Abstract
Metabolic profiling of host tissues and biofluids during parasitic infections can reveal new biomarker information and aid the elucidation of mechanisms of disease. The multicompartmental metabolic effects of an experimental Echinostoma caproni infection have been characterized in 12 outbred female mice infected orally with 30 E. caproni metacercariae each, using a further 12 uninfected animals as a control group. Mice were killed 36 days postinfection and brain, intestine (colon, ileum, jejeunum), kidney, liver, and spleen were removed. Metabolic profiles of tissue samples were measured using high-resolution magic angle spinning 1H NMR spectroscopy and biofluids measured by applying conventional 1H NMR spectroscopy. Spectral data were analyzed via principal component analysis, partial least-squares-derived methods and hierarchical projection analyses. Infection-induced metabolic changes in the tissues were correlated with altered metabolite concentrations in the biofluids (urine, plasma, fecal water) using hierarchical modeling and correlation analyses. Metabolic descriptors of infection were identified in liver, renal cortex, intestinal tissues but not in spleen, brain or renal medulla. The main physiological change observed in the mouse was malabsorption in the small intestine, which was evidenced by decreased levels of various amino acids in the ileum, for example, alanine, taurine, glutamine, and branched chain amino acids. Furthermore, altered gut microbial activity or composition was reflected by increased levels of trimethylamine in the colon. Our modeling approach facilitated in-depth appraisal of the covariation of the metabolic profiles of different biological matrices and found that urine and plasma most closely reflected changes in ileal compartments. In conclusion, an E. caproni infection not only results in direct localized (ileum and jejenum) effects, but also causes remote metabolic changes (colon and several peripheral organs), and therefore describes the panorganismal metabolic response of the infection.
Keywords: Biomarker, Cluster Analysis, Echinostoma caproni, Hierarchical Analysis, Magic Angle Spinning, Metabolic Profiling, NMR Spectroscopy, Systemic Effect
Short abstract
The intestinal fluke Echinostoma caproni represents an ideal model parasite for investigating subpathologic changes in the murine host. We present a multicompartmental assessment of tissues and biofluids using 1H NMR spectroscopy to obtain a holistic metabolic view of the infection. Application of a novel hierarchical cluster algorithm together with conventional multivariate approaches facilitates interpretation of the systemic biochemical changes demonstrating both localized and remote effects in the host.
Introduction
Parasitic infections induce metabolic changes in various physiological compartments of the host and impact upon both local and systemic tissue homeostasis. Protozoan parasites come into physical contact with multiple different tissues in the host organism, due to widespread distribution in the blood stages in their life cycle (e.g., Trypanosoma spp. and Plasmodium spp.).1,2 On the other hand, parasitic worms (i.e., helminths, consisting of cestodes, nematodes, and trematodes) are less likely to come into contact with multiple host tissues, as the worms are often restricted to a single organ once they have reached their place of maturation, such as the bile duct (Fasciola spp.), the lung (Paragonimus spp.) or the small intestinal tract (Echinostoma spp).(3) However, although the physical presence of the adult worm may be topographically limited, there are well-known situations where helminths can induce pathological states in remote organs, as for example in the case of neuroschistosomiasis, where parasite eggs are transported to the central nervous system (CNS) and cause local inflammation in the tissues in which they embed.(4) Another example is Fasciola hepatica-induced pancreatitis,(5) for which the mechanism has yet to be fully elucidated.
There is increasing awareness of the importance of the composition and function of the microbiota in many physiological and pathological processes.6,7 Several parasites have been shown to impact upon host microbial composition or activity.8−12 For example, Summers and colleagues (2003)(13) showed the benefits of a Trichuris suis infection on inflammatory bowel disease (IBD) and highlighted the importance of monitoring intestinal helminth-induced changes at the microenvironmental level, especially with respect to the interactions between gut microbiota, host, and parasite. Hence, the gut deserves special consideration when studying parasite-induced changes in physiology. This issue is substantiated by our own observations: approximately a third of all identified biomarkers in different parasite-rodent models carried out thus far indicated changes in the microbial composition of the host gut, and hence, a perturbed microbiome is a characteristic feature of parasitic infections.8−12,14,15
Gastrointestinal helminth infections represent a considerable public health and veterinary problem.16,17 Metabolic monitoring of physiological changes due to helminth infections can deepen our knowledge of direct and remote effects of a helminth infection on the host and contribute to the elucidation of mechanisms of action at the biochemical level, hence, leading to a better global understanding of host−parasite interactions. So far, conventional 1H NMR spectroscopy of biofluids has been conducted in various host−parasite models in order to characterize the metabolic signature of an infection.8−12,14,15 The additional use of high-resolution magic angle spinning (HR-MAS) delivers a means of nondestructively analyzing metabolic perturbations in tissues obtained from many different physiological and pathological states.18−22 Hence, HR-MAS is now an established approach for measuring semisolid tissues because of its advantages over conventional 1H NMR and magnetic resonance spectroscopy (MRS), which obtains in vivo spectra21,23,24 and carries complementary information to biofluid-derived spectral data, but is intrinsically less sensitive.
In the present study, a metabolic profiling strategy was employed to characterize the responses of female NMRI mice to an experimental infection with the intestinal fluke Echinostoma caproni. Direct local changes on the small intestine were investigated, as well as more subtle metabolic changes on the remote systemic environment of the mouse, for example, brain, kidney, liver, and spleen. The metabolic signatures of the E. caproni infection in mice tissue compartments were then co-analyzed and integrated with existing data from metabolic changes in biofluids (urine, plasma, and fecal water).(11) The ultimate aim of this work is to provide a more complete global picture of infection-induced metabolic changes, to model compartmental interactions, and to further our understanding of panorganismal metabolic responses of an experimental infection. Of note, E. caproni represents a suitable model trematode for study because of its close physical proximity to the host microbiota and its known and localized impact on the gut architecture and biochemistry (e.g., fused and eroded villi and villous atrophy combined with biochemical changes such as higher concentrations of free fatty acids in the host mucosa).25,26
Methods
Host−Parasite Model
The study was approved by the cantonal veterinary authority of Basel (license no. 2081). Twenty-four female out-bred NMRI mice were purchased from RCC (Füllinsdorf, Switzerland), housed in groups of 4 in macrolon cages at the animal care facility of the Swiss Tropical Institute (Basel, Switzerland), and kept under environmentally controlled conditions (temperature, ∼25 °C; relative humidity, 60−70%; light-dark cycle, 12−12 h). Mice were acclimatized for 7 days prior to the start of the experiment, in order to limit potential stress-related deviation in metabolic profiles. Throughout the experiment, mice had free access to community tap water and rodent food purchased from Nafag (Gossau, Switzerland).
At the start of the experiment—designated as day 0—mice were aged 5−6 weeks and had an average weight of 25.5 g (standard deviation (SD) = 0.92 g). Mice were split into a control (n = 12) and a treatment group (n = 12), with animals in the latter group each infected orally with 30 E. caproni metacercariae (kindly provided by B. Fried; Lafayette College, PA). Mice were weighed at 7 sequential time points, namely, days 1, 5, 8, 12, 19, 26, and 33 postinfection, using a Mettler balance (model K7T; Greifensee, Switzerland). At the same time points, small amounts of tail blood were collected into sodium-heparin hematocrit capillaries (40−50 μL) and the packed cell volume (PCV) was determined as described elsewhere.(9) Mice were killed 36 days postinfection, using CO2. To assess the worm burden, E. caproni were recovered from the ileum and jejunum and counted. No worms were found in 4 out of 12 infected animals, and these mice were excluded from further analysis.
Collection of Tissue Samples
The distal part of the left lateral lobe of the liver was harvested into a 2 mL cryotube, immediately immersed in liquid nitrogen, and stored at −80 °C. The same procedure was followed for the left kidney, the spleen, and the left half of the brain. Small portions (∼0.5 cm) of tissue at 3 different locations of the intestine, (i.e., colon, jejunum, and ileum) were cut from the middle of each section to avoid regional overlap. Tissue samples were washed with phosphate buffered saline, then transferred into small cryotubes, immersed in liquid nitrogen, and stored at −80 °C. All tissues were forwarded to Imperial College (London, U.K.) on dry ice, and stored at −80 °C prior to preparation for HR-MAS 1H NMR spectroscopy.
Small samples of the 3 intestinal parts, the liver, the right kidney, the right half of the brain, and the spleen, were transferred into 5 mL tubes containing a solution of 4% buffered formalin for subsequent histological analyses via hematoxylin and eosin staining.
Collection of Biofluids
Details of the collection, processing and spectral analyses of biofluids have been presented elsewhere.(11) For the current study, only those biofluid samples (i.e., urine, plasma, and fecal water) collected on day 33 postinfection, which is the closest-matching time point to that when tissues were sampled (i.e., 36 days postinfection), were considered.
Preparation and NMR Spectroscopy of Tissues
Tissue samples of 15−20 mg were placed in 4-mm outer diameter zirconia rotors (Bruker Analytische Messtechnik GmbH; Rheinstetten, Germany), to which a drop (∼50 μL) of 0.9% saline D2O was added and enclosed with a spherical insert and a Kel-F cap.
