Abstract
In yeast, Tsc10p catalyzes reduction of 3-ketosphinganine to dihydrosphingosine. In mammals, it has been proposed that this reaction is catalyzed by FVT1, which despite limited homology and a different predicted topology, can replace Tsc10p in yeast. Silencing of FVT1 revealed a direct correlation between FVT1 levels and reductase activity, showing that FVT1 is the principal 3-ketosphinganine reductase in mammalian cells. Localization and topology studies identified an N-terminal membrane-spanning domain in FVT1 (absent in Tsc10p) oriented to place it in the endoplasmic reticulum (ER) lumen. In contrast, protease digestion studies showed that the N terminus of Tsc10p is cytoplasmic. Fusion of the N-terminal domain of FVT1 to green fluorescent protein directed the fusion protein to the ER, demonstrating that it is sufficient for targeting. Although both proteins have two predicted transmembrane domains C-terminal to a cytoplasmic catalytic domain, neither had an identifiable lumenal loop. Nevertheless, both Tsc10p and the residual fragment of FVT1 produced by removal of the N-terminal domain with factor Xa protease behave as integral membrane proteins. In addition to their topological differences, mutation of conserved catalytic residues had different effects on the activities of the two enzymes. Thus, while FVT1 can replace Tsc10p in yeast, there are substantial differences between the two enzymes that may be important for regulation of sphingolipid biosynthesis in higher eukaryotes.
Keywords: sphingolipids, 3-ketososphinganine reductase, membrane protein topology
Although the pathway for the biosynthesis of sphingolipids has been delineated, the structural organization of many of the enzymes that catalyze the individual reactions remains unclear. Since many of the intermediate products of the biosynthetic pathway are known to be highly bioactive, it seems reasonable to hypothesize that the intracellular levels of these intermediates and the localization of their syntheses would be tightly controlled (1). In particular, 3-ketosphinganine (3-KDS), the product of the rate-limiting and committed step of sphingolipid synthesis, is highly chemically reactive, suggesting that the enzymes responsible for its synthesis and reduction could be spatially coupled. Indeed, all of the enzymes involved in ceramide synthesis are localized in the endoplasmic reticulum (ER) membrane, and the catalytic domains of serine palmitoyltransferase (SPT) and the ceramide synthases are cytosolic (2). Thus, it is likely that the active site of 3-KDS reductase is also cytosolic and that the active sites of all of the enzymes are organized to allow the bioactive intermediates to be channeled from one enzyme to the next.
However, addition of 3-KDS to the growth medium supports the growth of yeast mutants deficient in SPT (3), indicating that 3-KDS crosses the cell membrane and either diffuses or is transported to the ER for incorporation into ceramide. Thus, either 3-KDS is not intrinsically toxic or the mechanism of its uptake and transport protects it from promiscuous reaction with other cellular components. In addition, it has recently been reported that the mammalian 3-KDS reductase FVT1 can replace yeast Tsc10p as the 3-KDS reductase in the biosynthetic pathway for dihydrosphinganine (DHS) (4). While comparison of the sequences of the two enzymes shows significant homology, particularly in the putative catalytic site, there are substantial differences between the sequences of the two proteins that are predicted to result in distinctly different topologies and structural organization. In particular, the N-terminal extension of FVT1 is predicted to contain a membrane-spanning domain not present in Tsc10p. In addition, the C-terminal region of Tsc10p contains a dilysine ER retrieval motif (5) not found in FVT1. Thus, it is difficult to see how the mammalian 3-KDS reductase could have identical interactions with the upstream and downstream yeast enzymes of the pathway.
Given the ability of FVT1 and the 3-KDS reductases of a variety of other organisms to functionally substitute for Tsc10p in a pathway that might be tightly regulated, we considered it important to compare the membrane topology of the yeast 3-KDS reductase with that of a 3-KDS reductase from higher eukaryotes. In addition, it has recently been reported that an autosomal recessive mutation (A175T) in the catalytic core of bovine FVT1 results in a form of spinal muscular atrophy (SMA) (6). Curiously, the mutant protein had sufficient residual activity to rescue a yeast tsc10 knockout but was reported to be inactive when expressed in Escherichia coli (6). This result was surprising because threonine is the naturally occurring residue in the yeast 3-KDS reductase. It was therefore also important to examine the effects of this mutation, as well as mutations in the residues that constitute the catalytic triad of the short-chain reductases (SDRs) (7), on the activities of the yeast and mammalian proteins.
To facilitate our comparison of the yeast and mammalian 3-KDS reductases, we developed a highly sensitive radiometric enzyme assay. Using this assay, and RNA interference, we demonstrate that FVT1 is likely the sole 3-KDS reductase in the mammalian sphingolipid biosynthetic pathway. In addition, using probes for membrane topology (8–10), we find evidence that Tsc10p contains only a single membrane-embedded domain, between residues 257 and 303, thereby placing the majority of the protein, including the active site and the ER retrieval signal, in the cytosol. In contrast, in addition to a C-terminal membrane-associated segment similar to that in Tsc10p, FVT1 also contains an N-terminal membrane-spanning domain that is both necessary and sufficient for ER localization. Thus, despite the fact that the two proteins catalyze the same reaction, their distinctly different topologies argue that the enzymes involved in sphingoid base synthesis are not part of a single multisubunit complex.
As expected, mutations in the catalytic triad of FVT1 and Tsc10p significantly compromised their ability to complement the tsc10Δ mutant yeast at 37°C. Interestingly, while the introduction of the bovine SMA mutation into human FVT1 had only a modest effect on enzyme activity, tsc10Δ yeast expressing this mutant protein also failed to grow at 37°C. Given that SPT is generally considered the rate-limiting enzyme of sphingolipid biosynthesis, this result raises interesting questions about the role of 3-KDS reductases in the regulation of sphingolipid synthesis.
MATERIALS AND METHODS
Antibodies and reagents
Rabbit antibodies were raised against human FVT1 using the peptide CVARNEDKLLQAKKEIE (Fig. 1) and against SPTLC2 using the peptides CGKYSRHRLVPLLRPF and CGDRPFDETTYEETED (Sigma-Genosystems, Woodlands, TX). Anti-green fluorescent protein (GFP), anti-SPTLC1, anti-Myc, horseradish peroxidase (HRP)-conjugated mouse anti-HA, anti-calnexin, HRP-conjugated goat anti-rabbit, and anti-mouse IgGs and Cy3-conjugated goat anti-mouse IgG were obtained from various commercial sources. Phytosphingosine (PHS) and DHS were obtained from Sigma-Aldrich (St. Louis, MO) and 3-KDS from Matreya (Pleasant Gap, PA).
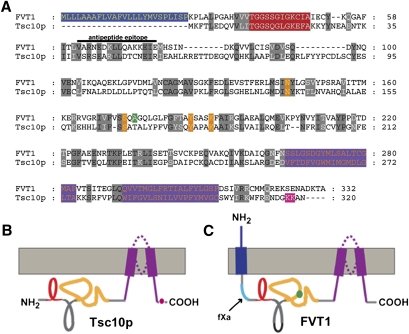
Fig. 1.
Alignment of FVT1 and Tsc10p. A: FVT1 was aligned with Tsc10p using Clustal (24% identity and 41% similarity). The N-terminal extension of FVT1 (blue), the Rossmann folds (red), conserved catalytic residues (orange), and the C-terminal hydrophobic domains (purple) are shown. In addition, the residue corresponding to that mutated in bovine SMA (green) and the dilysine motif presumed to be important for ER retention in Tsc10p (pink) are indicated. Residues (62–77) contained in the peptide used to generate the anti-FVT1 antibodies are indicated. B, C: Topological representation of Tsc10p and FVT1 are presented with relevant regions indicated as described in A. The fXa protease cleavage site inserted into FVT1 is denoted in aqua.
