Abstract
Proper partitioning of the contents of a cell between two daughters requires integration of spatial and temporal cues. The anaphase array of microtubules that self-organize at the spindle midzone contributes to positioning the cell division plane midway between the segregating chromosomes1. How this signaling occurs over micron length-scales, from the midzone to the cell cortex, is not known. Here we examine the anaphase dynamics of protein phosphorylation by Aurora B kinase, a key mitotic regulator, using FRET-based sensors in living cells and immunofluorescence of native Aurora B substrates. Quantitative analysis of phosphorylation dynamics, using chromosome and centromere targeted sensors, reveals that changes are due primarily to position along the division axis rather than time. These dynamics result in the formation of a spatial phosphorylation gradient early in anaphase that is centered at the spindle midzone. This gradient depends on Aurora B targeting to a subpopulation of microtubules that activate it. Aurora kinase activity organizes the targeted microtubules to generate a structure based feedback loop. We propose that feedback between Aurora B kinase activation and midzone microtubules generates a gradient of posttranslational marks that provides spatial information for events in anaphase and cytokinesis.
Keywords: anaphase, gradient, FRET, Aurora B, INCENP, histone H3, serine 10, mitotic spindle, Hesperadin, monopolar
It is believed that self-organizing systems position the cleavage furrow, since experimental displacement of the anaphase spindle results in repositioning of the cleavage furrow within minutes2. While mitotic chromosomes are thought to generate gradients of RanGTP that self-organize the prometaphase spindle3, this cannot be the only self-organizing signal in anaphase because cytokinesis can occur in the absence of chromatin4,5. Instead, the location of the cleavage furrow is coupled to the position of the spindle midzone where the Chromosome Passenger Complex (CPC) containing Aurora B kinase is localized. How signals are transmitted over micron length-scales between midzone microtubules and the cell cortex is unknown.
To examine spatial patterns of Aurora B signaling during anaphase, we developed a strategy using FRET-based sensors that report quantitative changes in substrate phosphorylation in living cells. We adapted a sensor design6 in which changes in intramolecular CFP-YFP FRET depend on changes in phosphorylation of an Aurora B substrate peptide, which is conserved among members of the kinesin-13 family7 (Fig. 1a). To mimic localizations of endogenous Aurora B substrates8, sensors were targeted to centromeres (CENP-B fusion), to chromatin (histone H2B fusion), or to cytosol (lacking targeting sequences) (Fig. S1a). To examine the sensor response to changes in Aurora B activity in living cells, we first imaged mitotic cells before and after kinase inhibition. Second, we imaged cells through anaphase, when endogenous Aurora B substrates are dephosphorylated9. For each sensor the YFP/CFP emission ratio increased both after inhibitor treatment and in anaphase, consistent with dephosphorylation for this sensor design6. The maximal increase in emission ratio following chemical inhibition is similar to the increase during anaphase for each sensor (Fig. S1b), indicating that the measured FRET changes correspond to full dephosphorylation of the sensor. To test specificity for Aurora B, the cytosolic sensor was treated with a Polo-like kinase (Plk) inhibitor, which did not cause an increase in the emission ratio (Fig. S2a). In addition, the cytosolic sensor was not phosphorylated in mitotic cells after Aurora B depletion by RNAi (Fig. S2b). Together, these data validate the sensors as reporters of Aurora B activity.
