Abstract
The N-terminal domain of Plasmodium falciparum merozoite surface protein-3 (PfMSP3) has been excluded from malaria vaccine development largely because of genetic diversity concerns. However, no study to date has followed N-terminal diversity over time. This study describes PfMSP3 variation in a hypoendemic longitudinal cohort in the Peruvian Amazon over the 2003-2006 transmission seasons. Polymerase chain reaction was used to amplify the N-terminal domain in 630 distinct P. falciparum infections, which were allele-typed by size and also screened for sequence variation using a new high-throughput technique, denaturing high performance liquid chromatography. PfMSP3 allele frequencies fluctuated significantly over the 4-year period, but sequence variation was very limited, with only 10 mutations being identified of 630 infections screened. The sequence of the PfMSP3 N-terminal domain is relatively stable over time in this setting, and further studies of its status as a vaccine candidate are therefore warranted.
INTRODUCTION
Plasmodium falciparum is responsible for between 300 and 500 million clinical episodes of malaria and between 1 and 3 million deaths annually, with ~90% of mortalities occurring in young children.1,2 With the continual threat of the emergence of drug-resistant P. falciparum strains, the need for a safe and effective malaria vaccine is more urgent than ever. Although a handful of P. falciparum vaccine candidates, such as merozoite surface protein 1 (MSP1),3-5 apical membrane antigen 1 (AMA1),6,7 and circumsporozoite protein (CSP),8-10 have been extensively studied for many years, there are many other promising vaccine candidates that have received much less attention. For many of these candidates, a pool that has radically increased with the recent rapid increase in genomic data,11 there are little, if any, supporting data, particularly regarding the candidate's sequence diversity or role in the generation of protective immunity in vivo. Given the problems of sequence diversity and poor antigenicity that can afflict even the most well-studied vaccine candidates, there is critical need for the rapid and early characterization of novel vaccine candidate potential and the adoption of high-throughput techniques to facilitate this type of analysis. It is with such concerns in mind that a collaborative malaria vaccine roadmap was recently released, which identifies key issues that need to be addressed for a vaccine candidate to effectively progress through the development pipeline (www.malariavaccineroadmap.net). With the increasing cost of vaccine clinical trials, this roadmap proposes that strict go/no-go criteria be enforced on candidate antigens (i.e., studies be conducted early that characterize the candidate's vaccine potential and either lay the groundwork for subsequent studies or remove it from the pool of viable candidates).
Plasmodium falciparum merozoite surface protein-3 (PfMSP3) is a major vaccine candidate that has not yet advanced to Phase II field trials. PfMSP3 is a highly immunogenic non-integral protein expressed on the surface of merozoites and has been suggested to be involved in erythrocyte binding, although its function remains unknown.12,13 Structurally, it possesses three blocks of heptad repeats that are proposed to form alpha-helical coil/coil domains, a hydrophobic glutamine-rich domain, and a putative leucine zipper domain at the extreme C terminus.14,15 Importantly, it is a major target of antibodies from malaria-immune African individuals, and immunization studies using full-length PfMSP3 elicited significant protection in a non-human primate model.16-18 PfMSP3 exists as two allele classes, termed K1 and 3D7 after the P. falciparum strains in which they were first identified. The majority of both intra- and interallele differences are localized to the heptad repeat region, which defines the N-terminal domain. Variation in the PfMSP3 N-terminal domain occurs largely between the blocks of heptad repeats and consists of both indels and SNPs. Conversely, the C-terminal domain, comprised of the glutamine-rich region and the leucine zipper motif, is almost entirely conserved.19 Although the presence of variation in the PfMSP3 N terminus has been established for some time using laboratory-adapted P. falciparum isolates,19 only one study has previously looked at natural variation in PfMSP3 sequences, with 48 and 50 PfMSP3 genotypes sequenced from samples in Nigeria and Thailand, respectively.20 PfMSP3 is therefore a strong vaccine candidate with limited epidemiologic data; data that are needed to support its continued development along the proposed malaria vaccine roadmap.
This study investigates genetic variation in the PfMSP3 N-terminal domain in samples collected from the Malaria Immunology and Genetics in the Amazon (MIGIA) cohort study, a longitudinal study of P. falciparum transmission in Zungarococha, which is a cluster of four villages located in the Peruvian Amazon. In this community, P. falciparum transmission is hypoendemic, with inoculation rates of < 0.04 infections per person per month during the 7-month transmission season, with most infections consisting of a single genetic type.21 Although transmission rates are low, there is still a significant level of overall genetic diversity, with at least five different genetic types defined for PfMSP1 Block 2 (P Sutton and OH Branch, unpublished data).
To date, no study has yet looked at the extent of PfMSP3 variation in hypoendemic settings such as in South America. Such data are important because the increased frequency of simple infections in such a setting enables us to look at changes in allele frequency over time, which might provide evidence for or against the presence of allele- and variant-specific immune responses. To characterize genetic variation within PfMSP3 in a hypoendemic setting, we initiated a retrospective study using blood samples obtained from P. falciparum-infected individuals enrolled in the MIGIA cohort study. To significantly increase our power and enable us to screen hundreds of samples, we adapted the recently developed high throughput denaturing high performance liquid chromatography (dHPLC) genotyping technology to screen for PfMSP3 variation.22-25 The existence of extensive clinical data generated by the MIGIA study also allowed us to test for correlates of protection with specific PfMSP3 genotypes. This study is therefore a comprehensive analysis of PfMSP3 genotypes across multiple transmission seasons and is an important window into the genetic diversity dynamics of this important vaccine candidate in a hypoendemic setting.
