Abstract
Lung cancer is the leading cause of cancer related deaths in the United States. It is estimated that in 2008 there were 215,000 new diagnoses of lung cancer and 163, 000 deaths. Despite emerging technologies for potential early diagnosis and discovery of novel targeted therapies, the overall five year survival remains a disappointing 15%. Explanations for the poor survival include late presentation of disease, a lack of markers for early detection and both phenotypic and genotypic heterogeneity within patients of similar histological classification. In order to further understand this heterogeneity and thus complexity of lung cancer, investigators have applied various technologies including high throughput analysis of both the genome and proteome. Such approaches have been successful in identifying signatures that may clarify molecular differences in tumors, identify new targets and improve prognostication. In the last decade, investigators have identified a new mode of gene regulation in the form of non-coding RNAs termed microRNAs (miRNAs or miRs). First determined to be of importance in larval development, microRNAs are ~19–22 nucleotide single stranded RNAs that regulate genes by either inducing mRNA degradation or inhibiting translation. MiRNAs have been implicated in several cellular processes including apoptosis, development, proliferation and differentiation. By regulating hundreds of genes simultaneously, miRNAs have the capacity for regulation of biological networks. Global alterations in miRNA expression in both solid organ and haematological malignancies suggest their importance in the pathogenesis of disease. To date, both in vivo and in vitro studies in lung cancer demonstrate a dysregulation of miRNA expression. Furthermore, investigators are beginning to identify individual targets and pathways of miRNAs relevant to lung tumorigenesis. Thus, miRNAs may identify critical targets and be important in the pathogenesis of lung cancer.
Introduction
Lung cancer remains the number one cause of cancer related deaths in the United States1. The consistent poor five year survival underscores the need for novel modalities for early detection, prognostication and targeted therapies. Successful application of targeted therapies such as epidermal growth factor receptor inhibition further supports the importance of understanding the molecular heterogeneity in lung cancer. Analysis of the genome has partially clarified this heterogeneity by identifying “signatures” for diagnosis and prognosis as well as biological targets. Given that patterns of mRNA expression may not reflect post translational modifications, evaluations of the proteome are equally critical to understanding disease pathogenesis and heterogeneity. Until recently, studies that rely on high throughput platforms have been limited by lack of reproducibility and small cohort size. 2–4 The molecular events that lead to lung cancer development and progression may be dependent upon multiple biological pathways. Therefore, approaches integrating multiple platforms to detect pathways or biological network dependence in lung cancer are important.
Approximately 15 years ago, investigators first determined in Caenorhabditis elegans that non-coding RNAs were relevant to larval development. 5 Now referred to as microRNAs (miRNAs or miRs), these non-coding RNAs have the capacity for simultaneous regulation of hundreds to thousands of genes. In fact, in silico analysis suggests that miRNAs may be responsible for the regulation of up to one third of the genome.6 MiRNAs are crucial to several biological processes including proliferation, apoptosis, development and cellular differentiation.7 To date, over 800 miRNAs have been identified in mammals, yet their biological relevance and functional targets remain largely unknown. Deregulation in miRNA expression in several malignancies compared to normal tissue as well as their frequent location in fragile chromosomal regions supports their relevance to malignancy.7 MiRNAs have the capacity to function as either oncogenes or tumor suppressors.8 Currently, miRNAs are being investigated across several solid and hematological malignancies as biomarkers for diagnosis and prognosis of disease.
We are now witnessing a growing body of literature focused on the role of miRNAs in lung cancer. Studies that both examine global changes in miRNA within lung tumors as well as those that have focused on the effects of individual miRNAs on lung cancer cell phenotype suggest miRNAs are involved in lung tumor development and progression and may potentially serve as biomarkers for diagnosis and prognosis. The review presented will provide an overview of miRNA biogenesis, targeting and regulation and offer a perspective on the emerging role of miRNAs in lung cancer.
miRNA Biogenesis/Targeting
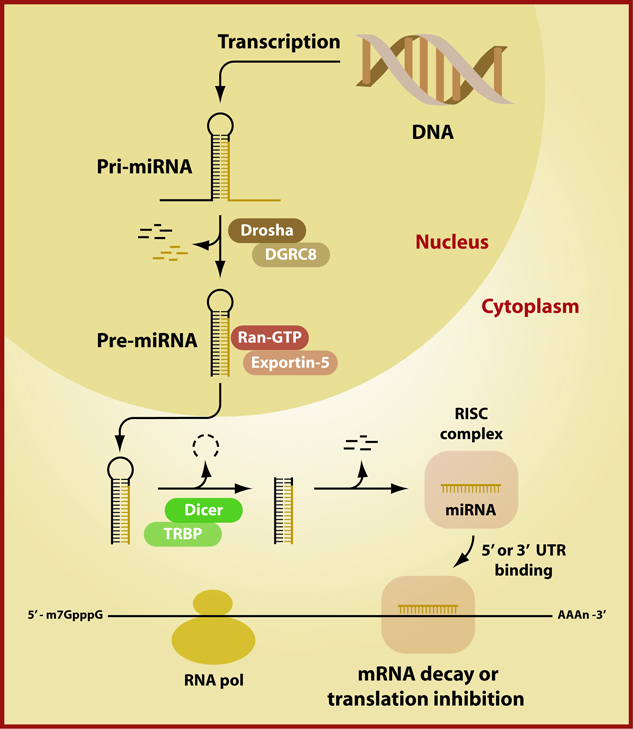
Generation of mature miRNAs requires multiple steps. (Figure 1) Much of our knowledge of miRNA biogenesis and targeting derives from invertebrate models, however, many of the proteins are evolutionary conserved throughout evolution and have similar functions in mammals. 9 Initially, a large primary (pri)-miRNA approximately 100 nucleotides (nt) in length is transcribed by RNA polymerase II.10 Within the nucleus, the pri-miRNAs are bound to double-stranded RNA-binding domain (dsRBD) protein. In invertebrates, Pasha or Partner of Drosha, while in humans DiGeorge syndrome critical region gene 8 (DGCR8) binds to the pri-miRNA. A complex containing DGCR8, the pri-miRNA and an universal RNase III endonuclease, RNASEN, (Drosha in invertebrates) facilitates the processing of the pri-miRNA to smaller ~70 nt precursor miRNA (pre-miRNA) molecules.9,11–13 The pre-miRNAs form an imperfect stem loop structure. These are then transported to the cytoplasm by a protein complex consisting of Exportin 5 and Ran GTPase. 