Yeast strains and media
The isolation and growth of yeast tsc10 mutants have been described previously (3). Media were prepared and cells were grown using standard procedures (11).
Cell culture and transfection
HEK293 or Chinese hamster ovary-K1 (CHO-K1) cells were maintained in DMEM containing 4.5 gm/l glucose (Mediatech, Herndon, VA) supplemented with 10% fetal bovine serum (JRH Biosciences, Lenexa, KS) and 2 mM glutamine as previously described (9). Cells were transfected with expression plasmids or small interfering RNAs (siRNAs ) using Lipofectamine 2000 (Invitrogen, Carlsbad, CA).
Plasmid construction
pFVT1 was prepared by PCR amplification of the FVT1 coding sequence from pOTB7 plasmid (Open Biosystems, Huntsville, AL). The amplified product was digested with BamHI and SpeI (at sites introduced on the ends of the PCR primers) and ligated into pcDNA3.1 (+). For C-terminal tagging, a SpeI-ended 3×-HA, a SpeI-ended 3×-MYC, or an XbaI-ended GFP cassette was ligated into an XbaI site introduced by Quikchange mutagenesis (QCM) (Stratagene, La Jolla, CA) just proximal to the stop codon in pFVT, thereby generating pFVT1-HA, pFVT1-MYC, and pFVT1-GFP. Construction of pADH-HA-TSC10 was described previously (3). The MYC-Tsc10p-expressing plasmid, pMYC-TSC10, was constructed by ligating a SalI fragment extending from approximately 1,300 bp upstream to 900 bp downstream of the TSC10 open reading frame into pRS316 and inserting a SpeI-ended 3×-MYC fragment into an AvrII site introduced by QCM after the start codon. pADH-HA-TSC10-MYC was constructed by inserting a SpeI-ended 3×-MYC cassette into an NheI site introduced by QCM between codons 313 and 314 of TSC10 in pADH1-HA-TSC10. pADH-HA-FVT1 was constructed by ligating a XhoI-ended PCR fragment encoding FVT1 into the SalI site of pADH (12). Topology reporters containing TSC10 and FVT1 gene fusion alleles with the invertase glycosylation cassette (GC) inserted at various positions were constructed as previously described (9). For construction of pFVT1(1-25)-GFP and pFVT1(Δ4-26)-GFP, a second XbaI site was introduced between codons 25 and 26 in the pFVT1 plasmid already containing an XbaI site preceding the stop codon (described above). Following digestion with XbaI to release the fragment encoding residues 26 to the end of FVT1, an XbaI-ended GFP cassette (without a stop codon) was ligated to the resulting vector fragment to generate pFVT1(1-25)-GFP. pFVT1(Δ4-26)-GFP plasmid was constructed by deleting the fragment encoding amino acids 4–26 from pFVT1-GFP by QCM. pFVT1-GFP(2-11)-fXa@25 was constructed by annealing two overlapping primers to generate a fragment encoding amino acids 2–11 of GFP followed by two factor Xa (fXa) protease sites that could be ligated into an XbaI site introduced by QCM after codon 25 in pFVT1. pADH1-FVT1-GFP, pADH1-FVT1(1-25)-GFP, and pADH1-FVT1(Δ4-26)-GFP were constructed by PCR amplification of the corresponding inserts from pFVT1-GFP, pFVT1(1-25)-GFP, and pFVT1(Δ4-26)-GFP using upstream primers flanked with an EcoRI site and downstream primers flanked with a SalI site and inserting the resulting amplicons into EcoRI/SalI-digested pADH1.
Construction of the FVT1 and Tsc10p catalytic mutations
The mutations in the putative catalytic residues of FVT1 and Tsc10p were introduced by QCM using the pADH-HA-FVT1 or pADH-HA-TSC10 plasmid.
Preparation and solubilization of membranes
Following transfection and incubation for 24–48 h, CHO-K1 or HEK cells were harvested, and microsomes were prepared as previously described (9). Microsomes were prepared from mid-log phase yeast cells, grown in minimal medium with selection to maintain the plasmid, as described (13). Membranes (1 mg/ml) were solubilized using 0.1% sucrose monolaurate (SML; Roche Diagnostics, Indianapolis, IN) for 10 min at 4°C, and the high speed (135,000 g, 30 min) supernatant collected. For protease protection assays, yeast right-side-out vesicles were prepared according to Romano and Michaelis (14) and right-side-out CHO vesicles were prepared as described by Feramisco, Goldstein, and Brown (15).
3-KDS reductase assay
The 3-KDS reductase was assayed as previously described (3) with the exception that [3H]3-KDS or [14C]3-KDS was prepared using soluble 6-His-tagged Sphingomonas paucimobilis SPT expressed in E. coli and purified by nickel-nitrilotriacetic acid (Ni-NTA) chromatography, palmitoyl-CoA, and either [3H]serine or [14C]serine. Extracted long-chain bases were dried, suspended in CHCl3:methanol (2:1, v/v), and resolved on a Silica Gel G TLC plate developed with CHCl3:methanol:2 M NH4OH (40:10:1). [3H]3-KDS and [3H]DHS (identified by ninhydrin-stained unlabeled standards resolved in an adjacent lane) were scraped and radioactivity determined by liquid scintillation spectrometry. [14C]3-KDS and [14C]DHS were quantified using a STORM 860 (GE Healthcare, Piscataway, NJ) phosphor imager. For determination of the specific activities of the enzymes, the amount of 3×-HA-tagged wild-type and A175T mutant FVT1 was determined by comparison to purified 3×-HA-tagged S. paucimobilis SPT using scanning densitometry of immunoblots.
Gel electrophoresis and immunoblotting
Denaturing gel electrophoresis was performed as previously described (9). Native gel electrophoresis was performed using 4–20% Tris-glycine nondenaturing gels (Invitrogen). Immunoblotting was performed as previously described (10) using HRP-conjugated secondary antibodies and ECL (GE Healthcare) detection.
Immunoprecipitation
Immunoadsorption was performed by incubating solubilized membranes with appropriate antibodies for 1 h at 4°C followed by incubation with protein A-Sepharose beads for an additional 1 h at 4°C. Beads were pelleted at 10,000 g and washed three times with 1 ml of 20 mM Tris-HCl (pH 7.5) containing 1 mM EGTA, 1 mM β-mercaptoethanol (β-ME), and 0.1% SML. Bound proteins were eluted by boiling in 100 µl of NuPAGE SDS loading buffer.
Determination of the glycosylation status of the Tsc10p-GC or FVT1-GC topology reporter fusion proteins
For analysis of the glycosylation of the GC-tagged proteins, microsomes were subjected to endoglycosidase H (EndoH) treatment as previously described (9).
Protease protection assay
Protease protection assays with yeast membranes were performed using proteinase K in the presence or absence of 0.4% Triton X-100 as described (14). Protease protection experiments using CHO cells were performed essentially as described for the yeast membranes except that the vesicles were suspended in 20 mM HEPES (pH 7.5) containing 250 mM sucrose, 1 mM sodium EDTA, 1 mM sodium EGTA, and 1 mM β-ME and digestion performed in the presence or absence of 1% Triton X-100. For digestion with fXa protease, right-side-out vesicles were prepared from CHO cells as described above and digested in the absence or presence of 0.2% Nonidet P-40 as described (9). The reaction was stopped by addition of PMSF (final concentration of 1 mM), and half the reaction mix was centrifuged at 100,000 g for 30 min. Total, soluble, and pellet fractions were separated by SDS-PAGE and analyzed by immunoblotting.