MATERIALS AND METHODS
Study location and malaria transmission
A detailed description of the four villages that comprise the longitudinal cohort study has been previously published.21 Briefly, the study enrolled residents from four villages of the Zungarococha community: Zungarococha village, Puerto Almendra, Ninarumi, and Llanchama. The community is located South of Iquitos in the Peruvian Amazon and is supported by the Nanay River, a tributary of the Amazon River. The village of Ninarumi contains a port that brings in boats from outside the community. The village residents have homogenous housing construction, income levels, and access to healthcare, provided by the MIGIA cohort physicians. The women generally work in or near their home, and the majority of men work as either community fishermen or in local agriculture. Importantly, travel outside the community is rare, with the most frequent travel being to the city of Iquitos, where malaria transmission is nonexistent.
The Zungarococha community was chosen as the focus of the MIGIA cohort because of the presence of continuing stable hypoendemic transmission of both P. vivax and P. falciparum, introduced during an epidemic in 1992. The annual malaria season lasts 7 months and closely follows the rainy season, which occurs between January and July. The major vector of malaria transmission is Anopheles darlingi, an anthropophilic species that has proliferated in the area since the early 1990s.26
Sample collection and extraction
All individuals enrolled in the MIGIA study have access to healthcare provided at a local health post located in Zungarococha village. On presentation to the health post, symptomatic individuals were tested for malaria parasites by Geimsa-stained microscopy. If positive, and consent was given, patients submitted a 0.5-mL blood sample by finger venipuncture and underwent a comprehensive medical evaluation. PCR verification of the microscopy result was subsequently performed using species-specific primers. Plasmodium infection was immediately treated with a combination of mefloquine (0.5 mg/kg for 7 days) and artesunate (4 mg/kg daily for 3 days).
Plasmodium falciparum infections were considered asymptomatic if the medical history and physical exam showed no malaria-related signs or symptoms, and the patient had not presented to the clinic. Clinical presentation or symptomatic infections identified in the community were labeled symptomatic infections.
The submitted blood sample was separated into a serum and packed red blood cell (RBC) fraction by centrifugation, and each was cataloged and stored at -80°C. P. falciparum DNA was extracted using a commercially available kit (Qiagen) and stored at -80°C until needed.
Only P. falciparum infections were included in this study, which spanned 4 years, from 2003 to 2006. Samples were chosen at random, but any infection occurring within 60 days of the initial infection date was excluded to minimize the risk of parasite recrudescence caused by incomplete drug clearance.
Nested PCR and allele-typing
Nested PCR was used to amplify the N-terminal region of PfMSP3 using the external PCR primers 5′-ATAATGTTGCTAGTAAAGAAATTG-3′ and 5′-AATACATCATCATTTTCCTTAG-3′ and the internal primers 5′-ATAATCTTAACTTAAGAAATGC-3′ and 5′-CGGCGGGGGCGATAAGCATTTTTTGCC-3′. The bold annotation identifies the CG clamp, which was added to the 5′ end of the internal reverse primer to facilitate the subsequent analysis by dHPLC. P. falciparum- infected patient extracted DNA (1.5 μL) was amplified using ChoiceTaq (Denville, South Plainfield, NJ) and underwent 35 cycles of 95°C for 30 seconds, 50°C for 30 seconds, and 65°C for 1 minute, followed by a final elongation step of 5 minutes at 65°C. The nested PCR reaction involved the same conditions except that 1 μL of the external PCR reaction was used as the template. Classification of the PfMSP3 allele was performed by size migration using 2% aga-rose gel electrophoresis supplemented with ethidium bromide.
dHPLC and sequencing
dHPLC analysis was performed using a WAVE column (Transgenomics, Omaha, NE) and standard protocols. Before screening, optimal WAVE temperatures for each PfMSP3 allele were determined using Wavemaker software (Transgenomics). The nested PCR mixture (2 μL) was diluted 1:10 in ddH20 and submitted for dHPLC analysis. The submitted sample was mixed with a control sequence from the homologous allele, and heterologous strand formation was performed by denaturation and subsequent reannealing of the mixture. The sample was bound to the WAVE column and eluted using a linear acetonitrile gradient. The presence of a second elution peak on the elution chromatograph was indicative of a mutation, and all detected mutants were immediately sequenced to identify the mutation. As positive controls, 3D7 and Borneo were complexed with the Peruvian alleles HB3 and K1, respectively. As an additional quality control step and to define the sensitivity and specificity of the technique, 10% of all samples that exhibited no mutations by dHPLC were randomly sequenced.
Ethics committee approval
This study was approved by review boards of the University of Alabama at Birmingham, Universidad Peruana Cayetano Heredia, and the Peruvian Ministerio de Salud, Instituto Naccional de Salud. Written consent was obtained from all participants before study enrollment.
Statistical analysis
Descriptive statistics, such as frequencies, percentages, means, and SDs, were used to summarize all study variables of interest. Comparisons between proportions of alleles (K1 and 3D7) for communities, mutant, sex, symptom status, subsequent allele, subsequent P. falciparum infections, year of infection, age group (< 15 and ≥ 15 years of age), time group to next P. falciparum infection (0-6 and ≥ 6 months), and comparisons between all other proportions of interest were performed using the two-group cχ2 test or Fisher exact test when the assumptions on the χ2 were not tenable. Comparisons between means of actual age and days to next P. falciparum infection were performed using the usual two-group t test or the two-group t test for unequal variances when needed. All statistical tests were two-sided and were performed using a 5% significance level (i.e., α = 0.05). SAS software (version 9.1.3; SAS Institute, Cary, NC) was used to perform all statistical analyses.
