Abstract
High resolution localized surface plasmon resonance (HR-LSPR) sensors were combined with matrix assisted laser desorption ionization mass spectrometry (MALDI-MS) for the first time. LSPR sensors provide real-time label-free detection of molecular adsorption. Subsequent MALDI-MS analysis enables identification of the adsorbed molecules. This synergistic LSPR-MS approach was applied to the detection and identification of amyloid beta oligomers which play an important role in the molecular pathogenesis of Alzheimer’s Disease.
Introduction
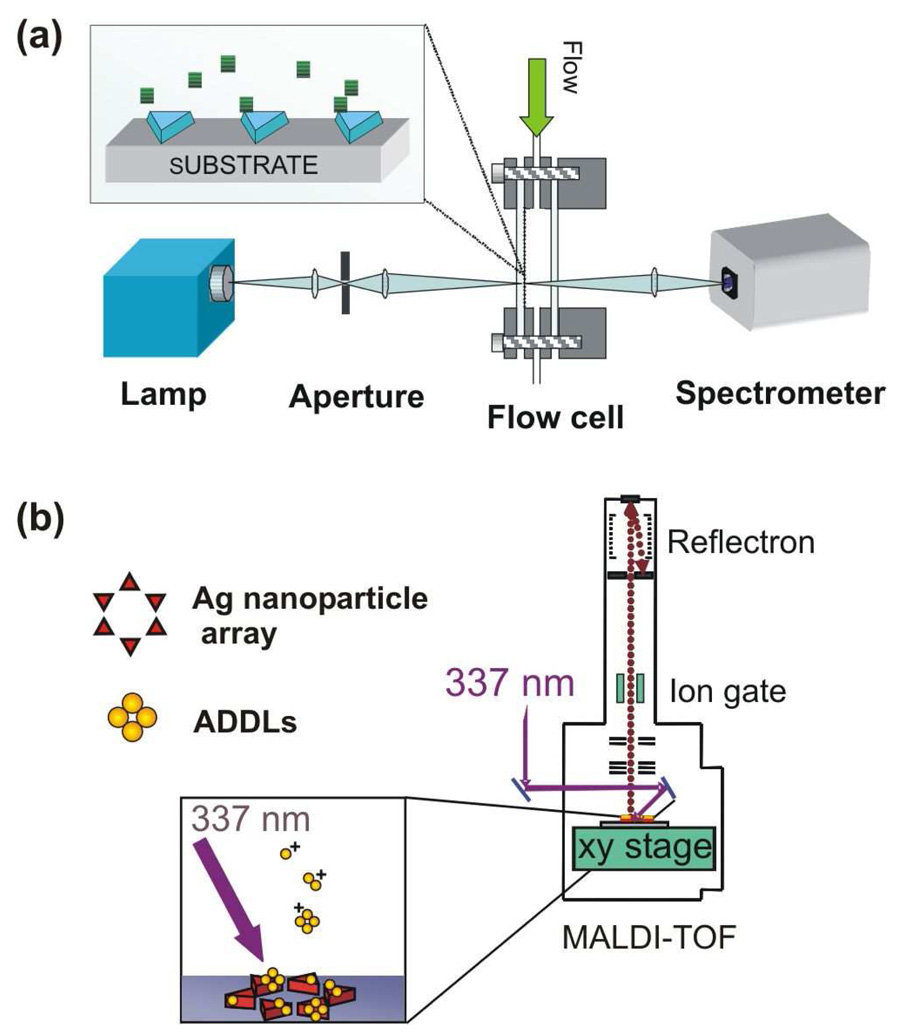
The ability to detect and identify small quantities of biomolecules is essential for characterizing ligand-receptor interactions and ultimately elucidating the molecular basis of disease. Real-time, label-free detection techniques based on quartz crystal microbalances (QCM), microcantilevers, optical cavity resonators, propagating surface plasmon resonance (SPR),1 and localized surface plasmon resonance (LSPR) spectroscopy2–4 are widely used to measure the thermodynamics and kinetics of binding. These techniques are analyte general and require receptor functionalized surfaces for specificity. In order to identify unknown molecules bound to the surface, a complementary analytical technique is needed, such as matrix assisted laser desorption ionization mass spectrometry (MALDI-MS). The advantages of integrating propagating SPR sensors with MALDI have been previously demonstrated.5,6 We report here the first experiments combining real-time, high resolution LSPR (HR-LSPR) and MALDI (See Figure 1). The techniques were applied to the detection of amyloid-beta derived diffusible ligands (ADDLs) which play an important role in the molecular pathogenesis of Alzheimer’s Disease.7
Figure 1.
