Abstract
Recent studies indicate that extracellular mechanical properties, including rigidity, profoundly affect cellular morphology, growth, migration, and differentiation [R. J. Pelham, Jr. and Y. Wang, Proc. Natl. Acad. Sci. U. S. A., 1997, 94(25), 13661–13665; H. B. Wang, M. Dembo and Y. L. Wang, Am. J. Physiol. Cell Physiol., 2000, 279(5), C1345–C1350; P. C. Georges, and P. A. Janmey, J. Appl. Physiol., 2005, 98(4), 1547–1553; C. M. Lo, H. B. Wang, M. Dembo and Y. L. Wang, Biophys. J., 2000. 79(1), 144–152; D. E. Discher, P. Janmey and Y. L. Wang, Science, 2005, 310(5751), 1139–1143; A. J. Engler, M. A. Griffin, S. Sen, C. G. Bonnemann, H. L. Sweeney and D. E. Discher, J. Cell Biol., 2004, 166(6), 877–887]. However, most studies involving rigidity sensing have been performed by comparing cells on separate substrata of fixed stiffness. To allow spatial and/or temporal manipulation of mechanical properties, we have developed a modulatable hydrogel by reacting linear polyacrylamide (PA) with a photosensitive crosslinker. This material allows UV-mediated control of rigidity, softening by 20–30% upon irradiation at a dose tolerated by live cells. Global UV irradiation induces an immediate recoiling of 3T3 fibroblasts and a reduced spread area at steady state. Furthermore, localized softening of the posterior substratum of polarized cells causes no apparent effect, while softening of the anterior substratum elicits pronounced retraction, indicating that rigidity sensing is localized to the frontal region. This type of material allows precise spatial and temporal control of mechanical signals for both basic research and regenerative medicine.
Introduction
Mechanosensing is believed to affect cancerous invasion, wound healing, stem cell differentiation, and many other physiological and pathological processes.1–8 The rigidity of adhesive substrata plays a particularly important role in cell regulation.9–11 Due to their favorable optical and tunable elastic properties, polyacrylamide (PA) gels have been used widely as adhesive substrata for manipulating mechanical cues.12 For example, stiff PA gels promote spreading and scattering of adherent cells,6,13,14 while soft PA gels promote soft tissue differentiation and tissue-like cell-cell associations.3,7,8,13,15 In addition, adherent cells migrate preferentially toward stiffer regions,1,16 a process known as durotaxis that may play a role in guiding cell migration in conjunction with chemical signals.
Most applications of PA gels in cell mechanics have involved substrata of constant rigidity,12 which limits the ability to study dynamic cellular responses. The large variability among cells further adds to the difficulty. Thus, the spatial and temporal mechanisms of mechanosensing remain elusive. These limitations motivated us to create gels with a rigidity that can be modulated in situ under conditions compatible with live cells. In particular, light-induced modulation allows a high degree of versatility while avoiding direct mechanical perturbations or cellular displacements that may confound the effects.
While gels that stiffen upon UV illumination have been reported,17 the typical use of photo-activated free radical generators raises serious concerns over toxic side effects to live cells. Therefore, we chose to develop a substratum that softens upon UV illumination, by crosslinking polymers with a photocleavable crosslinking reagent. Cell spreading is promoted by coating the surface with a thin layer of polyacrylamide-based material conjugated to fibronectin (FN). These gels can be softened at a specific time in an area that encompasses either the entire cell (global illumination) or a specific region (localized illumination), to allow observations of cellular response to spatially and temporally controlled substrate softening. Application of this approach to migrating 3T3 cells suggests that cells maintain an internal homeostatic tension in response to substratum rigidity and such a response is localized to the cell anterior.