14 Once in the cytoplasm, a pre-miRNA processing complex containing a second RNase III endonuclease, Dicer, and transactivating response RNA-binding protein (TRBP) is formed to cleave the pre-miRNA to double stranded ~22 nt miRNA molecules.13 The miRNA is then separated into two single stranded molecules; the anti-sense strand is incorporated in the RNA-induced silencing complex (RISC) through its interaction with Argonaute (AGO) proteins, while the other strand is degraded. Ago proteins bind to the 3´ end of the RNA and serve to align and stabilize the miRNA:mRNA duplex. It appears that there is a strong bias for the association of specific AGO proteins and small RNA subclasses. In plants, the asymmetry of the 5´ end of the miRNA dictates which Ago protein associates with the small RNA molecule, whereas in invertebrates structural features of the pre-miRNA dictate the binding of specific AGO proteins.15 The precise mechanisms for AGO-mediated miRNA loading to RISC in mammals have yet to be determined in mammals and remain the subject of intense investigation.
Figure 1.
miRNA Biogenesis: miRNA are transcribed as a large primary (pri)-miRNA which are approximately 100 nucleotides (nt) in length. Pri-miRNAs are bound to double-stranded RNA-binding domain (dsRBD) protein. In invertebrates, Pasha or Partner of Drosha, while in humans DiGeorge syndrome critical region gene 8 (DGCR8) binds to the pri-miRNA. DGCR8, the pri-miRNA and a universal RNase III endonuclease, RNASEN, (Drosha in invertebrates) facilitates the processing of the pri-miRNA to a smaller ~70 nt precursor miRNA (pre-miRNA) molecules These are then transported to the cytoplasm by the nuclear transmembrane Ran GTP-dependent transported Exportin 5. Once in the cytoplasm, a pre-miRNA processing complex containing a second RNase III endonuclease, Dicer, and a transactivating response RNA-binding protein (TRBP) is formed to cleave the pre-miRNA to double stranded ~22 nt miRNA molecules. The miRNA is then separated into two single stranded molecules; the anti-sense strand is incorporated in the RNA-induced silencing complex (RISC) through its interaction with Argonaute (AGO) proteins, while the other strand is degraded. Ago proteins bind to the 3´ end of the RNA and serve to align and stabilize the miRNA:mRNA duplex. The mature miRNA then targets either the 5’ or 3’UTR based on complimentartity
Although simplistic in concept that the AGO proteins and additional components of RISC are important in the formation of miRNA:mRNA duplex, this alone is not sufficient to predict whether translational repression or degradation of mRNA targets occur. Conserved base-pairing between the 3´ untranslated region (UTR) of the mRNA and the “seed sequence” located in the 5´ end of the miRNA also have an important function.16 Through evaluation of the minimal requirements for miRNA:mRNA interactions, Brennecke and colleagues identified two categories of target sites; the 5´-dominant sites that require little to no support from the 3´ miRNA end for function due to a high degree of complimentary and the 3´-compensatory sites that generally have sufficient complimentarity to the 5´ end of the miRNA.17 Interestingly, 3´-compensatory sites may not necessarily increase efficacy of interaction and function.18 There are four types of 5´-dominant interaction sites: 6mer, 7mer-m8, 7mer-A1, and 8mer.18 The 6-mer site matches perfectly to the seed sequence (2–7 nt), while the 7mer-m8 also matches these nucleotides with additional nucleotide in the 8 position. The 7mer-A1 matches the seed sequence which is augmented by an Ala in the first position of the miRNA. Lastly, the 8mer is a combination of the two 7mer sites and is comprised of a match seed sequence with the Ala in the first position and the matched 8th position of the miRNA. When the 8mer is the sole site present in the 3´ UTR of an mRNA target, deregulation of the mRNA target is optimal. Notably, multiple sites in the 3´ UTR of the targeted mRNA are better than one.17–20 However cooperation and spacing between these sites are also important factors .18 The number of mismatches between the miRNA and it target dictate the end result. If perfect complimentarity exists, then generally the target is degraded through ubiquitination. Imperfect matches mainly predict inhibition of protein translation. In some cases, the perfect match of one miRNA is not sufficient to degrade its target but rather multiple miRNAs are required to bind the 3´ UTR for a full effect. Given the ubiquitous nature of miRNA/mRNA targeting, several prediction databases have been developed based on in silico analysis. Many of these score the free-energy generated between 5´ and 3´ base-pairing, consider phylogentic conservation, and/or limit target identification by specific 5´ –interaction sites. For example, TargetScan (http://www.targetscan.org/) considers matches in the 3´ UTR for only 7mer and 8mer interactions sites.18,21,22 The Sanger Database (http://microrna.sanger.ac.uk/targets/v5) uses the miRanda algorithm which weighs 5´ miRNA complementary in the seed sequence, thermodynamic calculations for the RNA duplex, and conservation across species.23,24 Recently, the miRDB database/MirTarget2 (http://mirdb.org/miRDB) was developed using a machine learning approach to analyze miRNA targets that are conserved and non-conserved across species. Comparison of these databases using a specific dataset indicated that the MirTarget2 performed similarly to TargetScan.25 These bioinformatic tools are very useful in providing cues on the function of a particular miRNA. The predicted targets generated from these or other databases should then be validated experimentally. Using luciferase assays, by cloning the 3´-UTR of target gene into a luciferase reporter gene downstream and co-transfection with miRNA to observe the inhibition, is the standard method to confirm the direct interaction between miRNA and target genes.26 Western blotting or in situ hybridization can be used to confirm the interaction at protein expression level for miRNA and target protein localization.
