Abstract
The Niemann-Pick C1 Like 1 (NPC1L1) is a predicted polytopic membrane protein that is critical for cholesterol absorption. NPC1L1 takes up free cholesterol into cells through vesicular endocytosis. Ezetimibe, a clinically used cholesterol absorption inhibitor, blocks the endocytosis of NPC1L1 thereby inhibiting cholesterol uptake. Human NPC1L1 is a 1,332-amino acid protein with a putative sterol-sensing domain (SSD) that shows sequence homology to HMG-CoA reductase (HMGCR), Niemann-Pick C1 (NPC1), and SREBP cleavage-activating protein (SCAP). Here, we use protease protection and immunofluorescence in selectively permeabilized cells to study the topology of human NPC1L1. Our data indicate that NPC1L1 contains 13 transmembrane helices. The NH2-terminus of NPC1L1 is in the lumen while the COOH-terminus projects to the cytosol. human NPC1L1 contains seven small cytoplasmic loops—four small and three large luminal loops—one of which has been reported to bind ezetimibe. Ezetimibe-glucuronide, the major metabolite of ezetimibe in vivo, can block the internalization of NPC1L1 and cholesterol. The membrane topology of NPC1L1 is similar to that of NPC1, and the putative SSD of NPC1L1 is oriented in the same manner as those of HMGCR, NPC1, and SCAP. The defined topology of NPC1L1 provides necessary information for further dissecting the functions of the different domains of NPC1L1.
Keywords: ezetimibe, NPC1, Niemann-Pick C1 Like 1, SSD
The cholesterol homeostasis in the whole body is maintained by regulating the balance of de novo synthesis, absorption, and clearance (1). Excessive cholesterol uptake is becoming a major risk factor for hypercholesterolemia, which leads to severe pathological conditions such as coronary heart disease (2). Although cholesterol biosynthesis and clearance are well studied (3, 4), free cholesterol absorption from intestine is less documented.
Niemann-Pick C1 Like 1 (NPC1L1) has been recently identified as a critical protein for enterohepatic free cholesterol absorption (5–9). NPC1L1 is highly expressed in small intestine and liver (6, 7, 10). Genetic or pharmaceutical inactivation of NPC1L1 dramatically inhibits sterol absorption, decreases blood cholesterol level, and prevents atherosclerosis (6, 7, 9, 11). In contrast, hepatic overexpression of NPC1L1 in mice significantly decreases biliary cholesterol concentration and increases plasma cholesterol (5). Ezetimibe, a clinically used cholesterol absorption inhibitor, has been demonstrated to bind NPC1L1 directly and inhibit cholesterol uptake (12, 13).
Our previous studies have shown that NPC1L1 mediates cholesterol uptake through vesicular endocytosis (14, 15). The shuttle of NPC1L1 between endocytic recycling compartment (ERC) and plasma membrane (PM) is subjected to the regulation of cholesterol level changes. When cellular cholesterol is low, NPC1L1 relocates to PM from ERC (14, 16). After the cholesterol loading in the cell membrane, NPC1L1 and cholesterol are internalized together and transported back to ERC (14). Plant sterols, including β-sitosterol, campesterol, stigmasterol, and brassicasterol, cannot promote NPC1L1 endocytosis and are barely taken up (14). This sterol-specificity for NPC1L1 endocytosis partly explains the well-known phenomena that only cholesterol is efficiently absorbed in digestive system. The recycling of NPC1L1 requires microfilament network and clathrin/AP2 complex (14). Notably, ezetimibe prevents the association between NPC1L1 and clathrin/AP2, blocks the endocytosis of NPC1L1, and therefore decreases cholesterol uptake (14).
Human NPC1L1 protein is an integral membrane protein with 1,332 residues (6, 8). It shows approximately 50% sequence homology to Niemann-Pick C1 (NPC1), dysfunction of which causes cholesterol storage disease Niemann-Pick type C (17). Human NPC1 is reported to have 13 transmembrane domains, resides in late endosome and lysosome, and functions in LDL-derived cholesterol transport (18). Sequences analysis suggests that NPC1L1 contains a signal peptide, multiple N-linked glycosylation sites, and a sterol-sensing domain (SSD) that is conserved in HMG-CoA reductase, NPC1, SCAP, Patched-1, and translocation in renal carcinoma chromosome 8 gene (TRC8) (19–22).
In the current studies, we tested a model for the membrane topology of NPC1L1 through protease protection assays and immunofluorescence in selective permeabilized cells. These data support a model in which NPC1L1 contains 13 membrane-spanning helices. In addition, we demonstrated that ezetimibe-glucuronide, a major metabolite of ezetimibe in vivo, can inhibit the internalization of NPC1L1 and cholesterol. Determining the membrane topology of NPC1L1 provides a map to better explore the mechanism of NPC1L1-mediated cholesterol absorption.
EXPERIMENTAL PROCEDURES
Materials
We obtained horseradish peroxidase-conjugated donkey anti-mouse and anti-rabbit IgG from Jackson ImmunoResearch Laboratories; Alexa Fluor 555 donkey anti-mouse and anti-rabbit IgG from Invitrogen; mouse monoclonal antibody anti-Myc IgG-9E10 from Roche; mouse monoclonal antibody anti-KDEL and digitonin from Calbiochem; Rabbit polyclonal antibody anti-cathepsin D from Santa Cruz; Soybean trypsin inhibitor, filipin and Triton X-100 from Sigma-Aldrich, Inc.; methyl-β-cyclodextron (CDX) from Cyclodextrin Technologies Development, Inc.; Ezetimibe-glucuronide (ezetimibe-Gluc) was a generous gift from Schering-Plough and Merck; and other reagents from previously described sources (14, 23).
Plasmids
The plasmid expressing the EGFP-tagged full-length human NPC1L1 was described previously (14). The Myc tagged NPC1L1 was generated by two steps. The Asc I recognition site was first introduced into the sequences encoding the hydrophilic loops of NPC1L1 at the position as indicated in Fig. 1B. Then the DNA encoding 3 or 6 tandem copies of Myc epitopes (EQKLISEEDL) with AscI site at both ends was PCR amplified and inserted into the NPC1L1 cDNA. The plasmids encoding the truncated versions of NPC1L1 were generated by site-directed mutagenesis based on full-length human NPC1L1. All the constructs were verified by DNA sequencing.