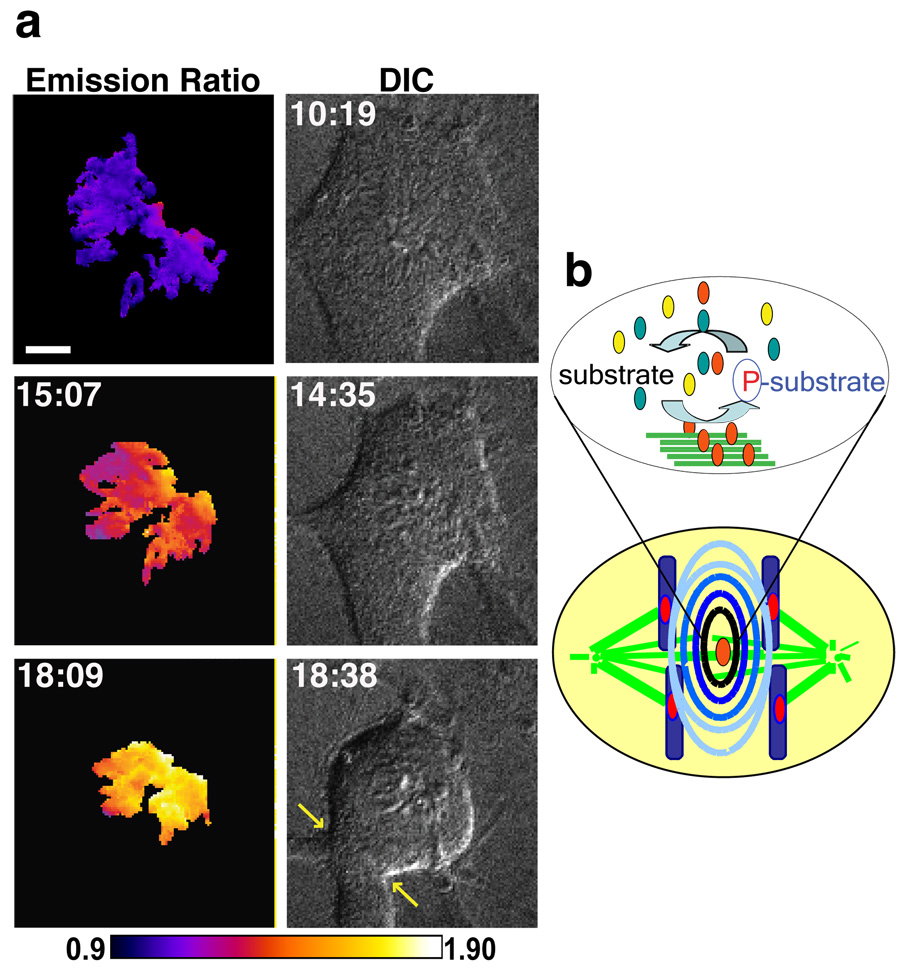
Figure 1. A FRET-based sensor of Aurora B kinase activity demonstrates a spatial phosphorylation gradient during anaphase.
a, Sensor design: phosphorylated threonine underlined, targeting sequences from histone H2B (chromatin) or CENP-B (centromere). b, Cells expressing cytosolic (untargeted), chromatin-targeted, or centromere-targeted sensors were imaged live through anaphase. The YFP/CFP emission ratio at each timepoint was normalized to vary from 0–100% and averaged over multiple cells (N≥4). c–e, Cells expressing the centromere-targeted sensor were imaged through anaphase, either with an intact checkpoint (c) or with Mad2 depleted by RNAi (d–e). Left panels (c,d): unprocessed YFP images, right panels: color-coded images of the emission ratio, timestamps (min) relative to anaphase onset, scale bars 5 um. In a plot of all timepoints (e), each circle represents an individual centromere characterized by time after anaphase onset, position along the division axis, and emission ratio (color scale). Dashed lines indicate data points plotted in Fig. S4.
To map Aurora B kinase activity during anaphase, we examined the kinetics of changes in sensor phosphorylation at different sites. Dephosphorylation of all three Aurora B sensors begins immediately after sister chromosome separation and is complete within 8 min for the centromere and chromatin targeted sensors, compared to 30 min for the cytosolic sensor (Fig. 1b). This analysis indicates that dephosphorylation kinetics of Aurora B substrates in anaphase depend on substrate localization. Mutation of the substrate threonine to alanine, using the chromosomal sensor, eliminated the change in emission ratio (Fig. S3).
The rapid dephosphorylation kinetics of the chromosome-targeted sensors are remarkably similar to the kinetics of chromosome segregation, suggesting that phosphorylation changes may be linked to chromosome position during anaphase. To test this possibility we calculated both the YFP/CFP emission ratio at each centromere (Fig. 1c) and its position along the division axis in cells expressing the centromere-targeted sensor. Analysis of single time points early in anaphase, when variance in centromere position is maximal, consistently revealed a correlation between position and sensor phosphorylation (Fig. S4a, Table S1). These results indicate that while dephosphorylation occurs at all centromeres over time, phosphorylation differences between individual centromeres depend on centromere position.
To determine the length scale over which position influences sensor phosphorylation, we depleted Mad2 by RNAi to inhibit the spindle checkpoint and increase the variance in centromere positions during anaphase. The sensor is dephosphorylated within 8 minutes of anaphase onset on centromeres that segregate normally in Mad2-depleted cells, but remains phosphorylated for up to 10 minutes on centromeres that remain in the center (Fig. 1d–e). Quantitative analyses demonstrate that changes in sensor phosphorylation depend primarily on centromere position along the division axis, over ~6 µm distance from the center rather than on time (Fig. 1e, Fig. S4, Table S1).