Results and discussion
Preparation of a UV-modulatable substratum
A gel is formed by crosslinking functionalized linear polyacrylamide (PA) with a UV-cleavable agent. PA was first activated with hydrazine hydrate to create polyacrylamide acryl hydrate (PAAH) with reactive amine groups (Fig. 1a).18 The linear polymers were then crosslinked with 4-bromomethyl-3-nitrobenzoic acid (BNBA).19 The bromomethyl group of BNBA undergoes nucleophilic reaction with the reactive amines on PAAH at elevated pH. The carboxyl group on BNBA was then linked with unmodified amines using 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDC), creating a gel (Fig. 1a). By systematically varying the reaction conditions, we created gels with an initial rigidity of ~7 kPa, where adherent fibroblasts were found to be highly sensitive to changes in substrate rigidity.20 This material contained on average ~0.08 mmol nitrobenzyl groups/g PAAH.
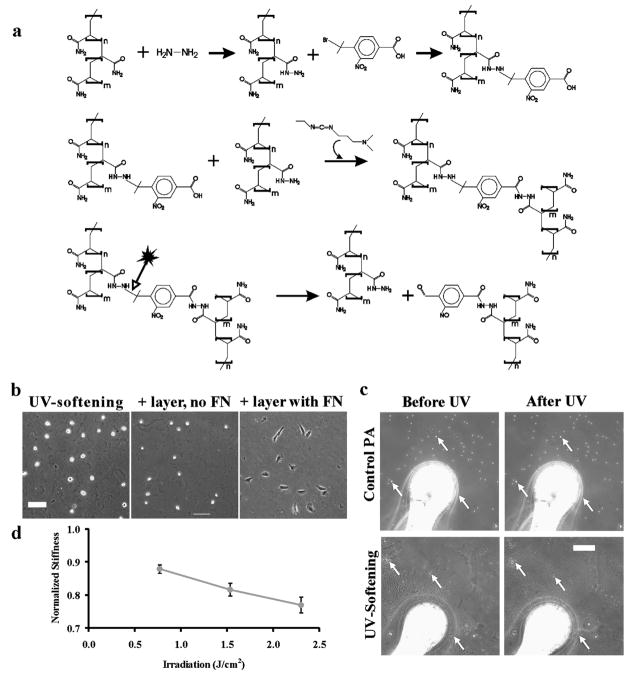
Fig. 1.
Preparation of a UV-softening, cell-adhesive gel. Linear polyacrylamide is functionalized with hydrazine hydrate followed by reaction with a UV-cleavable reagent, BNBA (a, upper reactions). Subsequent crosslinking of the polymers with EDC creates a gel (a, middle reaction). UV exposure cleaves near the nitrophenyl group and causes the gel to soften (a, lower reaction). While the gel is non-adhesive to cells (b, left panel), coating with a thin layer of PA gel with conjugated fibronectin allows cell adhesion (b, right panel; scale bar, 100 μm). UV irradiation softens the gel, as indicated by increased probe indentation that is reflected in the embedded beads going out of focus in the UV-softening gel only (c, arrows; scale bar, 20 μm; see also Movie 1, ESI†) that is not seen on light-insensitive control PA gels. Softening is dose-dependent (d, mean ± s.e.m., n = 10 from multiple gels). Scale bar, 100 μm.
Since crosslinked PAAH, like PA, is poorly adhesive for cells (Fig. 1a), its surface was further coated with a thin layer of FN-conjugated PA, prepared by copolymerizing acrylamide, bisacrylamide, and an NHS-ester derivative of acrylamide in the presence of FN (Fig. 1b),21 such that cells can sense rigidity changes in the underlying UV-sensitive gel. This versatile coating process also allows adhesions to be targeted at specific matrix proteins at controlled concentrations, independent of the underlying UV-sensitive gel. In addition, no small molecular byproducts capable of entering cells are released as a result of photolysis, as both sides of the cleaved crosslinkage remain associated with the polymer gel (Fig. 1a).
Response of modulatable substratum to UV irradiation
The nitrobenzyl group of BNBA used to crosslink the UV-softening gels is prone to efficient photolysis with 365 nm UV light, at an intensity similar to that used for the photo-activation of reagents caged with a nitrophenyl group. Changes in rigidity upon irradiation were probed with a microsphere indenter mounted on a micromanipulator22,23 (Fig. 1c and 1d). Irradiation softened the gel instantaneously, causing the probe to sink into the gel and beads sprinkled on the surface of the gel to jump out of focus (Fig. 1c, and Movie 1, ESI†). The amount of softening was dependent upon the energy of illumination (Fig. 1d). At maximal energy (see Experimental), we were able to soften the gels ~22% (Fig. 1d) from 7.2 ± 0.8 kPa to 5.5 ± 0.1 kPa (n = 10 each). In contrast, control PA gels, prepared with only the photo-insensitive FN-containing gel, were insensitive to UV exposure (E= 11.1 ± 1.0 kPa, n = 10; Fig. 1c.).