The 1H NMR spectra of all tissues were acquired on a Bruker DRX600 spectrometer operating at 600.13 MHz (14.1 T) and spun at a MAS rate of 5000 Hz. A constant temperature of 283 K was maintained during spectral acquisition by cooling the inlet gas, which is responsible for sample spinning and surveyed by the thermocouple system. One-dimensional (1D) 600 MHz 1H NMR spectra were acquired using the standard solvent suppression pulse sequence [recycle delay (RD)-90°-t1-90°-tm-90°-acquire free induction decay (FID)](27) to suppress baseline distortions caused by the large water signal. In this pulse program, a weak irradiation pulse was applied at the water resonance frequency during the RD of 3 s and during the mixing period tm (100 ms), with t1 fixed at 3 μs. Typically, 128 transients were collected into ∼32 600 data points, with a spectral width of 20 ppm and an acquisition time per scan of 1.36 s.
Two additional NMR experiments were used to analyze tissue samples. The 1D Carr−Purcell−Meiboom−Gill (CPMG) spin−echo experiment [RD-90°-(τ-180°-τ)n-acquire FID] takes advantage of the short transverse relaxation times (spin−spin relaxation, T2) of macromolecules, and hence can be used to edit out interfering broad resonances.(28) The diffusion-edited experiment using the bipolar-pair longitudinal-eddy-current delay (BPP-LED) pulse sequence [RD-90°-G1-τ-180°-G2-τ-90°-Δ-90°-G3-τ-180°-G4-τ-90°-te-90°-acquire FID](29) selectively attenuates signals from small molecules which have fast translational diffusion rate, and hence enhances the contribution from high molecular weight molecules.(30) Assignment of resonance peaks was achieved via 2-dimensional (2D) 1H−1H correlation spectroscopy (COSY)(31) and total correlation spectroscopy (TOCSY)(32) for selected samples and by statistical total correlation spectroscopy (STOCSY) performed on the whole 1D spectral data set.(33)
Data acquisition of biofluids was performed as described in our previous work.(11)
Data Processing of NMR Spectra
The 1H NMR spectra of the tissues and biofluids were phased and baseline-corrected with an in-house developed script and were referenced to the chemical shift of the β-CH3 resonance of alanine at δ 1.48 in XwinNMR 3.5 (Bruker Analytische Messtechnik GmbH; Rheinstetten, Germany). The signal intensity in each spectrum was calculated by the integration of 0.00025 ppm wide sections. The region containing the water resonance (δ 4.7−5.1) was removed and the spectra were converted into ∼28 000 data points over the range of δ 0.5−8.0 using MATLAB (Version 7; The Mathworks Inc.; Natwick, MA). Before pattern recognition analysis was performed, each spectrum was normalized to the total remaining spectral area in a MATLAB environment.
Multivariate Analysis of NMR Data
Principal component analysis (PCA) was applied to the 1H NMR spectral data sets for individual tissues in order to give an overview of the data distribution and intersample similarities (e.g., groupings and outliers) for each tissue.34,35 The descriptive components of the PCA models R2X and Q2cumX indicate the explanatory and the predictive power of each model, respectively. Partial least-squares analysis (PLS) was subsequently applied to each matrix to optimize biochemical discrimination of the tissues from uninfected control and E. caproni-infected mice. This method quantitatively relates the data matrix X to the Y-matrix containing additional information regarding a measure of response, in this case the number of worms recovered, and thus correlates the worm burden to the metabolic profiles. PLS can also be applied as qualitative measure in discriminant analyses (PLS-DA), whereby the second set of information (Y) contains a 0 or 1 coded matrix representing classification values, such as gender, presence or absence of infection, and so forth.(36)
Further modification of the PLS-DA algorithm has been made using an in-built orthogonal filter to split the variation in the NMR data into 3 parts, that is, (i) real intermatrix correlation, (ii) systematic variation relating to the Y-matrix only (i.e., class of sample), and (iii) residual variation related to noise and/or baseline imperfections.(36) This method of deconvolving the NMR data enables the exclusion of the latter 2 sources of variation from the modeling process and optimizes interpretation with respect to elucidating the effects that are directly related to infection. Further to this initial analysis on individual matrices, hierarchical PLS-DA (H-PLS-DA) was applied as a statistical aid for facilitating visualization and information-recovery from this complex multidimensional data set with emphasis on interrogating similarities and dissimilarities in response to infection across locally distinct tissues and organs. This method has previously been applied to NMR-spectral data sets to elucidate biological events on molecular, compartmental, and systems levels.(37) The sublevel of hierarchical analysis was built initially using PCA models for each of the investigated compartments, referred to as independent blocks and the derived scores tB for each tissue matrix was subsequently combined to deliver a new analytical template or ‘superblock’ of all significant principal component (PC) scores for all tissues that can be modeled by PCA or PLS.(38) The number of PCs included in the superblock was determined according to the optimal Q2X values (representing the predictivity of each model). A total of 34 independent blocks were chosen from the first analytical level and are comprised of the biological compartments liver, ileum, colon, renal cortex, plasma, and urine acquired by up to 3 different pulse programs (i.e., 1D standard, CPMG, diffusion-edited). The explanatory power of each data set (defined by a particular type of NMR pulse program as applied to an individual tissue type or compartment, e.g., CPMG spectra for liver, etc.), that is, Q2X values and visual differentiation between the infected and controls in the scores, was used to select the scores tB for building a new analytical template for hierarchical analysis. H-PLS-DA analysis delivers new compound scores tT based on the new descriptor variables (e.g., PC scores for each tissue) and the loadings pT, generated for this supralevel show the relative contribution of each tissue or biofluid compartment to groupings observed in the scores plot.39,40
Correlation Analysis of Selected Physiological Compartments
To assess the degree of interaction between biological compartments, which showed an E. caproni-induced alteration of the metabolic profile, an O-PLS-based correlation analysis was performed. For each compartment, metabolites that differentiated between control and infected groups, with a cutoff correlation coefficient of 0.55 for a group size of 8 animals, were selected and integrated over a nonoverlapped, representative peak region using the trapezium rule(41) for further correlation analysis. This procedure was repeated for 8 selected compartments (urine, plasma, fecal water, liver, kidney, ileum, jejunum, and colon) demonstrating differentiation between infected and control profiles and pairwise correlations were calculated for each integrated metabolite. The pairwise correlations between changes in metabolite concentrations, taking all differentiating biological matrices into account, were calculated using the Pearson correlation coefficient measure. The p-values for testing the null hypothesis (i.e., no correlations between tissue localized metabolites) were computed by t-statistics using n − 2 degrees of freedom. Each p-value is the probability of observing a correlation between the assessed metabolic concentrations, that is, integrated area, as large as the observed value by random chance, when the true correlation is zero. The correlations between metabolite concentration changes with p < 0.05 were used in the analysis. The metabolite correlation matrix was clustered using average linkage hierarchical clustering coupled with the correlation dissimilarity measure to identify subsets of highly correlated changes in metabolite concentrations relating to the infection induced response.
Results
Physiological Monitoring of Mice, Gross Parasite Burden, and Histology
Mice that were infected with E. caproni for up to 36 days showed no visible signs of ill-health. Moreover, their mean weight and mean PCV showed no significant difference to the uninfected control group at any time point, indicating that the mice had not become anemic as a result of the infection. Worms were recovered and counted after dissection of mice, in order to confirm and assess the level of infection. Worms were present in 8 out of the 12 mice initially infected (average worm count = 26.5; SD = 12.0; range = 10−44 worms). Only animals with an established infection were included in subsequent analyses. Histological analysis of the spleen, kidney, liver, brain, ileum, jejunum, and colon did not show any infection-induced abnormalities in the cellular architecture.
Analysis of Biofluids from E. caproni-Infected Mice
Time-induced changes associated with an E. caproni infection in the metabolic profiles of mouse urine, plasma, and fecal water have been reported elsewhere.(11) In brief, the dominant infection-related changes in the biofluids collected at the final time point (day 33 postinfection) included decreased hippurate and increased levels of trimethylamine in urine, evidence of increased lipids in the plasma, and higher excretion of several amino acids in the feces.
Tissue-Specific Metabolic Composition in Healthy Mice
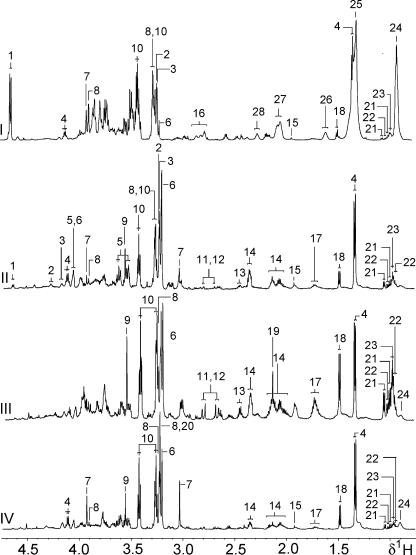
The biochemical composition of the various organs and tissues analyzed in the noninfected cohort was consistent with the extant literature.20,22,23 Lactate, choline, and alanine, the branched chain amino acids (BCAAs), glutamine, glutamate, betaine, and taurine were observed in all tissue samples (Figure 1), while other metabolites were more apparent in specific tissues only. For example, liver spectra contained generally higher glucose levels than the kidney and the small intestinal tissues (ileum and jejunum), while the spleen and colon showed practically no visible glucose resonances. Furthermore, the assessed liver and ileum regions contained the highest concentration of lipid components compared to the other tissues, whereas ileum and jejunum showed the highest levels of tyrosine and phenylalanine of all measured tissues. The spleen and kidney spectra contained particularly low relative levels of both these amino acids. As expected, comparison of kidney medulla with cortical tissue showed higher concentrations of osmotically active substances, such as betaine, myo- and scyllo-inositol, and taurine, together with higher levels of glycerophosphocholine (GPC), choline, creatine, and glucose in the medulla.