RESULTS
PfMSP3 allele frequency varies significantly between subsequent years and between villages
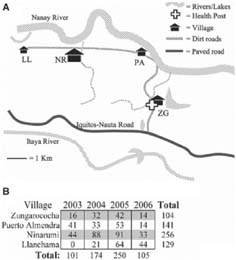
The Zungarococha community consists of four villages each ~2-3 km apart and supported by tributaries of the Amazon River: Zungarococha village (ZG, N = 805), Puerto Almendra (PA, N = 272), Ninarumi (NR, N = 590), and Llanchama (LL, N = 203) (Figure 1A). P. falciparum transmission is hypoendemic at this study site, transmitted largely by the anthropophilic vector An. darlingi. P. falciparum transmission is seasonal, occurring largely between January and August of each year, with entomologic bite rates described as high as 10-24 bites/person/h between April and July. This corresponds to an entomological inoculation rate (EIR) of 0.13 infections/person/mo during the 7-month transmission season.21
FIGURE 1.
P. falciparum transmission in Zungarococha from 2003 to 2006. A, Schema of Zungarococha, a small community of four villages fed by the Nanay and Itaya river systems: Zungarococha village (ZG, N = 805), Puerto Almendra (PA, N = 272), Ninarumi (NR, N = 590), and Llanchama (LL, N = 203). The community health post is located in Zungarococha village. B, Zungarococha community P. falciparum infection demographics from 2003 to 2006. A total of 630 P. falciparum infections, spanning all villages, were analyzed in this study.
A total of 630 P. falciparum infections were randomly chosen for inclusion in this study: 101 from 2003, 174 from 2004, 250 from 2005, and 105 from 2006 (Figure 1B). Samples were chosen to give the widest possible distribution across all four villages, although numbers for each village were not equal because the villages do not carry an equal burden of infections: although Zungarococha village is the largest village in the community, it had the least number of infections included in this study. The highest number of infections sampled was from Ninarumi, the second largest village.
Nested PCR was used to amplify and genotype PfMSP3 from P. falciparum-infected individuals. The primers amplified the PfMSP3 N-terminal domain, where the majority of genetic diversity has been shown to occur19 (nucleotides 117-507 in the 3D7 strain, excluding primer sequence) and the allele class identified using agarose gel electrophoresis. Size differences confirmed that both 3D7 and K1 allele classes of PfMSP3 are present in the Zungarococha community (Figure 2A), and eight samples from each allele classes were sequenced to identify the strain. The sequence of the smaller, 3D7 allele class samples were all identical to the published PfMSP3 HB3 reference strain (GenBank accession no. AF001137), whereas the larger, K1 allele class samples were all identical to PfMSP3 from the K1 reference strain (GenBank accession no. AF001151).
FIGURE 2.
PfMSP3 allelic diversity in the Zungarococha community. A, Agarose gel of nested PCR amplicons from P. falciparum DNA extracted from eight infected Peruvian individuals shows both PfMSP3 K1 (top arrow) and 3D7 (bottom arrow) allele classes are present in the Zungarococha community. B, PfMSP3 allele class frequency exhibits statistically significant fluctuations, by χ2, between subsequent years. *P < 0.001. PfMSP3 allele classes are significantly associated with community (C), with K1 infections being significantly elevated and decreased in Puerto Almendra and Llanchama, respectively (D).
Genotyping all 630 samples based on size established that, between 2003 and 2006, a significant majority, 570 (90.3%), of infections were of the 3D7 allele class, and only 57 infections were of the K1 allele class. Only three infections were complex infections, containing both allele classes, in keeping with the generally “simple” infections observed in hypoendemic transmission areas. Interestingly, the allele frequency distribution fluctuated significantly between subsequent years, with the K1 allele class frequency exhibited a decreasing trend; K1 class frequencies ranged from 19.1% in 2004 to 1.0% in 2006 (Figure 2B). The change in allele class frequency between 2004 and 2005 was statistically significant by χ2 analysis (P < 0.001). Additionally, in comparison with the 2003 allele frequency, the decrease in K1 class frequency in both 2005 and 2006 was also statistically significant (P < 0.001).
To assess if any correlation might be identified that could explain the significant allele class frequency variation, we looked at the village level for any correlation that exists between PfMSP3 allele and age, sex, community, patient symptom status, and subsequent infection. As shown in Figure 2C, only community was significantly associated with PfMSP3 allele class status. Interestingly, Puerto Almendra showed a significant increase in K1 class infections (> 3-fold, P < 0.0001) compared with the other villages. Additionally, Llanchama, the smallest of the villages, was associated with a significant decrease (> 16-fold, P = 0.002) in K1 class infections compared with the other villages, with only a single K1 class infection being detected across all four transmission season.
dHPLC can detect single nucleotide differences in PfMSP3 sequence
Because of the large number of samples allele-typed in this study, we adopted a high-throughput approach to screen for single nucleotide polymorphisms (SNPs) and indels within each amplicon: dHPLC. To our knowledge, this is the first time that dHPLC has been used to screen for SNPs in P. falciparum genes. As described in the Materials and Methods section, the technique detects mutations through the formation of heteroduplex bubbles. Briefly, a test amplicon is denatured and reannealed in the presence of a reference amplicon of known sequence. If the test and reference sequences are identical, only homoduplexes form, but if there are SNPs or indels in the test sequence that are not found in the reference sequence, heteroduplexes will form. The reannealed fragments are bound to a WAVE column (Transgenomics) and eluted with a linear acetonitrile gradient. Homoduplexes elute from the column as a single peak. However, if a heteroduplex bubble is present, it will elute at a different acetonitrile concentration and therefore form a second peak distinct from the homoduplex peak. Any samples that elute as two peaks from the WAVE column would be sequenced to confirm the presence and identify of the SNP(s) and/or indels. dHPLC is therefore a rapid method to screen for deviations from a reference sequence, and because the cost is significantly lower (10% or less) than sequencing, dHPLC is of particular use when the expected SNP frequency is low—it allows the user to rapidly screen large numbers of samples and only sequence those that are likely to contain SNPs.