(a) Schematic of the experimental setup for LSPR extinction spectroscopy. (b). Schematic of the experimental setup for for MALDI-MS on the LSPR substrates.
ADDLs are neurotoxic oligomers of amyloid beta-protein (Aβ), intermediate in size between monomers and the fibrils found in the senile plaques of Alzheimer’s patients. ADDLs specifically target synapses8 and selectively block long term potentiation.9 In transgenic Alzheimer’s mouse models, 12-mers of Aβ develop when memory failure first occurs; injection of these isolated 12-mers into normal mice causes memory deficiency.10 In human patients, there is a strong correlation between the presence of Alzheimer’s Disease and the concentration of ADDLs in cerebrospinal fluid and cerebral cortex tissue.11–13 Measuring Aβ oligomer concentrations and studying their aggregation rates is therefore important for understanding pathology and developing diagnostics and therapeutics.
LSPR shift assays detect molecular adsorption to noble metal nanoparticles. These nanoparticles intensely absorb and scatter light at wavelengths that depend on the nanoparticle size, shape, composition and local refractive index.2–4 Molecular adsoption to the nanoparticles increases the local refractive index, and leads to a red-shift in the extinction spectrum. With improvements in instrumentation and software, real-time LSPR shifts can now be monitored in solution with picometer spectral resolution.3,4 Indeed the sensitivity is sufficiently high to detect the conformational changes that accompany calcium binding to calmodulin.4 LSPR shift assays have also been used to detect ADDL concentrations down to 100 fM by measuring the LSPR wavelength in nitrogen before and after ADDL binding.13
To demonstrate the combination of HR-LSPR and MALDI-MS, we first used HR-LSPR to monitor binding of ADDLs to an anti-ADDL antibody (IgG, NU-1). Next, we acquired MALDI spectra from ADDLs on a stainless steel plate. Finally, we acquired MALDI spectra from an LSPR substrate after first recording real-time LSPR.
Experimental Methods
Preparation of Amyloid ADDLs
ADDLs were prepared according to previously published protocols.7 Aβ1–42 peptide (California Peptide Research, Napa, CA) was dissolved in hexafluoro-2-propanol (HFIP) and aliquoted to microcentrifuge tubes. HFIP was allowed to evaporate overnight in a fume hood, after which residual traces of HFIP were removed by drying for 15 min in a SpeedVac (Savant Instruments) at 6 mTorr. The tubes were stored desiccated at −80°C. To prepare the soluble oligomer solution, an aliquot of Aβ1–42 was dissolved in neat, cold dimethylsulfoxide (DMSO; freshly opened vial) to make a 5 mM solution. The DMSO stock was immediately diluted into cold phenol-free F12 medium (Life Technologies, Gaithersburg, MD) to make a 100-µM Aβ solution. This solution was then incubated at 4 °C for 24–30 hours, centrifuged at 14,000×g for 10 min, and the supernatant was collected. Protein concentration was determined using the Coomassie blue assay, with the monomer Aβ1–42 peptide as the molar reference. SDS-PAGE separation of these ADDLs shows a series of oligomers including higher order oliogmers > 50 kDa.7 SDS-PAGE studies with cross-linked ADDLs suggest that pentamers and hexamers are more stable configurations.14
Nanosphere Lithography (NSL)
Silver nanoprisms attached to a glass substrate were prepared using nanosphere lithography (NSL).15 Nanospheres (Interfacial Dynamics Corporation, Portland OR, d=390 nm) were drop coated onto a #2 18 mm glass coverslide to create a close-packed hexagonal monolayer. Silver was then deposited through the triangular spaces in the sphere mask to a height of 80 nm. The spheres were removed by attaching a layer of tape to the sample and then peeling it off, followed by sonicating the sample in ethanol for 10 seconds. The sample was then functionalized with a self-assembled monolayer (SAM) by incubation in an ethanolic solution of 0.75 mM octanethiol and 0.25 mM 11-mercaptoundecanoic acid (MUA) for at least 24–48 hrs. The SAM protects the nanoprisms by reducing the effect of solvent annealing as well as providing carboxyl groups onto which biomolecules can be covalently linked.