Indirect immunofluorescence
CHO cells were plated on coverslips, grown to >50% confluence, and transfected with pFVT1-GFP and pCHO-SPTLC1 (9) or pDsRed2-ER (Clontech, Mountain View, CA). Forty-eight hours after transfection, cells were washed with PBS, fixed with 4% (w/v) formaldehyde, and permeabilized with 0.2% Triton X-100 in PBS containing 1% BSA. For immunofluorescent detection, cells were incubated with a 1:5,000 dilution of an anti-SPTLC1 monoclonal antibody followed by incubation with a 1:10,000 dilution of Cy3-conjugated goat anti-mouse IgG (Molecular Probes, Eugene, OR). For detection of GFP fusion proteins or DsRed2-ER protein, cells were fixed and mounted as above. Fluorescence microscopy was performed using an Olympus IX70 inverted fluorescence microscope equipped with a HiQ fluorescein filter set. The excitation and emission wavelengths were 460–490 nm and 510 nm for detection of GFP and 510–550 nm and 590 nm for detection of Cy3. Images were collected with a Princeton Instruments 5-MHz MicroMax cooled CCD camera, a shutter and controller unit, and IPLab software (version 3.6; Scanalytic, Fairfax, VA). Alternatively, confocal fluorescent images were acquired using a Zeiss LSM 5 PASCAL confocal laser scanning microscope. Cells transected with pDsRed2-ER were observed using a helium/neon (543 nm) laser, and fluorescent images were collected using a 560-nm long-pass filter. GFP fusion proteins were observed with an argon (488 nm) laser and a 500–550 nm band-pass filter.
RESULTS
FVT1 is the major 3-KDS reductase in mammalian cells
Conversion of 3-KDS to DHS is catalyzed by Tsc10p in yeast. In mammalian cells, FVT1 is the closest homolog of Tsc10p, and it has been shown to rescue tsc10Δ mutants (4). Comparison of the two enzymes reveals that both contain a Rossman fold predicted to be involved in NADPH binding and a conserved catalytic triad that is present in other SDRs (7). However, Tsc10p contains a dilysine motif in its C-terminal region that is believed to be important for ER retention (5) as well as two predicted transmembrane domains in the C-terminal end (Fig. 1). In contrast, FVT1 has no C-terminal dilysine motif but contains an N-terminal extension with a predicted transmembrane domain (Fig. 1). Deletion of TSC10 abolishes all 3-KDS reductase activity in yeast, demonstrating that it is the major, if not only, 3-KDS reductase (3). However, given the overall lack of homology between Tsc10p and FVT1, the apparent differences in their structural organization, and the presence of several orphan SDRs in the mammalian genome with unknown substrates, it cannot be concluded that FVT1 is the only, or even the major, 3-KDS reductase in mammalian cells. We therefore sought to determine whether the level of 3-KDS reductase activity in mammalian cells is tightly correlated with the level of FVT1 expression. To do this, purified soluble S. paucimobilis SPT was used to synthesize reagent quantities of [3H]3-KDS with high specific activity. When added to microsomal membranes prepared from CHO cells, [3H]3-KDS was converted to [3H]3-DHS in a nearly linear fashion over a period of 20 min (Fig. 2A). In contrast, microsomes prepared from cells overexpressing FVT1 (Fig. 2B) showed significantly greater 3-KDS reductase activity (Fig. 2A). These results are consistent with those of Kihara and Igarashi (4) and confirm that FVT1 is indeed a 3-KDS reductase.
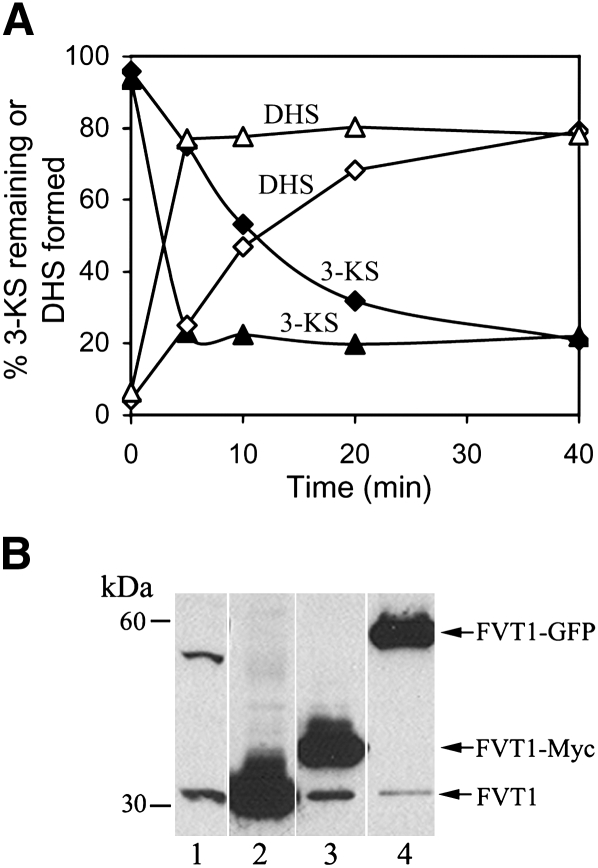
Fig. 2.
Overexpression of FVT1 increases 3-KDS reductase activity. A: FVT1 activity was measured by the conversion of 3-KDS (closed symbols) to DHS (open symbols) in cells transfected with pcDNA3.1 (closed and open diamonds) or pFVT1 (closed and open triangles). B: Solubilized microsomal protein prepared from cells transfected with pcDNA3.1 (lane 1), pFVT1 (lane 2), pFVT1-MYC (lane 3), or pFVT1-GFP (lane 4) was analyzed by immuno blotting using anti-FVT1 antibodies (see Materials and Methods).
The enhanced sensitivity of this assay allowed us to measure not only the activity of transfected FVT1, but endogenous 3-KDS reductase activity as well. We therefore determined to what extent siRNA directed against FVT1 would reduce endogenous levels of 3-KDS reductase activity. To identify a suitable siRNA, several candidates were cotransfected into CHO cells with pFVT1 and their relative abilities to reduce FVT1 mRNA determined. The most potent of these, Si-1, reduced expression of transfected FVT1 mRNA and protein by at least 70% (Fig. 3A, B). More importantly, Si-1 inhibited endogenous FVT1 expression in HEK cells without altering the level of expression of either subunit of SPT (Fig. 3C) and elicited a corresponding reduction in endogenous 3-KDS reductase activity (Fig. 3D). We therefore conclude that the major source of endogenous 3-KDS reductase activity must be FVT1.
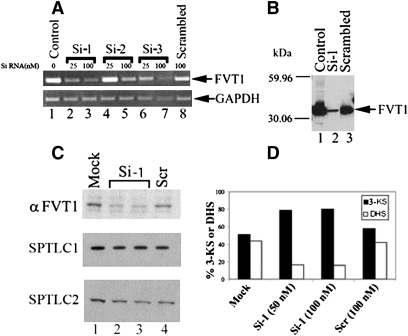
Fig. 3.
Reduction of FVT1 expression and 3-KDS reductase activity with siRNA. A: CHO cells were transfected with pFVT1 (lane 1), pFVT1 and 25 or 100 nM siRNAs directed against nucleotides 135–156 (Si-1, lanes 2 and 3); 592–613 (Si-2, lanes 4 and 5), or 926–946 (Si-3, lanes 6 and 7) of FVT1 or pFVT1 and a nonspecific scrambled siRNA control (lane 8). Twenty-four hours after transfection, total RNA was isolated and RT-PCR was performed with FVT1 (upper) and GAPDH (lower) specific primers using the SuperScript first-strand synthesis system from Invitrogen. B: Microsomal protein (8 μg) prepared from CHO cells transfected with pFVT1 (lane 1), pFVT1 and 50 nM Si-1 RNA (lane 2), or pFVT1 and 50 nM scrambled siRNA (lane 3) were separated by SDS-PAGE and immunoblotted using anti-FVT1. C: Twenty-five micrograms of microsomal protein from untransfected HEK cells (lane 1) or cells transfected with Si-1 RNA (50 nM, lane 2; 100 nM, lane 3) or nonspecific Scr RNA-transfected (100 nM, lane 4) was resolved by SDS-PAGE and immunoblotted using anti-FVT1, anti-SPTLC1, and anti-SPTLC2 antibodies. D: 3-KDS reductase was assayed in microsomes prepared from HEK cells as described in Materials and Methods. The specific activity of FVT1 in microsomes prepared from mock-transfected HEK cells was 40 pmol DHS formed/mg/min. Results are expressed as percentage of DHS formed and 3-KDS remaining.