There are several high-throughput approaches to quantify miRNAs in tissue samples but the gold standard is unclear. cDNA oligonucleotide arrays have become a standard global scale technique for miRNA profiling.27 MiRNA microarrays hybridize at a lower melting temperature compared with traditional cDNA arrays, therefore, probe design is critical. PCR arrays also provide another high-throughput approach to detect miRNA in a plate form.28 Cloning of all miRNAs is another technique mainly used to identify miRNAs and counting the number of sequenced miRNAs in a library also provides estimation of miRNA distribution in a specific tissue.29
Northern blotting using high percentage of urea-acrylamide gels was initially one of the primary modalities used to detect individual small RNAs. An advantage of northern blotting is that it can simultaneously identify both observe precursor and mature miRNA. Real time PCR has been successfully used for detecting low copy number precursor and mature miRNA with high sensitivity and specificity. Stem-loop RT-PCR which provides primers and probe binding sites by incorporating elongated stem-loop RT primer can be utilized for accurate miRNA quantification.30 In situ hybridization can provide the quantification and the distribution of miRNA simultaneously.31(Figure 2) Alternatively, bead-based flow cytometric miRNA expression profiling methods combining xMAP with Locked Nucleic Acids (LNAs) technology is just emerging. The principle in this approach is that biotin label total RNA is hybridized with LNA-modified capture probes coupled to xMAP beads, then a streptavidin-phycoerythrin reporter molecule is for detection on the Luminex analyser.32
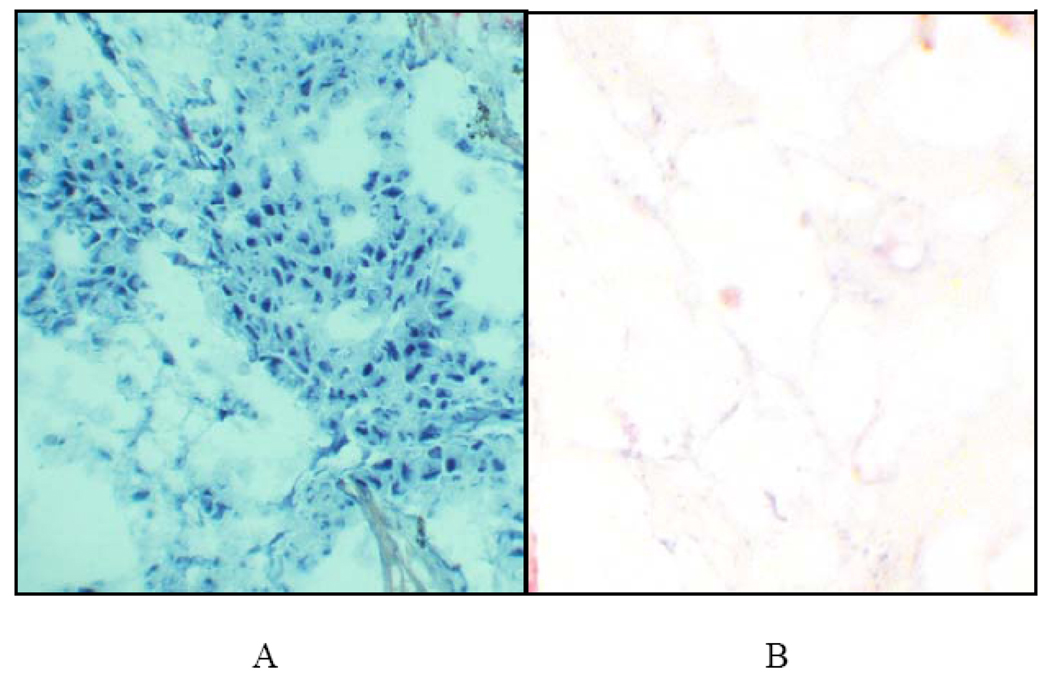
Figure 2.
In situ detection of miR-222 in squamous cell cancer of the lung. MiR-221 and 222 are commonly found in cancers, and can impact similar targets. MiR-222 was detected in most of the malignant cells of this squamous cell cancer of the lung using a digoxigenin-tagged LNA probe (panel A); the signal is blue and the counterstain pink. In comparison, miR-222 was not evident in the cells of this section of the adjacent normal lung (panel B).