We initially tested the sensitivity of dHPLC using PfMSP3-specific primers containing a 3′-CG clamp by mixing the HB3 allele either with itself or the same fragment amplified from genomic DNA from the 3D7 strain. Both HB3 and 3D7 are of the same allele class, but 3D7 has previously been shown to contain a single SNP relative to HB3. dHPLC analysis showed a single peak when HB3 was reannealed only to itself but two elution peaks when 3D7 and HB3 alleles were reannealed (Figure 3A). Sequencing the 3D7 amplicon confirmed the single SNP, showing that dHPLC has the sensitivity to detect single nucleotide changes in PfMSP3. Mixing K1 sequences with those from a different K1-class allele, genomic DNA from the Borneo P. falciparum strain (a kind gift from John Barnwell) also showed the presence of a heteroduplex. Sequencing the Borneo fragment showed that K1 and Borneo differ by 37 nucleotides, establishing a range of detection for the PfMSP3-specific dHPLC assay of 1-37 nucleotide differences. The sensitivity of dHPLC withstood wide changes in either reference or unknown amplicon concentrations (data not shown) as has been previously noted, increasing its utility with field P. falciparum samples where DNA concentrations can vary.
FIGURE 3.
PfMSP3 sequence variation in the Zungarococha community. A, dHPLC elution chromatograms show that adapting dHPLC to screen for PfMSP3 variation can detect sequence differences of between 1 and 37 nucleotide differences, as evident by the secondary elution peak (arrows). B, Screening 630 distinct P. falciparum infections yielded only 10 mutations in PfMSP3. A total of 86 dHPLC-negative samples were sequenced to ensure adequate sensitivity.
PfMSP3 coil/coil domain sequence variation is limited
To characterize the level of sequence variation within the N-terminal domain of PfMSP3, all 630 samples that had been allele-typed were subjected to dHPLC analysis. All 3D7 allele class samples were mixed with HB3 sequences as a reference, because, as mentioned above, initial sequencing of eight 3D7 class alleles showed all were identical in sequence to the HB3 allele. K1 allele class samples were mixed with K1 sequences as a reference, again because our initial sequencing had identified this as the predominant K1 class allele. Of the 630 samples screened, dHPLC detected heteroduplexes in 14 samples, all in the 3D7 allele class. All 14 samples were sequenced: 3 were false positives with no detectable mutation, whereas 11 samples had mutations that were different to the HB3 reference sequence (10 true mutations and 1 mutation that had double overlapping peaks at one position on the sequencing chromatogram and therefore presumably resulted from Taq polymerase error). Eighty-six samples that were mutation negative by dHPLC were also submitted to direct sequencing to test for sensitivity, and all 86 samples were found to be identical to the reference sequence, as predicted by dHPLC. We therefore estimate that using dHPLC to screen for PfMSP3 variation has a sensitivity of 100% and a specificity of 96.6%. Sensitivity is calculated by the equation A/(A + C), where A is the number of true positives in the dHPLC-mutant positive group (11) and C is the number of true positives in the dHPLC-mutant negative group (0). Specificity is calculated by the equation D/(B + D), where D is the number of true negatives in the dHPLC-mutant negative group (86) and B is the number of true negatives in the dHPLC-mutant positive group (3).
In 630 samples screened, we therefore found SNPs in only 10 samples (Figure 3B), all of which were 3D7-class alleles. Direct sequencing of these 10 mutants established that all were identical and contained the same SNP: a single C→T nucleotide substitution at position 203, which resulted in a non-synonymous mutation L68S. Interestingly, this is the SNP that differentiates the HB3 and 3D7 PfMSP3 sequences. All 10 samples were collected during the 2003 transmission season, but they were distributed widely across the community: 3 occurred in Zungarococha village, 5 in Ninarumi, and 2 in Puerto Almendra. No significant correlation was detected between these samples and age, sex, symptom status, or village (data not shown). To rule out the possibility that the detected mutations were caused by the introduction of a completely new strain into the Zungarococha community, another polymorphic loci, PfMSP2, was amplified and sequenced from 6 of these 10 samples. Three of the PfMSP2 genes were identical to the HB3 PfMSP2, but three PfMSP2 genes were significantly different (data not shown), suggesting that not all PfMSP3 mutant strains shared the same origin.
Assessing for evidence of protection for PfMSP3 alleles
The longitudinal MIGIA cohort study uses active case detection in addition to passive case detection. This allows for the identification of not only symptomatic infections but also asymptomatic infections that remain undiagnosed in the absence of active detection. Because premunition is characterized by the absence of malaria symptoms in combination with an inability to clear P. falciparum parasites, asymptomatic infections in Peruvian individuals might indicate protection against P. falciparum. We therefore compared our PfMSP3 dHPLC genotyping data, which identified all infections as either HB3, 3D7, or K1 alleles, with the available epidemiologic data to analyze whether there was any propensity for a specific allele type to be associated with a specific clinical outcome.