We next examined the chromatin-targeted and cytoplasmic sensors. Spatial phosphorylation patterns were not detected using the cytoplasmic sensor, possibly because rapid diffusion of cytosolic proteins may degrade any spatial patterns so that they are not detected by our methods. The chromatin-targeted sensor revealed a clear phosphorylation gradient. Early in anaphase sensor phosphorylation is highest on chromatin near the spindle midzone and lowest near the spindle poles (Fig. 2a–b). Chromosomes segregated normally in these experiments, indicating that microtubule attachments are not perturbed. As the phosphorylation gradient is not restricted to a few individual chromosomes, it is unlikely to reflect differences in chromosome-spindle attachments. A Plk sensor did not reveal spatial phosphorylation patterns in anaphase (Fig. S5), which indicates that the phosphorylation gradient is specific for Aurora B substrates.
Figure 2. The anaphase phosphorylation gradient is observed for multiple Aurora B substrates.
a, A cell expressing the chromatin-targeted Aurora B sensor was imaged live through anaphase. Top panels: unprocessed YFP images, bottom panels: color-coded images of the YFP/CFP emission ratio, timestamp (min) relative to anaphase onset, scale bar 5 µm. b, Average emission ratio (±stdev) plotted vs. position, at time=2.1 min, for pixels binned according to position along the division axis. c, HeLa cell fixed and stained to label chromosomes (DAPI, blue), H3(S10) phosphorylation (green), and Aurora B (red). d, Xenopus S3 cell fixed and stained for DAPI (blue), MCAK (green), and phospho-MCAK(S196) (red). e–f, Intensity profiles of phospho-H3(S10) (green) and DAPI (blue), or MCAK (green) and phospho-MCAK(S196) (red), measured along lines in merged images (c,d). The ratio of phospho-MCAK(S196) to MCAK is calculated at the indicated points (numbers in parenthesis).
We next examined phosphorylation of endogenous Aurora B substrates by immunofluorescence, using phospho-specific antibodies. First we analyzed histone H3 Ser-10 (H3(S10)) phosphorylation, which was highest towards the spindle midzone and lower towards the poles (Fig. 2c,e). H3(S10) phosphorylation increased 1.5–2.6 fold from pole to midzone in 78% of anaphase cells (60–120 cells per experiment, n=6). This anaphase H3(S10) phosphorylation gradient was verified in multiple cell types and using a second phospho-specific antibody (Fig. S6a–d). A similar result was reported in drosophila syncitial embryos10. Second, we analyzed another Aurora B substrate, MCAK Ser-1967. During anaphase MCAK localizes throughout the cell with highest concentrations at the spindle poles, while phospho-MCAK(S196) appears highest in the spindle midzone (Fig. 2d,f). Together, these data demonstrate phosphorylation gradients for endogenous and exogenous (FRET sensor) Aurora B substrates on chromosomes or the cytoskeleton during anaphase.
To determine whether Aurora B localization contributes to formation of the phosphorylation gradient, we used three different perturbations. First, brief (8 min) nocodazole treatment led to microtubule disassembly, spindle midzone disorganization, and dispersion of Aurora B throughout the cytoplasm11 (Fig. 3a, S7b–c). We observed loss of the normal H3(S10) phosphorylation gradient in 76% of nocodazole-treated HeLa cells (n=110) (Fig. 3a,d; S8d). Slight increases in H3(S10) phosphorylation were sometimes apparent on chromatin near the spindle midzone, most likely reflecting incomplete microtubule disruption (Fig. S7c). Second, we depleted the kinesin MKLP-2 with shRNAi (Fig. S8b–c), leading to loss of midzone localization of Aurora B in 63% of anaphase cells (n=27)12 and absence of the H3(S10) phosphorylation gradient in 70% of these cells (Fig. 3b,e). Third, following expression of non-degradable cyclin B, Aurora B remained on chromosome arms in anaphase11,13, and the H3(S10) phosphorylation gradient was disrupted (Fig. S7e). Together, these findings indicate that the anaphase phosphorylation gradient depends on Aurora B localization to the spindle midzone.
Figure 3. The phosphorylation gradient requires Aurora B localization to the midzone.
a–f, HeLa cells treated with nocodazole for 8 min (a,d), shRNA against MKLP2 (b,e), or mock transfected (c,f) were fixed and stained for chromosomes (DAPI, blue), H3(S10) phosphorylation (green), and Aurora B (red). Intensity profiles (d–f) show H3(S10) phosphorylation (green) and DAPI (blue) measured along lines in merged images, with distance increasing away from the midzone. g–k, Xenopus S3 cells treated for 8 min with either Hesperadin (g), nocodazole (h), or DMSO (i) were fixed and stained for chromosomes (DAPI, blue), tubulin (green), and phospho-INCENP(S850) (red). Total cellular INCENP and phospho-INCENP(S850) in anaphase were measured by quantitative confocal microscopy (j–k) (mean±sem, n≥10, *p<0.005). I, Antibodies against tubulin and Aurora B were used in a proximity ligation in situ assay (P-LISA) in an anaphase Xenopus S3 cell: PLISA product (red), tubulin (blue), kinetochores (NDC80, light blue). Scale bars 5 µm.
