have been enhanced under prolonged hypoxia. Accordingly, effects of DNKu70 expression under hypoxic conditions, such as reduced repair, reduced cell proliferation, and increased cell death, might have contributed to the increased enhancement of radiation response of hypoxic DNKu70 cells.
It is well known that the OER depends on, for example, oxygen concentrations, cell lines, and survival levels. The present study suggests that OER value is dynamic. The value is influenced by factors that occur during the course of fractionated treatment, such as cell proliferation and cell death. That of DNKu70-7 cells decreased to 2.1 and that of DNKu70-11 cells to a surprising value of 1.0 because of DNKu70-enhanced cell kill under hypoxic conditions. On the other hands, the OER of Rat-1 cells increased to 3.5 because of cell proliferation. The small OER for DNKu70-11 cells after single doses may also contribute to small OER after multiple doses, although the cause of the small OER after single doses is unknown.
Gene-radiotherapy has been studied by many research groups (22, 37–42). A major objective of this therapy is to enhance the radiation response of malignant cells by transfecting gene(s) that are involved in radiosensitization or repair inhibition of radiation-induced damage (22, 39–41). Other gene-radiotherapy projects may aim to obtain an additive effect, such as enhancing host-immunity, increasing the effect of chemotherapeutic agents, or decreasing tumorigenicity of cancer cells (37, 38, 40, 42).
Our research project is to investigate the feasibility of using adenovirus-mediated expression of the DNKu70 fragment in gene-radiotherapy. The present study showed that over-expression of DNKu70 can enhance radiation response and that this enhancement can be obtained after a small radiation dose and repeatedly obtained after multiple small doses, such as 3 Gy per fraction. An encouraging result may be that this enhancement was stronger under hypoxic than under oxic conditions. Furthermore, the over-expression of DNKu70 seemed to inhibit PLD repair in addition to its inhibitory effect on the DNA-DSB repair and to enhance hypoxia-induced cell kill. These results not only justify but also encourage the use of adenovirus-mediated expression of the DNKu70 fragment antisense Ku70 in gene-radiotherapy.
Footnotes
Conflict of interest: none.
References
- 1.Collis SJ, DeWeese TL. Enhanced radiation response through directed molecular targeting approaches. Cancer Metastasis Rev. 2004;23:277–292. doi: 10.1023/B:CANC.0000031767.30730.d1. [DOI] [PubMed] [Google Scholar]
- 2.Hoeijmakers JH. Genome maintenance mechanisms for preventing cancer. Nature. 2001;411:366–374. doi: 10.1038/35077232. [DOI] [PubMed] [Google Scholar]
- 3.Jackson SP. Detecting, signalling and repairing DNA double-strand breaks. Biochem Soc Trans. 2001;29:655–661. doi: 10.1042/0300-5127:0290655. [DOI] [PubMed] [Google Scholar]
- 4.Shiloh Y. ATM and related protein kinases: Safeguarding genome integrity. Nat Rev Cancer. 2003;3:155–168. doi: 10.1038/nrc1011. [DOI] [PubMed] [Google Scholar]
- 5.Petrini JH, Stracker TH. The cellular response to DNA double-strand breaks: Defining the sensors and mediators. Trends Cell Biol. 2003;13:458–462. doi: 10.1016/s0962-8924(03)00170-3. [DOI] [PubMed] [Google Scholar]
- 6.Valerie K, Povirk LF. Regulation and mechanisms of mammalian double-strand break repair. Oncogene. 2003;22:5792–5812. doi: 10.1038/sj.onc.1206679. [DOI] [PubMed] [Google Scholar]
- 7.Yang J, Yu Y, Hamrick HE, et al. ATM, ATR and DNA-PK: Initiators of the cellular genotoxic stress responses. Carcinogenesis. 2003;24:1571–1580. doi: 10.1093/carcin/bgg137. [DOI] [PubMed] [Google Scholar]
- 8.Iliakis G, Wang Y, Guan J, et al. DNA damage checkpoint control in cells exposed to ionizing radiation. Oncogene. 2003;22:5834–5847. doi: 10.1038/sj.onc.1206682. [DOI] [PubMed] [Google Scholar]