Because of the hypoendemic transmission and low rate of mixed infections at this study site, it is possible to follow subsequent infections in single individuals that are separated by time. We therefore assessed whether the PfMSP3 allele frequency in these subsequent infections in the same individual was any different to the population level allele frequency. If there is strong allele-specific immunity to the PfMSP3 N-terminal domain, one would expect that allele frequencies would change in subsequent infections—that is, an HB3 infection would be more likely to be followed by a K1 infection or vice versa. There were 106 successive infection pairs (individuals with two P. falciparum infections in a < 500-day period) in the 630 infections for which we had genotyped PfMSP3. Comparing the genotypes for the first and second infections in these individuals, we found no statistically significant deviation from whole population allele frequencies— that is, there was no trend for an infection of one allele to be followed by an infection of another allele, as would be expected if protection was largely allele specific (Table 1A; P = 0.93). Additionally, we tested for the possibility that infection with either allele class would lead to a decrease in subsequent P. falciparum infections, but this relationship was not significant (P =0.17).
Table 1.
Assessing for allele-specific protection in the Zungarococha community: (A) assessing for an association between PfMSP3 allele and subsequent allele, the potential to experience a subsequent P. falciparum infection, and the time-to-next-infection failed to identify a significant association; and (B) comparison of PfMSP3 allele-typed infection and subsequent infection symptom status
| A | |||
|---|---|---|---|
| HB3 | K1 | P | |
| Subsequent allele | 0.9309 | ||
| HB3 | 86 | 13 | |
| K1 | 6 | 1 | |
| Subsequent Pf infection | 0.1651 | ||
| No | 412 | 36 | |
| Yes | 161 | 21 | |
| Next infection (days) | 0.5179 | ||
| Mean | 263 | 237 | |
| N | 73 | 12 | |
| B | ||||
|---|---|---|---|---|
| Asym. | Sym. | %Asym | P | |
| Heterologous | 0.4335 | |||
| HB3:HB3 | 26 | 42 | 38.2% | |
| K1:K1 | 0 | 1 | 0.0% | |
| Homologous | 0.9126 | |||
| HB3:K1 | 1 | 2 | 33.3% | |
| K1:K1 | 3 | 7 | 30.0% | |
| Total Homologous | 26 | 42 | 38.2% | |
| Total Heterologous | 4 | 9 | 30.8% | 0.6095 |
No significant association was found that suggested any shift in the asymptomatic rate.
The propensity of individuals to experience another P. falciparum infection within a 6-month period was also used to assess for possible protection, as well as assessing for an increase in the “time to next infection.” Because the infection rate has remained relatively steady at 0.13 infections/person/mo during the 7-month transmission season,21 any significant decrease in infection rates in individuals infected with either allele would be suggestive of immune-mediated protection. As shown in Table 1A, no significant delay in time-to-next-infection was detected for either homologous or heterologous allele infections. There was, however, a non-significant trend that K1-infected individuals had a decreased “time to next infection” value (on average 26 days earlier, P > 0.05). However, because we showed above that the K1 allele class frequency is significantly associated with community, any difference is most likely caused by transmission differences between the villages.
We also tested for any association that might exist between the presence of either PfMSP3 allele and the rate of asymptomatic infections in subsequent infections. If the asymptomatic rate is higher in subsequent infections with the homologous antigen (i.e., subsequent HB3 infection after a primary HB3 infection), immune selection might be a significant factor in mediating this phenomenon. The rate of asymptomatic infections in subsequent heterologous infection is also immensely informative because, if rates are similar to homologous infections, it suggests some degree of cross-protection. Conversely, a significant increase in asymptomatic infections found in homologous infections but absent in asymptomatic heterologous infections would suggest a significant strain-specific immune response. Compared with primary infections, no statistical significant difference in HB3 or K1 asymptomatic rates was detected for either homologous or heterologous subsequent infections (Table 1B). Even after pooling the infections into homologous/heterologous subsequent infections, we were unable to detect a significant increase in asymptomatic infections for either group.
DISCUSSION
In this study, we described the use of dHPLC to screen for variation within the malaria vaccine candidate PfMSP3 in a hypoendemic setting. Because dHPLC has been shown to exhibit a sensitivity and specificity of up to 100% in some tests, possesses a high-throughput capacity, and is considerably more cost-efficient compared with direct sequencing, we decided to adapt it to screen for PfMSP3 N-terminal domain variation. Even with our resequencing of both mutation positive and negative samples for specificity and sensitivity analysis, our overall costs were only 25% of the cost of direct sequencing all 630 isolates. We therefore believe that dHPLC is an effective high-throughput approach for P. falciparum genotyping, particularly in hypoendemic transmission regions where genetic variation is suspected to be low. Although we applied dHPLC here to study a vaccine candidate antigen, another obvious application would be for drug resistance screening, where SNPs associated with drug resistance in PfCRT, PfDHFR, and PfDHPS are well established.
Using nested PCR, we allele typed 630 individual P. falciparum infections and found that both 3D7 and K1 alleles classes of PfMSP3 were present in the Zungarococha community. Direct sequencing of those samples identified 3D7 class alleles as identical to the previously published sequences for HB3 and K1 class alleles as identical to the K1 sequence itself. Interestingly, 3D7 class alleles were dominant throughout the community and, over time, became increasingly more dominant. Between 2003 and 2006, the allele frequency shifted significantly, with 3D7 class percent increasing from 80.9% to 99% of all infections. Although the possibility exists that the observed changes in allelic frequency are caused by extraneous factors such as genetic drift, which is a major contributor to allele variation in small populations, we believe our study population is sufficiently large that the effects of genetic drift are minimal. Such large-scale and directional changes in allele frequencies over such a short time frame are unlikely to be caused by genetic drift, but this remains a formal possibility. Thus, it seems most likely that an as of yet unidentified selection pressure is responsible for driving the observed allele frequency variation. Importantly, if this selection pressure is immunologically mediated, the target of that response is most likely against the N-terminal domain, because the C-terminal domain is almost entirely conserved between both alleles.