HR-LSPR
Silver nanoprism substrates, prepared by NSL, were placed in a home built flow cell. Unpolarized light from a BPS100 lamp (BWTek Inc., Newark, DE) was focused on the substrate (1 mm spot size), and transmitted light was analyzed with a BRC711E photodiode array spectrometer (BWTek Inc., Newark, DE). A clear region on the substrate, made by masking with Kapton tape during silver deposition, was used as a spectral reference. Transmittance at each wavelength was calculated as T=(sample-dark)/(reference-dark), and extinction was calculated as −log10(T). A program written in Labview (National Instruments, Austin, TX) acquired spectra at 1 s intervals, calculated the maximum wavelength by fitting a 100 nm spectral region around the peak to a fourth order polynomial, and displayed λmax in real-time. Solutions were injected into the flow cell with a syringe. To switch solutions, the solution within the flow cell was aspirated with the syringe, and a new solution was added; spectral acquisition was paused during the roughly 10 s that this process took.
The binding curves for each step were fit to a single exponential with a linear drift correction term: y(t)= ysat – b exp(-t/τ) – c t. The linear correction term accounted for the particle shape annealing in aqueous solvent, and was less than 0.001 nm/s in all cases. The total LSPR shift due to adsorption of biomolecules to the LSPR surface varies from sample to sample by +/− 30% due to differences in nanoparticle morphology and initial λmax, which is known to affect the sensitivity of nanoparticles to refractive index changes.
MALDI on stainless steel plate
The ADDLs were spotted onto a MALDI plate using the sandwich method with sinapinic acid as the matrix. A sinapinic acid stock solution (SA) was prepared as 10 mg/mL sinapinic acid in a solvent of 50% (v/v) acetonitrile, 49.95% water, 0.05% trifluoroacetic acid. 0.5 µL SA was pipetted onto the stainless steel plate and aspirated to leave a thin film. After drying, 0.5 µL of ADDLs solution was added, and 0.5 µL of SA was added to the droplet and allowed to dry. The mass spectra were acquired on a MALDI time of flight (TOF) instrument, Axima CFR (Shimadzu, Tokyo, Japan/Kratos, Manchester, UK) which uses a 337 nm nitrogen laser. The Instrument was calibrated using mass calibration standards of bovine insulin and insulin oxidized B chain.
MALDI on an LSPR sample
The LSPR sample consisted of silver nanoprisms on a glass slide prepared by NSL. The nanoprisms had been previously exposed to ADDLs, and the adsorption was monitored in real-time using HR-LSPR. The sample was then spotted with 1 µL SA and allowed to dry. On some spots, we also co-deposited 5 pmols of bovine insulin and insulin oxidized B chain as calibration standards. Subsequently, the sample was attached to a stainless steel MALDI plate using kapton tape and introduced into the MALDI instrument.