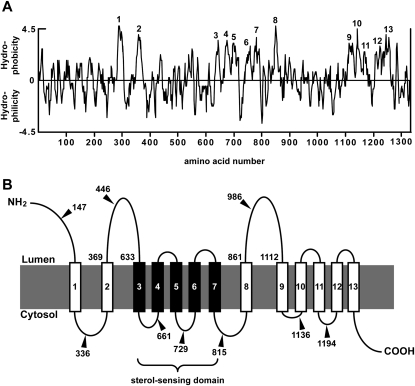
Fig. 1.
Hydropathy plot and predicted membrane topology of human NPC1L1. A: Hydropathy plot of human NPC1L1. The hydropathy profile of NPC1L1 was analyzed using the software of DNASTAR. The putative transmembrane helices were numbered 1–13. B: Predicted membrane topology of human NPC1L1. The solid lines denote the hydrophilic loops; the rectangles numbered 1–13 denote the membrane-spanning regions with a sterol-sensing domain indicated by the black rectangles 3–7. The numbers pointed by triangles indicate the residues behind which Myc tags were inserted, respectively. NPC1L1, Niemann-Pick C1 Like 1.
Cell culture and transient transfection
CRL-1601 (McArdle RH7777 rat hepatoma cell), and CRL-1601/NPC1L1-EGFP that stably expresses NPC1L1-EGFP were grown in monolayer at 37°C in 5% CO2. The cells were maintained in medium A (Dulbecco's modified Eagle's medium containing 100 units/ml penicillin and 100 μg/ml streptomycin sulfate) supplemented with 10% fetal bovine serum (FBS). Cholesterol-depleting medium is medium A supplemented with 5% lipoprotein deficient serum (LPDS), 10 µM compactin, 50 µM mevalonate, and 1.5% CDX. Cholesterol-replenishing medium contains medium A supplemented with 5% LPDS, 10 µM compactin, 50 µM mevalonate, and 15 μg/ml cholesterol/CDX. The cholesterol/CDX inclusion complexes were prepared as described before (14). Transfection of cells with Fugene HD (Roche) was performed according to the manufacture's manual.
Trypsin proteolysis
Cells were harvested, lysed in 0.5 ml buffer C (10 mM Hepes-KOH at pH7.4, 10 mM KCl, 1.5 mM MgCl2, 5 mM sodium EDTA, 5 mM sodium EGTA, 250 mM sucrose), and clarified by centrifugation at 1,000 g for 7 min at 4°C. The pellet was discarded, and the supernatant was centrifuged at 12,000 g for 15 min at 4°C. Then, the pellet was resuspended in 70 μl buffer A2 (buffer C plus 0.1 M NaCl). Aliquots (52 μl) were taken out and supplemented with either 3 μl buffer A2 (for no detergent condition) or 3 μl 10% Triton X-100 (final concentration 0.5%) following addition of various amounts of trypsin as indicated to a final volume of 58 μl. The samples were incubated at 30°C for 25 min. The reaction was stopped by addition of 400 units of soybean trypsin inhibitor (2 μl) and 20 μl 4×loading buffer. After boiling for 5 min at 95°C, the samples were subjected to SDS-PAGE and immunoblot analysis.
Immunofluorescence assay in selectively permeabilized cells
Cells grown on glass coverslips were fixed with 4% paraformaldehyde for 15 min at room temperature and washed with PBS. For selective permeabilization, cells were incubated in buffer S (10 mM Hepes-KOH at pH7.4, 0.3 M sucrose, 0.1 M KCl, 2.5 mM MgCl2, 1 mM sodium EDTA) containing 50 μg/ml digitonin for 3 min on ice. For complete permeabilization, cells were incubated in PBS containing 0.2% Triton X-100 for 5 min at room temperature. After permeabilization, cells were incubated in blocking buffer (PBS plus 1% BSA) for 30 min at 4°C. Myc or EGFP was detected by incubating cells in blocking buffer containing 1.3 μg/ml anti-Myc antibody or 0.5 μg/ml anti-EGFP antibody for 1 h at 4°C, followed by washing with PBS and additional incubating in blocking buffer containing fluorescent secondary antibodies for 40 min at 4°C. Immunofluorescence microscopy was carried out using an Olympus BX51 microscope. In each experiment, images of the same channel were acquired at identical laser output, gain, and offset.
Filipin staining and fluorescence quantification
Cells were fixed with 4% paraformaldehyde for 30 min at room temperature after each treatment and washed three times with PBS. A fresh 5 mg/ml filipin stock solution was prepared in ethanol and diluted to a concentration of 50 μg/ml with PBS containing 10% FBS. Cells were stained with 50 μg/ml filipin in the dark for 30 min at room temperature followed by three washes with PBS. Filipin signals of stained cells were analyzed with a Leica TCS SP5 confocal microscope equipped with a two-photon laser using an excitatory wavelength of 720 nm. Red pseudocolor was assigned to show the filipin signal. In each experiment, images of the same channel were acquired at identical laser output, gain and offset. Fluorescence quantification was carried out as previously described (14).
RESULTS
Predicted membrane topology of human NPC1L1
To predict the potential transmembrane domains of human NPC1L1, we used the software DNASTAR to calculate its hydropathy plot. Based on the computational result, NPC1L1 was predicted to contain 13 transmembrane segments (Fig. 1A). According to this analysis, the membrane topology graph for NPC1L1 was generated (Fig. 1B). The transmembrane helices corresponded to the hydrophobic sequences 1-13 in the hydropathy plot. The NH2-terminus of NPC1L1 was proposed to extend into the lumen, while the COOH-terminus projected to the cytosol. In addition, NPC1L1 was predicted to contain three large luminal loops (a.a. 1-287, a.a. 369-633 and a.a. 861-1112), the second of which has been reported to bind ezetimibe (13). The sterol-sensing domain spanning transmembrane domains 3–7 showed similarity to the five other SSD-containing proteins: HMG-CoA reductase, NPC1, SCAP, Patched-1, and TRC8.