Comparison of catalytic properties of Tsc10p and FVT1 and analysis of key catalytic and putative disease-causing residues
To compare the catalytic properties of Tsc10p and FVT1, both proteins were expressed in yeast and their activities assayed. The results showed that the affinity of FVT1 for 3-KDS was approximately an order of magnitude greater than that of Tsc10p and that the enzymes had similar Vmax values (data not shown). Mutation of any of the three residues that constitute the catalytic triad of FVT1 did not completely inactivate the enzyme, as these mutant proteins were able to support growth of tsc10Δ mutant cells at 26°C. This is consistent with our previous observation that very little 3-KDS reductase activity is required for growth at this temperature (3). However, none of the mutants in the catalytic triad supported growth at 37°C (Fig. 4). Comparable results were obtained for Tsc10p, with the exception that mutation of Y180 was less deleterious at 37°C. In some SDRs, a conserved asparagine has also been shown to participate in catalysis (16, 17). We therefore investigated the effects of mutating the corresponding residues, N145 in FVT1 and N140 in Tsc10p. Our results (Fig. 4) suggest that, in contrast to the tyrosine residue, which was more critical for FVT1 activity, this conserved asparagine is more important for the activity of Tsc10p.
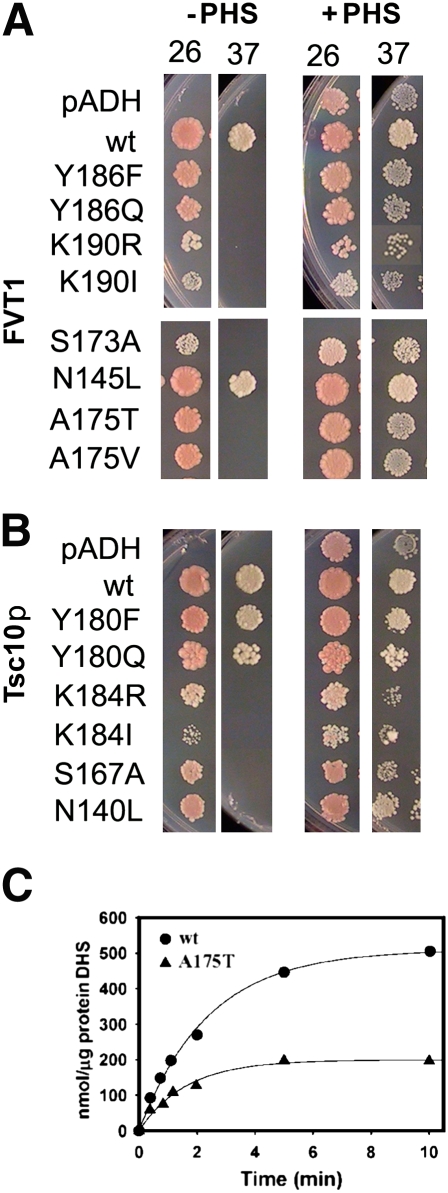
Fig. 4.
Mutations in the conserved residues of the catalytic domain of FVT1 and Tsc10p differentially affect function. Wild-type and mutant FVT1 (A) and Tsc10p (B) were expressed in yeast tsc10Δ mutant cells and their ability to support growth at 26°C or 37°C in the absence or presence of PHS determined. C: 3-KDS reductase activity of the wild-type and A175T mutant FVT1 proteins was assayed at approximately 10 μM 3-KDS as described in Materials and Methods.
Based on the recent report that bovine SMA is the result of a substitution of threonine for alanine at residue 175 in FVT1 (6), we investigated the effects of this mutation on human FVT1. Our results show that the mutant protein complemented the growth of tsc10Δ mutant yeast at 26°C, but not at 37°C (Fig. 4A), and was less active than wild-type FVT1 when assayed in microsomes prepared from the tsc10Δ mutants (Fig. 4C). In these experiments, N-terminally HA-tagged mutant and wild-type FVT1 were used, and the amount of each enzyme determined by comparison to purified HA-tagged S. paucimobilis SPT after immunoblotting with anti-HA antibodies as described in Materials and Methods. Mutant and wild-type FVT1 were expressed at similar levels, indicating that although no comparable mutation has been identified in human FVT1, the A175T mutation is clearly deleterious to the human enzyme.
Tsc10p and FVT1 are both multimeric enzymes
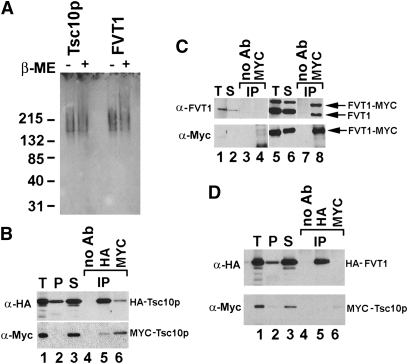
Members of the SDR family of enzymes are generally multimeric, but the organization of the 3-KDS reductases has not been investigated. Native gel electrophoresis showed that both FVT1 and Tsc10p migrated with approximate molecular masses of 150 kDa, suggesting that both proteins may be tetrameric (Fig. 5A). Direct evidence that the enzymes are oligomeric was provided by coimmunoprecipitation of solubilized microsomes prepared from cells expressing both HA- and Myc-tagged Tsc10p. Immunoprecipitation with anti-HA antibody followed by immunoblotting with an anti-Myc antibody or immunoprecipitation with an anti-Myc antibody followed by immunoblotting with an anti-HA antibody demonstrated association of the Myc- and HA-tagged proteins (Fig. 5B). Similarly, when FVT1-Myc and untagged FVT1 were both expressed in CHO cells, immunoprecipitation of solubilized microsomes with anti-Myc antibodies resulted in the coimmunoprecipitation of the untagged FVT1 (Fig. 5C). However, FVT1 and Tsc10p do not interact with one another as demonstrated by their failure to coimmunoprecipitate (Fig. 5D).
Fig. 5.
FVT1 and Tsc10p are both multimeric enzymes but do not interact with each other. A: Microsomal protein prepared from yeast expressing HA-Tsc10p or CHO cells expressing FVT1-HA was resolved by native gel electrophoresis before or after treatment with β-ME and immunoblotted with anti-HA antibodies. B: Microsomes were prepared from yeast cells coexpressing HA-Tsc10p and MYC-Tsc10p. Ten micrograms of total microsomal protein (lane 1, T) were solubilized with SML yielding a pellet (lane 2, P) or supernatant (lane 3, S) after centrifugation at 100,000 g. The supernatant was incubated in the absence (lane 4) or presence of anti-HA (lane 5) or anti-MYC (lane 6) antibodies followed by adsorption to protein A-Sepharose beads as described in Materials and Methods. Immunoprecipitated proteins were subjected to immunoblot analysis using either anti-HA (upper) or anti-MYC (lower) antibodies as described in Materials and Methods. C: Microsomes were prepared from CHO cells transfected with pcDNA3.1 (lanes 1–4) or pFVT1 and pFVT1-MYC (lanes 5–8). Following solubilization with SML, the 100,000 g supernatants (lanes 2 and 6) were incubated in the absence (lanes 3 and 7) or presence (lanes 4 and 8) of anti-MYC antibodies and FVT1 resolved by SDS-PAGE after adsorption to protein A-Sepharose beads as described in Materials and Methods. FVT1 was visualized with anti-FVT1 (upper) or anti-MYC (lower) antibodies. D: Microsomes were prepared from yeast cells coexpressing HA-FVT1 and MYC-Tsc10p. Ten micrograms of total microsomal protein (lane 1, T) was solubilized with SML, and the 100,000 g supernatant (lane 3, S) was incubated in the absence (lane 4) or presence of anti-HA (lane 5) or anti-MYC (lane 6) antibodies. HA-FVT1 and MYC-Tsc10p were resolved by SDS-PAGE after adsorption to protein A-Sepharose beads and visualized with anti-HA (upper) or anti-MYC (lower) antibodies.