We next addressed how a gradient might be established. One of the best examples occurs during development, when morphogen gradients are produced by self-organizing systems that require localization of an activator and positive feedback14. To determine where Aurora B is activated during anaphase, we analyzed INCENP Ser-850 phosphorylation in Xenopus cells (Table S2) and Aurora B Thr-232 phosphorylation in HeLa cells. Both modifications are associated with full Aurora B activation15,16. Using phospho-specific antibodies, we find that both INCENP(S850) and Aurora B(T232) phosphorylation are limited to the spindle midzone (Fig. 3i, Fig. S6d), indicating that Aurora B activation is restricted to this site. Brief (8 min) treatment with an Aurora B inhibitor, Hesperadin17, led to disruption of midzone microtubule organization (Fig. 3g, Fig. S9a) and reduction of total phospho-INCENP(S850) staining by 88% (Fig. 3g, 3k, Table S3). Loss of phospho-INCENP(S850) is not caused by a decrease in INCENP protein, as Hesperadin treatment increased total INCENP staining during anaphase by over 70% (Fig. 3j, Fig. S9a, S9d–e). Together, these data suggest that Aurora B must be continuously activated during anaphase, and that active kinase localizes to the spindle midzone.
To test the possibility that Aurora B activation depends on microtubule association in anaphase, INCENP(S850) phosphorylation was examined following nocodazole treatment, which led to 85% reduction in phospho-INCENP(S850) (Fig. 3h, 3k). Brief nocodazole treatment did not depolymerize all microtubules, and residual phospho-INCENP(S850) was confined to the remaining midzone microtubules. Nocodazole treatment also reduced anaphase H3(S10) phosphorylation by approximately 50% (Table S4). Microtubules can directly stimulate Aurora B kinase activity in vitro (Fig. S10a–b), consistent with previous results18. To determine if Aurora B directly contacts microtubules during anaphase, we performed a proximity ligation in situ assay19 (P-LISA). The P-LISA product was detected primarily within the spindle midzone, consistent with a direct interaction between midzone microtubules and Aurora B (Fig. 3i). This signal co-localized with both markers of Aurora B activation, phospho-INCENP(S850) and phospho-Aurora B(T232), but not the bulk of tubulin (Fig. S10c–d). Together, these data indicate that Aurora kinase activity at the spindle midzone is continuously maintained through local interactions with microtubules.
Formation of a phosphorylation gradient centered at the spindle midzone suggests a mechanism to communicate the position of the midzone to the cortex. Although inhibition of Aurora B or of midzone components such as MKLP-2 perturbs cytokinesis, it is difficult to separate the function of the gradient from other functions of these proteins. To test whether the gradient may provide spatial information to position the cleavage furrow, we changed the shape of the gradient by perturbing the spatial organization of the anaphase spindle. In the presence of a kinesin-5 inhibitor, spindles are monopolar but anaphase still occurs if the spindle checkpoint is inhibited. Chromosomes are pulled to one side of the cell, followed by microtubule stabilization and cell cleavage on the opposite side20. This assay introduces a dramatic spatial change without directly inhibiting Aurora B or other midzone or furrow components. We observe a phosphorylation gradient within 1.5 minutes (±0.5 sem, N=6) of chromosome movement in monopolar anaphase. The gradient is oriented with maximal phosphorylation opposite the direction of chromosome movement (N=9 out of 12 cells examined) (Fig 4a–b). This result demonstrates that gradient formation is robust to changes in spindle geometry. Although we do not always observe a cleavage furrow in monopolar anaphase, furrows that do form are positioned in the direction of maximal phosphorylation in the gradient (Fig. S11). We also find that Aurora B disappears from centromeres in a monopolar anaphase and subsequently redistributes to the cortex where the cleavage furrow forms (Fig 4c–d), beginning 3.1 min (±0.2 sem, N=5) after chromosome movement. As the gradient precedes both cortical Aurora B localization and furrow ingression, these data suggest that the anaphase phosphorylation gradient provides spatial information to position the cleavage furrow.
Figure 4. The phosphorylation gradient in a monopolar anaphase predicts the cleavage site.
a–d, Cells expressing either the chromatin-targeted sensor (a–b) or Aurora B-GFP (c–d) were depleted of Mad2 by RNAi and imaged through anaphase in the presence of the kinesin-5 inhibitor monastrol. Unprocessed YFP images (a) show chromosome movement relative to the cell perimeter (outlined in white), and color coded images show the YFP/CFP emission ratio (b). Aurora B-GFP (c) redistributes to the cortex, with chromosomes and cleavage furrow visualized by DIC (d). Timestamps min:sec. Scale bar 5 um. e, Model: Following activation on midzone microtubules, Aurora B remains active until dephosphorylation by cytosolic phosphatases. The resulting phosphorylation gradient (contour lines) extends from the midzone to the cortex (yellow ovals -Aurora B complex, yellow ovals with star - active Aurora B complex, teal ovals-phosphatase).