Results and Discussion
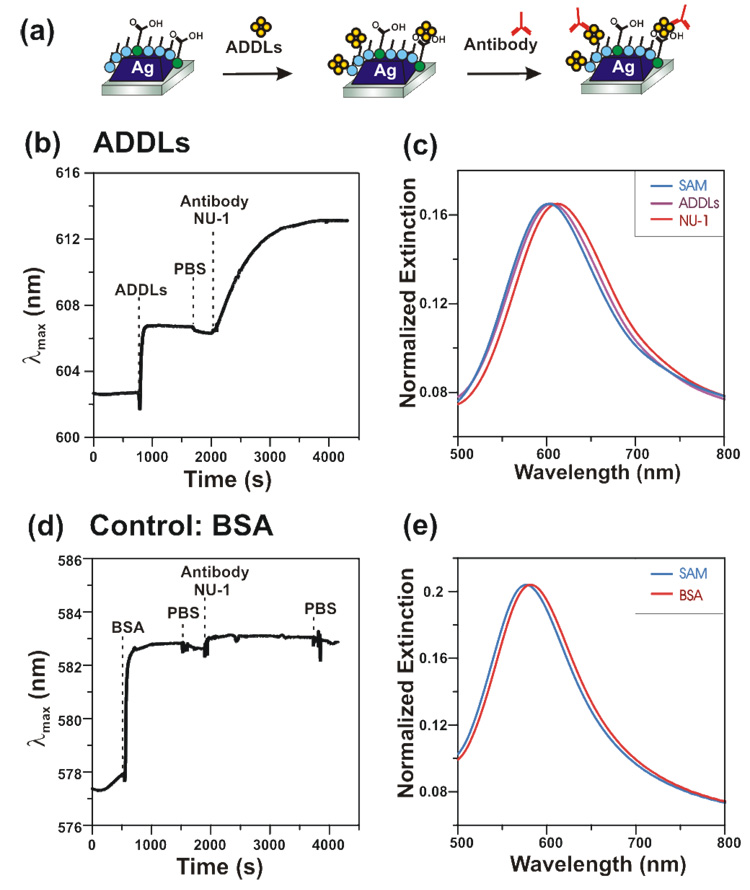
Figure 2 shows an HR-LSPR sensorgram of ADDLs binding to silver nanoprisms fabricated using nanosphere lithography. The nanoprism substrate was incubated in 10 mM phosphate buffered saline (PBS) at pH 7.4, followed by ADDLs (10 µM in Aβ), a PBS rinse, and a final incubation in 200 nM anti-ADDLs. All binding curves were fit to a sum of an exponential plus a linear drift term: y(t)= ysat – b exp(-t/τ) – c t. Upon incubation with ADDLs, the LSPR extinction shifted by 5.0 nm with a 24 s exponential time constant (τ) due to adsorption. Rinsing in PBS resulted in a small blue-shift (0.4 nm) due to removal of weakly bound ADDLs. Subsequent adsorption of antibody resulted in an additional 6.8 nm shift (τ=725 s). Figure 2b shows the real-time tracking of the peak wavelength. Figure 2c shows extinction spectra at each stage of ADDLs binding. A control experiment using bovine serum albumin (BSA, MW 66 kDa) resulted in a similar initial shift of 5.0 nm (τ =19 s), but minimal antibody adsorption (0.52 nm), see Figure 2d and 2e. These results indicate that a similar amount of BSA and ADDLs adsorb to the nanoprisms, and at a similar rate, but that the antibody binds specifically to the ADDLs functionalized surface. The signal to noise ratio of these measurements is high (~500) and measurements can be made over many hours with no photobleaching. Although previous researchers have used propagating SPR to examine Aβ oligomerization,16 this marks the first time that LSPR sensors detected real-time binding of ADDLs and antibody.
Figure 2.