miRNA Regulation
MiRNAs are often conserved across species. Not only do miRNAs serve as important regulators of gene expression, but their expression is also regulated by multiple factors. The genomic organization of miRNAs often dictates translational control. MiRNAs tend to be located in fragile chromosomal regions that are susceptible to translocations, microdeletions and amplifications.33 Allelic loss is a frequent genetic alteration in human lung cancers. Cytogenetic and molecular analyses have revealed chromosomal deletion on 3p, 8p, 9p, 11p, 13q, 17p, 18q and 22q.34 In fact, several miRNAs located in these regions are currently being studied. Most miRNAs are located in genomic regions distant from annotated genes and encoded in independent transcription units.10 Approximately one quarter of human miRNAs are within introns of mRNAs in the same orientation as these miRNAs, suggesting that they are not transcribed from their own promoters but processed from introns. Other miRNAs are clustered in the genome and transcribed as a polycistronic primary transcript, suggesting their functional relationships. To date, 148 miRNAs have been identified within 51 clusters. 35 Each of these clusters contains an upstream promoter region. For example, the miR-17~92 cluster encodes seven miRNAs and is regulated by promoter elements for c-Myc, and E2F1-3 transcription factors.36,37 Dews and colleagues reported that the c-Myc transcription factor upregulates this cluster leading to the promotion of angiogenesis in solid tumors .36 This finding has led to the hypothesis that increased expression of c-Myc in malignancies would augment the expression of miRNAs. Surprisingly, over-expression of c-Myc in B-cell lymphomas leads to global decrease in miRNAs expression.38 It is predicted that c-Myc binds promoter elements to inhibit translation. Another important cluster in tumorigenesis is the let-7 family which is decreased in many malignancies. 39 Despite these clusters being regulated by a promoter, not all the miRNAs contained within the cluster are similarly expressed, suggesting a second level of post-transcriptional regulation.40 During embryogenesis, the pri-miRNA for the let-7 cluster is abundant whereas the pre-miRNAs and mature miRNAs from this cluster are absent. 41 Newman and colleagues identified highly conserved nucleotides in the loop regions of the primary transcript for let-7 cluster to which a Drosha inhibitor binds preventing the processing of the pri-miRNA.42
Tumor suppressors within lung cancer are often susceptible to epigenetic silencing by methylation.43 The miR-29 family functionally targets DNA methytransferases (DNMT) 3A and −3B, two key enzymes involved in DNA methylation, and often up-regulated in several malignancies including lung cancer.44 Re-expression of miR-29s in lung cancer cells restores the normal DNA methylation patterns and reduced tumor development.45 MiRNA expression profiling comparing DNA methyltransferase deficient cells with normal HCT116 cells found that miR-124a underwent inactivation by CpG island hypermethylation in human tumors including lung cancers.46
Investigators have determined that polymorphisms within the 3´UTR of a target mRNA may affect miRNA functional targeting as well. MiRNAs may also be regulated by other miRNAs and environmental factors such as hypoxia and cigarette smoke. For example, miRNAs appear to be globally altered following cigarette smoke exposure.47,48 However, the mechanisms by which this occurs and biological implications for lung tumorigenesis remain to be explored.
miRNA and Lung Cancer Cell Phenotype
Several miRNAs have been implicated in lung cancer by altering fundamental cellular processes (Table 1) thus making them attractive candidates as therapeutic targets. Both miR-221 and miR-222 are up-regulated in TNF-alpha related apoptosis-inducing ligand cell lines. Silencing of these miRNAs sensitized resistant cell lines to TRAIL agents.49 MiR-200c may enhance the initiation of an invasive phenotype through decreasing the expression of zinc finger transcription factor transcription factor 8 (TCF8), which is an inhibitor of E-cadherin that is a crucial event in epithelial-to-mesenchymal transition.50 Nasser et al recently showed that the muscle enriched miR-1 inhibited both in vitro growth and survival in NSCLC cell lines through functional targeting of several oncogenes including MET, Pim-1, FoxP1 and HDAC4. In addition, miR-1 sensitized cells to chemotherapeutic mediated apoptosis.51 MiR-21 is over-expressed in several solid malignancies including glioblastoma, breast cancer and lung cancer. MiR-21 targets several tumor suppressors including PDCD4 and has anti-apoptotic properties.52 In lung cancer, increased tumor expression of miR-21 carried a poor overall survival but did not correlate with histological features.53 MiR-126 alters both tumor growth and metastatic potential in breast cancer.54 A recent investigation suggested that miR-126 may alter lung cancer cell migratory and invasive capacity through functional targeting of Crk.55
Table 1.
MiRNA implicated in lung cancer
| miRNA | Function | Target gene | Reference |
|---|---|---|---|
| miR-1 | proliferation apoptosis | MET, Pim-1, FoxP1 and HDAC4 | 50 |
| miR-7 | EGFR | 72 | |
| Let-7 | proliferation | Cdk6, CDC25, NRAS, KRAS, HMGA2 | 61–64, |
| miR-17-92 | angiogenesis proliferation | Tsp1, CTGF HIF1α | 36 |
| miR-21 | proliferation | PDCD4 | 52 |
| miR-29 | demethylation | DNMT 3A and 3B | 45 |
| miR-126 | invasion | CRK | 55 |
| miR-128b | EGFR | 71 | |
| miR-200c | Epithelial mesenchymal transition | TCF 8 | 50 |
| miR-221,miR-222 | apoptosis | p27 (kip1), Kit | 49 |
miRNA Biogenesis and Lung Cancer
Global repression of miRNA maturation by targeting three key components of miRNA processing machinery decreased miRNA expression in steady-state miRNA levels and caused a pronounced transformed phenotype in lung cancer cells. Impaired processing through in vitro targeting of Drosha, DGCR8 and Dicer resulted in accelerated colony formation and growth in soft agar and in vivo tumor development.56 Karube et al, demonstrated in a cohort of 67 lung cancer patients that reduced tumor expression of Dicer predicted a poor post resection prognosis independent of disease stage.57 Chiosea et al reported a transient increase in Dicer expression in both atypical adenomatous hyperplasia and bronchoalveolar carcinoma followed by a steady decrease in expression with more advanced invasive adenocarcinoma.58 This would indicate that abrogation of global miRNA processing promotes tumorigenesis.