FVT1 and Tsc10p have distinct membrane topologies
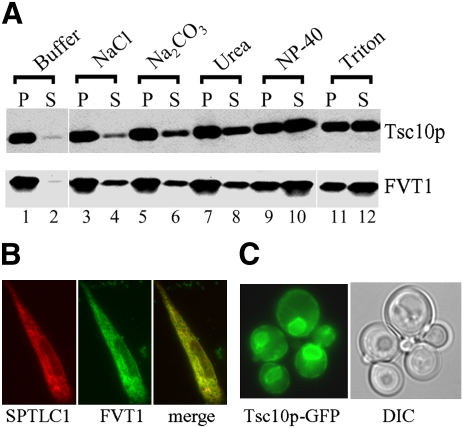
Given the fact that both Tsc10p and FVT1 catalyze the same reaction and are both multimeric, yet appear to have significant structural differences and are predicted by a variety of algorithms to have different membrane topologies (Table 1), it was important to experimentally examine their membrane association and topological organization. Treatment of microsomes prepared from yeast or CHO cells with NaCl, Na2CO3, urea, Nonidet P-40, and Triton X-100 showed that efficient extraction of both enzymes required detergent (Fig. 6A). In addition, FVT1-GFP expressed in CHO cells (Fig. 2B) showed the same ER localization as seen for the SPTLC1 subunit of SPT (Fig. 6B). Similarly, Tsc10p-GFP localizes to the yeast ER membrane (Fig. 6C). These results essentially confirm those of Kihara and Igarashi (4) and suggest that despite differences in their predicted membrane topologies, both are tightly associated with the ER membrane.
TABLE 1.
Predicted transmembrane domains
| Algorithm (Ref.) | Tsc10p | FVT1 |
|---|---|---|
| HMMTop2 (23) | 162-181 257-275 286-305 | 6-23 |
| TMPred (24) | 161-178 283-303 | 2-19 39-55 165-185 265-283 285-309 |
| TopPred (25) | 160-180 256-276 285-305 | 1-21 27-47 168-188 271-291 |
| MEMSAT (26) | 286-305 | 5-24 |
| TMHMM (27) | 285-307 | 2-24 |
| PHDhtm (28) | 162-179 281-300 | 2-20 40-57 171-188 292-309 |
| ConPred II (29) | 159-179 255-275 284-304 | 3-23 |
Fig. 6.
FVT1 and Tsc10p are both integral ER membrane proteins. A: Microsomal protein (100 μg) prepared from yeast cells expressing HA-Tsc10p (upper) or CHO cells overexpressing FVT1 (lower) was incubated with 0.5 M NaCl, 0.1 M Na2CO3, 2.5 M urea, 0.2% Nonidet P-40, or 1% Triton X-100 at room temperature for 1 h. Following centrifugation, the proteins from the supernatants (S) and pellets (P) were resolved by SDS-PAGE and analyzed by immunoblotting with anti-HA (upper) or anti-FVT1 (lower) antibodies. B: Intracellular localization of FVT1-GFP was determined by fluorescence microscopy and compared with the localization of SPTLC1 as described in Materials and Methods. C: Localization of Tsc10p-GFP was determined by fluorescence and differential interference contrast phase microscopy.
To more thoroughly investigate the topologies of Tsc10p and FVT1, two sets of fusion proteins, containing glycosylation reporter cassettes (GCs) inserted in-frame at various positions along the lengths of the proteins, were constructed. The GC consisted of a 53-amino acid domain comprising residues 80–133 of yeast invertase (Suc2p) that contains three NXS/T sites for N-linked glycosylation. This reporter cassette has been successfully used by us and others to determine the topology of a variety of ER-localized proteins, including Lcb1p (8–10).
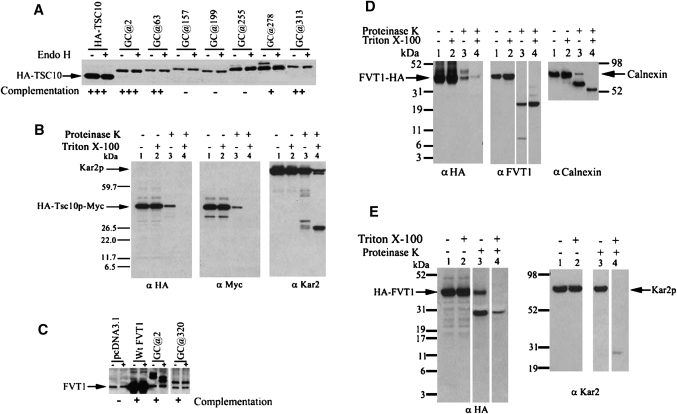
In the case of Tsc10p, some glycosylation of the GC inserted at position 278 was observed, indicating that at least a fraction of the cassette inserted at this site is in the lumen of the ER (Fig. 7A). This region of Tsc10p lies between two hydrophobic stretches predicted by several of the algorithms to be membrane-spanning domains (Fig. 1, Table 1). With the exception of the GC at 255 that was weakly glycosylated, none of the other GC insertions were glycosylated. Two of the fusion proteins that showed no glycosylation also failed to complement the tsc10Δ mutant, and it is therefore not possible to definitively conclude that they report the native topology. However, GCs inserted at the N and C termini did not disrupt protein function and were not glycosylated, clearly indicating that both ends of the protein are cytoplasmic. This is consistent with the predicted cytosolic location of the C-terminal dilysine motif and suggests that Tsc10p may contain two closely spaced membrane-spanning domains in the C-terminal region of the protein. An alternative and perhaps more likely possibility is that Tsc10p contains a single hydrophobic domain embedded in the membrane (Fig. 1B).
Fig. 7.
Tsc10p and FVT1 have distinct membrane topologies. A: HA-Tsc10p-GC fusion proteins with GC inserted at the indicated positions were expressed in yeast tsc10Δ cells. Microsomal proteins (50 μg) were fractionated with or without EndoH treatment by SDS-PAGE, and the HA-Tsc10p-GC proteins were visualized by immunoblotting using anti-HA antibodies. The ability of each HA-Tsc10p-GC fusion protein to complement the PHS requirement of the tsc10Δ mutant cells is indicated. B: Right-side-out sealed vesicles prepared from tsc10Δ cells expressing HA-Tsc10p-MYC were prepared and digested with or without proteinase K in the absence or presence of Triton X-100 as described in Materials and Methods. Following TCA precipitation, 15 μg of protein was resolved by SDS-PAGE and visualized by immunoblotting with anti-HA and anti-Myc antibodies. The integrity of the membrane vesicles was assessed by immunoblotting with antibodies to the ER lumenal protein, Kar2p. C: FVT1-GC fusion proteins with GC inserted at the indicated positions were expressed in CHO cells and analyzed as in A. The ability of the FVT1-GC fusion proteins to complement the PHS requirement of the yeast tsc10Δ cells is indicated. D: Right-side-out vesicles prepared from CHO cells transfected with pFVT1-HA were incubated with and without proteinase K in the absence or presence of Triton X-100. Following TCA precipitation, 15 μg of protein was analyzed by immunoblotting with anti-HA (left) or anti-FVT1 (middle). The integrity of the membrane vesicles was assessed by immunoblotting with antibodies to the ER lumenal protein, calnexin. E: pADH1-HA-FVT1 was expressed in the yeast tsc10Δ mutant, and right-side-out sealed vesicles were prepared and treated with proteinase K in the absence or presence of Triton X-100 and analyzed by immunoblotting with anti-HA antibodies. The integrity of the membranes vesicles was assessed by immunoblotting with antibodies to the ER lumenal protein, Kar2p.