(a) Schematic of the experimental protocol for binding of ADDLs and antibody. (b) Sensorgram showing λmax as a function of time during binding of ADDLs, rinsing, and addition of antibody (1 spectrum/s collection rate). (c) Extinction spectra for SAM functionalized substrate in solution before ADDLs adsorption (λmax = 602.70 nm), after ADDLs adsorption (λmax = 606.67 nm), and after NU-1 antibody binding (λmax = 613.13 nm). (d) Control experiment showing binding of BSA but minimal antibody binding. (e) Extinction spectra for SAM functionalized substrate in solution before BSA adsorption (λmax = 577.90 nm), and after BSA adsorption (λmax = 582.82 nm); the spectrum after NU-1 antibody incubation largely overlaps with the spectrum after BSA and is not shown.
ADDLs were spotted onto a stainless steel MALDI plate along with the MALDI matrix, sinapinic acid (SA). Approximately 35 pmol of Aβ was deposited per spot. MALDI spectra were acquired on an Axima CFR instrument (Shimadzu, Tokyo, Japan). In addition to the Aβ peak, we observed a second set of peaks shifted by 16 mass units (see supplemental Figure S1). Previous researchers have also observed this peak, and attributed it to oxidation of the methionine.17 It is reported that high concentrations of the oxidized species of Aβ are found in senile amyloid plaques, although in vitro, it is less prone to oligomerization and fibrillization.17,18 The oxidized ADDLs are reported to have decreased toxicity over periods of less than 24 hours, with restored toxicity after 96 hours.17,18
The MALDI spectrum also displays a series of oligomers up to at least 30-mers (see supplemental Figure S2). The observed ions are likely a combination of intact molecular ions, fragments from larger oligomers, and gas phase aggregates. Indeed, post source decay of the dimer into monomer was observed using reflectron mode (see supplemental Figure S3). Hung and coworkers published the first MALDI spectra of Aβ oligomers up to octomers.19 Their studies of Aβ peptides and oligomers adsorbed to hydrophobic C-6 modified surfaces showed that the GxxxG peptide repeat sequence of Aβ is important in oligomerization. Replacing any of the glycines in those repeat sequencies with leucine residues reduced both the amount of oligomerization and toxicity of the Aβ. The present results demonstrate that that even larger oligomers can also be observed with MALDI. Work is ongoing to acquire MALDI from electrophoretically separated ADDLs as well cross-linked ADDLs and isotope labeled ADDLs to elucidate the aggregation and dissociation mechanisms in the MALDI spectra.
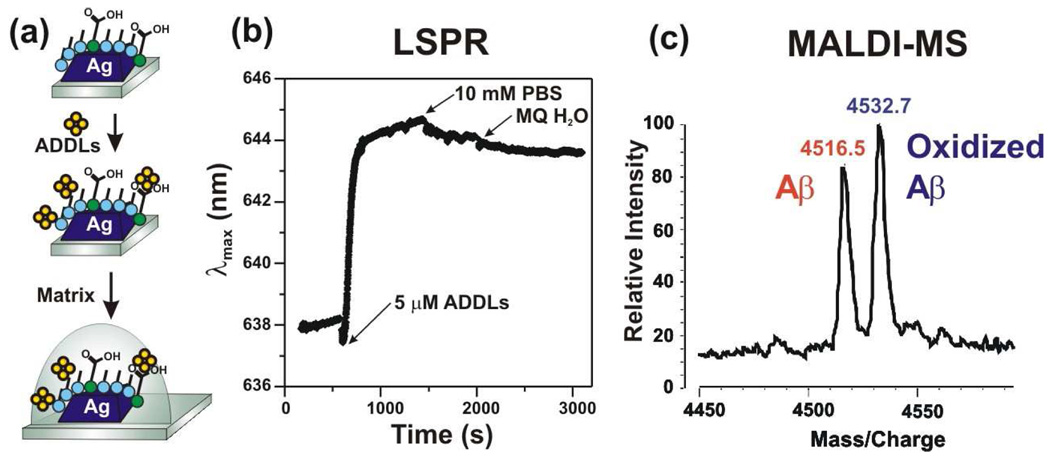
We next performed MALDI directly on an LSPR sample after adsorption of ADDLs (Figure 3). Figure 3a shows real-time binding of 5 µM ADDLs to an LSPR substrate. The shift is ~6.5 nm (τ=49 s); the reduced adsorption rate compared to Figure 2 may be due to lower concentration, while the similar total shift indicates that the surface saturates after long incubation times at 5 µM concentration. Next, we pipetted 1 µL SA onto the substrate and allowed it to dry. The number of moles of ADDLs on the nanoprisms spotted with matrix is estimated to be approximately 4 orders of magnitude less concentrated than on the samples directly drop coated onto the MALDI plate (Figure S2). As a result, oligomers are obscured by background. The monomer peak is clearly observed and resolved into two peaks due to normal and oxidized Aβ. Such posttranscriptional modifications could not be resolved using LSPR alone.