Membrane orientation of a.a. 369-633 and a.a. 861-1112 of NPC1L1
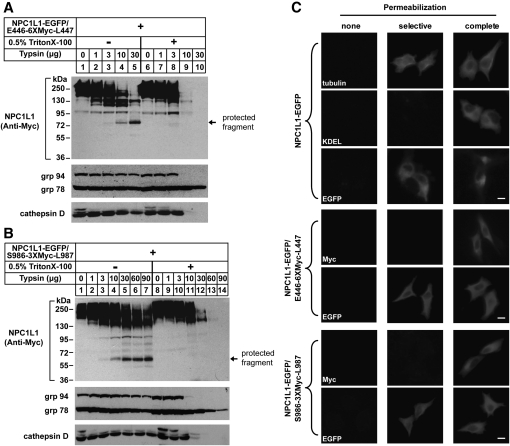
To determine the membrane orientation of the a.a. 369-633 fragment (predicted as the second large luminal loop) of NPC1L1, we constructed a vector encoding NPC1L1 with a COOH-terminal EGFP tag and a 6-copy of Myc tag (6×Myc) inserted between 446 and 447 residue (NPC1L1-EGFP/E446-6×Myc-L447), which we used to perform trypsin protection assay (Fig. 1B). CRL-1601 was transiently transfected with NPC1L1-EGFP/E446-6×Myc-L447 expression plasmid. The intact membrane vesicles were isolated and treated with increasing amounts of trypsin in the absence or presence of Triton X-100. The samples were then subjected to SDS-PAGE followed by immunoblot with indicated antibodies. In the absence of Triton X-100, NPC1L1 showed resistance to trypsin digestion (Fig. 2A, upper panel, lanes 1–5) and a protected fragment of ∼75 kDa was yielded (Fig. 2A, upper panel, lanes 4 and 5). In the presence of Triton X-100, NPC1L1 was destroyed by trypsin and no protected fragment was generated (Fig. 2A, upper panel, lanes 9 and 10). These results indicated that the Myc epitope was located within the lumen. As controls, we utilized the antibody against KDEL to detect two ER luminal proteins, grp78 and grp94, both of which have the common KDEL sequences (Fig. 2A, middle panel). Grp94 was resistant to trypsin proteolysis in the absence of detergent (Fig. 2A, middle panel, lanes 1–5). But in the presence of Triton X-100, the KDEL signal of grp94 was digested at high dosage of trypsin (Fig. 2A, middle panel, lanes 9 and 10). This pattern was identical to that of NPC1L1 detected by anti-Myc antibody. Unlike grp94, grp78 was not digested by trypsin either in the absence or presence of detergent, as grp78 is a intrinsic trypsin-resistant protein (21, 24). Cathepsin D, a protein in lysosome, was used as another control to validate the integrity of vesicles (Fig. 2A, lower panel). Cathepsin D showed a similar pattern to that of NPC1L1 (Fig. 2A, lower panel, lanes 9 and 10). These data indicated that the a.a. 369-633 loop of NPC1L1 was located in the lumen.
Fig. 2.
Membrane orientation of the a.a. 369-633 and a.a. 861-1112 of NPC1L1 as determined by trypsin proteolysis and immunofluorescence in selectively permeabilized cells. A: Membrane orientation of the a.a. 369-633 of NPC1L1. On day 0, CRL-1601 were set up at 7 × 105 per 60-mm dish. On day 2, cells were transfected with construct of NPC1L1-EGFP/E446-6×Myc-L447. On day 3, cells were harvested, fractionated, and trypsin proteolysis was performed as described under Experimental Procedures. The samples were then mixed with 4× SDS loading buffer, boiled for 5 min at 95°C, subjected to SDS-PAGE, and blotted with antibodies against Myc, cathepsin D, or anti-KDEL (against grp94 and grp78). B: Membrane orientation of the a.a. 861-1112 of NPC1L1. CRL-1601 cells were set up and transfected with construct of NPC1L1-EGFP/S986-3×Myc-L987. After 24 h, the cells were treated as described in (A). The samples were then mixed with 4× SDS loading buffer, boiled for 5 min at 95°C, subjected to SDS-PAGE, and blotted with antibodies against Myc, cathepsin D or anti-KDEL (against grp94 and grp78). C: Immunofluorescence in selective permeabilized cells. CRL-1601 transiently transfected with constructs of NPC1L1-EGFP or NPC1L1-EGFP/E446-6×Myc-L447 or NPC1L1-EGFP/S986-3×Myc-L987 were grown on glass coverslips, fixed with 4% paraformaldehyde, and permeabilized with either 50 μg/ml digitonin (selective permeabilization) or 0.2% Triton X-100 (complete permeabilization) as described under Experimental Procedures. Then cells were blocked with 1% BSA in PBS and labeled with primary antibodies and fluorescent secondary antibodies as indicated in the figure. Immunofluorescence microscopy was performed using an Olympus BX51 microscope. None, no permeabilization; selective, selective permeabilization; complete, complete permeabilization. Scale bar = 10 μm.
We next used NPC1L1-EGFP/S986-3×Myc-L987 to test the membrane orientation of the a.a. 861-1112 loop (the predicted third large luminal loop) of NPC1L1 (Fig. 1B). CRL-1601 cells were transfected with NPC1L1-EGFP/S986-3×Myc-L987 expression construct and trypsin protection assay was carried out as in Fig. 2A. In the absence of Triton X-100, NPC1L1 was resistant to trypsin and a protected fragment of ∼60 kDa was observed (Fig. 2B, upper panel, lanes 4-7). At high dosage of trypsin, NPC1L1 was completely digested in the presence of Triton X-100 (Fig. 2B, upper panel, lanes 13 and 14). The patterns of grp94 and cathepsin D were similar to that of NPC1L1 (Fig. 2B, middle and lower panels), indicating that the orientation of the a.a. 861-1112 fragment of NPC1L1 was luminal.