Regardless of whether there are two membrane-spanning domains or a single membrane-embedded domain, both ends of the protein would be predicted to be cytoplasmic. However, several algorithms predict alternative topologies in which either the N or C termini are in the lumen of the ER (Table 1). Limited proteolysis of right-side-out membrane vesicles containing Tsc10p tagged at the N terminus with a 3×-HA epitope and at the C terminus with a 3×-MYC epitope was therefore used to definitively establish the locations of the N and C termini of Tsc10p. In the absence or presence of detergent, epitopes at either end of Tsc10p were cleaved by treatment with proteinase K consistent with both ends being cytoplasmic (Fig. 7B). The integrity of vesicles was verified by showing that the lumenal Kar2p protein was resistant to proteolysis in the absence of detergent (Fig. 7B). If, as predicted by some of the hydropathy analyses, the N terminus had been lumenal, a protected HA-tagged fragment of at least 25 kDa should have been seen after digestion in the absence of detergent. Similarly, if the C terminus had been in the lumen, a protected MYC-tagged fragment of at least 8 kDa should have been seen after the same treatment. As neither fragment was detected, we conclude that the interpretation of the GC reporter data (Fig. 7A) is correct and that both ends of Tsc10p are cytoplasmic (Fig. 1B).
Insertion of GC cassettes into FVT1 was less informative because none of the internal insertions was stably expressed when transfected into CHO cells (data not shown). However, insertion of the GC cassette at position 320 yielded a stable protein that was not glycosylated (Fig. 7C), indicating that like the C terminus of Tsc10p, the C terminus of FVT1 is cytoplasmic. To confirm the location of the C terminus of FVT1, right-side-out vesicles prepared from CHO cells expressing C-terminally HA-tagged FVT1 were digested with proteinase K in the absence or presence of detergent. The results showed that FVT1-HA was sensitive to proteinase K digestion in the absence of detergent (Fig. 7D), thereby placing the C terminus of the protein in the cytosol. In this respect, FVT1 and Tsc10p appear similar. In these experiments, the integrity of the vesicles was established by digestion of calnexin in the absence or presence of detergent (18). Interestingly, the anti-FVT1 immunoblot revealed the presence of a proteinase K-resistant fragment of approximately 23 kDa (Fig. 7D), which, based on the localization of the epitope against which the anti-FVT1 antibody was raised (Fig. 1), should contain the catalytic domain of the 3-KDS reductase. This result is distinctly different from those reported by Kihara and Igarashi, who found no such protease resistant fragment (4).
In contrast to the cytoplasmic localization of the N terminus of Tsc10p, the N terminus of FVT1 is lumenal. FVT1 containing a GC cassette at its N terminus was clearly glycosylated (Fig. 7C) when expressed in yeast. Treatment of solubilized membranes with EndoH increased the electrophoretic mobility of FVT1, indicating that unlike the N terminus of Tsc10p, the N terminus of FVT1 is lumenal. Additional evidence for the lumenal location of the N terminus of FVT1 was obtained from limited proteinase K digestion, in the absence and presence of detergent, of yeast microsomes containing N-terminally HA-tagged FVT1. In the absence of detergent, a protected fragment of approximately 27 kDa was observed whose presence was greatly diminished if digestion was performed in the presence of detergent (Fig. 7E). It was not possible to perform the analogous experiment in mammalian cells because of the difficulty in obtaining adequate expression of an N-terminally HA-tagged protein.
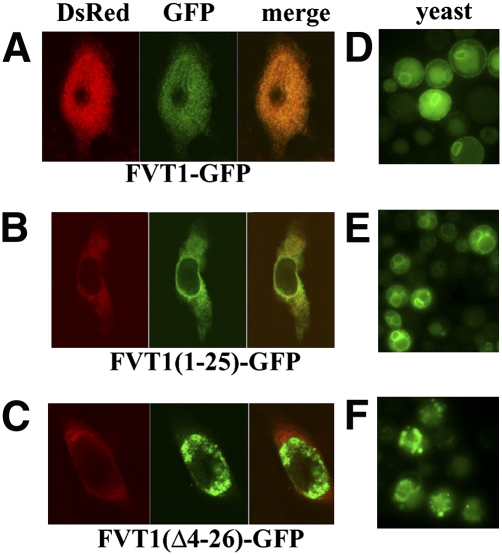
If, indeed, the N terminus of FVT1 is in the lumen of the ER, then there must be a membrane-spanning domain not found in Tsc10p, most likely the hydrophobic N-terminal extension that is missing from Tsc10p (Fig. 1). To test this possibility, the first 25 residues of FVT1 were fused to GFP, and the localization of this protein was compared with that of the full-length FVT1-GFP when expressed in CHO cells. Both proteins showed a typical ER localization pattern and colocalization with a DsRed2-ER marker (Fig. 8A, B). In contrast, deletion of residues 4–26 from the FVT1-GFP protein abolished ER localization (Fig. 8C). This indicates that the N-terminal hydrophobic extension of FVT1 is both necessary and sufficient for proper ER targeting in mammalian cells. Thus, while the C-terminal regions of FVT1 and Tsc10p are topologically similar, the N-terminal regions of the two proteins are clearly distinct. Surprisingly, when either FVT1-GFP or FVT1(1-25)-GFP was expressed in yeast, each protein was found in the ER membrane (Fig. 8D, E). Furthermore, deletion of the N-terminal membrane-spanning domain abolished ER localization (Fig. 8F). Thus, although neither Tsc10p nor any other yeast protein has a sequence homologous to the ER targeting sequence of FVT1, this sequence is recognized by the yeast ER targeting machinery.
Fig. 8.
The N-terminal 25-amino acid domain of FVT1 is sufficient to target GFP to ER membranes. CHO cells were transfected with pFVT1-GFP (A), pFVT1(1-25)-GFP (B), and pFVT1(Δ4-26)-GFP (C) and pDsRed2-ER. Proteins were visualized by confocal microscopy 36 h after transfection. Yeast cells (mat a tsc10Δ::TRP ura3-52 trp1Δ ade 2-101 his3Δ leu2Δ) were transformed with pADH1-FVT1-GFP (D), pADH1-FVT1 (1–25)-GFP (E), or pADH1-FVT1(Δ4-26)-GFP (F). Transformed cells were grown in SD medium without leucine and tryptophan (with PHS) for several generations, and proteins were visualized by fluorescence microscopy.