Formation of the cleavage furrow depends on signals from the spindle midzone, but how the midzone is initially established in unknown. We propose that release of active Aurora B from centromeres establishes a phosphorylation gradient early in anaphase (Fig. 2a), so that substrates known to regulate microtubule organization1 are preferentially phosphorylated at the center of the anaphase spindle. The phosphorylation gradient is maintained through a positive feedback loop in which Aurora B activity organizes midzone microtubules, and the midzone catalyzes local Aurora B auto-phosphorylation of its own regulatory sites. Active Aurora B diffuses away from the midzone where it is inactivated by cytosolic phosphatases (Fig. 4e). Many Aurora B substrates are localized to structures (chromosomes or the cytoskeleton) that would limit diffusion and maintain gradient information. While we favor this model, we cannot exclude alternatives, for example involving spatial patterns of phosphatase activity.
We have shown that perturbations that block cytokinesis (nocodazole21, hesperadin17, MKLP-2 siRNA22, non-degradable cyclinB13) also inhibit gradient formation (Fig. 3a–b,d–e; Fig. S7c–e,g; Fig. S8d). Moreover, the relationship between gradient direction and furrow location persists in monopolar anaphase. We propose that the anaphase phosphorylation gradient, which extends over micron length-scales, provides a signaling mechanism to communicate the location and orientation of the spindle midzone to the cell cortex to position the cleavage furrow. Our molecular dissection has uncovered the underlying regulatory basis for an anaphase phosphorylation gradient, and our quantitative analysis of phosphorylation dynamics will lend itself to future mathematical modeling of spatial patterning in anaphase.
Methods Summary
The Aurora B phosphorylation sensor is designed so that the efficiency of intramolecular energy transfer between CFP and YFP depends on the phosphorylation state of the substrate peptide, through reversible binding to an FHA2 phospho-threonine binding domain6. The substrate sequence was selected to minimize phosphorylation by other kinases23 and optimized for binding to the FHA2 domain24. Further details of the sensor designs are provided in Methods.
For centromere and chromatin targeted sensors, live imaging was performed with a spinning disk confocal (Yokogawa). CFP was excited at 440 nm, and CFP and YFP emissions were acquired simultaneously with a beamsplitter (Dual-View, Optical Insights). Maximal intensity projections are shown for YFP emissions to show sensor localization.
Custom software was written in Matlab (Mathworks) for image analysis. For the centromere-targeted sensor, we designed image analysis algorithms to identify individual centromeres in three dimensions from confocal image stacks and calculate the YFP/CFP emission ratio at each centromere. The sensor response at individual centromeres is then described by a multi-dimensional data set consisting of each centromere’s spatial coordinates, sensor phosphorylation state as represented by the YFP/CFP emission ratio, and time. The projection of centromere position onto the division axis was calculated to collapse the data set to three dimensions: position as distance from the center of the separating chromosomes, time after anaphase onset, and emission ratio. Further details of the image analysis are provided in Methods.
For the P-LISA assay, oligonucleotides were directly conjugated to antibodies against anti-Xenopus Aurora B and anti-tubulin. The close proximity of these two antibodies is detected by adding two additional oligonucleotides that can form a template for rolling-circle replication after ligation. The rolling-circle product is then detected by hybridization of fluorescent probes. Further details of the P-LISA techniques are provided in Supplementary Information.
Full Methods
Sensor construction
The Aurora B sensor was generated by modifying the protein kinase C sensor CKAR, a gift from Alexandra Newton (UCSD). The PKC substrate sequence was replaced with KVNKIVKNRRTVAI. This sequence is from HsKif2, residues 57–70, with an Ile inserted at position +3 relative to the Thr to promote binding to the FHA2 domain24. Analysis of this sequence with Scansite23 does not predict phosphorylation of this sequence by any other kinases, even at low stringency. The CyPet-YPet variants of CFP-YFP, which have been optimized for FRET25, were used to maximize sensitivity and dynamic range of the sensor. Truncation of the substrate sequence or further optimization for FHA2 binding did not improve the sensor response (data not shown).
For targeting to centromeres, residues 1–167 from human CENP-B (Invitrogen, clone ID 6470289) were amplified by PCR and fused to the N-terminus of the sensor. For targeting to chromatin, human histone H2B was amplified from pBOS-H2BGFP (BD Pharmingen) and inserted in place of CENP-B. These sequences have been previously shown to target GFP to the centromere or to chromatin26. The Plk sensor was constructed by replacing the substrate sequence in the Aurora sensor with LLLDSTLSINWD. This sequence is from Myt1, residues 421–432. The Plk substrate, Ser-426, is replaced with Thr, and an Ile is inserted at 429 to promote FHA2 binding.