Screening for sequence variation identified only 10 mutations in PfMSP3 in 630 infections. All mutations occurred within the 2003 transmission season and were identical, a C T non-synonymous substitution at position 203. It is interesting to note that this is the same SNP that was most commonly observed in other global studies, being present in 48% (20 of 42 total) of 3D7 allele class variants in Nigeria and Thailand.20 The frequency and global distribution of this SNP suggests a globally conserved function, perhaps in immune evasion. Although the possibility exists that the lack of PfMSP3 variation that we observed in the Zungarococha community is caused by an absence of genetic diversity in this setting, work by Sutton and others (unpublished data) has shown that MSP1-block 2 variation in the Zungarococha community is significant and is similar in spectrum, although not in scale, to diversity seen in a hyperendemic environment (P Sutton and OH Branch, unpublished data). Additionally, microsatellite studies by Branch and others suggested extensive diversity within the community (OH Branch, personal communication). This implies that the lack of variation in PfMSP3 is likely not caused by a lack of genome-wide diversity in the community but by a combination of the relative stability of the N-terminal domain antigen and the relatively recent introduction of P. falciparum in the Zungarococha community. Subsequent studies of PfMSP3 mutants by sequencing the highly polymorphic PfMSP2 showed that three of those infections contained PfMSP2 sequences that were significantly different than HB3, suggesting that the introduction of new 3D7 class strains into the community can explain some, but perhaps not all, PfMSP3 mutations. It is likely that the introduction of new P. falciparum strains occurred through the naval port located in Ninarumi. In support of this, all three PfMSP2-variant infections occurred in Ninarumi (data not shown).
A significant strength of the MIGIA cohort study is the use of active case detection in addition to passive detection of P. falciparum infections. Such infections, marked by a lack of malaria-related symptoms in the presence of active parasitemia, mimic protection in malaria-immune individuals, a phenomenon termed premunition, and might predict malaria-immune status in these individuals. Thus, the use of asymptomatic individuals, detected through active case detection, allows us to assess for correlates of protection for P. falciparum alleles.
To identify correlates of protection for PfMSP3 alleles, we combined our PfMSP3 allele data with the available epidemiologic data. Evidence of protection was assessed by testing whether the presence of either PfMSP3 allele correlated with an increased frequency of subsequent allele infections, the rate of subsequent infections, or an increase in the rate of subsequent asymptomatic infections. Although we were unable to find significant correlations of protection for either PfMSP3 allele, this outcome does not rule out the potential for allele-specific protection. Such correlates of protection for PfMSP3 alleles, although important to assess for, are indirect outcomes of host protection and infrequent P. falciparum infections, which define the hypoendemic environment, make the identification of significant correlations unfavorable because of a lack of statistical power. More direct tests, such as directly testing antibody levels, will be critical to clearly identifying the role that PfMSP3 plays in mediating immunity in the hypoendemic setting.
In terms of PfMSP3 vaccine development, our central finding of genetic stability in the PfMSP3 N-terminal domain supports a reassessment of the inclusion of this domain in a PfMSP3-based P. falciparum vaccine, especially because genetic variation has been the major factor excluding N-terminal domain-based vaccine development. Because a large population of the world lives in hypoendemic environments, the observed stable nature of the N-terminal domain of PfMSP3 in our study is significant. Previous reports have shown more extensive PfMSP3 N-terminal domain variation in the hyperendemic setting, but this variation may simply represent a much larger pool of population-level genetic diversity and does not necessarily mean that the N-terminal domain should be considered hypervariable. In such hyperendemic environments, the high inoculation rate makes the differentiation of new polymorphisms from infections with heterologous strains difficult to distinguish. In contrast, the hypoendemic environment removes the confounding of heterologous/variant strain inoculation, allowing the dynamics of candidate polymorphisms to be easily studied.
Recent studies have suggested the N-terminal domain of PfMSP3 is significantly more immunogenic than the C-terminal domain,20 supporting our recommendation that the N-terminal domain be reassessed for future vaccine development. Importantly, because PfMSP3 N-terminal domain variation occurs largely between the heptad repeat motifs, which are almost entirely conserved, concerns about N-terminal domain variation can be largely alleviated if antibody responses against the N-terminal domain are not allele specific but instead provide cross-protection between variants. Our genotyping data certainly show no evidence for allele-specific responses, although this clearly needs to be followed up with immuno-logic data. The potential of antibodies to mediate cross-protection between N-terminal sequence variants therefore needs to be studied to either support the current C-terminal vaccine constituent or alternatively to provide evidence that supports a novel PfMSP3 N-terminal domain-based subunit vaccine. Such studies are ongoing in our laboratory.
Acknowledgments
The authors thank Dr. Michael Crowley for technical expertise and assistance in using dHPLC to screen for PfMSP3 sequence variation; Gino Nivarro Anderson, Jean Hernandez, and Patrick Sutton for help with sample processing and obtaining the corresponding epidemiologic data; and the residents of the Zungarococha community for their willingness to participate in the MIGIA study.