Figure 1.
Typical 600 MHz 1H MAS NMR 1D spectra of different tissues from an uninfected control mouse, aged 8 weeks, (I) liver, (II) kidney cortex, (III) spleen, and (IV) ileum. Key: 1, β-glucose; 2, glycerophosphocholine; 3, phosphocholine; 4; lactate; 5, myo-inositol; 6, choline; 7, creatine; 8, betaine; 9, glycine; 10, taurine; 11, aspartate; 12, asparagine; 13, glutamine; 14, glutamate; 15, acetate; 16, triglyceride (CH=CH); 17, lysine; 18, alanine; 19, methionine; 20, trimethylamine-N-oxide (TMAO); 21, valine; 22, isoleucine; 23, leucine; 24, triglyceride (CH)3; 25, triglyceride (CH)2; 26, triglyceride (CH2CH2CO); 27, triglyceride (CH=CHCH2); 28, triglyceride (CH2CO).
The spectral comparison of the colon with the 2 small intestinal tissues jejunum and ileum indicated that the main histological difference between these regions was the short chain fatty acid (SCFA) levels. For example, butyrate and propionate were only present in appreciable concentrations in the colon (data not shown). Lipid signals were strongest in ileum and weakest in colon. Acetate was found in all 3 intestinal regions, with highest levels in the colon. The various metabolites detected in different tissue samples are summarized in Table 1.
Table 1. Metabolites Found in Different Organs of NMRI Female Mice, Aged 7−8 Weeksa.
| metabolite | coupling pattern (Hz) | δ (ppm) | infection induced changes |
|---|---|---|---|
| 2-ketoisocaproate | CH2, CH, 2xCH3 | 2.61 (d), 2.10 (m), 0.94 (d) | (▼)U |
| 5-aminovalerate | 5-CH2, 2-CH2, 3,4-CH2 | 3.02 (t), 2.24 (t), 1.65 (m) | (▲)F |
| acetate | CH3 | 1.91 (s) | (▼)P |
| alanine | α-CH, β-CH3 | 3.81 (q), 1.48 (d) | (▲)Kc, (▼)I |
| ascorbate | CH, α-CH, β-CH2 | 4.52 (d), 4.03 (ddd), 3.75 (dd) | (▼)I |
| asparagine | α-CH, half β-CH2, half β-CH2 | 3.90 (dd), 2.81 (dd), 2.69 (dd) | |
| aspartate | α-CH, half β-CH2, half β-CH2 | 3.90 (m), 2.81 (dd), 2.68 (dd) | (▲)Kc, (▼)I |
| betaine | CH2, CH3 | 3.90 (s), 3.27 (s) | (▲)L, (▼b)Km |
| butyrate | α-CH2, β-CH2, γ-CH3 | 2.16 (t), 1.56 (m), 0.90 (t) | |
| choline | 3xCH3, α-CH2, β-CH2 | 3.21 (s), 4.07 (m), 3.52 (m) | (▼)P |
| creatine | CH3, CH2 | 3.04 (s), 3.93 (s) | (▼)P |
| formate | CH | 8.45 (s) | (▲)P |
| fumarate | CH | 6.53 (s) | (▼)I |
| glutamate | α-CH, β-CH2, γ-CH2 | 3.78 (m), 2.06 (m), 2.36 (m) | (▲)Kc, (▲b)L |
| glutamine | α-CH, β-CH2, γ-CH2 | 3.78 (m), 2.15 (m), 2.46 (m) | (▼)I |
| glycerophosphocholine | 3xCH3, half α-CH2, half α-CH2, half β-CH2, | 3.23 (s), 4.32 (t), 3.60 (dd), 3.68 (t), | (▼)Kc, (▲)C, (▲)I, |
| half β-CH2, γ-CH2 | 3.89 (m), 3.72 (dd) | (▼)P | |
| glycine | CH2 | 3.55 (s) | (▼)I |
| hippurate | CH2, 2,6-CH, 3,5-CH, 4-CH | 3.97 (d), 7.84 (d), 7.55 (t), 7.64 (t) | (▼)U |
| isoleucine | α-CH, β-CH, half γ-CH2, half γ-CH2, δ-CH3, β-CH3 | 3.68 (d), 1.93 (m), 1.25 (m), 1.47 (m), 0.99 (d), 1.02 (d) | (▼)I, (▼)L, (▼)P |
| lactate | CH, CH3 | 4.12 (q), 1.33 (d) | |
| leucine | α-CH, β-CH2, γ-CH, δ-CH3, δ-CH3 | 3.72 (t), 1.96 (m), 1.63 (m), 1.69 (m), 0.91 (d), 0.94 (d) | (▼)L, (▼)P |
| lysine | α-CH, β-CH2, γ-CH2, δ-CH2, ε-CH2 | 3.77 (t), 1.92 (m), 1.73 (m), 1.47 (m), 3.05 (t) | |
| myo-inositol | 1,3-CH, 2-CH, 5-CH, 4,6-CH | 3.53 (dd), 4.06 (t), 3.28 (t), 3.63 (t) | (▼)J |
| phenylacetylglycine | 2,6-CH, 3,5-CH, Ph−CH2, N−CH2 | 7.43 (m), 7.37 (m), 3.75 (d), 3.68 (s) | (▲)U |
| phenylalanine | 2,6-CH, 3,5-CH, 4-CH, CH2, CH | 7.44 (m), 7.39 (m), 7.33 (m), 3.17 (dd), 3.30 (dd), 3.99 (dd) | (▲)J |
| phosphocholine | CH3, α-CH2, β-CH2 | 3.24 (s), 3.60 (m), 4.18 (m) | (▲)L |
| propionate | CH2, CH3 | 2.19 (q), 1.06 (t) | |
| scyllo-inositol | 6xCH | 3.35 (s) | (▲)Kc |
| taurine | CH2N, CH2S | 3.27 (t), 3.43 (t) | (▼)I, (▼)J |
| trimethylamine | 3xCH3 | 2.88 (s) | (▲)C, (▲)U |
| trimethylamine-N-oxide | 3xCH3 | 3.27 (s) | |
| tyrosine | 2,6-CH, 3,5-CH, CH2, α-CH | 7.20 (d), 6.91 (d), 3.95 (dd), 3.20 (dd), 3.06 (dd) | (▲)J |
| uracil | 5-CH, 6-CH | 5.81 (d), 7.59 (d) | (▼)I |
| valine | α-CH, β-CH, γ-CH3, γ′-CH3 | 3.62 (d), 2.28 (m), 0.98 (d), 1.03 (d) | (▼)I, (▼)L, (▼)P |
| α-glucose | 1-CH, 2-CH, 3-CH, 4-CH, 5-CH, half 6-CH2, half 6-CH2 | 5.24 (d), 3.56 (dd), 3.70 (t), 3.40 (t), 3.83 (m), 3.72 (dd), 3.85 (m) | (▲)I, (▲)P |
| β-glucose | 1-CH, 2-CH, 3-CH, 4-CH, 5-CH, half 6-CH2, half 6-CH2 | 4.65 (d), 3.25 (dd), 3.47 (t), 3.40 (t), 3.47 (ddd), 3.78 (dd), 3.90 (dd) | (▲)I, (▲)P |
| lipid fractions | CH3 | 0.90 (t) | (▲)P, (▲b)Km |
| (CH2)n | 1.20 (m) | ||
| CH2C=C | 1.97 (m), 2.00 (m) | ||
| CH2CO | 2.23 (m) | ||
| C=CCH2C=C | 2.72 (m) | ||
| CH=CH | 5.29 (m) |
The arrows indicate the direction of the metabolic change, associated with an E. caproni infection, e.g., significantly increased (▲) or decreased (▼) in infected animals compared to noninfected control animals. Key: C, colon; F, feces, Kc, renal cortex; Km, renal medulla; L, liver; I, ileum; J, jejenum; P, plasma; U, urine. Multiplicity: s, singlet; d, doublet; dd, doublet of doublets, ddd, doublet of doublets of doublets; t, triplet; m, multiplet; q, quadruplet.
Correlation with infection intensity.