Cellular responses to global illumination of substrata
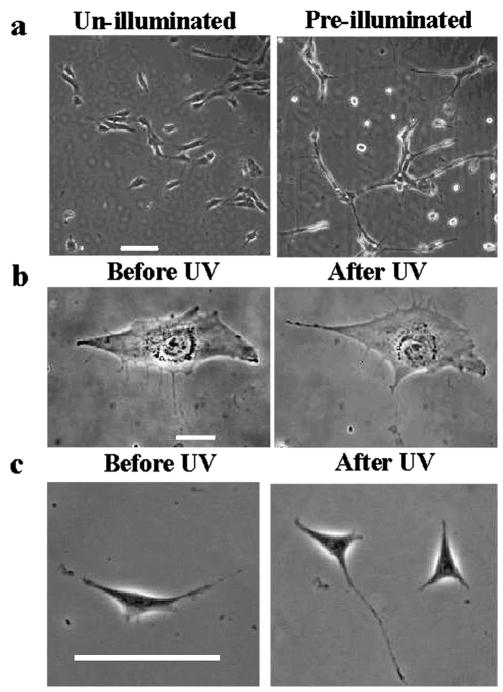
NIH 3T3 fibroblasts plated on un-illuminated and globally pre-illuminated UV-sensitive gels showed similar differences in morphology to cells on photo-insensitive stiff and soft PA gels, respectively12,21 (Fig. 2a). Equivalent illumination, when applied to cells on control PA gels or on FN-coated coverslips, caused no detectable response in spread area or protrusive activities (Fig. 2b and ESI Movie 2†), nor were there significant changes in migration speed (0.29 ± 0.03 and 0.25 ± 0.03 μm/min calculated over 2 h before and after UV, respectively, n = 8 each, p-value = 0.1126) or directional persistence24 (0.59 ± 0.10 and 0.66 ± 0.08 before and after UV, respectively, n = 8 each, p-value = 0.4890 see Experimental for the definition). Additionally, cell division proceeded normally after illumination (n = 6; Fig. 2c). The density of the illumination energy was estimated to be 2.3 J/cm2, which was much lower than that reported to cause radiation damage.25
Fig. 2.
Irradiation affects morphology of cells on UV-softening gels but not on control PA gels. Bulk irradiation of the gel surface inhibits spreading of NIH 3T3 cells on UV-softening substrates (a, right), as compared to cells on substrata without irradiation (a, left; scale bar, 100 μm). Cells on control PA gel also appear unaffected by the irradiation, showing similar morphology just before and 2 h after UV exposure (b; scale bar, 20 μm; see also ESI Movie 2†). An irradiated cell on an FN-coated coverslip went through normal mitosis, creating two daughter cells (c, right panel; scale bar, 100 μm).