Let-7
Let-7 was the first miRNA reported to be aberrantly expressed in human lung cancer.59 The RAS 3´-UTR contains multiple let-7 complementary sites making it a functional target.60 HMGA2, another oncogene in a variety of tumors including lung cancers and mesenchymal tumors, is also a predicted target of let-7.61 Although both RAS and HMGA2 are let-7 targets, K-Ras seems to be stronger than HMGA2 in rescuing let-7g mediated tumor suppression.62 Over-expression of let-7 in the A549 cell line inhibited cell growth and reduced cell cycle progression and division.63 In a murine model of K-Ras(G12D) induced lung cancer let-7g significantly reduced the number, size and volume of murine tumors and human non-small cell lung xenografts.62 In addition, intranasal let-7 administration reduced tumor formation in a similar K-Ras (G12D) model.64 These findings have translated to human application. For example, analysis of a small cohort of early in situ bronchioloalveolar carcinomas, well-differentiated adenocarcinomas and invasive adenocarcinomas revealed that let-7 is important to early occurrence in carcinogenesis but is not related to prognosis.65 Furthermore, these observations are supported clinically by the correlation between reduced let-7 expression in 143 resected lung cancer cases and poor clinical outcome.59
miR-17~92 Cluster
Although Dicer affects miRNA maturation, the miRNA cluster miR-17~92 is important in lung development and homeostasis. MiR-17~92 cluster expression is high in embryonic development and steadily declines throughout development and into adulthood.66 The miR-17~92 cluster was first identified as a potential oncogene in B-cell lymphoma.67 This Myc-activated miRNA cluster can increase tumor vasculature and form larger, better-perfused tumors by functionally targeting anti-angiogenic thrombosopndin-1 (Tsp1) and connective tissue growth factor (CTGF) genes.36 In human lung cancers, particularly small cell carcinoma, miR-17~92 is also over-expressed and in vitro introduction enhanced cell proliferation.66 Antisense oligonucletide mediated inhibition of both miR-17-5p and miR-20a induced apoptosis selectively in lung cancer cells over-expressing miR-17~92, suggesting oncogenic potential.68
miRNA and Epidermal Growth Factor
The presence of abnormal levels of growth factor and its receptor can deregulate the normal cell growth process and initiate cancer. Epidermal growth factor receptor (EGFR) has increased expression in several malignancies and has become the focus of targeted therapeutics.69 EGFR induces cancer in three ways: overexpression of EGFR ligands, amplification of EGFR, and mutational activation of EGFR.69 Mutations in the EGFR tyrosine kinase in NSCLC can cause oncogenic transformation and change the sensitivity to tyrosine kinase inhibitors such as gefitinib.70 Two recent studies demonstrated that EGFR may be a functional target of miRNAs. MiRNA-128b, located on chromosome 3p which is frequently deleted in lung cancers, directly down-regulated EGFR.71 However, EGFR expression and mutation status did not correlate with survival outcome, suggesting a minor effect of this miRNA on EGFR regulation. Webster et al determined in vitro that EGFR was a functional target of let-7.72 Let-7 also altered expression patterns of several other signaling effectors including protein kinase B (Akt) and extracellular signal regulated kinase 1/2 (ERK 1/2). 72
Polymorphisms and miRNA expression
Single-nucleotide polymorphisms (SNPs), are major determinants of variations in disease susceptibility, medication response and toxicity.73 Thirty-seven SNPs have been identified in the p53 gene alone.74 The presence of polymorphisms in the 3´UTR of the target gene may also compromise the miRNA binding and regulation.75 In addition, SNPs in pre-miRNAs can alter miRNA processing, expression, and binding to target mRNA. The homozygous CC at SNP rs11614913 in miR-196a2 correlated with survival in individuals with NSCLC.76 This mutation enhanced the processing of pre-miRNA to its mature form and affected the miRNA binding ability to its target mRNA. Sequence regions within the 3´UTR of K-RAS which is a functional target of let-7 contain SNPs as well. Variant alleles within this SNP caused a 2.3 fold increased risk for NSCLC and in vitro suppression of let-7 mediated regulation of K-RAS.77
miRNAs Signatures and Lung Cancer
Platforms for global miRNA detection have been used to identify their involvement in lung carcinogenesis. Yanaihara et al compared miRNA patterns of expression in tumor versus adjacent uninvolved lung in 104 cases of lung cancer.78 They identified 43 differentially expressed miRNAs between lung tumors and adjacent uninvolved lung. In addition, five miRNAs (miR-155, 17-3p, let-7a-2, 145 and 21) predicted prognosis among patients with lung cancer.
Using an RT-PCR platform for miRNA analysis, Yu at al recently showed that a five miRNA signature correlated with overall disease free survival in a cohort of 122 patients with NSCLC.39 In vitro allied studies identified three high-risk miRNAs (miR-137, miR-182*, miR-372) that increased the invasive capacity in cancer cells while the protective miR-221 decreased invasion.