Pairwise Comparison Between E. caproni-Infected and Uninfected Control Mice for Each Biocompartment and Correlation of Metabolite Profiles with Worm Burden
The R2 and Q2 values for all the sublevel mathematical models are summarized in Table 2. Differentiation between control and infected groups was observed for all tissues examined, except renal medulla, brain, and spleen. A candidate set of biomarkers, representing the impact of an E. caproni infection on mouse tissues, was recovered using O-PLS-DA for all discriminatory data sets. Table 2 shows the Q2Y values, indicating the quality of each of the models corresponding to individual tissues. In the liver, betaine and phosphocholine levels were significantly higher in E. caproni-infected compared to uninfected control mice (Figure 2), while the BCAA concentrations were decreased in several matrices (i.e., liver, plasma, and ileum) after infection, with a corresponding increase of these metabolites observed in fecal water (Table 1). Assessment of the correlation of worm burden with the spectral changes achieved via O-PLS modeling revealed an association of hepatic glutamate with worm burden (R2Y = 0.92, Q2Y = 0.43), in addition to those metabolites simply associated with presence or absence of infection.
Table 2. Q2 and R2 Values Are Provided for All Created Analytical Models, Whereas Q2 Is an Indicator of Model-Predictability and Represents the Quality of the Modela.
| PCA |
PLS-DA |
O-PLS-DA |
||||||
|---|---|---|---|---|---|---|---|---|
| Q2X | R2cumX | Q2Y | R2X | R2Y | Q2Y | R2Y | ||
| Brain | standard 1D | 0.64 | 0.71 | −0.21 | 0.24 | 0.72 | −0.03 | 0.74 |
| Colon | standard 1D | 0.93 | 0.96 | 0.31 | 0.81 | 0.68 | 0.15 | 0.64 |
| CPMG | 0.89 | 0.94 | 0.42 | 0.78 | 0.78 | 0.13 | 0.84 | |
| diff. editedb | 0.40 | 0.58 | 0.13 | 0.31 | 0.75 | 0.03 | 0.78 | |
| Ileum | standard 1D | 0.74 | 0.84 | 0.70 | 0.50 | 0.88 | 0.54 | 0.85 |
| CPMG | 0.72 | 0.80 | 0.66 | 0.53 | 0.88 | 0.32 | 0.75 | |
| diff. edited | 0.83 | 0.90 | 0.05 | 0.50 | 0.76 | 0.20 | 0.77 | |
| Jejenum | standard 1Db | 0.86 | 0.88 | 0.25 | 0.64 | 0.53 | 0.46 | 0.80 |
| CPMGb | 0.71 | 0.83 | 0.29 | 0.43 | 0.67 | 0.51 | 0.78 | |
| diff. editedb | 0.90 | 0.93 | 0.46 | 0.70 | 0.63 | 0.35 | 0.75 | |
| Liver | standard 1D | 0.85 | 0.91 | 0.66 | 0.71 | 0.79 | 0.73 | 0.87 |
| CPMG | 0.81 | 0.88 | 0.57 | 0.69 | 0.78 | 0.52 | 0.94 | |
| diff. edited | 0.86 | 0.90 | 0.60 | 0.58 | 0.78 | 0.57 | 0.91 | |
| Plasma | standard 1D | 0.76 | 0.83 | 0.80 | 0.68 | 0.87 | 0.73 | 0.98 |
| CPMG | 0.62 | 0.77 | 0.78 | 0.42 | 0.92 | 0.54 | 0.96 | |
| diff. editedb | −0.60 | 0.25 | −0.19 | 0.16 | 91.00 | 0.63 | 0.94 | |
| Renal cortex | standard 1D | 0.70 | 0.83 | 0.15 | 0.49 | 0.68 | 0.25 | 0.72 |
| CPMGb | 0.44 | 0.71 | 0.38 | 0.38 | 0.85 | 0.50 | 0.88 | |
| diff. editedb | 0.50 | 0.64 | −0.21 | 0.22 | 0.83 | 0.28 | 0.89 | |
| Renal medulla | standard 1Db | 0.74 | 0.85 | −0.21 | 0.33 | 0.65 | −0.20 | 0.51 |
| CPMGb | 0.53 | 0.71 | 0.29 | 0.55 | 0.86 | 0.14 | 0.77 | |
| diff. editedb | 0.50 | 0.72 | 0.14 | 0.34 | 0.74 | 0.05 | 0.79 | |
| Spleen | standard 1Db | 0.89 | 0.93 | −0.21 | 0.72 | 0.58 | −0.70 | 0.57 |
| CPMGb | 0.70 | 0.85 | 0.04 | 0.50 | 0.80 | 0.06 | 0.74 | |
| Urine | standard 1D | 0.24 | 0.62 | 0.37 | 0.42 | 0.77 | 0.73 | 0.96 |
R2 shows how much of the variation in X and Y, respectively, is explained.
Models were excluded from hierarchical analysis due to lack of discrimination of infected and control group.
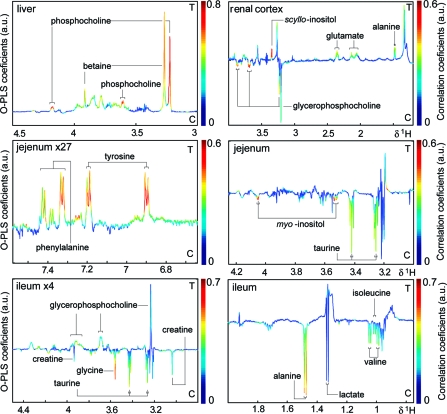
Figure 2.
Selected HR-MAS 1H NMR spectral regions in liver, renal cortex, jejunum, and ileum O-PLS-DA analysis. For illustrative purposes, a standard 1D pulse program was chosen for spectral analysis in liver, kidney and the jejunum aromatic region, whereas the CPMG data expressed the low molecular weight components, which mainly accounted for class differentiation in the ileum and the aliphatic region of the jejunum. The color bars indicate maximal contribution of the class separating metabolites in arbitrary units (a.u.).
GPC was found to be decreased in the renal cortex of E. caproni-infected mice, whereas alanine, aspartate, scyllo-inositol, and glutamate were increased in the infected group (Figure 2).
The renal medulla, brain, and spleen showed no infection-related metabolic changes at all based on PCA modeling, which is underscored by the low Q2Y values as shown in Table 2 for these models. However, O-PLS analysis of the renal medulla using worm burden as the Y or response variable data gave an overall model with an R2Y value of 0.55 and a Q2Y value of 0.29 and identified an inverse correlation of betaine with the worm burden, whereas the lipid components were positively correlated with the infection intensity. Since the PCA model for the renal medulla was not robust, this matrix was not further analyzed in the hierarchical models.
The intestinal tissues showed significant changes with worm burden in all 3 assessed regions. The relative concentrations of 2 aromatic amino acids (e.g., phenylalanine and tyrosine) were higher in the jejunum of infected mice (Figure 2), compared to the uninfected control group, and also positively correlated with worm burden (R2Y = 0.76, Q2Y = 0.55). Conversely myo-inositol and taurine decreased with infection, but showed no obvious correlation with worm burden. Infection with E. caproni also induced various metabolic changes in the ileum, such as increased glucose and decreased concentrations of glycine, fumarate, uracil, taurine, alanine, aspartate, valine, glutamine, ascorbate, and isoleucine (Figure 2). A negative correlation to the worm burden was confirmed for fumarate, ascorbate, glycine, taurine, and alanine (overall model statistics: R2Y = 0.74, Q2Y = 0.24). In the colon, higher levels of trimethylamine and GPC were observed in the E. caproni-infected mice.
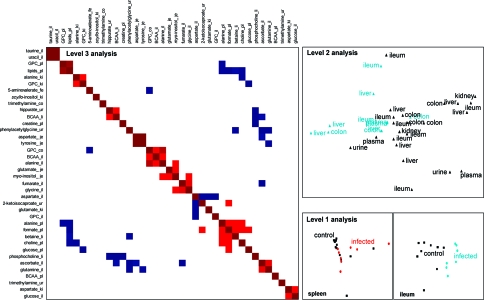
Multivariate Sublevel and Hierarchical Systems Analysis
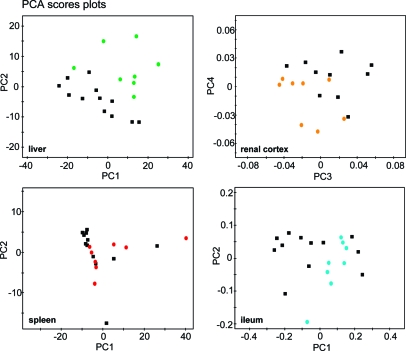
On the basis of the PCA models for each biocompartment, a second level or hierarchical analysis was performed to assess intercompartmental correlations characteristic of an E. caproni infection. Those models that resulted in positive Q2X values (Table 2) when comparing infected and controls, and achieved clear visual separation in the corresponding scores plot after the first sublevel PCA analysis, were chosen for incorporation into the hierarchical analysis. In the liver, separation between the metabolic profiles of E. caproni-infected and uninfected control mice was achieved for each of the applied pulse programs with PCA (Figure 3) and PLS-DA indicating that both low molecular weight molecules and the macromolecular compounds were discriminatory with regard to infection status. Visual differentiation of renal cortical samples between infected and uninfected mice were obtained in PCA of the CPMG-acquired data (Figure 3), but not in the standard or diffusion edited data, indicating that the key discriminatory molecules were low molecular weight components. In contrast, the renal medulla, brain, and spleen showed no infection-related separation. All 3 intestinal tissues revealed clear infection-induced differences. In the ileum, PCA models for standard 1D and CPMG (Figure 3), but not diffusion edited spectra were significant, and the PLS-DA, via CPMG-acquired spectra delivered a good discriminatory model, again implicating the relatively stronger role of the lower molecular weight molecules in comparison with the lipidic or membrane components. Similarly, the class differences in colon were obvious when employing standard 1D and CPMG-acquired spectra in PCA. In the case of the jejunal portion of the small intestine, only the CPMG spectra showed class-separation in PCA. One animal (denoted by ‘t2’) appeared as an outlier in the scores plots of the ileum analysis, which might be explained by the fact that the highest worm burden was observed in this animal (i.e., 44 adult E. caproni worms recovered upon mouse dissection).