Figure 3.
(a) Experimental protocol. (b) Real-time detection of ADDLs binding to octanethiol/11-MUA functionalized nanoprisms. (c) MALDI mass spectrum of Aβ monomers bound to the nanoprisms, using a layer of sinapinic acid as the matrix.
Conclusion
The LSPR-MS approach offers detection and identification capabilities similar to the established SPR-MALDI approach.5,6 In addition, LSPR systems display less interference from bulk refractive index changes,20 observation of protein conformational changes,4 and higher spatial resolution down to single particles.2 Nanoparticles also provide advantages for mass spectrometry because nanoparticles may also be suspended in solution and concentrated through centrifugation.21 In addition, nanoparticles have been reported to enhance desorption and ionization processes in MALDI imaging.22,23
In conclusion, we have demonstrated the first use of MALDI-MS on an LSPR sample. The LSPR provided real-time binding data, while the MALDI allowed the identification of normal and oxidized Aβ in ADDLs. Integrating LSPR with MS therefore represents an important advance in the analytical capability of LSPR. We expect complementary LSPR and MS analysis will be important in the in vitro analysis of Aβ aggregation and interaction with biomolecules, as well as the characterization and analysis of Aβ in tissue samples and cerebral spinal fluid.13,24
Supplementary Material
Additional figures are available free of charge via the internet at http://pubs.acs.org. Figure S1 is a high resolution MALDI spectra of amyloid beta including its oxidized form. Figure S2 is a MALDI spectrum amyloid beta oligomeres. Figure S3 shows the postsource decay MALDI spectrum of amyloid beta dimers.
Acknowledgement
This research was supported by the National Science Foundation (Grants EEC-0647560, CHE-0414554, DMR-0520513, and BES-0507036), The National Institutes of Health (Grants R01 AG029460 and R01 AG022547), The International Institute for Nanotechnology, Northwestern University through a grant from the State of Illinois Department of Commerce and Economic Opportunity (#06-205022), a Ruth L. Kirschstein National Research Service Award (5 F32 GM077020) to J.N.A., and a Ryan Fellowship to W.P.H.
Contributor Information
Milan Mrksich, Email: mmrksich@uchicago.edu.
William L. Klein, Email: wklein@northwestern.edu.
Richard P. Van Duyne, Email: vanduyne@northwestern.edu.