Financial support: This work was supported by National Institute of Health Grant R21 AI072421 and the UAB Sparkman Center for Global Health.
REFERENCES
- 1.Breman JG. The ears of the hippopotamus: manifestations, determinants, and estimates of the malaria burden. Am J Trop Med Hyg. 2001;64:1–11. doi: 10.4269/ajtmh.2001.64.1. [DOI] [PubMed] [Google Scholar]
- 2.Marsh K. Malaria—a neglected disease? Parasitology. 1992;104(Suppl):S53–S69. doi: 10.1017/s0031182000075247. [DOI] [PubMed] [Google Scholar]
- 3.Blackman MJ, Heidrich HG, Donachie S, McBride JS, Holder AA. A single fragment of a malaria merozoite surface protein remains on the parasite during red cell invasion and is the target of invasion-inhibiting antibodies. J Exp Med. 1990;172:379–382. doi: 10.1084/jem.172.1.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Malkin E, Long CA, Stowers AW, Zou L, Singh S, Macdonald NJ, Narum DL, Miles AP, Orcutt AC, Muratova O, Moretz SE, Zhou H, Diouf A, Fay M, Tierney E, Leese P, Mahanty S, Miller LH, Saul A, Martin LB. Phase 1 study of two merozoite surface protein 1 (MSP1(42)) vaccines for Plasmodium falciparum malaria. PLoS Clin Trials. 2007;2:e12. doi: 10.1371/journal.pctr.0020012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yuen D, Leung WH, Cheung R, Hashimoto C, Ng SF, Ho W, Hui G. Antigenicity and immunogenicity of the N-terminal 33-kDa processing fragment of the Plasmodium falciparum merozoite surface protein 1, MSP1: Implications for vaccine development. Vaccine. 2006;25:490–499. doi: 10.1016/j.vaccine.2006.07.053. [DOI] [PubMed] [Google Scholar]
- 6.Malkin EM, Diemert DJ, McArthur JH, Perreault JR, Miles AP, Giersing BK, Mullen GE, Orcutt A, Muratova O, Awkal M, Zhou H, Wang J, Stowers A, Long CA, Mahanty S, Miller LH, Saul A, Durbin AP. Phase 1 clinical trial of apical membrane antigen 1: an asexual blood-stage vaccine for Plasmodium falciparum malaria. Infect Immun. 2005;73:3677–3685. doi: 10.1128/IAI.73.6.3677-3685.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Remarque EJ, Faber BW, Kocken CH, Thomas AW. Apical membrane antigen 1: a malaria vaccine candidate in review. Trends Parasitol. 2008;24:74–84. doi: 10.1016/j.pt.2007.12.002. [DOI] [PubMed] [Google Scholar]
- 8.Frevert U, Sinnis P, Cerami C, Shreffler W, Takacs B, Nussenzweig V. Malaria circumsporozoite protein binds to heparan sulfate proteoglycans associated with the surface membrane of hepatocytes. J Exp Med. 1993;177:1287–1298. doi: 10.1084/jem.177.5.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doherty JF, Pinder M, Tornieporth N, Carton C, Vigneron L, Milligan P, Ballou WR, Holland CA, Kester KE, Voss G, Momin P, Greenwood BM, McAdam KP, Cohen J. A phase I safety and immunogenicity trial with the candidate malaria vaccine RTS,S/SBAS2 in semi-immune adults in The Gambia. Am J Trop Med Hyg. 1999;61:865–868. doi: 10.4269/ajtmh.1999.61.865. [DOI] [PubMed] [Google Scholar]
- 10.Heppner DGJ, Kester KE, Ockenhouse CF, Tornieporth N, Ofori O, Lyon JA, Stewart VA, Dubois P, Lanar DE, Krzych U, Moris P, Angov E, Cummings JF, Leach A, Hall BT, Dutta S, Schwenk R, Hillier C, Barbosa A, Ware LA, Nair L, Darko CA, Withers MR, Ogutu B, Polhemus ME, Fukuda M, Pichyangkul S, Gettyacamin M, Diggs C, Soisson L, Milman J, Dubois MC, Garcon N, Tucker K, Wittes J, Plowe CV, Thera MA, Duombo OK, Pau MG, Goudsmit J, Ballou WR, Cohen J. Towards an RTS,S-based, multi-stage, multi-antigen vaccine against falciparum malaria: progress at the Walter Reed Army Institute of Research. Vaccine. 2005;23:2243–2250. doi: 10.1016/j.vaccine.2005.01.142. [DOI] [PubMed] [Google Scholar]
- 11.Gardner MJ, Hall N, Fung E, White O, Berriman M, Hyman RW, Carlton JM, Pain A, Nelson KE, Bowman S, Paulsen IT, James K, Eisen JA, Rutherford K, Salzberg SL, Craig A, Kyes S, Chan MS, Nene V, Shallom SJ, Suh B, Peterson J, Angiuoli S, Pertea M, Allen J, Selengut J, Haft D, Mather MW, Vaidya AB, Martin DM, Fairlamb AH, Fraunholz MJ, Roos DS, Ralph SA, McFadden GI, Cummings LM, Subramanian GM, Mungall C, Venter JC, Carucci DJ, Hoffman SL, Newbold C, Davis RW, Fraser CM, Barrell B. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature. 2002;419:498–511. doi: 10.1038/nature01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mills KE, Pearce JA, Crabb BS, Cowman AF. Truncation of merozoite surface protein 3 disrupts its trafficking and that of acidic-basic repeat protein to the surface of Plasmodium falciparum merozoites. Mol Microbiol. 2002;43:1401–1411. doi: 10.1046/j.1365-2958.2002.02834.x. [DOI] [PubMed] [Google Scholar]