Another novel approach to identifying miRNA signatures involves examining target genes of lung enriched miRNAs. In one such study, investigators identified a 17 gene signature of genes targeted by miR-34b/34c/449 that accurately distinguished adenocarcinomas from squamous cell and carried a 90% sensitivity in detecting lung cancer in the airway when combined with cytopathology.79
Conclusion
Lung cancer remains a deadly disease that carries a poor five year prognosis. However, studies focused on elucidating the molecular heterogeneity are proving successful in identifying novel targets and subcategorizing clinical phenotypes. MiRNAs represent newly identified molecules that have the capacity of multiple gene and thus pathways regulation. To date, several studies have suggested that miRNAs demonstrate discrete patterns of expression in lung cancer and are likely critical to the pathogenesis of lung cancer. Therefore, the study of miRNAs in lung cancer may complement other more established platforms. We still have only a preliminary understanding of miRNAs and how targeting both in vitro and murine studies may be translated to human disease. Several questions remain: What genes regulate miRNAs? Which miRNAs regulate which genes? What are the mechanisms of miRNA function? Given the multitude of genes that may be targeted by a single miRNA, how can miRNAs be applied as targeted therapies in vivo while avoiding deleterious effects? With the development of techniques of miRNA cloning, more functional miRNAs related to lung cancer will be discovered. In addition, the identification of miRNAs by non-invasive means ( bronchoalveolar lavage and peripheral blood) are being studied. MiRNA offer a promising platform that has the potential to eventually influence the prevention, diagnosis, prognosis, and therapy of lung cancer.
Acknowledgments
This work was supported by National Institutes of Health Grants #HL077717 (S.P.N.) and Chest/LUNGevity Foundation Grant (S.P.N.).
Reference List
- 1.Jemal A, Siegel R, Ward E, Murray T, Xu J, Thun MJ. Cancer statistics. 2007 10 CA Cancer J Clin. 2007;57:43–66. doi: 10.3322/canjclin.57.1.43. [DOI] [PubMed] [Google Scholar]
- 2.Beer DG, Kardia SL, Huang CC, et al. Gene-expression profiles predict survival of patients with lung adenocarcinoma. Nat Med. 2002;8:816–824. doi: 10.1038/nm733. [DOI] [PubMed] [Google Scholar]
- 3.Shedden K, Taylor JM, Enkemann SA, et al. Gene expression-based survival prediction in lung adenocarcinoma: a multi-site, blinded validation study. Nat Med. 2008;14:822–827. doi: 10.1038/nm.1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Meyerson M, Carbone D. Genomic and proteomic profiling of lung cancers: lung cancer classification in the age of targeted therapy. J Clin Oncol. 2005;23:3219–3226. doi: 10.1200/JCO.2005.15.511. [DOI] [PubMed] [Google Scholar]
- 5.Lee RC, Feinbaum RL, Ambros V. The C.elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 6.Chen CZ. MicroRNAs as oncogenes and tumor suppressors. N Engl J Med. 2005;353:1768–1771. doi: 10.1056/NEJMp058190. [DOI] [PubMed] [Google Scholar]
- 7.Calin GA, Dumitru CD, Shimizu M, et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 9.Murphy D, Dancis B, Brown JR. The evolution of core proteins involved in microRNA biogenesis. BMC Evol Biol. 2008;8:92. doi: 10.1186/1471-2148-8-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 11.Gregory RI, Yan KP, Amuthan G, et al. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- 12.Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18:3016–3027. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gregory RI, Shiekhattar R. MicroRNA biogenesis and cancer. Cancer Res. 2005;65:3509–3512. doi: 10.1158/0008-5472.CAN-05-0298. [DOI] [PubMed] [Google Scholar]
- 14.Lund E, Guttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- 15.Mi S, Cai T, Hu Y, et al. Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5’ terminal nucleotide. Cell. 2008;133:116–127. doi: 10.1016/j.cell.2008.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 17.Brennecke J, Stark A, Russell RB, Cohen SM. Principles of microRNA-target recognition. PLoS Biol. 2005;3:e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing 1. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Doench JG, Sharp PA. Specificity of microRNA target selection in translational repression. Genes Dev. 2004;18:504–511. doi: 10.1101/gad.1184404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lai EC, Wiel C, Rubin GM. Complementary miRNA pairs suggest a regulatory role for miRNA:miRNA duplexes. RNA. 2004;10:171–175. doi: 10.1261/rna.5191904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 22.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 23.John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human MicroRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Enright AJ, John B, Gaul U, Tuschl T, Sander C, Marks DS. MicroRNA targets in Drosophila. Genome Biol. 2003;5:R1. doi: 10.1186/gb-2003-5-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang X, El N I. Prediction of both conserved and nonconserved microRNA targets in animals. Bioinformatics. 2008;24:325–332. doi: 10.1093/bioinformatics/btm595. [DOI] [PubMed] [Google Scholar]