- 9.Featherstone C, Jackson SP. Ku, a DNA repair protein with multiple cellular functions? Mutat Res. 1999;434:3–15. doi: 10.1016/s0921-8777(99)00006-3. [DOI] [PubMed] [Google Scholar]
- 10.Tuteja R, Tuteja N. Ku autoantigen: A multifunctional DNA-binding protein. Crit Rev Biochem Mol Biol. 2000;35:1–33. doi: 10.1080/10409230091169177. [DOI] [PubMed] [Google Scholar]
- 11.Kurimasa A, Ouyang H, Dong L-J, et al. Catalytic subunit of DNA-dependent protein kinase: Impact on lymphocyte development and tumorigenesis. Proc Natl Acad Sci U S A. 1999;96:1403–1408. doi: 10.1073/pnas.96.4.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nussenzweig A, Chen C, da Costa Soares V, et al. Requirement for Ku80 in growth and immunoglobulin V(D)J recombination. Nature (London) 1996;382:551–555. doi: 10.1038/382551a0. [DOI] [PubMed] [Google Scholar]
- 13.Ouyang H, Nussenzweig A, Kurimasa A, et al. Ku70 is required for DNA repair but not for TCR gene recombination in vivo. J Exp Med. 1997;186:921–929. doi: 10.1084/jem.186.6.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li GC, He F, Shao X, et al. Adenoviral-mediated heat-activated antisense Ku70 RNA radiosensitizes tumor cells in vitro and in vivo. Cancer Res. 2003;63:3268–3274. [PubMed] [Google Scholar]
- 15.Marangoni E, Le Romancer M, Foray N, et al. Transfer of Ku86 RNA antisense decreases the radioresistance of human fibroblasts. Cancer Gene Ther. 2000;7:339–346. doi: 10.1038/sj.cgt.7700111. [DOI] [PubMed] [Google Scholar]
- 16.Jin S, Weaver DT. Double-strand break repair by Ku70 requires heterodimerization with Ku80 and DNA binding functions. EMBO J. 1997;16:6874–6885. doi: 10.1093/emboj/16.22.6874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang J, Dong X, Myung K, et al. Identification of two domains of the p70 Ku protein mediating dimerization with p80 and DNA binding. J Biol Chem. 1998;273:842–848. doi: 10.1074/jbc.273.2.842. [DOI] [PubMed] [Google Scholar]
- 18.Wang J, Dong X, Reeves WH. A model for Ku heterodimer assembly and interaction with DNA. J Biol Chem. 1998;273:31068–31074. doi: 10.1074/jbc.273.47.31068. [DOI] [PubMed] [Google Scholar]
- 19.Cary RB, Chen F, Shen Z, et al. A central region of Ku80 mediates interaction with Ku70 in vivo. Nucleic Acids Res. 1998;26:974–979. doi: 10.1093/nar/26.4.974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Z, Zhu L, Lin D, et al. The three-dimensional structure of the C-terminal DNA-binding domain of human Ku70. J Biol Chem. 2001;276:38231–38236. doi: 10.1074/jbc.M105238200. [DOI] [PubMed] [Google Scholar]
- 21.Walker JR, Corpina RA, Goldberg J. Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature (London) 2001;412:607–614. doi: 10.1038/35088000. [DOI] [PubMed] [Google Scholar]
- 22.He F, Li L, Kim D, et al. Adenovirus-mediated expression of a dominant negative Ku70 fragment radiosensitizes human glioma U-87MG cells under aerobic and hypoxic conditions. Cancer Res. 2007:634–642. doi: 10.1158/0008-5472.CAN-06-1860. [DOI] [PubMed] [Google Scholar]
- 23.Phillips RA, Tolmach LJ. Repair of potentially lethal damage in x-irradiated HeLa cells. Radiat Res. 1966;29:413–432. [PubMed] [Google Scholar]
- 24.Zininger GF, Little JB. Fractionated radiation response of human cells in stationary and exponential phases of growth. Radiology. 1973;108:423–428. doi: 10.1148/108.2.423. [DOI] [PubMed] [Google Scholar]
- 25.Rasey JS, Nelson NJ. Repair of potentially lethal damage following irradiation with X rays or cyclotron neutrons: response of the EMT-6/uw tumor system treated under various growth conditions in vitro and in vivo. Radiat Res. 1981;85:69–84. [PubMed] [Google Scholar]
- 26.Urano M, Koike S. Comparison of the effects of neutron and/or photon irradiation on spontaneous squamous-cell carcinoma in mice. Radiology. 1980;134:219–225. doi: 10.1148/radiology.134.1.6444252. [DOI] [PubMed] [Google Scholar]