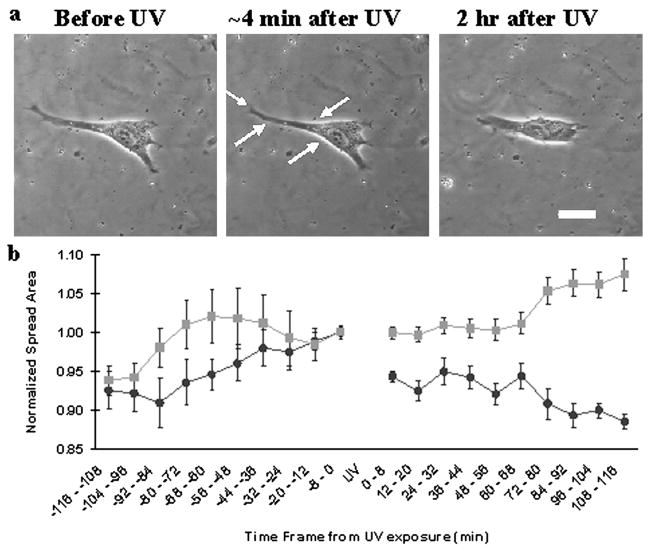
We examined the behavior of NIH 3T3 cells on UV-softening substrates before and after UV exposure that encompassed the entire cell. The spread area of cells decreased substantially upon whole-cell exposure to UV (n = 8, Fig. 3a and 3b; see also ESI Movie 3†). The response showed a rapid retraction (6 ± 1% reduction in area immediately after UV exposure, p-value <0.0001; ESI Movie 4†) followed by a gradual, further decline in spread area to reach a steady state (12 ± 1% reduction in area compared to just before UV, p-value <0.0001, Fig. 3a and 3b). The initial retraction may, in part, reflect recoiling as a result of lowered substrate resistance against cellular traction forces, as indicated by the inward movements of fluorescent beads embedded in the substrata (i.e., increase in strain; Fig. 3; see also ESI Movie 5†). The beads were released subsequently while the cell remained in a retracted state, consistent with a decrease of traction forces and release of focal adhesions as cells reached a steady state on the softened substratum.1
Fig. 3.
Responses of cells on UV-softening substrata to irradiation-induced substrate softening. Response to substrate softening includes an initial rapid retraction (a, middle; see also ESI Movie 4†) and a slower phase of reduction in spread area (a, right; scale bar, 20 μm; see also ESI Movie 3†). Measurements of cell area before and after irradiation at t = 0 shows the time course of retraction upon substrate softening (b, ●), while cells on control gels show no such response (b, ■). Spread area was normalized to the area just before UV and values are mean ± s.e.m. (n = 8).
Cellular responses to localized softening in posterior or anterior regions
By closing down the illumination field diaphragm to generate a UV beam ~50 μm in diameter, we compared the sensitivity of different regions of the cell to changes in substrate rigidity. Polarized cells with well-defined posterior and anterior regions were illuminated over either region, such that less than 50% of the nucleus was exposed in either case. Illumination of regions posterior to the nucleus caused no detectable response (Fig. 4 and ESI Movie 6†), nor did illuminations of cells on control PA gels in either the anterior or posterior region (ESI Movie 7† and not shown). The cell area increased by 3 ± 9% after illumination of the posterior region (n = 8; ESI Movie 6†). Similar results were obtained following the illumination of cells on control PA gels (13 ± 3% and 9 ± 2% 2 h following frontal and rear illumination respectively, n = 8 each; ESI Movie 7† and not shown). The slight increase in area was due to the selection of cells that were not undergoing spontaneous retraction prior to UV irradiation.
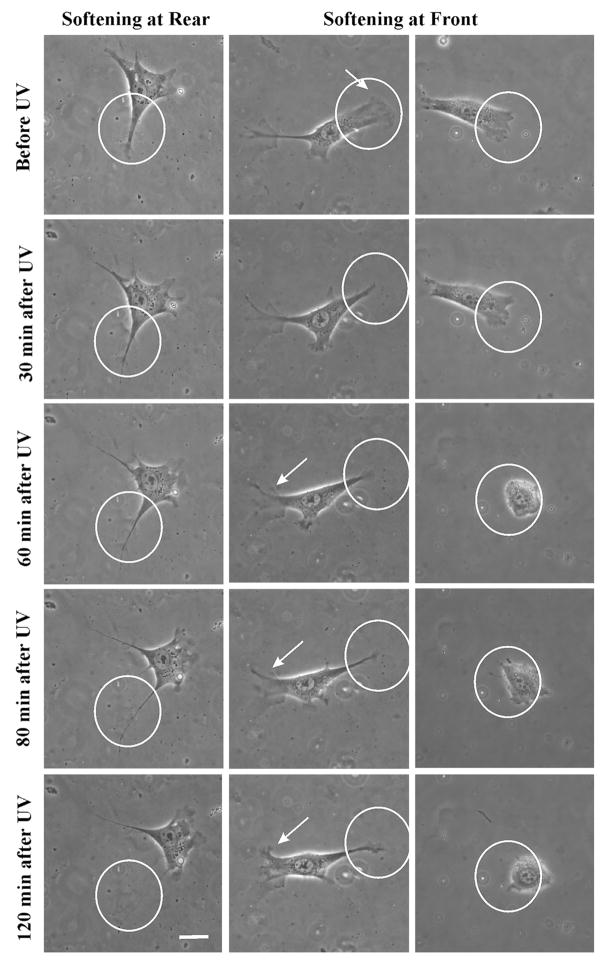
Fig. 4.