Cell culture, transfection, and live imaging
Xenopus S3 cells were maintained in 66% L-15 medium containing10% FBS, 50 IU/ml penicillin, 50 mg/ml streptomycin, and 1 mM sodium pyruvate at room temperature. HeLa, Du145 and DLD21 cells were cultured in growth medium, DMEM (Invitrogen) with 10% fetal bovine serum (Sigma) and penicillin-streptomcyin (100 U/ml and 100 µg/ml respectively, Invitrogen), at 37°C in a humidified atmosphere with 5% CO2.
For live cell studies, cells were transfected with plasmid DNA using Fugene (Roche Diagnostics) followed in some cases by a second transfection with an siRNA duplex targeting Mad2 (5’-AAGAGUCGGGACCACAGUUUA-3’, Dharmacon) using Oligofectamine (Invitrogen). Transfection of plasmid DNA and siRNA targeting Aurora B (5’- AACGCGGCACUUCACAAUUGA-3’, Dharmacon) was performed simultaneously using Lipofectamine 2000 (Invitrogen) to increase the probability of co-transfection. Aurora B knock-down was verified by immunostaining using a monoclonal antibody (BD Transduction Laboratories) to show that cells expressing the sensor were also depleted of Aurora B.
One day after transfection cells were plated on 22×22 mm No. 1.5 glass coverslips (Fisher Scientific) coated with Poly-D-lysine (Sigma) and used for imaging the following day. Coverslips were mounted in Rose chambers for live imaging, using L-15 medium without phenol-red (Invitrogen). Temperature was maintained at 35–37°C, using either a temperature controlled chamber (Solent Scientific) or an air stream incubator (ASI 400, Nevtek). The kinesin-5 inhibitor monastrol was used at 100 uM to induce monopolar spindles.
For Plk and Aurora B inhibition experiments, cells were first incubated with 0.5 µg/ml nocodazole (Sigma) to prevent mitotic exit. Cells were imaged live before and after addition of the Aurora B inhibitor hesperadin17 (50 nM) or the Plk inhibitor BTO-1 (20 µM)27. The YFP/CFP emission ratio was calculated from images acquired at each timepoint. Cells were followed until the maximal increase in emission ratio was achieved.
For live imaging of sensors without targeting domains, images were acquired on a Carl Zeiss Axiovert 200M microscope with a 63× 1.4 NA objective, a cooled, back-thinned electron multiplier CCD camera (Cascade II 512B, Photometrics), and Metamorph software (Universal Imaging). CFP and YFP emissions were acquired sequentially with CFP excitation. CFP and YFP emissions were summed over an entire cell following background subtraction, and the YFP/CFP emission ratio was calculated.
For centromere and chromatin targeted sensors, images were aquired on a Carl Zeiss Axiovert 200M microscope equipped with a z-motor, a 100× 1.4 NA objective, and a Yokogawa spinning disk confocal QLC100 unit. CFP was excited at 440 nm, and both CFP and YFP emissions were acquired simultaneously using a beamsplitter (Dual-View, Optical Insights), a cooled, back-thinned electron multiplier CCD camera (Hamamatsu, C9100-12), and Metamorph software (Universal Imaging). The pixel size in this configuration is 0.16 µm. Confocal image stacks were acquired with 0.5 µm spacing, typically 12 sections per stack.
Image analysis of targeted FRET sensors
Custom software was written in Matlab (Mathworks) for image analysis. CFP and YFP emissions were aligned by minimizing the correlation coefficient between the two images. Background intensities were calculated either locally around each centromere (for the centromere-targeted sensor) or globally around the entire spindle (for the chromatin-targeted sensor). For the chromatin-targeted sensor, intensity thresholds were selected manually, and mean CFP and YFP intensities were calculated over a 5×5 pixel square centered on each pixel within the thresholded area. The YFP/CFP emission ratio was calculated from these local means and used to create a ratio image, while pixels outside the thresholded area were set to zero. Projections of the ratio images were calculated as the average over the z-dimension of all non-zero pixels at each (x,y) coordinate. The projections were color-coded for graphical representation of the sensor phosphorylation state at each pixel. The color scale was set to incorporate the entire range of emission ratio during anaphase. To plot the change in emission ratio vs. position, pixels were binned by distance from the center of the separating chromosomes, in increments of 1.6 µm, and the average emission ratio calculated for each bin.