- 26.Eubank TD, Roberts R, Galloway M, Wang Y, Cohn DE, Marsh CB. GM-CSF induces expression of soluble VEGF receptor-1 from human monocytes and inhibits angiogenesis in mice. Immunity. 2004;21:831–842. doi: 10.1016/j.immuni.2004.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu CG, Spizzo R, Calin GA, Croce CM. Expression profiling of microRNA using oligo DNA arrays. Methods. 2008;44:22–30. doi: 10.1016/j.ymeth.2007.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiang J, Lee EJ, Gusev Y, Schmittgen TD. Real-time expression profiling of microRNA precursors in human cancer cell lines. Nucleic Acids Res. 2005;33:5394–5403. doi: 10.1093/nar/gki863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Landgraf P, Rusu M, Sheridan R, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen C, Ridzon DA, Broomer AJ, et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nuovo GJ. In situ detection of precursor and mature microRNAs in paraffin embedded, formalin fixed tissues and cell preparations. Methods. 2008;44:39–46. doi: 10.1016/j.ymeth.2007.10.008. [DOI] [PubMed] [Google Scholar]
- 32.Becker JL, Steigbigel RT, Nuovo GJ. In situ detection of PCR-amplified HIV-1 and EBV nucleic acids in hyperplastic lymph nodes and in AIDS-related lymphoma. J Histochem Cytochem. 1996;44:1085–1089. doi: 10.1177/44.10.8813072. [DOI] [PubMed] [Google Scholar]
- 33.Di Leva G, Calin GA, Croce CM. MicroRNAs: Fundamental facts and involvement in human diseases. Birth Defects Res C Embryo Today. 2006;78:180–189. doi: 10.1002/bdrc.20073. [DOI] [PubMed] [Google Scholar]
- 34.Osada H, Takahashi T. Genetic alterations of multiple tumor suppressors and oncogenes in the carcinogenesis and progression of lung cancer. Oncogene. 2002;21:7421–7434. doi: 10.1038/sj.onc.1205802. [DOI] [PubMed] [Google Scholar]
- 35.Yu J, Wang F, Yang GH, et al. Human microRNA clusters: genomic organization and expression profile in leukemia cell lines. Biochem Biophys Res Commun. 2006;349:59–68. doi: 10.1016/j.bbrc.2006.07.207. [DOI] [PubMed] [Google Scholar]
- 36.Dews M, Homayouni A, Yu D, et al. Augmentation of tumor angiogenesis by a Myc-activated microRNA cluster. Nat Genet. 2006;38:1060–1065. doi: 10.1038/ng1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sylvestre Y, De G V, Querido E, et al. An E2F/miR-20a autoregulatory feedback loop. J Biol Chem. 2007;282:2135–2143. doi: 10.1074/jbc.M608939200. [DOI] [PubMed] [Google Scholar]
- 38.Chang TC, Yu D, Lee YS, et al. Widespread microRNA repression by Myc contributes to tumorigenesis. Nat Genet. 2008;40:43–50. doi: 10.1038/ng.2007.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu SL, Chen HY, Chang GC, et al. MicroRNA signature predicts survival and relapse in lung cancer 4. Cancer Cell. 2008;13:48–57. doi: 10.1016/j.ccr.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 40.He L, He X, Lim LP, et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thomson JM, Newman M, Parker JS, Morin-Kensicki EM, Wright T, Hammond SM. Extensive post-transcriptional regulation of microRNAs and its implications for cancer. Genes Dev. 2006;20:2202–2207. doi: 10.1101/gad.1444406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Newman MA, Thomson JM, Hammond SM. Lin-28 interaction with the Let-7 precursor loop mediates regulated microRNA processing. RNA. 2008;14:1539–1549. doi: 10.1261/rna.1155108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Esteller M. Epigenetics in cancer. N Engl J Med. 2008;358:1148–1159. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- 44.Kim H, Kwon YM, Kim JS, et al. Elevated mRNA levels of DNA methyltransferase-1 as an independent prognostic factor in primary nonsmall cell lung cancer. Cancer. 2006;107:1042–1049. doi: 10.1002/cncr.22087. [DOI] [PubMed] [Google Scholar]
- 45.Fabbri M, Garzon R, Cimmino A, et al. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc Natl Acad Sci U S A. 2007;104:15805–15810. doi: 10.1073/pnas.0707628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lujambio A, Esteller M. CpG island hypermethylation of tumor suppressor microRNAs in human cancer. Cell Cycle. 2007;6:1455–1459. [PubMed] [Google Scholar]
- 47.Izzotti A, Calin GA, Arrigo P, Steele VE, Croce CM, De FS. Downregulation of microRNA expression in the lungs of rats exposed to cigarette smoke. FASEB J. 2008 doi: 10.1096/fj.08-121384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schembri F, Sridhar S, Perdomo C, et al. MicroRNAs as modulators of smoking-induced gene expression changes in human airway epithelium. Proc Natl Acad Sci U S A. 2009 doi: 10.1073/pnas.0806383106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Garofalo M, Quintavalle C, Di LG, et al. MicroRNA signatures of TRAIL resistance in human non-small cell lung cancer. Oncogene. 2008;27:3845–3855. doi: 10.1038/onc.2008.6. [DOI] [PubMed] [Google Scholar]
- 50.Hurteau GJ, Carlson JA, Spivack SD, Brock GJ. Overexpression of the microRNA hsa-miR-200c leads to reduced expression of transcription factor 8 and increased expression of E-cadherin. Cancer Res. 2007;67:7972–7976. doi: 10.1158/0008-5472.CAN-07-1058. [DOI] [PubMed] [Google Scholar]
- 51.Nasser MW, Datta J, Nuovo G, et al. Down-regulation of micro-RNA-1 (miR-1) inlung cancer. Suppression of tumorigenic property of lung cancer cells and their sensitization to doxorubicin-induced apoptosis by miR-1. J Biol Chem. 2008;283:33394–33405. doi: 10.1074/jbc.M804788200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 52.Chan JA, Krichevsky AM, Kosik KS. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65:6029–6033. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- 53.Markou A, Tsaroucha EG, Kaklamanis L, Fotinou M, Georgoulias V, Lianidou ES. Prognostic value of mature microRNA-21 and microRNA-205 overexpression in non-small cell lung cancer by quantitative real-time RT-PCR. Clin Chem. 2008;54:1696–1704. doi: 10.1373/clinchem.2007.101741. [DOI] [PubMed] [Google Scholar]