Localization of rigidity sensing to the cell anterior or posterior region. Softening of posterior substratum produces no change in spread area (left column; see also ESI Movie 6†). The cell often migrates away from the softened area (ESI Movie 5†). In contrast, softening of the anterior substratum elicits a dramatic response, whereby the cell either reverses polarity (central column, arrows; see also ESI Movie 8†) or becomes trapped in the softened region (right column; see also ESI Movie 9†). Circles indicate irradiated areas. Scale bar, 20 μm.
In contrast, softening the substratum region anterior to the nucleus caused a striking reduction in spread area (−16 ± 7%, 2 h following the irradiation, n = 8; see also ESI Movies 8 and 9†). The response to frontal illumination was more striking than that following global illumination, likely due to the stronger imbalance of forces when the softening was limited to part of the cell. There were two distinct types of response, in which the cell either reversed its direction of migration, typical of durotaxis (ESI Movie 8†), or migrated randomly and became trapped within/around the softened region (Fig. 4 and ESI Movie 9†). Cells that reversed their direction had a more branched morphology with secondary protrusions outside exposed areas, which expanded into a leading edge after illumination. These cells also showed strong recoil upon substrate softening, reminiscent of retraction induced spreading26 (ESI Movie 8†). In contrast, cells that became trapped had no secondary protrusions and showed weak frontal retraction upon illumination. These results indicate that rigidity sensing is largely confined to the anterior region, consistent with the idea that active traction forces, which are concentrated in the frontal region, are involved in probing substrate rigidity.27,28
Observations of subcellular structures at a high magnification require the use of thinner gels to accommodate the short working distance of the objective lens, which may in turn affect the cell’s ability to deform the material due to the tethering to the underlying rigid glass surface at a short distance. However, we found that the morphological responses were qualitatively similar to those seen on thicker substrata following either global or localized exposure. In addition, exposure to the UV beam caused no noticeable bleaching to GFP or RFP probes. Thus the material should allow direct observations of the responses of subcellular structures including focal adhesions to mechanical signals.
Conclusions
We have created a UV-softening hydrogel substratum by cross-linking linear functionalized PA with a photosensitive reagent, and demonstrated that the material is suitable for probing the spatial and temporal responses of adherent cells to substrate rigidity. Responses to localized softening indicated that mechanosensing is largely localized to the anterior of polarized cells. The same chemical principles may be applied to other linear polymers to obtain a wide variety of UV-softening materials. In addition to probing cellular rigidity sensing, such materials may find wide applications in tissue engineering and regenerative medicine. Potential applications include the generation of micropatterned rigidity to differentially regulate cell polarity, growth and differentiation, as well as controlled timing for triggering these events at the optimal stage of tissue formation.
Experimental
Unless otherwise specified, all chemicals were purchased from Sigma.
Preparation of UV-softening substrates and control PA gels
Linear polyacrylamide (MW 600 000–1 000 000, 10% in water; Polysciences, Warrington, PA) was dried in a Savant DNA 110 Speed Vac (Global Medical Instrumentation, Inc., Ramsey, MN) and resuspended in hydrazine hydrate (24–26%) at a concentration of 26.4 mg/ml.18 Following incubation for 3 h at 50 °C with frequent vortexing, the solution was cooled on ice and mixed with 3 volumes of cold methanol to precipitate the PA, which was collected by centrifugation. After several rinses with methanol, the pellet of polyacrylamide acyl hydrazide (PAAH) was dried with nitrogen and stored dessicated at 4 °C.