- 27.Iliakis G. Radiation-induced potentially lethal damage: DNA lesions susceptible to fixation. Int J Radiat Biol Relat Stud Phys Chem Med. 1988;53:541–584. doi: 10.1080/09553008814550901. [DOI] [PubMed] [Google Scholar]
- 28.Thames HD, Jr, Withers HR, Peters LJ, et al. Changes in early and late radiation responses with altered dose fractionation: implications for dose-survival relationships. Int J Radiat Oncol Biol Phys. 1982;8:219–226. doi: 10.1016/0360-3016(82)90517-x. [DOI] [PubMed] [Google Scholar]
- 29.Omori S, Takiguchi Y, Suda A, et al. Suppression of a DNA double-strand break repair gene, Ku70, increases radio- and chemosensitivity in a human lung cancer cell lines. DNA Repair (Amst) 2002;1:299–310. doi: 10.1016/s1568-7864(02)00006-x. [DOI] [PubMed] [Google Scholar]
- 30.Hahn GM, Bagshaw MA, Evans RG, et al. Repair of potentially lethal lesions in x-irradiated, density-inhibited Chinese hamster cells: Metabolic effects and hypoxia. Radiat Res. 1973;55:280–290. [PubMed] [Google Scholar]
- 31.Urano M, Nesumi N, Ando K, et al. Repair of potentially lethal radiation damage in acute and chronically hypoxic tumor cells in vivo. Radiology. 1976;118:447–451. doi: 10.1148/118.2.447. [DOI] [PubMed] [Google Scholar]
- 32.Spiro IJ, Rice GC, Durand RE, et al. Cell killing, radiosensitization and cell cycle redistribution induced by chronic hypoxia. Int J Radiat Oncol Biol Phys. 1984;10:1275–1280. doi: 10.1016/0360-3016(84)90332-8. [DOI] [PubMed] [Google Scholar]
- 33.Durand RE, Sham E. The lifetime of hypoxic human tumor cells. Int J Radiat Oncol Biol Phys. 1998;42:711–715. doi: 10.1016/s0360-3016(98)00305-8. [DOI] [PubMed] [Google Scholar]
- 34.Lim JW, Kim H, Kim KH. Expression of Ku70 and Ku80 mediated by NF-kB and cyclooxygenase-2 is related to proliferation of human gastric cancer cells. J Biol Chem. 2002;277:46093–46100. doi: 10.1074/jbc.M206603200. [DOI] [PubMed] [Google Scholar]
- 35.Phillips TL, Hanks GE. Apparent absence of recovery in endogenous colony-forming cells after irradiation under hypoxic conditions. Radiat Res. 1968;33:517–532. [PubMed] [Google Scholar]
- 36.Hall EJ. The effect of hypoxia on the repair of sublethal radiation damage in cultured mammalian cells. Radiat Res. 1972;49:405–415. [PubMed] [Google Scholar]
- 37.Urano M, Kuroda M, Reynolds R, et al. Expression of manganese superoxide dismutase reduces tumor control radiation dose: Gene-radiotherapy. Cancer Res. 1995;55:2490–2493. [PubMed] [Google Scholar]
- 38.Quigg M, Mairs RJ, Brown SM, et al. Assessment in vitro of a novel therapeutic strategy for glioma, combining herpes simplex virus HSV1716-mediated oncolysis with gene transfer and targeted radiotherapy. Med Chem. 2005;1:423–429. doi: 10.2174/1573406054864124. [DOI] [PubMed] [Google Scholar]
- 39.Sun Y, Huang YC, Xu QZ, et al. HIV-1 Tat depresses DNA-PK(CS) expression and DNA repair, and sensitizes cells to ionizing radiation. Int J Radiat Oncol Biol Phys. 2006;65:842–850. doi: 10.1016/j.ijrobp.2006.02.040. [DOI] [PubMed] [Google Scholar]
- 40.Goblirsch M, Zwolak P, Ramnaraine ML, et al. Novel cytosine deaminase fusion gene enhances the effect of radiation on breast cancer in bone by reducing tumor burden, osteolysis, and skeletal fracture. Clin Cancer Res. 2006;12:3168–3176. doi: 10.1158/1078-0432.CCR-05-2729. [DOI] [PubMed] [Google Scholar]
- 41.Kim JH, Kim SH, Kolozsvary A, et al. Selective enhancement of radiation response of herpes simplex virus thymidine kinase transduced 9L gliosarcoma cells in vitro and in vivo by antiviral agents. Int J Radiat Oncol Biol Phys. 1995;33:861–868. doi: 10.1016/0360-3016(95)00134-9. [DOI] [PubMed] [Google Scholar]
- 42.Weichselbaum RR, Hallahan DE, Beckett MA, et al. Gene therapy targeted by radiation preferentially radiosensitizes tumor cells. Cancer Res. 1994;54:4266–4269. [PubMed] [Google Scholar]