References
- 1.Schuck P. Use of Surface Plasmon Resonance to Probe the Equilibrium and Dynamic Aspects of Interactions Between Biological Macromolecules. Annu. Rev. Biophys. Biomol. Struct. 1997;26:541–566. doi: 10.1146/annurev.biophys.26.1.541. [DOI] [PubMed] [Google Scholar]
- 2.Anker JN, Hall WP, Lyandres O, Shah NC, Zhao J, Van Duyne RP. Biosensing with plasmonic nanosensors. Nat. Mater. 2008;7:442–453. doi: 10.1038/nmat2162. [DOI] [PubMed] [Google Scholar]
- 3.Dahlin AB, Tegenfeldt JO, Hook F. Improving the instrumental resolution of sensors based on localized surface plasmon resonance. Anal. Chem. 2006;78:4416–4423. doi: 10.1021/ac0601967. [DOI] [PubMed] [Google Scholar]
- 4.Hall WP, Anker JN, Lin Y, Modica J, Mrksich M, Van Duyne RP. A Calcium-Modulated Plasmonic Switch. J. Am. Chem. Soc. 2008;130:5836–5837. doi: 10.1021/ja7109037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grote J, Dankbar N, Gedig E, Koenig S. Surface plasmon resonance/mass spectrometry interface. Anal. Chem. 2005;77:1157–1162. doi: 10.1021/ac049033d. [DOI] [PubMed] [Google Scholar]
- 6.Gurard-Levin ZA, Mrksich M. Combining Self-Assembled Monolayers and Mass Spectrometry for Applications in Biochips. Annu. Rev. Anal. Chem. 2008;1:767–800. doi: 10.1146/annurev.anchem.1.031207.112903. [DOI] [PubMed] [Google Scholar]
- 7.Klein WL. Aβ toxicity in Alzheimer’s disease: globular oligomers (ADDLs) as new vaccine and drug targets. Neurochem. Int. 2002;41:345–352. doi: 10.1016/s0197-0186(02)00050-5. [DOI] [PubMed] [Google Scholar]
- 8.Lacor PN, Buniel MC, Chang L, Fernandez SJ, Gong Y, Viola KL, Lambert MP, Velasco PT, Bigio EH, Finch CE. Synaptic Targeting by Alzheimer's-Related Amyloid β Oligomers. J. Neurosci. 2004;24:10191. doi: 10.1523/JNEUROSCI.3432-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang HW, Pasternak JF, Kuo H, Ristic H, Lambert MP, Chromy B, Viola KL, Klein WL, Stine WB, Krafft GA. Soluble oligomers of β amyloid (1–42) inhibit long-term potentiation but not long-term depression in rat dentate gyrus. Brain Research. 2002;924:133–140. doi: 10.1016/s0006-8993(01)03058-x. [DOI] [PubMed] [Google Scholar]
- 10.Lesne S, Koh MT, Kotilinek L, Kayed R, Glabe CG, Yang A, Gallagher M, Ashe KH. A specific amyloid-b protein assembly in the brain impairs memory. Nature. 2006;440:352–357. doi: 10.1038/nature04533. [DOI] [PubMed] [Google Scholar]
- 11.Gong Y, Chang L, Viola KL, Lacor PN, Lambert MP, Finch CE, Krafft GA, Klein WL. Alzheimer's Disease-affected brain: Presence of oligomeric Aβ ligands (ADDLs) suggests a molecular basis for reversible memory loss. Proc. Nat. Acad. Sci. U.S.A. 2003;100:10417–10422. doi: 10.1073/pnas.1834302100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Georganopoulou DG, Chang L, Nam JM, Thaxton CS, Mufson EJ, Klein WL, Mirkin CA. Nanoparticle-based detection in cerebral spinal fluid of a soluble pathogenic biomarker for Alzheimer's Disease. Proc. Nat. Acad. Sci. U.S.A. 2005;102:2273–2276. doi: 10.1073/pnas.0409336102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haes AJ, Chang L, Klein WL, Van Duyne RP. Detection of a biomarker for Alzheimer's Disease from synthetic and clinical samples using a nanoscale optical biosensor. J. Am. Chem. Soc. 2005;127:2264–2271. doi: 10.1021/ja044087q. [DOI] [PubMed] [Google Scholar]