4-Bromomethyl-3-nitrobenzoic acid (BNBA) was handled under reduced illumination throughout the procedure. A fresh solution of 20% (w/v) BNBA in dimethyl sulfoxide was added to 45 mg/ml of PAAH in 10 mM boric acid (pH 12.5), at a volume ratio of 0.072.19 After stirring for 2 h at room temperature, the solution was mixed with an equal volume of 100 mM 2-(N morpholino) ethane sulfonic acid (pH 6.0; Research Organics, Cleveland, OH), and additional dry PAAH at 0.5 weight relative to the initial weight. The solution was degassed for 20 min to avoid the formation of bubbles in subsequent steps, then mixed with 0.42 M EDC at a volume ratio of 21:4. The mixture was immediately plated onto glutaraldehyde activated coverslips,29 and weighted with a 7/8 inch diameter × 1 inch stainless steel cylinder atop a 7/8 inch diameter thin Teflon disk (<1/16 inch thick virgin electrical grade Teflon (PTFE), McMaster Carr, Elmhurst, IL). The crosslinking reaction was allowed to proceed for 30 min at room temperature to form a thin sheet. The gel was then soaked in HEPES (pH 8.5) for 10 min and the stainless steel cylinder and Teflon sheet carefully removed. The gel was then washed extensively with phosphate buffer solution (PBS). The surface of the gel was then covered with a thin cell-adhesive layer of PA gel.21 First, 200 μl acrylamide (40%; BioRad), 20 μl bis-acrylamide (2%, BioRad), 100 μl 10X PBS, 2 μl TEMED (EMD), and 455 μl distilled water were mixed and pH adjusted to 7.2–7.6. After addition of 50 μg fibronectin (1 mg/ml), 1 μl of acrylic acid N-hydroxy succinimide ester (10 mg/ml), and 10 μl fluorescent beads (0.5 μm diameter; Polysciences, 19507), the volume was adjusted to 995 μl with distilled water. The solution was degassed for 20 min, and the polymerization initiated by adding 5 μl of ammonium persulfate (BioRad) at 100 mg/ml. The solution was layered immediately atop the UV-sensitive gel (10 μl per substratum of 7/8 inch diameter), and covered using the Teflon disk and weight for 30 min prior to soaking and rinsing with HEPES as described above.
Substrata may be stored in the dark at 4 °C in PBS. Prior to experiments, the substrata were incubated with complete media for 45 min. Control PA substrates were prepared by placing 42 μl of the polymerizing solution for the top layer directly onto activated coverslips, omitting the bottom UV-sensitive layer.
Measurement of BNBA concentration
The extent of modification of PAAH by BNBA, in mmol BNBA/g PAAH, was determined by spectrophotometry (Ultrospec 2100 pro, Cambridge, UK). Following the 2 h reaction of BNBA with PAAH, and neutralization of the solution with MES (n = 2), solutions were dialyzed extensively against 10 mM boric acid, pH 12.5, then measured for absorbance at 350 nm in triplicates. The average of these values were then compared against a standard curve obtained from measuring known concentrations of BNBA in the same buffer (n = 9 from 3 preparations).
UV light source and power measurement
Substrata were irradiated using a 105W mercury arc lamp (HBO 105; Carl Zeiss, Thornwood, NY) filtered with a 365 nm interference filter and a BG38 heat filter. All experiments were performed using an exposure energy density of 2.3 J/cm2, except for the sequential illumination measurements where the intensity was reduced to one-third of the maximum (Fig. 1d). The illumination power was measured with a power meter (LaserMate-Q A/D, Coherent, Inc.) at the output of the objective lens at each experiment, and the exposure time was adjusted accordingly to obtain a consistent total energy density. To perform proof of principle experiments on softening, pre-illuminated substrates were exposed for 3 min at a distance of 2 1/4 inch from two 15 W UVB bulbs.