To further study the membrane orientation of NPC1L1, we established an immunofluorescence assay in which the cells were treated using one of the following conditions: (1) no permeabilization (absence of detergent); (2) selective permeabilization of the plasma membrane (50 μg/ml of digitonin); or (3) complete permeabilization (0.2% Triton X-100). As shown in the first set of panels (Fig. 2C), tubulin, a cytoplasmic protein was detected in both selective and complete permeabilization conditions. However, anti-KDEL immunofluorescent signal was observed in the complete permeabilized but not in selective permeabilized cells. EGFP that was fused to the C-terminal of NPC1L1 was stained in both selective and complete permeabilization conditions (Fig. 2C, the first set of panels). Altogether, these data demonstrated the specificity of the cell permeabilization protocol and indicated the COOH-terminus of NPC1L1 projected to cytosol.
To further confirm the luminal orientation of a.a. 369-633 fragment and a.a. 861-1112 fragment of NPC1L1, CRL-1601 cells were transfected with different expression constructs, and immunofluorescence assay was carried out following the indicated permeabilization conditions. NPC1L1 protein was detected with anti-Myc antibody and a fluorescein-conjugated secondary antibody. As a control, the anti-EGFP staining was also performed to measure the orientation of the COOH-terminus of NPC1L1 in the following individual experiment. The Myc tag was detected in the complete permeabilized cells but not in the selective permeabilized cells, when either NPC1L1-EGFP/E446-6×Myc-L447 or NPC1L1-EGFP/S986-3×Myc-L987 was transfected (Fig. 2C, the second and third sets of panels). Consistent with the protease protection results (Fig. 2A and 2B), these immunofluorescence data showed that the a.a. 369-633 and a.a. 861-1112 fragments of NPC1L1 resided in lumen.
Localization of epitopes in full-length NPC1L1
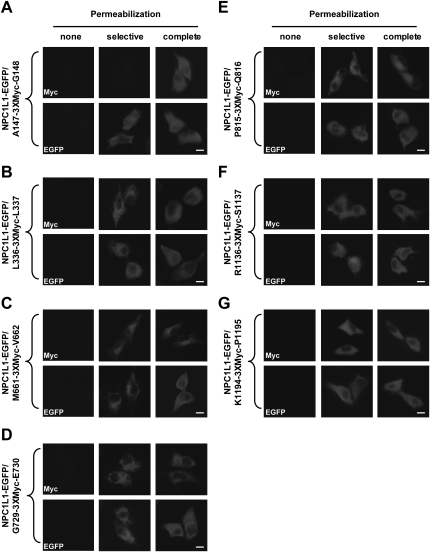
To further dissect the membrane orientation of the hydrophilic domains of NPC1L1, Myc tag (3×Myc) was inserted into the different positions of NPC1L1-EGFP (Fig. 1B). Next, the cells were transfected with the indicated constructs, permeabilized under different conditions, and stained with anti-Myc or anti-EGFP antibodies. As shown in Fig. 3A, Myc tag between A147 and G148 was detected in complete, but not selective, permeabilization. These data suggested that the NH2-terminus of NPC1L1 resided in lumen.
Fig. 3.
Membrane orientation of Myc epitopes in NPC1L1 as determined by immunofluorescence in selectively permeabilized cells. CRL-1601 grown on glass coverslips were transfected with constructs as shown in A–G. None, selective or complete permeabilization were performed as in Figure 2C. After permeabilization, cells were labeled with antibodies against Myc or EGFP followed by fluorescent secondary antibody. Immunofluorescence microscopy was performed using an Olympus BX51 microscope. Scale bar = 10 μm.
When Myc tag was inserted after L336, M661, G729, P815, R1136, and K1194, anti-Myc staining could be observed in both selective and complete permeabilization conditions (Fig. 3B–G). These results illustrated that at these positions, Myc tags were located in cytosol. For every Myc-epitoped protein, anti-EGFP staining showed that their COOH-termini resided in cytosol (Fig. 3B–G), suggesting that the Myc insertion did not alter the topology of NPC1L1 protein.
Localization of EGFP epitopes in truncated NPC1L1
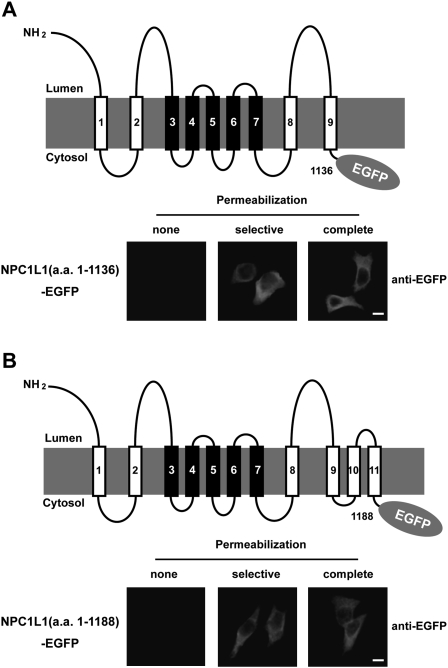
To verify the orientation of the loop between the ninth and tenth transmembrane domain (TM) and the loop between TM 11 and 12, two truncated NPC1L1 constructs, which encode the NH2-terminus to TM 9 and the NH2-terminus to TM 11 fragments with a fused COOH-terminal EGFP, were expressed and analyzed as described above. In both cases, EGFP tags were undetected in nonpermeabilized cells but were strongly stained in selective or complete permeabilization conditions (Fig. 4A and 4B). These observations were consistent with the immunofluorescence results of NPC1L1-EGFP/R1136-3×Myc-S1137 and NPC1L1-EGFP/K1194-3×Myc-P1195 (Fig. 3F and 3G), showing that these domains were located in the cytosol.
Fig. 4.