- 54.Tavazoie SF, Alarcon C, Oskarsson T, et al. Endogenous human microRNAs that suppress breast cancer metastasis. Nature. 2008;451:147–152. doi: 10.1038/nature06487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Crawford M, Brawner E, Batte K, et al. MicroRNA-126 inhibits invasion in non-small cell lung carcinoma cell lines. Biochem Biophys Res Commun. 2008;373:607–612. doi: 10.1016/j.bbrc.2008.06.090. [DOI] [PubMed] [Google Scholar]
- 56.Kumar MS, Lu J, Mercer KL, Golub TR, Jacks T. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat Genet. 2007;39:673–677. doi: 10.1038/ng2003. [DOI] [PubMed] [Google Scholar]
- 57.Karube Y, Tanaka H, Osada H, et al. Reduced expression of Dicer associated with poor prognosis in lung cancer patients. Cancer Sci. 2005;96:111–115. doi: 10.1111/j.1349-7006.2005.00015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chiosea S, Jelezcova E, Chandran U, et al. Overexpression of Dicer in precursor lesions of lung adenocarcinoma. Cancer Res. 2007;67:2345–2350. doi: 10.1158/0008-5472.CAN-06-3533. [DOI] [PubMed] [Google Scholar]
- 59.Takamizawa J, Konishi H, Yanagisawa K, et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64:3753–3756. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- 60.Johnson SM, Grosshans H, Shingara J, et al. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 61.Lee YS, Dutta A. The tumor suppressor microRNA let-7 represses the HMGA2 oncogene. Genes Dev. 2007;21:1025–1030. doi: 10.1101/gad.1540407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kumar MS, Erkeland SJ, Pester RE, et al. Suppression of non-small cell lung tumor development by the let-7 microRNA family. Proc Natl Acad Sci U S A. 2008;105:3903–3908. doi: 10.1073/pnas.0712321105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Johnson CD, Esquela-Kerscher A, Stefani G, et al. The let-7 microRNA represses cell proliferation pathways in human cells. Cancer Res. 2007;67:7713–7722. doi: 10.1158/0008-5472.CAN-07-1083. [DOI] [PubMed] [Google Scholar]
- 64.Esquela-Kerscher A, Trang P, Wiggins JF, et al. The let-7 microRNA reduces tumor growth in mouse models of lung cancer. Cell Cycle. 2008;7:759–764. doi: 10.4161/cc.7.6.5834. [DOI] [PubMed] [Google Scholar]
- 65.Inamura K, Togashi Y, Nomura K, et al. let-7 microRNA expression is reduced in bronchioloalveolar carcinoma, a non-invasive carcinoma, and is not correlated with prognosis. Lung Cancer. 2007;58:392–396. doi: 10.1016/j.lungcan.2007.07.013. [DOI] [PubMed] [Google Scholar]
- 66.Hayashita Y, Osada H, Tatematsu Y, et al. A polycistronic microRNA cluster, miR-17-92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res. 2005;65:9628–9632. doi: 10.1158/0008-5472.CAN-05-2352. [DOI] [PubMed] [Google Scholar]
- 67.He L, Thomson JM, Hemann MT, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Matsubara H, Takeuchi T, Nishikawa E, et al. Apoptosis induction by antisense oligonucleotides against miR-17-5p and miR-20a in lung cancers overexpressing miR-17-92. Oncogene. 2007;26:6099–6105. doi: 10.1038/sj.onc.1210425. [DOI] [PubMed] [Google Scholar]
- 69.Pao W, Miller VA. Epidermal growth factor receptor mutations, small-molecule kinase inhibitors, and non-small-cell lung cancer: current knowledge and future directions. J Clin Oncol. 2005;23:2556–2568. doi: 10.1200/JCO.2005.07.799. [DOI] [PubMed] [Google Scholar]
- 70.Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib 6. N Engl J Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 71.Weiss GJ, Bemis LT, Nakajima E, et al. EGFR regulation by microRNA in lung cancer: correlation with clinical response and survival to gefitinib and EGFR expression in cell lines. Ann Oncol. 2008;19:1053–1059. doi: 10.1093/annonc/mdn006. [DOI] [PubMed] [Google Scholar]
- 72.Webster RJ, Giles KM, Price KJ, Zhang PM, Mattick JS, Leedman PJ. Regulation of epidermal growth factor receptor signaling in human cancer cells by microRNA-7. J Biol Chem. 2008 doi: 10.1074/jbc.M804280200. [DOI] [PubMed] [Google Scholar]
- 73.Sauna ZE, Kimchi-Sarfaty C, Ambudkar SV, Gottesman MM. Silent polymorphisms speak: how they affect pharmacogenomics and the treatment of cancer. Cancer Res. 2007;67:9609–9612. doi: 10.1158/0008-5472.CAN-07-2377. [DOI] [PubMed] [Google Scholar]
- 74.Soussi T, Wiman KG. Shaping genetic alterations in human cancer: the p53 mutation paradigm. Cancer Cell. 2007;12:303–312. doi: 10.1016/j.ccr.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 75.Tan Z, Randall G, Fan J, et al. Allele-specific targeting of microRNAs to HLA-G and risk of asthma. Am J Hum Genet. 2007;81:829–834. doi: 10.1086/521200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hu Z, Chen J, Tian T, et al. Genetic variants of miRNA sequences and non-small cell lung cancer survival. J Clin Invest. 2008;118:2600–2608. doi: 10.1172/JCI34934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chin LJ, Ratner E, Leng S, et al. A SNP in a let-7 microRNA complementary site in the KRAS 3’ untranslated region increases non-small cell lung cancer risk. Cancer Res. 2008;68:8535–8540. doi: 10.1158/0008-5472.CAN-08-2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yanaihara N, Caplen N, Bowman E, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 79.Liang Y. An expression meta-analysis of predicted microRNA targets identifies a diagnostic signature for lung cancer. BMC Med Genomics. 2008;1:61. doi: 10.1186/1755-8794-1-61. [DOI] [PMC free article] [PubMed] [Google Scholar]