- 14.Bitan G, Kirkitadze MD, Lomakin A, Vollers SS, Benedek GB, Teplow DB. Amyloid beta -protein (Abeta) assembly: Abeta 40 and Abeta 42 oligomerize through distinct pathways. Proc. Nat. Acad. Sci. U.S.A. 2003;100:330–335. doi: 10.1073/pnas.222681699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haynes CL, Van Duyne RP. Nanosphere lithography: A versatile nanofabrication tool for studies of size-dependent nanoparticle optics. J. Phys. Chem. B. 2001;105:5599–5611. [Google Scholar]
- 16.Ryu J, Joung HA, Kim MG, Park CB. Surface Plasmon Resonance Analysis of Alzheimer's-Amyloid Aggregation on a Solid Surface: From Monomers to Fully-Grown Fibrils. Anal. Chem. 2008;80:2400–2407. doi: 10.1021/ac7019514. [DOI] [PubMed] [Google Scholar]
- 17.Butterfield DA, Bush AI. Alzheimer’s amyloid β-peptide (1–42): involvement of methionine residue 35 in the oxidative stress and neurotoxicity properties of this peptide. Neurobiol. Aging. 2004;25:563–568. doi: 10.1016/j.neurobiolaging.2003.12.027. [DOI] [PubMed] [Google Scholar]
- 18.Barnham KJ, Ciccotosto GD, Tickler AK, Ali FE, Smith DG, Williamson NA, Lam YH, Carrington D, Tew D, Kocak G. Neurotoxic, Redox-competent Alzheimer's β-Amyloid Is Released from Lipid Membrane by Methionine Oxidation. J. Biol. Chem. 2003;278:42959–42965. doi: 10.1074/jbc.M305494200. [DOI] [PubMed] [Google Scholar]
- 19.Hung LW, Ciccotosto GD, Giannakis E, Tew DJ, Perez K, Masters CL, Cappai R, Wade JD, Barnham KJ. Amyloid-β Peptide (A β) Neurotoxicity Is Modulated by the Rate of Peptide Aggregation: A β Dimers and Trimers Correlate with Neurotoxicity. J. Neurosci. 2008;28:11950. doi: 10.1523/JNEUROSCI.3916-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yonzon CR, Jeoung E, Zou S, Schatz GC, Mrksich M, Van Duyne RP. A comparative analysis of localized and propagating surface plasmon resonance sensors: The binding of concanavalin A to a monosaccharide functionalized self-assembled monolayer. J. Am. Chem. Soc. 2004;126:12669–12676. doi: 10.1021/ja047118q. [DOI] [PubMed] [Google Scholar]
- 21.Huang YF, Chang HT. Nile Red-adsorbed gold nanoparticle matrixes for determining aminothiols through surface-assisted laser desorption/ionization mass spectrometry. Anal. Chem. 2006;78:1485–1493. doi: 10.1021/ac0517646. [DOI] [PubMed] [Google Scholar]
- 22.Altelaar AFM, Klinkert I, Jalink K, de Lange RP, Adan RAH, Heeren RMA, Piersma SR. Gold-enhanced biomolecular surface imaging of cells and tissue by SIMS and MALDI mass spectrometry. Anal. Chem. 2006;78:734–742. doi: 10.1021/ac0513111. [DOI] [PubMed] [Google Scholar]
- 23.Taira S, Sugiura Y, Moritake S, Shimma S, Ichiyanagi Y, Setou M. Nanoparticle-Assisted Laser Desorption/Ionization Based Mass Imaging with Cellular Resolution. Anal. Chem. 2008;80:4761–4766. doi: 10.1021/ac800081z. [DOI] [PubMed] [Google Scholar]
- 24.Portelius E, Zetterberg H, Andreasson U, Brinkmalm G, Andreasen N, Wallin A, Westman-Brinkmalm A, Blennow K. An Alzheimer's disease-specific β-amyloid fragment signature in cerebrospinal fluid. Neurosci. Lett. 2006;409:215–219. doi: 10.1016/j.neulet.2006.09.044. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional figures are available free of charge via the internet at http://pubs.acs.org. Figure S1 is a high resolution MALDI spectra of amyloid beta including its oxidized form. Figure S2 is a MALDI spectrum amyloid beta oligomeres. Figure S3 shows the postsource decay MALDI spectrum of amyloid beta dimers.