Membrane orientation of EGFP epitopes in truncated NPC1L1 as determined by immunofluorescence in selectively permeabilized cells. A: Membrane localization of EGFP in NPC1L1(a.a. 1-1136)-EGFP. A schematic illustration of NPC1L1(a.a. 1-1136)-EGFP is shown in the upper panel. In the lower panel, CRL-1601 cells transiently transfected with the construct of NPC1L1-EGFP(a.a. 1-1136) were permeabilized as described in Figure 3C. Cells were labeled with primary antibody against EGFP and fluorescent secondary antibody. Immunofluorescence microscopy was performed using an Olympus BX51 microscope. Scale bar = 10 μm. B: Membrane localization of EGFP in NPC1L1(a.a. 1-1188)-EGFP. A schematic illustration of NPC1L1-EGFP(a.a. 1-1188) is shown in the upper panel, and the immunofluorescence experiments were performed as described in (A). Scale bar = 10 μm.
Sterol-regulated recycling of epitope-tagged NPC1L1
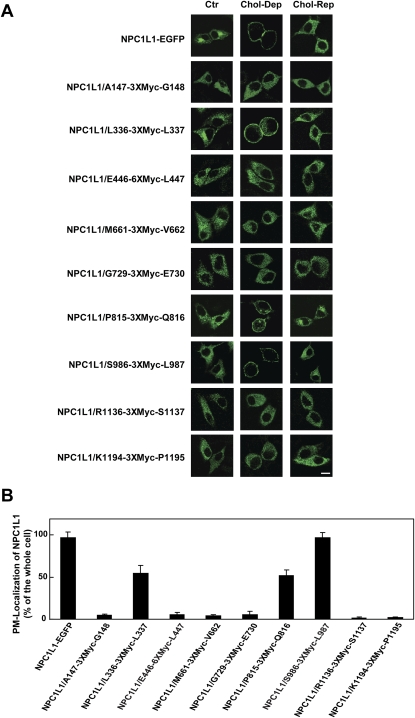
In previous studies, we have shown that the subcellular localization of NPC1L1 protein is regulated by cholesterol (14). Cholesterol depletion induces the translocation of NPC1L1 to PM, and cholesterol replenishing causes the internalization of NPC1L1 and cholesterol (14). To analyze whether the epitope insertion changed the function of NPC1L1, we transfected CRL-1601 cells with the plasmids, encoding different epitope-tagged forms of NPC1L1-EGFP. When cells were depleted of cholesterol for 60 min, the majority of NPC1L1-EGFP and NPC1L1-EGFP/S986-3×Myc-L987 were relocated to PM, and cholesterol replenishing caused their internalization (Fig. 5A, panels 1 and 8). The NPC1L1-EGFP forms with the insertion of the 3×Myc epitope after L336 and P815 showed partial regulation by cholesterol (Fig. 5A, panels 3 and 7). However, epitope insertion at other positions (A147, E446, M661, G729, R1136, and K1194) of NPC1L1 severely impaired the sterol-regulated recycling of NPC1L1 (Fig. 5A, panels 2, 4, 5, 6, 9, and 10).
Fig. 5.
Sterol-regulated recycling of epitope-tagged NPC1L1. A: Translocation of epitope-tagged NPC1L1-EGFP regulated by cholesterol alterations. Cells transfected with indicated plasmids were incubated in cholesterol-depleting medium for 60 min (Chol-Dep). Then the cells were replenished with 15 μg/ml of cholesterol/CDX for 60 min (Chol-Rep). The cells were fixed and examined by confocal microscopy. Ctr, control. Scale bar = 10 μm. B: Quantification of plasma membrane-located NPC1L1 of the cells in (A) at the cholesterol-depleting condition. Error bars represent standard deviations.
Inhibition of the internalization of NPC1L1 and cholesterol by Glucuronidated ezetimibe
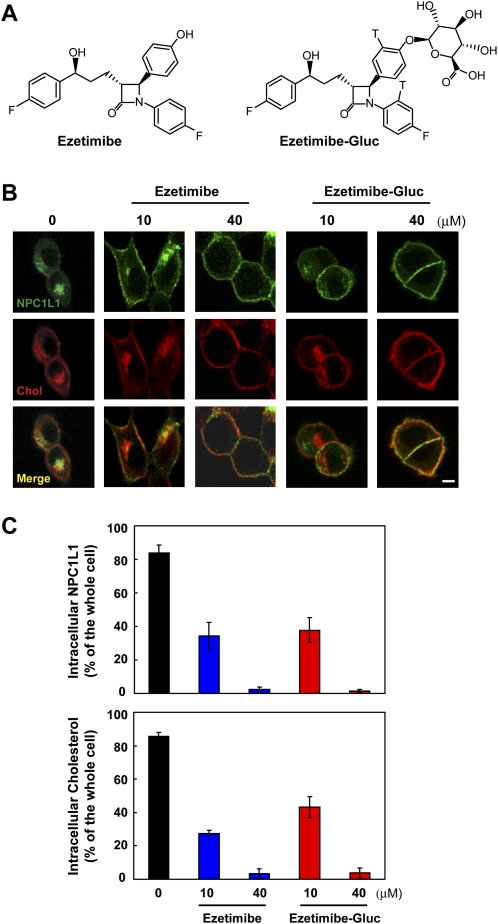
After oral taken-up, ezetimibe is rapidly glucuronidated. The ezetimibe-glucuronide (ezetimibe-Gluc) can inhibit cholesterol absorption as potently as ezetimibe (25, 26). Our previous studies have demonstrated that ezetimibe inhibits cholesterol uptake by blocking the endocytosis of NPC1L1 (14). To test whether ezetimibe-Gluc acts by the similar mechanism, we measured cholesterol uptake by NPC1L1 in our established cellular system (14). Cholesterol-depleted cells were treated with increasing concentrations of ezetimibe or ezetimibe-Gluc, followed by cholesterol replenishment. Both ezetimibe and ezetimibe-Gluc inhibited NPC1L1 endocytosis in a concentration-dependent manner (Fig. 6B). Meanwhile, the inward transport of cholesterol was greatly diminished by both compounds (Fig. 6B and 6C).
Fig. 6.