Measurements of gel rigidity and observations of cell responses
The rigidity of the gel was measured with a calibrated glass microneedle with a spherical tip.22,23 Using beads placed on the surface and embedded in the bottom UV-sensitive gel, we obtained a typical thickness of 50 μm for the UV-sensitive layer and submicron thickness (beyond the detection limit) for the top layer. Assuming a thick and homogenous gel, the elastic moduli were calculated using the equation for Hertz contact,22,30 with a probe radius, R, and probe stiffness, k, of ~30 μm and 0.08 N/m respectively. Indentation depths between 0.3 < R < 0.6 probed sufficiently deep into the bulk of the UV-sensitive gel, and similar results were obtained with or without the top layer (data not shown).
NIH 3T3 fibroblasts were cultured in DMEM (Invitrogen) containing 10% donor calf serum (Hyclone) and 100 units/ml penicillin, 100 μg/ml streptomycin and 4 mM supplemental glutamine (Invitrogen) for 12–36 h before experiments. Cellular responses to substrate softening were studied by irradiating cells plated on UV-softening gels and comparing against those on control PA. Cells were imaged with phase contrast optics under red illumination (Axiovert-S100, Zeiss, Thornwood, NY, with a custom stage incubator) using a 10× (Achromat, N.A. 0.25) 40× (Plan-NeoFluar, N.A. 0.75) objective lens for either 2 h 20 min before UV illumination (for global and localized irradi ation, respectively). All experiments were performed with motile interphase cells well separated from one another. To distinguish between illumination-induced and spontaneous retraction and eliminate potentially mitotic cells, cells showing a decreasing trend in area before UV illumination were rejected. After UV illumination, the cells were imaged for 2 h. The spread area was computed, normalized, and averaged in 8 min intervals suppress noise, using custom software. Directional persistence was determined as a ratio of total distance travelled to the net displacement24 and speed was calculated as the distance travelled per unit time.
Statistical analysis
Values before and after illumination (n = 24 from 8 cells for area measurements; n = 8 for speed measurements) were compared using paired students t-tests performed with GraphPad© soft ware after normality was verified using the Lillifors test. All data are represented as mean ± standard error of the mean (s.e.m.).
Supplementary Material
Appendix.
abbreviations list
| FN | fibronectin |
| BNBA | 4-bromomethyl-3-nitrobenzoic acid |
| EDC | 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide |
| PBS | phosphate buffer solution |
| PAAH | polyacrylamide acryl hydrate |
| PA | polyacrylamide |
| UV | ultraviolet light |
| DMEM | Dulbecco’s modified eagle medium |
Acknowledgments
This work was supported by a grant from the NIH GM32476.
The authors wish to thank Angela Throm for helpful discussions and Corrie Painter for her assistance with ChemDraw.
Footnotes
Electronic supplementary information (ESI) available: Movies 1–9.
References
- 1.Lo CM, Wang HB, Dembo M, Wang YL. Biophys J. 2000;79(1):144–152. doi: 10.1016/S0006-3495(00)76279-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wozniak MA, Desai R, Solski PA, Der CJ, Keely PJ. J Cell Biol. 2003;163(3):583–595. doi: 10.1083/jcb.200305010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Paszek MJ, Zahir N, Johnson KR, Lakins JN, Rozenberg GI, Gefen A, Reinhart-King CA, Margulies SS, Dembo M, Boettiger D, Hammerand DA, Weaver VM. Cancer Cell. 2005;8(3):241–254. doi: 10.1016/j.ccr.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 4.Huang S, Ingber DE. Cancer Cell. 2005;8(3):175–176. doi: 10.1016/j.ccr.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 5.Engler AJ, Sweeney HL, Discher DE, Schwarzbauer JE. J Musculoskelet Neuronal Interact. 2007;7(4):335. [PubMed] [Google Scholar]