Figure 3.
PCA scores plots for liver, renal cortex, spleen, and ileum spectra acquired by a CPMG pulse sequence, showing E. caproni infection-related differentiation in all assessed host tissues apart from the spleen, which did not show any changes induced by the infection. The renal cortex spectra of two uninfected control mice (c3 and c6) were excluded from the PCA model due to unsatisfactory water suppression.
For hierarchical modeling of the different tissue compartments, 34 scores vectors tB from previous individual PCA models were used to form a new analytical template and included 3 PCs (scores vectors tB) for all of the included compartments and applied pulse programs with the exception of the plasma spectra acquired via the standard pulse sequence and diffusion-edited liver spectra, where 2 PCs were sufficient to explain the optimal variation in the data set. For liver and ileum, spectral data from all 3 applied pulse programs could be further used for hierarchical analysis, whereas in colon, separation was gained in spectral data derived only from the 1D and CPMG pulse-sequences. Additionally, the renal cortical PCs from the CPMG acquisition were modeled. The PCA model for the spleen showed no visual separation in the sublevel PCA scores plot (Figure 3), but the explained variation in the CPMG-derived spectral analysis was good, and hence the scores tB were incorporated in the H-PLS-DA model.
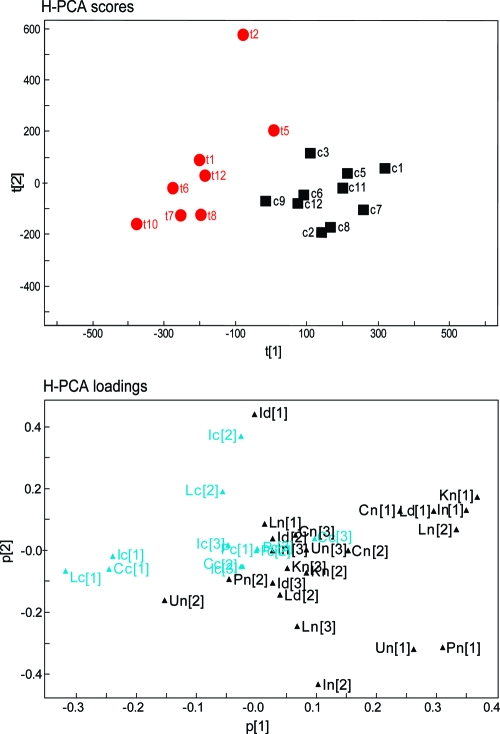
Among the multivariate statistical models for the biofluids, PCA models of the plasma and urine obtained at the ultimate sampling time point (i.e., day 33 postinfection) acquired by the standard 1D pulse sequence showed good separation in the scores plots and, additionally, the CPMG-acquired spectra for plasma were included in the hierarchical model. H-PLS-DA analyses were based on the new 34-descriptor template and were optimally modeled by 2 PCs (PLS-DA: R2X = 0.35, Q2Y = 0.76). In the H-PLS loadings plot (Figure 4), the main source of variation corresponding to the 2 separate classes (infected and noninfected control) in the scores plot lay in differentiation of the pulse programs rather than the biological compartments (Figure 4). Thus, the 1D and diffusion-edited experiments for all biological compartments (with the exception of the 1D plasma and urine and the diffusion-edited ileum spectral information) co-mapped with the control animals, whereas the spectra acquired by the CPMG pulse sequence predominantly influenced the animals infected with E. caproni, again indicating the relatively greater contribution of the low molecular weight components to the characterization of E. caproni infection.
Figure 4.
Hierarchical-PCA scores plot, assessing the weight of the different biological matrices due to an E. caproni infection. Initially, separate PCAs have been performed for all the biological matrices, and subsequently the PCs were extracted from those models with positive Q2X and/or R2cumX and which showed a visual separation in the scores plot (i.e., liver, renal cortex, ileum, colon, spleen, plasma, and urine). Two or 3 extracted PCs from each chosen compartment formed the sublevel of the hierarchical analysis. Only the closest time point to the dissection was chosen for the biofluids (i.e., day 33 postinfection), in order to minimize time-dependent variation compared to the time point of tissue-removal (i.e., day 36 postinfection). The loadings plot shows the distribution of the biomatrices in relation to the 2 groups in the corresponding scores plot, whereby ‘t’ indicates E. caproni-infected animals and ‘c’ the uninfected control mice. Of note, two uninfected control mice (c4 and c10) were excluded from the global hierarchical analysis due to unsatisfactory water suppression in the 1D spectra of ileum and urine, respectively. The H-PCA loadings are composed of the tissues (C, colon; I, ileum; K, renal cortex; L, liver; P, plasma; U, urine) and the corresponding pulse program that has been applied for 1H NMR data acquisition (c, CPMG; d, DOESY; n, 1D pulse sequence).
Calculation of Intercompartment Correlation Structures
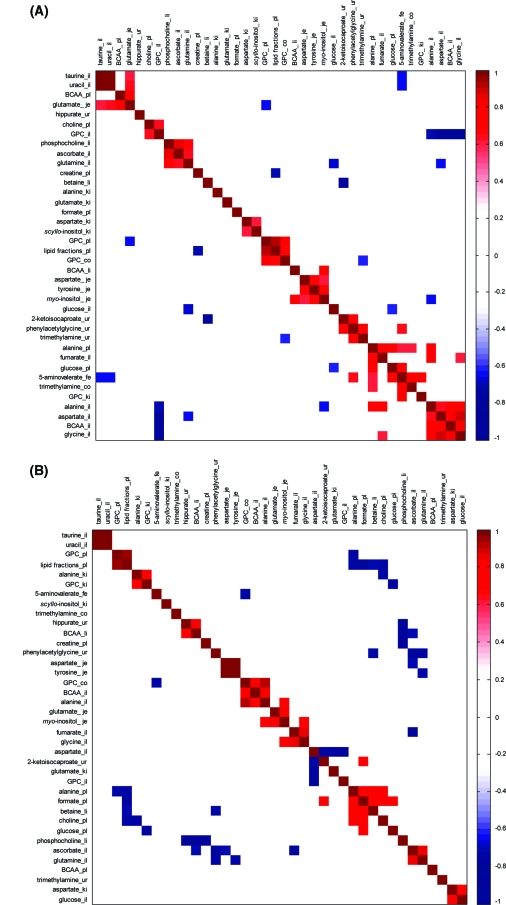
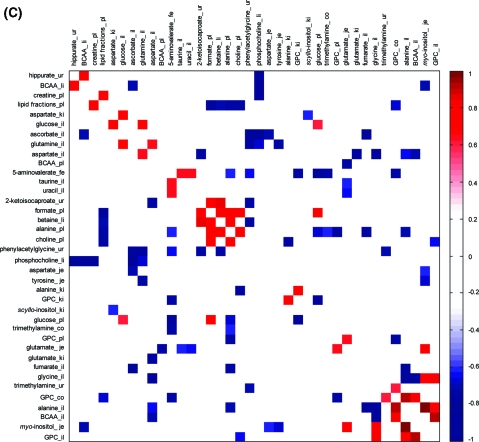
Three separate correlation matrices were calculated using integral values for all metabolites, which were identified as discriminatory between uninfected control and E. caproni-infected mice in particular tissues. These matrices were calculated for (i) control group only (Figure 5A); (ii) infected group only (Figure 5B); and (iii) the difference in metabolite pairwise correlations between control and infected groups (Figure 5C). The correlation matrix constructed for the control data set showed several positive and negative correlations between paired metabolites. The greatest number of infection-induced perturbed metabolite levels occurred in the ileum (n = 11), the plasma (n = 8) and kidney (n = 5), while only a single metabolite, that is, 5-aminovalerate, was statistically altered in the fecal water profile. The colon showed significant changes in the levels of just 2 metabolites after infection. In some matrices, such as urine and ileum, the discriminatory biomarkers, in general, co-varied (either positively or negatively) within that matrix. In other matrices such as the kidney, the key infection discriminating metabolites did not show strong within-matrix correlations. The correlation matrix was structured using a clustering algorithm indicating patterns of similarity and dissimilarity between metabolites regardless of their topographical location (Figure 5A−C). For the control group, several ileal amino acids (alanine, valine, leucine, aspartate, and glycine) demonstrate positive correlations. The GPC level in the colon was correlated with GPC in the plasma and was also positively associated with plasma lipids, whereas ileal GPC was negatively correlated with the ileal amino acids. The correlation matrix calculated for the infected group (Figure 5B) showed conservation of some pairwise correlations (e.g., ileal BCAAs and alanine) observed for the control data set, but the correlations for other metabolite pairs observed in the control case disintegrated (e.g., GPC levels in the plasma and colon) and new infection-dependent associations were formed, such as those between glutamate and myo-inositol in the jejunum or urinary hippurate and hepatic BCAAs. The differences in the underlying structure of metabolite correlations between the control and infected groups are further underscored in Figure 5C, which illustrates the differential cluster matrix of E. caproni-induced effects.