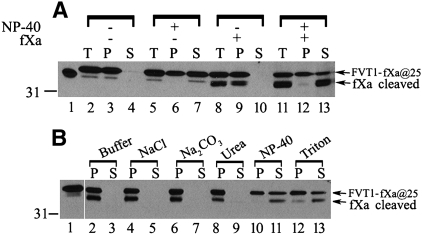
Based on hydropathy analysis, it had been suggested that FVT1 contains two additional membrane-spanning domains comparable to those in Tsc10p (4). Because of the difficulty in generating stably expressed GC fusion proteins that would accurately report the topology of this region of FVT1, an alternative approach for assessing the membrane association of this region of FVT1 was adopted. A short spacer consisting of the first 10 amino acids of GFP followed by tandem fXa protease cleavage sites was inserted between the end of the N-terminal transmembrane domain 1 and the epitope against which the anti-FVT1 antibodies had been raised (Fig. 1C). This allowed us to specifically cleave transmembrane domain 1 from the protein, thereby generating a C-terminal domain whose membrane association could be assessed. After fXa protease digestion of microsomes prepared from CHO cells expressing this construct, approximately 50% of the protein was cleaved (Fig. 9A). Moreover, the fragment displayed fractionation properties consistent with those expected of a tightly associated membrane protein (Fig. 9B); it could only be released by detergent treatment. Thus, while we have not precisely mapped the location of the putative membrane-associated domain(s) analogous to those in Tsc10p, it is clear that the C-terminal portion of the FVT1 protein is membrane associated (Fig. 1C).
Fig. 9.
FVT1 lacking the N-terminal transmembrane domain retains membrane association. A: Right-side-out vesicles (80 μg) were prepared from CHO cells expressing pFVT1-GFP(2-11)-fXa@25. Following treatment with or without fXa protease in the absence or presence of 0.2% Nonidet P-40, half of the samples were centrifuged at 100,000 g. Fractions of the total (T), the 100,000 g supernatant (S), and the pellet (P) were analyzed by SDS-PAGE and immunoblotted using anti-FVT1 antibodies. B: The pellet obtained from samples digested with fXa protease in the absence of Nonidet P-40 was incubated with buffer, 0.5 M NaCl, 0.1 M Na2CO3, 2.5 M urea, 0.2% Nonidet P-40, and 1% Triton X-100 on ice for 1 h and centrifuged at 100,000 g for 30 min. The pellet (P) and the soluble fractions (S) equivalent to 15 μg of original microsomes were analyzed by immunoblotting using anti-FVT1 antibodies.
DISCUSSION
In yeast, reduction of 3-KDS to DHS is catalyzed exclusively by Tsc10p, a member of a large family of SDRs. The most closely related human SDR (albeit only 24% identical, 42% similar) is FVT1, which Kihara and Igarashi (4) have shown complements a yeast tsc10Δ mutant. However, in higher eukaryotes, there are multiple isoforms of several enzymes involved in sphingolipid synthesis, including, for example, SPT (19). It therefore seemed possible that in higher eukaryotes, SDRs in addition to FVT1 might have 3-KDS reductase activity. Indeed, sequence comparisons of Tsc10p with mammalian SDRs identified significant regions of homology in a number of uncharacterized SDRs. Moreover, we have recently shown, experimentally, that Arabidopsis thaliana contains two SDRs that can reduce 3-KDS (K. Gable et al., unpublished observations). To investigate the possibility that more than one 3-KDS reductase exists in higher eukaryotes, RNA interference was used to reduce the expression of FVT1 and examine the effects on overall 3-KDS reductase activity in CHO cells. The results of these experiments showed that reduction of FVT1 expression resulted in a concomitant loss of 3-KDS reductase activity, indicating that FVT1 is the principal 3-KDS reductase in mammalian cells. This conclusion is consistent with the recent report that mutations in bovine FVT1 that have only a modest affect on enzyme activity result in a profound neurologic deficiency (6).
Based on hydropathy algorithms and limited proteolysis that placed the catalytic domain of FVT1 in the cytoplasm, Kihara and Igarashi (4) predicted that Tsc10p and FVT1 both contain two distally located transmembrane domains flanking a short lumenal loop. To test this prediction, GC cassettes were inserted into the regions Tsc10p and FVT1 predicted to reside in the lumen. For Tsc10p, the fusion protein with the GC inserted between the two predicted transmembrane domains, Tsc10p-GC@278, was both stable and functional. However, the limited extent of glycosylation raises the possibility that Tsc10p may not contain two distinct domains that span the entire membrane, but rather an extended hydrophobic region embedded in the membrane that confers properties typical of integral membrane proteins. This may be true for FVT1 as well, as it contains analogously positioned predicted transmembrane domains (Fig. 1), and its C-terminal half exhibits properties of an integral membrane protein. However, insertion of GCs into the region between these two predicted transmembrane domains of FVT1 rendered the protein unstable, and it was therefore not possible to directly test this possibility.
The difference in the N-terminal topologies of the yeast and mammalian 3-KDS reductases are clearly important for their correct targeting and insertion into the ER membrane. Fusion of the N-terminal extension of FVT1 to a heterologous protein revealed that it is both necessary and sufficient for ER targeting. Moreover, N-terminally truncated FVT1 failed to localize to the ER. As Tsc10p, which lacks an N-terminal transmembrane domain, properly localizes to the ER while FVT1 requires an additional transmembrane domain, we conclude that their distinctly different topologies reflect important functional differences in the mechanisms for their ER targeting and retention. This is consistent with the presence of a canonical ER retention signal (5) in the C-terminal portion of Tsc10p that is not found in FVT1. It is important to note that although the N-terminal membrane-spanning domain of FVT1 is necessary for targeting, once inserted, its removal leaves a C-terminal fragment that retains the properties of an integral membrane protein. Thus, although there is no direct evidence for the lumenal localization of any of the distal portion of FVT1, our data clearly support the hypothesis that it contains a membrane-associated domain. Nevertheless, despite the different topologies of FVT1 and Tsc10p, FVT1 functionally complements a tsc10Δ mutant, demonstrating that these differences do not prevent ER localization of FVT1 in yeast. Not only is the N-terminal transmembrane domain of FVT1 necessary and sufficient for ER targeting in mammalian cells, but it is also required for proper ER targeting when FVT1 is expressed in yeast. This result suggests that the same ER targeting mechanism used by FVT1 in mammalian cells is present in yeast and provides additional evidence that ER targeting is not determined by primary sequence (20).
The data presented here show that FVT1 and Tsc10p are both multimeric, a general property of SDRs (21). In addition, our results support the conclusion that the catalytic regions of the two proteins are cytoplasmic (Fig. 1). This is consistent with the cytoplasmic localization of the catalytic site of SPT and suggests that SPT and the 3-KDS reductase could be part of a higher-order complex. However, coimmunoprecipitation experiments showed that FVT1 and Tsc10p do not heterodimerize, a result consonant with the relatively low sequence homology between the two proteins. Thus, although it was considered possible that the highly reactive 3-KDS intermediate might be channeled from SPT to the reductase, the ability of FVT1 to substitute for Tsc10p suggests that this is not necessary. Indeed, exogenously supplied 3-KDS can rescue yeast mutants lacking SPT (3). Moreover, prokaryotic SPT is a soluble enzyme whose expression can also rescue yeast mutants lacking SPT (G. Han et al., unpublished observations). Clearly, additional experiments will be required to resolve the organization of the sphingoid base biosynthetic pathway.
Both FVT1 and Tsc10p contain the highly conserved tyrosine, lysine, and serine residues that constitute the catalytic triad of most SDRs (22). Interestingly, while mutation of the serine or lysine residues in either protein results in a temperature-sensitive growth phenotype, mutation of the tyrosine in FVT1 appeared more deleterious than mutation of the corresponding residue of Tsc10p. Conversely, mutation of a conserved asparagine, suggested to also be important for catalysis, was more deleterious in Tsc10p. Thus, in addition to their topological differences, there are also differences in the catalytic sites of the two reductases. While it is tempting to speculate that these differences are responsible for the higher affinity of FVT1 for 3-KDS, this remains to be proven.