Glucuronidated ezetimibe inhibits the internalization of NPC1L1 and cholesterol. A: Chemical structures of ezetimibe and ezetimibe-glucuronide (Ezetimibe-Gluc). B: Ezetimibe and Ezetimibe-Gluc inhibit NPC1L1-mediated cholesterol uptake. CRL-1601/NPC1L1-EGFP cells were incubated in cholesterol-depleting medium for 60 min. Then the cells were treated with indicated com pound for 30 min followed by replenishing with 15 μg/ml of choles terol/CDX for 60 min. The cells were fixed, stained with filipin, and examined by two-photon confocal microscopy. Scale bar = 10 μm. C: Quantification of intracellular NPC1L1 and cholesterol of the cells in (B). Error bars represent standard deviations.
DISCUSSION
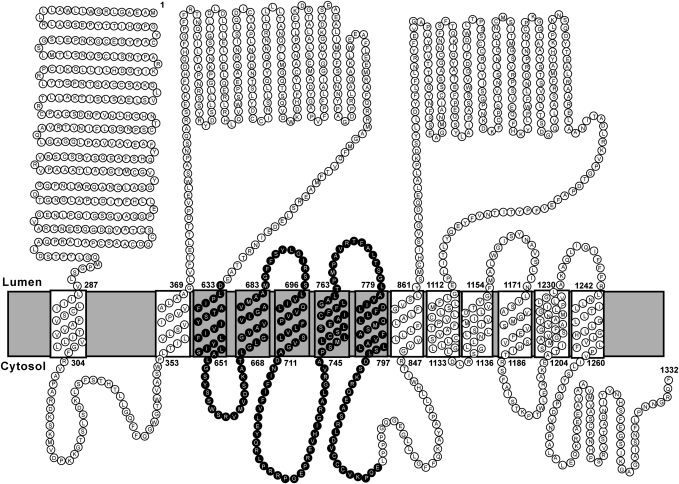
The current study defines the membrane topography of NPC1L1, an endocytic recycling protein that mediates biliary and dietary cholesterol uptake in the liver and intestine. As summarized in Fig. 7, NPC1L1 contains 13 membrane-spanning segments. The NH2-terminus of NPC1L1 with a predicted signal peptide faces the lumen or extracellular matrix, whereas the COOH-terminus projects to the cytosol. NPC1L1 contains 7 small cytoplasmic segments (four small and three large luminal loops).
Fig. 7.
The topological model of human NPC1L1. A topological model based on experimental data in these studies is shown. Each circle represents an amino acid. The sterol-sensing domain is highlighted.
The topologic model of NPC1L1 is based on the analysis of membrane orientation of the epitopes inserted at different positions of NPC1L1. To rule out the possibility that these epitope insertions changed the topology of NPC1L1, we analyzed the membrane orientation of the COOH-terminal EGFP tag in all of the immunofluorescence experiments and tested whether they are functional by measuring the cholesterol-regulated recycling. The results showed that the COOH-terminal EGFP of the tag-inserted NPC1L1-EGFP protein was always localized in cytosol, suggesting the epitopes insertion did not alter the topology of NPC1L1. NPC1L1 with insertions after L336, P815, and S986 can recycle between ERC and PM in response to cholesterol alterations, indicating these insertions do not change the topology and function of NPC1L1. However, epitopes insertion at other positions (A147, E446, M661, G729, R1136, and K1194) of NPC1L1 dramatically impaired the cholesterol-regulated recycling, suggesting the domains (loops) where the tags were inserted are essential for NPC1L1's function. Although these tag-inserted forms of NPC1L1-EGFP showed correct localization of EGFP, we cannot exclude the possibility that the insertion of tags at these positions affects the even numbers of TM regions, which changes the NPC1L1 topology but not the localization of the C-terminal EGFP tag.
The defined topological structure will aid the functional characterization of NPC1L1. Previous studies have shown that NPC1L1 is a multi-functional protein. Its subcellular localization is dynamically regulated by cholesterol (14, 16). The inward transport of NPC1L1 depends on clathrin/AP2 complex (14). The cytosolic portions of NPC1L1 may provide binding sites for the proteins that are responsible for vesicular trafficking. Ezetimibe is a clinically used cholesterol absorption inhibitor that physically interacts with NPC1L1 and inhibits its function. The ezetimibe-binding domain in NPC1L1 has been mapped to the predicted loop C, which corresponds to the second luminal loop in our model (13). Importantly, we have shown that ezetimibe prevents the binding of NPC1L1 to clathrin/AP2 (14). It will be interesting to study how the binding of ezetimibe in the extracellular loop prevents the intracellular association of NPC1L1 and clathrin/AP2.
We have shown that NPC1L1 facilitates cholesterol uptake via vesicular endocytosis, and ezetimibe acts by preventing the internalization of NPC1L1 in cultured cell system (14). After in vivo administration, ezetimibe is rapidly glucuronidated and recycled by the enterohepatic circulation (27). Here we provide experimental evidence that ezetimibe-Gluc can inhibit the internalization of NPC1L1 and cholesterol as potently as ezetimibe. These data indicate that ezetimibe inhibits cholesterol absorption by blocking NPC1L1's endocytosis in vivo.
The membrane topology of NPC1L1 is similar to that of NPC1, and the putative SSD of NPC1L1 is oriented in the same manner as those of NPC1, HMGCR, and SCAP (20–22). These results indicate that NPC1L1 may have similar function and is regulated similarly to NPC1, HMGCR, and SCAP. The SSDs are important for the functions of NPC1, HMGCR, and SCAP, all of which are involved in different aspects of cholesterol metabolism. High levels of sterols promote the binding of Insig proteins to the SSDs of HMGCR and SCAP that result in the ER-associated degradation of HMGCR and ER-retention of SCAP-SREBP complex (28–30). It has been demonstrated that cholesterol directly binds the SSD of SCAP, induces the conformational changes, and therefore promotes the interaction of SCAP with Insig (4). It is yet unknown whether the SSD of HMGCR binds sterols directly. However, NPC1 can bind cholesterol via its SSD that is important for its function in cholesterol transport (31). Recently, it was found that besides SSD, NPC1 can bind cholesterol and oxysterol through its luminal NH2-terminal domain although the sterol binding site on this domain is not essential for NPC1's function in fibroblasts (32, 33). We have shown that cholesterol induces the clathrin-dependent endocytosis of NPC1L1. The topological resemblances of NPC1L1 to NPC1 and other SSD-containing proteins inspires us to hypothesize that the free cholesterol may bind NPC1L1 via the first luminal loop or SSD and induce the conformational change that exposes the cytosolic binding site for clathrin/AP2. Then NPC1L1 and cholesterol can be internalized into the cells. More studies are needed to fully elucidate this hypothesis. The defined topological structure of NPC1L1 will greatly facilitate the future functional and mechanistic studies on NPC1L1.