Figure 5.
Cluster analysis based on E. caproni infection-induced metabolic interactions between 8 assessed biological matrices. (A) Correlation structure of control animals demonstrating natural variation due to genetic and experimental individual background; (B) correlation structure composed of inherent individual variation and the effects of an infection; (C) differential cluster matrix showing only the remaining effects of the E. caproni infection. Key: co, colon; fe, fecal water; il, ileum; je, jejunum; ki, kidney; li, liver; pl, plasma; ur, urine. Statistically significant pairwise correlations between metabolite levels with p < 0.05 were explored in the analysis.
Discussion
The systemic effect of an E. caproni infection in the mouse was reflected in 5 of the 8 host tissues examined and in all 3 biofluids (urine, plasma, and fecal water) investigated. Since no overt tissue damage was revealed by histology, the observed alterations in biochemistry associated with an E. caproni infection can be deemed to be subpathological. As expected, the tissue which was most affected in terms of the number of perturbed metabolites was the ileum, since the small intestine is the primary target location of E. caproni. The infection-induced metabolic changes suggested a malabsorption in the ileum reflected by low ileal concentrations of the BCAAs, taurine, ascorbate, alanine, uracil, and glutamine and may suggest an impairment of the Na+-coupled co-transporters, which are the main form of cross-luminal transport in the small intestinal tract.(42) A host of transporter molecules have been summarized previously,(43) whereby 3 main groups were identified: (i) B0AT1/SLC6A19 for neutral amino acids(44) (e.g., valine, alanine, and isoleucine), which cluster in the correlation map (Figure 5B); (ii) SLC1A1(45) for co-transport of anionic amino acid (e.g., aspartate); and (iii) B0,+ATB0,+/SLC6A14, which is responsible for neutral and cationic amino acid co-transport (e.g., glutamine).(46) Additionally, taurine, ascorbate, and uracil depletion in the small intestinal tissues can be explained by a disruption of Na+-coupled transport mechanisms.47,48 A potential intestinal malabsorption has already been suggested in our previous research focusing on biofluids obtained from E. caproni-infected mice,(11) whereby increased BCAA concentration was observed in the feces and subsequently decreased BCAA concentration in plasma. Here, the depletion of serum BCAAs was consistent with the relatively lower ileal and liver concentrations of these metabolites. The jejunal portion of the small intestine showed increased concentrations of the aromatic amino acids, tyrosine and phenylalanine, which are oxidized in the liver to phenylacetate and p-hydroxyphenylacetate, respectively, and are then converted into p-cresol by gut microbial species (e.g., Clostridium difficile and Clostridium scatologenes).(49) Since p-cresol undergoes phase II conjugation to the glucuronide or sulfate in the kidney prior to excretion via urine, this might explain the higher concentrations of p-cresol glucuronide found in the urine of E. caproni-infected mice, which was significantly elevated at time points prior to day 33 postinfection.(11)
E. caproni infection-induced changes in the mouse colon included relatively higher levels of trimethylamine (TMA), which again may reflect altered microbial activity, as dietary methyl components such as carnitine and choline are degraded to TMA by gut bacterial species.(50) TMA is bound to lipoproteins after transluminal transport and reaches the liver via blood circulation where it is oxidized to trimethylamine-N-oxide (TMAO).(51) Both choline degradation products (TMA and TMAO) exhibited a higher urinary excretion in the infected animals, which might be reflective of a systemic increase in these products. Furthermore, relatively higher levels of GPC were found in infected mice, which may be due to the reported E. caproni-induced degradation of host membranes.(25) This observation is further supported by the previously identified increase of lipid fractions (e.g., fatty acids, triacylglycerides, and lipoproteins) in the plasma of infected mice.(11) We hypothesize that the significantly higher levels of phosphocholine, with subsequent betaine production in the liver, and the increase of inositol and lipid fractions in the kidney, compensate for the intestinal lipid degradation by increased uptake and depletion of plasma choline.52−55 Hierarchical multivariate analysis revealed that the main source of variance in the combined data set, in connection to the response to infection, was mainly accounted for by changes in the low molecular weight tissue components (e.g., intestinal malabsorption of amino acids) as the spectral data acquired via the CPMG pulse sequence, which seems to drive the mapping position of the infected group in the metabolic space (see Figure 4, blue coordinates), are selective for small molecular weight components. Interestingly, one tissue component unrelated to CPMG descriptors (i.e., diffusion-edited spectra of ileum) co-mapped with the infected animal designated ‘t2’, which showed the highest worm burden. This may indicate an increased degradation of biological membranes in the small intestine,(56) hence, resulting in increased systemic lipid components. Although some groupings between biological matrices are evident, for example, the ileum, colon, and liver map to a similar location in the hierarchical loadings plot, the transparency of the individual metabolites contributing to the structure of the model (e.g., the discrimination between infected and control samples) is limited. Thus, an exploration of covariance between metabolites both within and across biological matrices can generate a clearer picture of potential mechanistic molecular connectivities.
For the differential candidate biomarkers selected on the basis of the O-PLS-DA models, correlation structures were assessed across the full range of tissues and biofluids separately for the control and infected groups and the differences between the two correlation matrices compared. While certain relationships across both control and infected correlation matrices were consistent, for example, the positive association between the various amino acids in the ileum, or plasma GPC with plasma lipids, for some metabolite pairs the direction of correlation was either not preserved (e.g., plasma and colonic GPC) or even reversed (jejunal myo-inositol and ileal alanine). Additional correlation analysis between the selected tissue compartments revealed interesting insight into the systems dynamics in response to the infection. The most diagnostic tissue in terms of the number of perturbed metabolites was the ileum, which is consistent with the localization of the maturing and adult trematode in the ileum. Urine was also strongly diagnostic of infection with several metabolites correlating directly with ileal metabolites, for example, a negative correlation of urinary phenylacetylglycine was found with ileal ascorbate and glutamine. Two previous studies using high-performance thin-layer chromatography for the analysis of urine of E. caproni-infected mice reported altered neutral lipid profile and amino acids in infected animals.26,57 The fecal water spectra showed the least degree of intra- or extra-matrix correlation, which can, at least partially, be explained by the metagenomic influence as the gut microbial species produce approximately 60% of the fecal mass and are intra- and interindividually highly dynamic. For these reasons, fecal samples are most variable in biochemical composition and difficult to interpret within a systems context, but can add important pathway information relating to the influence of the infection on the microbiota or vice versa. Surprisingly, the fecal mass profile showed little correlation with the colon, despite their close physical connection, but is most likely explained by the low number of compartment-specific infection-induced changes, which were the driving factor of the pairwise correlation analysis. The association between metabolite levels of control animals arises due to intrinsic variability of a metabolic system influenced by genetic and environmental factors. In turn, the effects of infection, disrupting regulatory activity of metabolic networks, may change the underlying structure of metabolite inter-relationships. The correlation analysis thus offers a way to explore additional information about the physiological or pathological state of an organism.
The clustered similarity matrix of pairwise correlations between discriminatory metabolites indicates that some metabolic changes are highly correlated with numerous metabolites, while others covary with only 1 or 2 other infection-induced changes in metabolite levels. The number of correlations between a given candidate biomarker and other metabolites may allow estimation of the specificity and utility of that metabolite as a biomarker of an E. caproni infection. For instance, it is likely that metabolites which are catholic in their correlation with other infection-related metabolite changes are more likely to inform on a global response to the infection at a systems level, while metabolites demonstrating discriminatory behavior between infected and control, but which are not widely correlated to other discriminatory metabolites are more likely to report on a pathway-specific or localized effect. Plasma alanine concentrations were increased in the E. caproni-infected mice and correlated with 10 other infection-related metabolite changes within and across compartments, including decreased plasma glucose concentrations, which may be indicative of an up-regulation of gluconeogenesis. Other metabolites that correlate only within 1 tissue may be more indicative of localized effects. The inverse correlation of urinary trimethylamine and phenylacetylglycine, for instance, both of which can be derived from gut microbial metabolism, suggests that an E. caproni infection causes disruption of the intestinal microbiota, which is consistent with observations made for several other host−parasite models.8−12
Conclusion
This study emphasizes the contribution of multilevel metabolic profiling of host−parasite systems to obtain information on the pathology and pathway perturbations, and to reveal information on a panorganismal metabolic level. Furthermore, a promising approach is utilized for visualizing compartmental interactions with a view to identifying the metabolic features (e.g., the candidate biomarkers) of the present infection, and hence, assess the degree of physiological involvement in disease mechanisms. The sensitivity of the approach is underlined by the additional histological analyses of all assessed tissues, which did not show any obvious infection-induced changes, thus, illustrating the ability of the applied spectroscopic tools for detecting sub- and prepathological states. The suitability of E. caproni as a model trematode has been confirmed in view of the subtle metabolic findings presented here. An infection with this intestinal fluke indeed appears to weaken the overall physiological state of the host by inducing an intestinal malabsorption of nutrients and affecting both direct and remote tissue compartments. While the impact of this parasite may be relatively harmless per se, in co-infection situations, it may exert a more deleterious effect. For example, the hepatic manifestation of the low BCAA status could have adverse effects in the case of a hepatic dysfunction, as the BCAAs are known to play a key role in hepatic regenerative processes,(58) which is of particular importance in Asian countries due to the high burden of hepato-biliary trematodes (e.g., Clonorchis sinensis and Opisthorchis viverrini), and which additionally account for the highest global rates of liver cancer and associated liver cirrhosis.59,60
Acknowledgments
We thank Dr. Olivier Cloarec for providing the Matlab script for O-PLS-DA and STOCSY analysis. Our investigation received financial support from Imperial College London, Biomedical Science Division and the Swiss National Science Foundation. J.S., J.V.L. and J.U. (project no. PPOOB-102883 and PPOOB-119129) and J.K. (project no. PPOOA-114941) are grateful to the Swiss National Science Foundation for financial support, and Y.W. acknowledges Nestlé and Chinese Academy of Science (KJCX-2YW-W11) for provision of funds. Additional support was provided to J.S. by the Wellcome Trust/Imperial College VIP award (PS1041). Authors’ contributions: J.S., Y.W., J.K., E.H. and J.U. designed the study. J.S., J.V.L., J.K., S.D. and J.U. carried out the experiments. J.S., J.V.L., Y.W., J.K., S.D., K.V., I.K.S.Y., J.K.N., E.H., and J.U. analyzed and interpreted the data. J.S. drafted the manuscript. All authors read and approved the final version of the manuscript before submission and contributed to the revision of the manuscript.