- 13.Rodriguez LE, Curtidor H, Ocampo M, Garcia J, Puentes A, Valbuena J, Vera R, Lopez R, Patarroyo ME. Identifying Plasmodium falciparum merozoite surface antigen 3 (MSP3) protein peptides that bind specifically to erythrocytes and inhibit merozoite invasion. Protein Sci. 2005;14:1778–1786. doi: 10.1110/ps.041304505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mulhern TD, Howlett GJ, Reid GE, Simpson RJ, McColl DJ, Anders RF, Norton RS. Solution structure of a polypep-tide containing four heptad repeat units from a merozoite surface antigen of Plasmodium falciparum. Biochemistry. 1995;34:3479–3491. doi: 10.1021/bi00011a001. [DOI] [PubMed] [Google Scholar]
- 15.Burgess BR, Schuck P, Garboczi DN. Dissection of merozoite surface protein 3, a representative of a family of Plasmodium falciparum surface proteins, reveals an oligomeric and highly elongated molecule. J Biol Chem. 2005;280:37236–37245. doi: 10.1074/jbc.M506753200. [DOI] [PubMed] [Google Scholar]
- 16.Oeuvray C, Bouharoun-Tayoun H, Grass-Masse H, Lepers JP, Ralamboranto L, Tartar A, Druilhe P. A novel merozoite surface antigen of Plasmodium falciparum (MSP-3) identified by cellular-antibody cooperative mechanism antigenicity and biological activity of antibodies. Mem Inst Oswaldo Cruz. 1994;89(Suppl 2):77–80. doi: 10.1590/s0074-02761994000600018. [DOI] [PubMed] [Google Scholar]
- 17.McColl DJ, Silva A, Foley M, Kun JF, Favaloro JM, Thompson JK, Marshall VM, Coppel RL, Kemp DJ, Anders RF. Molecular variation in a novel polymorphic antigen associated with Plasmodium falciparum merozoites. Mol Biochem Parasitol. 1994;68:53–67. doi: 10.1016/0166-6851(94)00149-9. [DOI] [PubMed] [Google Scholar]
- 18.Oeuvray C, Bouharoun-Tayoun H, Gras-Masse H, Bottius E, Kaidoh T, Aikawa M, Filgueira MC, Tartar A, Druilhe P. Merozoite surface protein-3: a malaria protein inducing antibodies that promote Plasmodium falciparum killing by cooperation with blood monocytes. Blood. 1994;84:1594–1602. [PubMed] [Google Scholar]
- 19.Huber W, Felger I, Matile H, Lipps HJ, Steiger S, Beck HP. Limited sequence polymorphism in the Plasmodium falciparum merozoite surface protein 3. Mol Biochem Parasitol. 1997;87:231–234. doi: 10.1016/s0166-6851(97)00067-4. [DOI] [PubMed] [Google Scholar]
- 20.Polley SD, Tetteh KK, Lloyd JM, Akpogheneta OJ, Greenwood BM, Bojang KA, Conway DJ. Plasmodium falciparum merozoite surface protein 3 is a target of allele-specific immunity and alleles are maintained by natural selection. J Infect Dis. 2007;195:279–287. doi: 10.1086/509806. [DOI] [PubMed] [Google Scholar]
- 21.Branch O, Casapia WM, Gamboa DV, Hernandez JN, Alava FF, Roncal N, Alvarez E, Perez EJ, Gotuzzo E. Clustered local transmission and asymptomatic Plasmodium falciparum and Plasmodium vivax malaria infections in a recently emerged, hypoendemic Peruvian Amazon community. Malar J. 2005;4:27. doi: 10.1186/1475-2875-4-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O'Donovan MC, Oefner PJ, Roberts SC, Austin J, Hoogendoorn B, Guy C, Speight G, Upadhyaya M, Sommer SS, McGuffin P. Blind analysis of denaturing high-performance liquid chromatography as a tool for mutation detection. Genomics. 1998;52:44–49. doi: 10.1006/geno.1998.5411. [DOI] [PubMed] [Google Scholar]
- 23.Balogh K, Patocs A, Majnik J, Racz K, Hunyady L. Genetic screening methods for the detection of mutations responsible for multiple endocrine neoplasia type 1. Mol Genet Metab. 2004;83:74–81. doi: 10.1016/j.ymgme.2004.08.013. [DOI] [PubMed] [Google Scholar]
- 24.Keller G, Hartmann A, Mueller J, Hofler H. Denaturing high pressure liquid chromatography (DHPLC) for the analysis of somatic p53 mutations. Lab Invest. 2001;81:1735–1737. doi: 10.1038/labinvest.3780387. [DOI] [PubMed] [Google Scholar]
- 25.Wolford JK, Blunt D, Ballecer C, Prochazka M. High-throughput SNP detection by using DNA pooling and denaturing high performance liquid chromatography (DHPLC) Hum Genet. 2000;107:483–487. doi: 10.1007/s004390000396. [DOI] [PubMed] [Google Scholar]
- 26.Vittor AY, Gilman RH, Tielsch J, Glass G, Shields T, Lozano WS, Pinedo-Cancino V, Patz JA. The effect of deforestation on the human-biting rate of Anopheles darlingi, the primary vector of Falciparum malaria in the Peruvian Amazon. Am J Trop Med Hyg. 2006;74:3–11. [PubMed] [Google Scholar]