Acknowledgments
The authors thank Dr. Harry R. Davis (Schering-Plough Research Institute) for helpful discussion; Hong-Hua Miao, Yu-Xiu Qu, Su-ZhePan, Qin Li, and Chun-Gen Yi for technical assistance; and Dr. Wei Qi for critical reading of the manuscript.
Footnotes
Abbreviations:
- a.a.
- amino acid
- CDX
- methyl-β-cyclodextron
- ERC
- endocytic recycling compartment
- ezetimibe-Gluc
- ezetimibe-glucuronide
- HMGCR
- HMG-CoA reductase
- LPDS
- lipoprotein deficient serum
- NPC1
- Niemann-Pick C1
- NPC1L1
- Niemann-Pick C1 Like 1
- PM
- plasma membrane
- SSD
- sterol-sensing domain
- TM
- transmembrane
This work was supported in part by a research grant from the Investigator-Initiated Studies Program of Merck and grants from the Ministry of Science and Technology of China (2009CB919000), National Natural Science Foundation of China (30670432), and Shanghai Science and Technology Committee (074119620, 08JC1421300 and 08431900500).
REFERENCES
- 1.Dietschy J. M., Turley S. D., Spady D. K. 1993. Role of liver in the maintenance of cholesterol and low density lipoprotein homeostasis in different animal species, including humans. J. Lipid Res. 34: 1637–1659 [PubMed] [Google Scholar]
- 2.Grundy S. M.1990. Cholesterol and coronary heart disease. Future directions. JAMA. 264: 3053–3059 [PubMed] [Google Scholar]
- 3.Lo S. G., Petruzzelli M., Moschetta A. 2008. A translational view on the biliary lipid secretory network. Biochim. Biophys. Acta. 1781: 79–96 [DOI] [PubMed] [Google Scholar]
- 4.Goldstein J. L., Bose-Boyd R. A., Brown M. S. 2006. Protein sensors for membrane sterols. Cell. 124: 35–46 [DOI] [PubMed] [Google Scholar]
- 5.Temel R. E., Tang W., Ma Y., Rudel L. L., Willingham M. C., Ioannou Y. A., Davies J. P., Nilsson L. M., Yu L. 2007. Hepatic Niemann-Pick C1-like 1 regulates biliary cholesterol concentration and is a target of ezetimibe. J. Clin. Invest. 117: 1968–1978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Altmann S. W., Davis H. R., Jr., Zhu L. J., Yao X., Hoos L. M., Tetzloff G., Iyer S. P., Maguire M., Golovko A., Zeng M., et al. 2004. Niemann-Pick C1 Like 1 protein is critical for intestinal cholesterol absorption. Science. 303: 1201–1204 [DOI] [PubMed] [Google Scholar]
- 7.Davis H. R., Jr., Zhu L. J., Hoos L. M., Tetzloff G., Maguire M., Liu J., Yao X., Iyer S. P., Lam M. H., Lund E. G., et al. 2004. Niemann-Pick C1 Like 1 (NPC1L1) is the intestinal phytosterol and cholesterol transporter and a key modulator of whole-body cholesterol homeostasis. J. Biol. Chem. 279: 33586–33592 [DOI] [PubMed] [Google Scholar]
- 8.Davies J. P., Levy B., Ioannou Y. A. 2000. Evidence for a Niemann-pick C (NPC) gene family: identification and characterization of NPC1L1. Genomics. 65: 137–145 [DOI] [PubMed] [Google Scholar]
- 9.Davies J. P., Scott C., Oishi K., Liapis A., Ioannou Y. A. 2005. Inactivation of NPC1L1 causes multiple lipid transport defects and protects against diet-induced hypercholesterolemia. J. Biol. Chem. 280: 12710–12720 [DOI] [PubMed] [Google Scholar]
- 10.Sane A. T., Sinnett D., Delvin E., Bendayan M., Marcil V., Menard D., Beaulieu J. F., Levy E. 2006. Localization and role of NPC1L1 in cholesterol absorption in human intestine. J. Lipid Res. 47: 2112–2120 [DOI] [PubMed] [Google Scholar]
- 11.Kuhlencordt P. J., Padmapriya P., Rutzel S., Schodel J., Hu K., Schafer A., Huang P. L., Ertl G., Bauersachs J. 2008. Ezetimibe potently reduces vascular inflammation and arteriosclerosis in eNOS-deficient ApoE ko mice. Atherosclerosis. 202: 48–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garcia-Calvo M., Lisnock J., Bull H. G., Hawes B. E., Burnett D. A., Braun M. P., Crona J. H., Davis H. R., Jr., Dean D. C., Detmers P. A., et al. 2005. The target of ezetimibe is Niemann-Pick C1-Like 1 (NPC1L1). Proc. Natl. Acad. Sci. USA. 102: 8132–8137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weinglass A. B., Kohler M., Schulte U., Liu J., Nketiah E. O., Thomas A., Schmalhofer W., Williams B., Bildl W., McMasters D. R., et al. 2008. Extracellular loop C of NPC1L1 is important for binding to ezetimibe. Proc. Natl. Acad. Sci. USA. 105: 11140–11145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ge L., Wang J., Qi W., Miao H. H., Cao J., Qu Y. X., Li B. L., Song B. L. 2008. The cholesterol absorption inhibitor ezetimibe acts by blocking the sterol-induced internalization of NPC1L1. Cell Metab. 7: 508–519 [DOI] [PubMed] [Google Scholar]
- 15.Chang T. Y., Chang C. 2008. Ezetimibe blocks internalization of the NPC1L1/cholesterol complex. Cell Metab. 7: 469–471 [DOI] [PubMed] [Google Scholar]