For the centromere targeted sensor, 3-dimensional (x,y,z) images were created from the confocal image stacks. The images were made binary using CFP and YFP intensity thresholds. Objects were defined from the binary images as connected pixels in 3 dimensions with a minimum size of 10 or 20 pixels. The CFP and YFP intensity thresholds used to create the binary images were initially selected manually, and all objects below 100 pixels in size were considered individual centromeres. For objects larger than 100 pixels, the intensity thresholds were locally increased incrementally until all objects were below a maximum size of 100 pixels. This algorithm ensures that centromeres that are close together are not merged into a single large object. Many, but not all centromeres are separated by this algorithm. Because the intensity threshold is determined locally, both dimmer and brighter centromeres are included in the analysis. For each object the CFP and YFP intensities were summed, and a single emission ratio was calculated to represent that object in a three-dimensional ratio image. By averaging over multiple pixels, the signal-to-noise ratio is improved dramatically over a pixel-by-pixel analysis. Pixels not included in any object were set to zero. Projections were calculated and color-coded as described above for the chromatin-targeted sensor.
Preparation of fixed cells, image acquisition and analysis for fixed cells, kinase assays and proximity ligation assays were performed using standard techniques essentially as described 7,18,19. These specific techniques are further described in Supplementary Methods.
Supplementary Material
Supplementary data: 11 Figures, 4 tables, 9 movies
Acknowledgements
Department of Biochemistry and Molecular Genetics for its support; Dan Burke for many discussions; Weijie Lan for his many contributions to this manuscript; Yu-Li Wang (University of Massachusetts Medical School) for the Aurora B-GFP plasmid; Hidewaki Nakagawa (University of Tokyo) for the anti-MKLP2 antibody; C. David Allis (Rockefeller University) for the anti-phospho-H3(S10) antibody; Alexandra Newton (UCSD) for the CKAR plasmid; Alison North and the Rockefeller University Bioimaging facility. This work was supported by American Lung foundation (PT); NIH grants to (TMK), (PTS), (DLB); Francis Goulet Fellowship at Rockefeller University (MAL); NICHD T32 Training Grant 'Cellular and Physiologic Mechanisms in Reproduction’ at the University of Virginia (BGF); and by the Pew Charitable Trust. EAF is a Robert Blount Family Fellow of the Damon Runyon Cancer Research Foundation; We thank Norbert Kraut and Boehringer Ingelheim for Hesparadin.
References
- 1.Glotzer M. The molecular requirements for cytokinesis. Science. 2005;307:1735–1739. doi: 10.1126/science.1096896. [DOI] [PubMed] [Google Scholar]
- 2.Bement WM, Benink HA, von Dassow G. A microtubule-dependent zone of active RhoA during cleavage plane specification. J Cell Biol. 2005;170:91–101. doi: 10.1083/jcb.200501131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kalab P, Pralle A, Isacoff EY, Heald R, Weis K. Analysis of a RanGTP-regulated gradient in mitotic somatic cells. Nature. 2006;440:697–701. doi: 10.1038/nature04589. *9- [DOI] [PubMed] [Google Scholar]
- 4.Rappaport R. Cytokinesis in Animal Cells. Cambridge: Cambridge University Press; 1996. [Google Scholar]
- 5.Alsop GB, Zhang D. Microtubules continuously dictate distribution of actin filaments and positioning of cell cleavage in grasshopper spermatocytes. J Cell Sci. 2004;117:1591–1602. doi: 10.1242/jcs.01007. [DOI] [PubMed] [Google Scholar]
- 6.Violin JD, et al. A genetically encoded fluorescent reporter reveals oscillatory phosphorylation by protein kinase C. J Cell Biol. 2003;161:899–909. doi: 10.1083/jcb.200302125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lan W, et al. Aurora B phosphorylates centromeric MCAK and regulates its localization and microtubule depolymerization activity. Curr Biol. 2004;14:273–286. doi: 10.1016/j.cub.2004.01.055. [DOI] [PubMed] [Google Scholar]
- 8.Ruchaud S, Carmena M, Earnshaw WC. Chromosomal passengers: conducting cell division. Nat Rev Mol Cell BiolOct. 2007;8(10):798–812. doi: 10.1038/nrm2257. Review. [DOI] [PubMed] [Google Scholar]
- 9.Zeitlin SG, et al. Differential regulation of CENP-A and histone H3 phosphorylation in G2/M. J Cell Sci. 2001;114:653–661. doi: 10.1242/jcs.114.4.653. [DOI] [PubMed] [Google Scholar]