- 6.Guo WH, Frey MT, Burnham NA, Wang YL. Biophys J. 2006;90(6):2213–2220. doi: 10.1529/biophysj.105.070144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Georges PC, Miller WJ, Meaney DF, Sawyer E, Janmey PA. Biophys J. 2006;90(8):3012–3018. doi: 10.1529/biophysj.105.073114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kass L, Erler JT, Dembo M, Weaver VM. Int J Biochem Cell Biol. 2007;39(11):1987–1994. doi: 10.1016/j.biocel.2007.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang HB, Dembo M, Wang YL. Am J Physiol Cell Physiol. 2000;279(5):C1345–C1350. doi: 10.1152/ajpcell.2000.279.5.C1345. [DOI] [PubMed] [Google Scholar]
- 10.Discher DE, Janmey P, Wang YL. Science. 2005;310(5751):1139–1143. doi: 10.1126/science.1116995. [DOI] [PubMed] [Google Scholar]
- 11.Ingber DE. Semin Cancer Biol. 2008;18(5):356–364. doi: 10.1016/j.semcancer.2008.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pelham RJ, Jr, Wang Y. Proc Natl Acad Sci U S A. 1997;94(25):13661–13665. doi: 10.1073/pnas.94.25.13661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Georgesand PC, Janmey PA. J Appl Physiol. 2005;98(4):1547–1553. doi: 10.1152/japplphysiol.01121.2004. [DOI] [PubMed] [Google Scholar]
- 14.Engler A, Bacakova L, Newman C, Hategan A, Griffin M, Discher D. Biophys J. 2004;86(1):617–628. doi: 10.1016/S0006-3495(04)74140-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Engler AJ, Griffin MA, Sen S, Bonnemann CG, Sweeney HL, Discher DE. J Cell Biol. 2004;166(6):877–887. doi: 10.1083/jcb.200405004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gray DS, Tien J, Chen CS. J Biomed Mater Res A. 2003;66(3):605–614. doi: 10.1002/jbm.a.10585. [DOI] [PubMed] [Google Scholar]
- 17.Bourke SL, Al-Khalili M, Briggs T, Michniak BB, Kohn J, Poole-Warren LA. AAPS PharmSci. 2003;5(4):E33. doi: 10.1208/ps050433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Freeman A. In: Methods in Enzymology. Part B. Mosbach K, editor. Vol. 135. Academic Press, Inc; Orlando: 1987. pp. 216–222. [Google Scholar]
- 19.Marriott G, Ottl J. In: Methods in Enzymology. Caged Compounds. Marriott G, editor. Vol. 291. Academic Press; San Diego: 1998. pp. 155–175. [DOI] [PubMed] [Google Scholar]
- 20.Solon J, Levental I, Sengupta K, Georges PC, Janmey PA. Biophys J. 2007;93(12):4453–4461. doi: 10.1529/biophysj.106.101386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rajagopalan P, Marganski WA, Brown XQ, Wong JY. Biophys J. 2004;87(4):2818–2827. doi: 10.1529/biophysj.103.037218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frey M, Engler A, Lee J, Wang Y, Discher D. Methods in Cell Biology. Vol. 83. Elsevier, Inc; 2007. pp. 45–63. [DOI] [PubMed] [Google Scholar]
- 23.Jacot JG, Dianis S, Schnall J, Wong JY. J Biomed Mater Res A. 2006;79(3):485–494. doi: 10.1002/jbm.a.30812. [DOI] [PubMed] [Google Scholar]
- 24.Frey MT, Tsai IY, Russell TP, Hanks SK, Wang YL. Biophys J. 2006;90(10):3774–3782. doi: 10.1529/biophysj.105.074526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Besaratinia A, Kim SI, Bates SE, Pfeifer GP. Proc Natl Acad Sci U S A. 2007;104(14):5953–5958. doi: 10.1073/pnas.0610534104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen WT. J Cell Biol. 1979;81(3):684–691. doi: 10.1083/jcb.81.3.684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Beningo KA, Dembo M, Kaverina I, Small JV, Wang YL. J Cell Biol. 2001;153(4):881–888. doi: 10.1083/jcb.153.4.881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dembo M, Wang YL. Biophys J. 1999;76(4):2307–2316. doi: 10.1016/S0006-3495(99)77386-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alpin J, Hughes C. Anal Biochem. 1981;113:144. doi: 10.1016/0003-2697(81)90057-9. [DOI] [PubMed] [Google Scholar]
- 30.Hertz HJ. J Reine Angew Mathematik. 1882;92:156–171. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.