- 16.Yu L., Bharadwaj S., Brown J. M., Ma Y., Du W., Davis M. A., Michaely P., Liu P., Willingham M. C., Rudel L. L. 2006. Cholesterol-regulated translocation of NPC1L1 to the cell surface facilitates free cholesterol uptake. J. Biol. Chem. 281: 6616–6624 [DOI] [PubMed] [Google Scholar]
- 17.Loftus S. K., Morris J. A., Carstea E. D., Gu J. Z., Cummings C., Brown A., Ellison J., Ohno K., Rosenfeld M. A., Tagle D. A., et al. 1997. Murine model of Niemann-Pick C disease: mutation in a cholesterol homeostasis gene. Science. 277: 232–235 [DOI] [PubMed] [Google Scholar]
- 18.Chang T. Y., Reid P. C., Sugii S., Ohgami N., Cruz J. C., Chang C. C. 2005. Niemann-Pick type C disease and intracellular cholesterol trafficking. J. Biol. Chem. 280: 20917–20920 [DOI] [PubMed] [Google Scholar]
- 19.Kuwabara P. E., Labouesse M. 2002. The sterol-sensing domain: multiple families, a unique role? Trends Genet. 18: 193–201 [DOI] [PubMed] [Google Scholar]
- 20.Davies J. P., Ioannou Y. A. 2000. Topological analysis of Niemann-Pick C1 protein reveals that the membrane orientation of the putative sterol-sensing domain is identical to those of 3-hydroxy-3-methylglutaryl-CoA reductase and sterol regulatory element binding protein cleavage-activating protein. J. Biol. Chem. 275: 24367–24374 [DOI] [PubMed] [Google Scholar]
- 21.Nohturfft A., Brown M. S., Goldstein J. L. 1998. Topology of SREBP cleavage-activating protein, a polytopic membrane protein with a sterol-sensing domain. J. Biol. Chem. 273: 17243–17250 [DOI] [PubMed] [Google Scholar]
- 22.Olender E. H., Simon R. D. 1992. The intracellular targeting and membrane topology of 3-hydroxy-3-methylglutaryl-CoA reductase. J. Biol. Chem. 267: 4223–4235 [PubMed] [Google Scholar]
- 23.Cao J., Wang J., Qi W., Miao H. H., Wang J., Ge L., Bose-Boyd R. A., Tang J. J., Li B. L., Song B. L. 2007. Ufd1 is a cofactor of gp78 and plays a key role in cholesterol metabolism by regulating the stability of HMG-CoA reductase. Cell Metab. 6: 115–128 [DOI] [PubMed] [Google Scholar]
- 24.Feramisco J. D., Goldstein J. L., Brown M. S. 2004. Membrane topology of human insig-1, a protein regulator of lipid synthesis. J. Biol. Chem. 279: 8487–8496 [DOI] [PubMed] [Google Scholar]
- 25.Clader J. W.2004. The discovery of ezetimibe: a view from outside the receptor. J. Med. Chem. 47: 1–9 [DOI] [PubMed] [Google Scholar]
- 26.Van Heek M., France C. F., Compton D. S., McLeod R. L., Yumibe N. P., Alton K. B., Sybertz E. J., Davis H. R., Jr 1997. In vivo metabolism-based discovery of a potent cholesterol absorption inhibitor, SCH58235, in the rat and rhesus monkey through the identification of the active metabolites of SCH48461. J. Pharmacol. Exp. Ther. 283: 157–163 [PubMed] [Google Scholar]
- 27.Harris M., Davis W., Brown W. V. 2003. Ezetimibe. Drugs Today (Barc.). 39: 229–247 [DOI] [PubMed] [Google Scholar]
- 28.Song B. L., Sever N., Bose-Boyd R. A. 2005. Gp78, a membrane-anchored ubiquitin ligase, associates with Insig-1 and couples sterol-regulated ubiquitination to degradation of HMG CoA reductase. Mol. Cell. 19: 829–840 [DOI] [PubMed] [Google Scholar]
- 29.Sever N., Yang T., Brown M. S., Goldstein J. L., Bose-Boyd R. A. 2003. Accelerated degradation of HMG CoA reductase mediated by binding of insig-1 to its sterol-sensing domain. Mol. Cell. 11: 25–33 [DOI] [PubMed] [Google Scholar]
- 30.Yang T., Espenshade P. J., Wright M. E., Yabe D., Gong Y., Aebersold R., Goldstein J. L., Brown M. S. 2002. Crucial step in cholesterol homeostasis: sterols promote binding of SCAP to INSIG-1, a membrane protein that facilitates retention of SREBPs in ER. Cell. 110: 489–500 [DOI] [PubMed] [Google Scholar]
- 31.Ohgami N., Ko D. C., Thomas M., Scott M. P., Chang C. C., Chang T. Y. 2004. Binding between the Niemann-Pick C1 protein and a photoactivatable cholesterol analog requires a functional sterol-sensing domain. Proc. Natl. Acad. Sci. USA. 101: 12473–12478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Infante R. E., Abi-Mosleh L., Radhakrishnan A., Dale J. D., Brown M. S., Goldstein J. L. 2008. Purified NPC1 protein. I. Binding of cholesterol and oxysterols to a 1278-amino acid membrane protein. J. Biol. Chem. 283: 1052–1063 [DOI] [PubMed] [Google Scholar]
- 33.Infante R. E., Radhakrishnan A., Abi-Mosleh L., Kinch L. N., Wang M. L., Grishin N. V., Goldstein J. L., Brown M. S. 2008. Purified NPC1 protein: II. Localization of sterol binding to a 240-amino acid soluble luminal loop. J. Biol. Chem. 283: 1064–1075 [DOI] [PubMed] [Google Scholar]