- 10.Su TT, Sprenger F, DiGregorio PJ, Campbell SD, O'Farrell PH. Exit from mitosis in Drosophila syncytial embryos requires proteolysis and cyclin degradation, and is associated with localized dephosphorylation. Genes Dev. 1998;21:495–503. doi: 10.1101/gad.12.10.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Murata-Hori M, Tatsuka M, Wang YL. Probing the dynamics and functions of aurora B kinase in living cells during mitosis and cytokinesis. Mol Biol Cell. 2002;4:1099–1108. doi: 10.1091/mbc.01-09-0467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gruneberg U, Neef R, Honda R, Nigg EA, Barr FA. Relocation of Aurora B from centromeres to the central spindle at the metaphase to anaphase transition requires MKLP2. J Cell Biol. 2004;166:167–172. doi: 10.1083/jcb.200403084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wheatley SP, Hinchcliffe EH, Glotzer M, Hyman AA, Sluder G, Wang Y. CDK1 inactivation regulates anaphase spindle dynamics and cytokinesis in vivo. J Cell Biol. 1997 Jul 28;138(2):385–393. doi: 10.1083/jcb.138.2.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meinhardt H, Greier A. Pattern formation by local self-activation and lateral inhibition. Bioessays. 2000;22:753–760. doi: 10.1002/1521-1878(200008)22:8<753::AID-BIES9>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 15.Bishop JD, Schumacher JM. Phosphorylation of the carboxyl terminus of inner centromere protein (INCENP) by the Aurora B Kinase stimulates Aurora B kinase activity. J Biol Chem. 2002;277:27577–27580. doi: 10.1074/jbc.C200307200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yasui Y, Urano T, Kawajiri A, Nagata K, Tatsuka M, Saya H, Furukawa K, Takahashi T, Izawa I, Inagaki M. Autophosphorylation of a newly identified site of Aurora-B is indispensable for cytokinesis. J Biol Chem. 2004 Mar 26;279(13):12997–12300. doi: 10.1074/jbc.M311128200. [DOI] [PubMed] [Google Scholar]
- 17.Hauf S, et al. The small molecule Hesperadin reveals a role for Aurora B in correcting kinetochore-microtubule attachment and in maintaining the spindle assembly checkpoint. J Cell Biol. 2003;161:281–294. doi: 10.1083/jcb.200208092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rosasco-Nitcher SE, Lan W, Khorasanizadeh S, Stukenberg PT. Centromeric Aurora-B actvation requires TD-60, microtubules and substrate priming phophorylation. Science. 2008 Jan 25;319(5862):469–472. doi: 10.1126/science.1148980. [DOI] [PubMed] [Google Scholar]
- 19.Söderberg O, Gullberg M, Jarvius M, Ridderstråle K, Leuchowius KJ, Jarvius J, Wester K, Hydbring P, Bahram F, Larsson LG, Landegren U. Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat Methods. 2006 Dec;3(12):995–1000. doi: 10.1038/nmeth947. [DOI] [PubMed] [Google Scholar]
- 20.Canman JC, et al. Determining the position of the cell division plane. Nature. 2003;424:1074–1078. doi: 10.1038/nature01860. [DOI] [PubMed] [Google Scholar]
- 21.Wheatley SP, Wang Y-L. Midzone microtubule bundles are continuously required for cytokinesis in cultured epithelial cells. J Cell Biol. 1996;135:981–989. doi: 10.1083/jcb.135.4.981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Neef R, Preisinger C, Sutcliffe J, Kopajtich R, Nigg EA, Mayer TU, Barr FA. Phosphorylation of mitotic kinesin-like protein 2 by polo-like kinase 1 is required for cytokinesis. J Cell Biol. 2003 Sep 1;162(5):863–875. doi: 10.1083/jcb.200306009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Obenauer JC, Cantley LC, Yaffe MB. Scansite 2.0: Proteome-wide prediction of cell signaling interactions using short sequence motifs. Nucleic Acids Res. 2003;31:3635–3641. doi: 10.1093/nar/gkg584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Durocher D, et al. The molecular basis of FHA domain: phosphopeptide binding specificity and implications for phospho-dependent signaling mechanisms. Mol Cell. 2000;6:1169–1182. doi: 10.1016/s1097-2765(00)00114-3. [DOI] [PubMed] [Google Scholar]
Methods References
- 25.Nguyen AW, Daugherty PS. Evolutionary optimization of fluorescent proteins for intracellular FRET. Nat Biotechnol. 2005;23:355–360. doi: 10.1038/nbt1066. [DOI] [PubMed] [Google Scholar]
- 26.Shelby RD, Hahn KM, Sullivan KF. Dynamic elastic behavior of alpha-satellite DNA domains visualized in situ in living human cells. J Cell Biol. 1996;135:545–557. doi: 10.1083/jcb.135.3.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peters U, et al. Probing cell-division phenotype space and Polo-like kinase function using small molecules. Nat Chem Biol. 2006;2:618–626. doi: 10.1038/nchembio826. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data: 11 Figures, 4 tables, 9 movies