References
- Kennedy P. G. The continuing problem of human African trypanosomiasis (sleeping sickness). Ann. Neurol. 2008, 64 (2), 116–126. [DOI] [PubMed] [Google Scholar]
- Winzeler E. A. Malaria research in the post-genomic era. Nature 2008, 455 (7214), 751–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keiser J.; Utzinger J. Chemotherapy for major food-borne trematodes: a review. Expert Opin. Pharmacother. 2004, 5 (8), 1711–1726. [DOI] [PubMed] [Google Scholar]
- Nascimento-Carvalho C. M.; Moreno-Carvalho O. A. Neuroschistosomiasis due to Schistosoma mansoni: a review of pathogenesis, clinical syndromes and diagnostic approaches. Rev. Inst. Med. Trop. Sao Paulo 2005, 47 (4), 179–184. [DOI] [PubMed] [Google Scholar]
- Echenique-Elizondo M.; Amondarain J.; Liron de Robles C. Fascioliasis: an exceptional cause of acute pancreatitis. JOP 2005, 6 (1), 36–39. [PubMed] [Google Scholar]
- Guarner F.; Malagelada J. R. Gut flora in health and disease. Lancet 2003, 361 (9356), 512–519. [DOI] [PubMed] [Google Scholar]
- Tuohy K. M.; Probert H. M.; Smejkal C. W.; Gibson G. R. Using probiotics and prebiotics to improve gut health. Drug Discovery Today 2003, 8 (15), 692–700. [DOI] [PubMed] [Google Scholar]
- Martin F. P. J.; Verdu E. F.; Wang Y.; Dumas M. E.; Yap I. K. S.; Cloarec O.; Bergonzelli G. E.; Corthesy-Theulaz I.; Kochhar S.; Holmes E.; Lindon J. C.; Collins S. M.; Nicholson J. K. Transgenomic metabolic interactions in a mouse disease model: interactions of Trichinella spiralis infection with dietary Lactobacillus paracasei supplementation. J. Proteome Res. 2006, 5 (9), 2185–2193. [DOI] [PubMed] [Google Scholar]
- Li J. V.; Wang Y.; Saric J.; Nicholson J. K.; Dirnhofer S.; Singer B. H.; Tanner M.; Wittlin S.; Holmes E.; Utzinger J. Global metabolic responses of NMRI mice to an experimental Plasmodium berghei infection. J. Proteome Res. 2008, 7 (9), 3948–3956. [DOI] [PubMed] [Google Scholar]
- Wang Y.; Holmes E.; Nicholson J. K.; Cloarec O.; Chollet J.; Tanner M.; Singer B. H.; Utzinger J. Metabonomic investigations in mice infected with Schistosoma mansoni: an approach for biomarker identification. Proc. Natl. Acad. Sci. U.S.A. 2004, 101 (34), 12676–12681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saric J.; Li J. V.; Wang Y.; Keiser J.; Bundy J. G.; Holmes E.; Utzinger J. Metabolic profiling of an Echinostoma caproni infection in the mouse for biomarker discovery. PLoS Negl. Trop. Dis. 2008, 2 (7), e254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.; Utzinger J.; Saric J.; Li J. V.; Burckhardt J.; Dirnhofer S.; Nicholson J. K.; Singer B. H.; Brun R.; Holmes E. Global metabolic responses of mice to Trypanosoma brucei brucei infection. Proc. Natl. Acad. Sci. U.S.A. 2008, 105 (16), 6127–6132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers R. W.; Elliott D. E.; Qadir K.; Urban J. F. Jr.; Thompson R.; Weinstock J. V. Trichuris suis seems to be safe and possibly effective in the treatment of inflammatory bowel disease. Am. J. Gastroenterol. 2003, 98 (9), 2034–2041. [DOI] [PubMed] [Google Scholar]
- Wang Y.; Utzinger J.; Xiao S. H.; Xue J.; Nicholson J. K.; Tanner M.; Singer B. H.; Holmes E. System level metabolic effects of a Schistosoma japonicum infection in the Syrian hamster. Mol. Biochem. Parasitol. 2006, 146 (1), 1–9. [DOI] [PubMed] [Google Scholar]
- Li J. V.; Holmes E.; Saric J.; Keiser J.; Dirnhofer S.; Utzinger J.; Wang Y. Metabolic profiling of a Schistosoma mansoni infection in mouse tissues using magic angle spinning-nuclear magnetic resonance spectroscopy. Int. J. Parasitol 2009, 39 (5), 547–558. [DOI] [PubMed] [Google Scholar]
- Hotez P. J.; Brindley P. J.; Bethony J. M.; King C. H.; Pearce E. J.; Jacobson J. Helminth infections: the great neglected tropical diseases. J. Clin. Invest. 2008, 118 (4), 1311–1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corwin R. M.Economics of gastrointestinal parasitism of cattle. Vet. Parasitol. 1997, 72 (3−4) 451−457, discussion 457−460. [DOI] [PubMed] [Google Scholar]
- Tsang T. M.; Griffin J. L.; Haselden J.; Fish C.; Holmes E. Metabolic characterization of distinct neuroanatomical regions in rats by magic angle spinning 1H nuclear magnetic resonance spectroscopy. Magn. Reson. Med. 2005, 53 (5), 1018–1024. [DOI] [PubMed] [Google Scholar]
- Griffin J. L.; Troke J.; Walker L. A.; Shore R. F.; Lindon J. C.; Nicholson J. K. The biochemical profile of rat testicular tissue as measured by magic angle spinning 1H NMR spectroscopy. FEBS Lett. 2000, 486 (3), 225–229. [DOI] [PubMed] [Google Scholar]
- Bollard M. E.; Garrod S.; Holmes E.; Lindon J. C.; Humpfer E.; Spraul M.; Nicholson J. K. High-resolution 1H and 1H-13C magic angle spinning NMR spectroscopy of rat liver. Magn. Reson. Med. 2000, 44 (2), 201–207. [DOI] [PubMed] [Google Scholar]
- Wilson M.; Davies N. P.; Grundy R. G.; Peet A. C. A quantitative comparison of metabolite signals as detected by in vivo MRS with ex vivo1H HR-MAS for childhood brain tumours. NMR Biomed. 2009, 22 (2), 213–219. [DOI] [PubMed] [Google Scholar]
- Wang Y.; Holmes E.; Comelli E. M.; Fotopoulos G.; Dorta G.; Tang H.; Rantalainen M. J.; Lindon J. C.; Corthesy-Theulaz I. E.; Fay L. B.; Kochhar S.; Nicholson J. K. Topographical variation in metabolic signatures of human gastrointestinal biopsies revealed by high-resolution magic-angle spinning 1H NMR spectroscopy. J. Proteome Res. 2007, 6 (10), 3944–3951. [DOI] [PubMed] [Google Scholar]
- Garrod S.; Humpfer E.; Spraul M.; Connor S. C.; Polley S.; Connelly J.; Lindon J. C.; Nicholson J. K.; Holmes E. High-resolution magic angle spinning 1H NMR spectroscopic studies on intact rat renal cortex and medulla. Magn. Reson. Med. 1999, 41 (6), 1108–1118. [DOI] [PubMed] [Google Scholar]
- Millis K. K.; Maas W. E.; Cory D. G.; Singer S. Gradient, high-resolution, magic-angle spinning nuclear magnetic resonance spectroscopy of human adipocyte tissue. Magn. Reson. Med. 1997, 38 (3), 399–403. [DOI] [PubMed] [Google Scholar]
- Fried B.; Huffman J. E. The biology of the intestinal trematode Echinostoma caproni. Adv. Parasitol. 1996, 38, 311–368. [DOI] [PubMed] [Google Scholar]
- Vasta J. D.; Fried B.; Sherma J. High performance thin layer chromatographic analysis of neutral lipids in the urine of BALB/c mice infected with Echinostoma caproni. Parasitol. Res. 2008, 102 (4), 625–629. [DOI] [PubMed] [Google Scholar]
- Nicholson J. K.; Foxall P. J.; Spraul M.; Farrant R. D.; Lindon J. C. 750 MHz 1H and 1H-13C NMR spectroscopy of human blood plasma. Anal. Chem. 1995, 67 (5), 793–811. [DOI] [PubMed] [Google Scholar]
- Meiboom S.; Gill D. Modified spin-echo method for measuring nuclear relaxation times. Rev. Sci. Instrum. 1958, 29, 688–691. [Google Scholar]
- Wu D. H.; Chen A. D.; Johnson C. S. An improved diffusion-ordered spectroscopy experiment incorporating bipolar-gradient pulses. J. Magn. Reson. Ser. A 1995, 115 (2), 260–264. [Google Scholar]
- James T. L.; McDonald G. G. Measurement of the self-diffusion coefficient of each component in a complex system using pulsed-gradient Fourier transform NMR. J. Magn. Reson. 1973, 11 (1), 58–61. [Google Scholar]
- Ernst R. R.; Bodenhauser G.; Wokaun A.. Principles of Nuclear Magnetic Resonance in One and Two Dimensions. Oxford University Press, London/New York, 1987. [Google Scholar]
- Bax A.; Lerner L.. Two-dimensional nuclear magnetic resonance spectroscopy. Science, 232 (4753), 960−967. [DOI] [PubMed]
- Cloarec O.; Dumas M. E.; Craig A.; Barton R. H.; Trygg J.; Hudson J.; Blancher C.; Gauguier D.; Lindon J. C.; Holmes E.; Nicholson J. Statistical total correlation spectroscopy: an exploratory approach for latent biomarker identification from metabolic 1H NMR data sets. Anal. Chem. 2005, 77 (5), 1282–1289. [DOI] [PubMed] [Google Scholar]
- Eriksson L.; Johansson E.; Kettaneh-Wold N.; Trygg J.; Wikstrom C.. Multi and Megavariate Data Analysis; Umetrics Academy: Umeå, Sweden, 2001. [Google Scholar]
- Eriksson L.; Antti H.; Gottfries J.; Holmes E.; Johansson E.; Lindgren F.; Long I.; Lundstedt T.; Trygg J.; Wold S. Using chemometrics for navigating in the large data sets of genomics, proteomics, and metabonomics (gpm). Anal. Bioanal. Chem. 2004, 380 (3), 419–429. [DOI] [PubMed] [Google Scholar]
- Trygg J.; Holmes E.; Lundstedt T. Chemometrics in metabonomics. J. Proteome Res. 2007, 6 (2), 469–479. [DOI] [PubMed] [Google Scholar]
- Martin F. P. J.; Wang Y.; Sprenger N.; Yap I. K. S.; Lundstedt T.; Lek P.; Rezzi S.; Ramadan Z.; van Bladeren P.; Fay L. B.; Kochhar S.; Lindon J. C.; Holmes E.; Nicholson J. K. Probiotic modulation of symbiotic gut microbial-host metabolic interactions in a humanized microbiome mouse model. Mol. Syst. Biol. 2008, 4, 157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergman R.; Johansson M.; Lundstedt T.; Seifert E.; Åberg J. Optimization of a tabletting process by sequential design and multivariate analysis. Chemom. Intell. Lab. Syst. 1998, 44 (1−2), 271–286. [Google Scholar]
- Lundstedt T.; Seifert E.; Abramo L.; Thelin B.; Nyström A.; Pettersen J.; Bergman R. Experimental design and optimization. Chemom. Intell. Lab. Syst. 1998, 42 (1−2), 3–40. [Google Scholar]
- Westerhuis J. A.; Kourti T.; MacGregor J. F. Analysis of multiblock and hierarchical PCA and PLS models. J. Chemometr. 1998, 12 (5), 301–321. [Google Scholar]
- Press W. H.; Flannery B. P.; Teukolsky S. A.; Vetterling W. T.. Numerical Recipes in Fortran 77 and Fortran 90; Cambridge University Press: Cambridge, U.K., 1997. [Google Scholar]
- O’Flaherty L.; Stapleton P. P.; Redmond H. P.; Bouchier-Hayes D. J. Intestinal taurine transport: a review. Eur. J. Clin. Invest. 1997, 27 (11), 873–880. [DOI] [PubMed] [Google Scholar]
- Thwaites D. T.; Anderson C. M. H+-coupled nutrient, micronutrient and drug transporters in the mammalian small intestine. Exp. Physiol. 2007, 92 (4), 603–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broer A.; Klingel K.; Kowalczuk S.; Rasko J. E.; Cavanaugh J.; Broer S. Molecular cloning of mouse amino acid transport system B0, a neutral amino acid transporter related to Hartnup disorder. J. Biol. Chem. 2004, 279 (23), 24467–24476. [DOI] [PubMed] [Google Scholar]
- Kanai Y.; Hediger M. A. The glutamate/neutral amino acid transporter family SLC1: molecular, physiological and pharmacological aspects. Pfluegers Arch. 2004, 447 (5), 469–479. [DOI] [PubMed] [Google Scholar]
- Hatanaka T.; Haramura M.; Fei Y. J.; Miyauchi S.; Bridges C. C.; Ganapathy P. S.; Smith S. B.; Ganapathy V.; Ganapathy M. E. Transport of amino acid-based prodrugs by the Na+- and Cl(-) -coupled amino acid transporter ATB0,+ and expression of the transporter in tissues amenable for drug delivery. J. Pharmacol. Exp. Ther. 2004, 308 (3), 1138–1147. [DOI] [PubMed] [Google Scholar]
- Chen N. H.; Reith M. E.; Quick M. W. Synaptic uptake and beyond: the sodium- and chloride-dependent neurotransmitter transporter family SLC6. Pfluegers Arch. 2004, 447 (5), 519–531. [DOI] [PubMed] [Google Scholar]
- Takanaga H.; Mackenzie B.; Hediger M. A. Sodium-dependent ascorbic acid transporter family SLC23. Pfluegers Arch. 2004, 447 (5), 677–682. [DOI] [PubMed] [Google Scholar]
- Yu L.; Blaser M.; Andrei P. I.; Pierik A. J.; Selmer T. 4-Hydroxyphenylacetate decarboxylases: properties of a novel subclass of glycyl radical enzyme systems. Biochemistry 2006, 45 (31), 9584–9592. [DOI] [PubMed] [Google Scholar]
- Seibel B. A.; Walsh P. J. Trimethylamine oxide accumulation in marine animals: relationship to acylglycerol storage. J. Exp. Biol. 2002, 205 (3), 297–306. [DOI] [PubMed] [Google Scholar]
- Smith J. L.; Wishnok J. S.; Deen W. M. Metabolism and excretion of methylamines in rats. Toxicol. Appl. Pharmacol. 1994, 125 (2), 296–308. [DOI] [PubMed] [Google Scholar]
- Zeisel S. H.; Blusztajn J. K. Choline and human nutrition. Annu. Rev. Nutr. 1994, 14, 269–296. [DOI] [PubMed] [Google Scholar]
- Zeisel S. H.; daCosta K. A.; Youssef M.; Hensey S. Conversion of dietary choline to trimethylamine and dimethylamine in rats: dose-response relationship. J. Nutr. 1989, 119 (5), 800–804. [DOI] [PubMed] [Google Scholar]
- Garcia-Perez A.; Burg M. B. Renal medullary organic osmolytes. Physiol. Rev. 1991, 71 (4), 1081–1115. [DOI] [PubMed] [Google Scholar]
- Troyer D. A.; Schwertz D. W.; Kreisberg J. I.; Venkatachalam M. A. Inositol phospholipid metabolism in the kidney. Annu. Rev. Physiol. 1986, 48, 51–71. [DOI] [PubMed] [Google Scholar]
- Horutz K.; Fried B. Effects of Echinostoma caproni infection on the neutral lipid content of the intestinal mucosa of ICR mice. Int. J. Parasitol. 1995, 25 (5), 653–655. [DOI] [PubMed] [Google Scholar]
- Vasta J. D.; Fried B.; Sherma J. Determination and quantification of amino acids in the urine of BALB/c mice infected with Echinostoma caproni by high performance thin-layer chromatography-densitometry. J. Liq. Chromatogr. Relat. Technol. 2009, 32 (9), 1210–1222. [Google Scholar]
- Urata Y.; Okita K.; Korenaga K.; Uchida K.; Yamasaki T.; Sakaida I. The effect of supplementation with branched-chain amino acids in patients with liver cirrhosis. Hepatol. Res. 2007, 37 (7), 510–516. [DOI] [PubMed] [Google Scholar]
- Lai E. C.; Lau W. Y. The continuing challenge of hepatic cancer in Asia. Surgeon 2005, 3 (3), 210–215. [DOI] [PubMed] [Google Scholar]
- Keiser J.; Utzinger J.. Food-borne trematodiases: a review. Clin. Microbiol. Rev. 2009, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]