Krebs et al. (6) have recently reported that an A175T mutation in bovine FVT1 is responsible for a form of spinal muscular atrophy. Mutation of this residue in human FVT1 had only a modest affect (about 2-fold) on enzymatic activity when assayed at a substrate concentration approximately equal to the Km of the wild-type enzyme. However, the actual concentration of 3-KDS in cells is unknown. Thus, if the local concentration of 3-KDS were substantially lower than the Km, this mutation could result in significant reduction of DHS synthesis, thereby accounting for the SMA phenotype. Although the decrease caused by the A175T mutation was significant, it was far less than that reported for the same mutation in bovine FVT1 (6). This could reflect a basic difference between the human and bovine enzymes. Alternatively, since assays of bovine FVT1 activity were performed using N-terminally truncated HIS-tagged enzyme purified from E. coli, the apparent difference could be the result of the different expression systems used. Based on the structures of other SDRs, A175 resides near the catalytic triad. However, it is not as highly conserved in SDRs as are the residues of the catalytic triad; indeed, in yeast Tsc10p, the corresponding residue is a threonine. Thus, it is not clear whether this mutation directly affects catalytic activity or acts less directly.
The results presented here demonstrate that FVT1 is the major 3-KDS reductase in mammalian cells. In addition, they show that although FVT1 can substitute for Tsc10p in yeast, there are significant topological differences that mediate the targeting and localization of the two enzymes. These results raise important questions about how SPT and 3-KDS reductase interact to transfer the highly reactive 3-KDS intermediate. Experiments to address these questions are in progress.
Footnotes
Abbreviations:
- CHO
- Chinese hamster ovary
- DHS
- dihydrosphingosine
- EndoH
- endoglycosidase H
- ER
- endoplasmic reticulum
- fXa
- factor Xa
- GC
- glycosylation cassette
- GFP
- green fluorescent protein
- HRP
- horseradish peroxidase
- 3-KDS
- 3-ketosphinganine
- β-ME
- β-mercaptoethanol
- PHS
- phytosphingosine
- QCM
- Quikchange mutagenesis
- SDR
- short-chain reductase
- siRNA
- short interfering RNA
- SMA
- spinal muscular atrophy
- SML
- sucrose monolaurate
- SPT
- serine palmitoyltransferase
This work was supported by United States Public Health Service Grant NS47717 awarded by the National Institute of Neurological Disorders and Stroke to T.M.D. and J.M.H.
REFERENCES
- 1.Merrill A. H., Jr2002. De novo sphingolipid biosynthesis: a necessary, but dangerous, pathway. J. Biol. Chem. 277: 25843–25846 [DOI] [PubMed] [Google Scholar]
- 2.Riezman H.2006. Organization and functions of sphingolipid biosynthesis in yeast. Biochem. Soc. Trans. 34: 367–369 [DOI] [PubMed] [Google Scholar]
- 3.Beeler T., Bacikova D., Gable K., Hopkins L., Johnson C., Slife H., Dunn T. 1998. The Saccharomyces cerevisiae TSC10/YBR265w gene encoding 3- ketosphinganine reductase is identified in a screen for temperature-sensitive suppressors of the Ca2+-sensitive csg2Δ mutant. J. Biol. Chem. 273: 30688–30694 [DOI] [PubMed] [Google Scholar]
- 4.Kihara A., Igarashi Y. 2004. FVT-1 is a mammalian 3-ketodihydrosphingosine reductase with an active site that faces the cytosolic side of the endoplasmic reticulum membrane. J. Biol. Chem. 279: 49243–49250 [DOI] [PubMed] [Google Scholar]
- 5.Jackson M. R., Nilsson T., Peterson P. A. 1993. Retrieval of transmembrane proteins to the endoplasmic reticulum. J. Cell Biol. 121: 317–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Krebs S., Medugorac I., Rother S., Strasser K., Forster M. 2007. A missense mutation in the 3-ketodihydrosphingosine reductase FVT1 as candidate causal mutation for bovine spinal muscular atrophy. Proc. Natl. Acad. Sci. USA. 104: 6746–6751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tanabe T., Tanaka N., Uchikawa K., Kabashima T., Ito K., Nonaka T., Mitsui Y., Tsuru M., Yoshimoto T. 1998. Roles of the Ser146, Tyr159, and Lys163 residues in the catalytic action of 7α-hydroxysteroid dehydrogenase from Escherichia coli. J. Biochem. 124: 634–641 [DOI] [PubMed] [Google Scholar]
- 8.Gilstring C. F., Ljungdahl P. O. 2000. A method for determining the in vivo topology of yeast polytopic membrane proteins demonstrates that Gap1p fully integrates into the membrane independently of Shr3p. J. Biol. Chem. 275: 31488–31495 [DOI] [PubMed] [Google Scholar]
- 9.Han G., Gable K., Yan L., Natarajan M., Krishnamurthy J., Gupta S. D., Borovitskaya A., Harmon J. M., Dunn T. M. 2004. The topology of the Lcb1p subunit of yeast serine palmitoyltransferase. J. Biol. Chem. 279: 53707–53716 [DOI] [PubMed] [Google Scholar]
- 10.Paul S., Gable K., Dunn T. M. 2007. A six-membrane-spanning topology for yeast and Arabidopsis Tsc13p, the enoyl reductases of the microsomal fatty acid elongating system. J. Biol. Chem. 282: 19237–19246 [DOI] [PubMed] [Google Scholar]
- 11.Sherman F., Fink G. R., Hicks J. B. 1986. Methods in Yeast Genetics: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 12.Kohlwein S. D., Eder S., Oh C. S., Martin C. E., Gable K., Bacikova D., Dunn T. 2001. Tsc13p is required for fatty acid elongation and localizes to a novel structure at the nuclear-vacuolar interface in Saccharomyces cerevisiae. Mol. Cell. Biol. 21: 109–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gable K., Slife H., Bacikova D., Monaghan E., Dunn T. M. 2000. Tsc3p is an 80-amino acid protein associated with serine palmitoyltransferase and required for optimal enzyme activity. J. Biol. Chem. 275: 7597–7603 [DOI] [PubMed] [Google Scholar]
- 14.Romano J. D., Michaelis S. 2001. Topological and mutational analysis of Saccharomyces cerevisiae Ste14p, founding member of the isoprenylcysteine carboxyl methyltransferase family. Mol. Biol. Cell. 12: 1957–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feramisco J. D., Goldstein J. L., Brown M. S. 2004. Membrane topology of human insig-1, a protein regulator of lipid synthesis. J. Biol. Chem. 279: 8487–8496 [DOI] [PubMed] [Google Scholar]
- 16.El-Kabbani O., Ishikura S., Darmanin C., Carbone V., Chung R. P., Usami N., Hara A. 2004. Crystal structure of human L-xylulose reductase holoenzyme: probing the role of Asn107 with site-directed mutagenesis. Proteins. 55: 724–732 [DOI] [PubMed] [Google Scholar]
- 17.Filling C., Berndt K. D., Benach J., Knapp S., Prozorovski T., Nordling E., Ladenstein R., Jornvall H., Oppermann U. 2002. Critical residues for structure and catalysis in short-chain dehydrogenases/reductases. J. Biol. Chem. 277: 25677–25684 [DOI] [PubMed] [Google Scholar]
- 18.Ou W. J., Cameron P. H., Thomas D. Y., Bergeron J. J. 1993. Association of folding intermediates of glycoproteins with calnexin during protein maturation. Nature. 364: 771–776 [DOI] [PubMed] [Google Scholar]
- 19.Hornemann T., Richard S., Rutti M. F., Wei Y., von Eckardstein A. 2006. Cloning and initial characterization of a new subunit for mammalian serine-palmitoyltransferase. J. Biol. Chem. 281: 37275–37281 [DOI] [PubMed] [Google Scholar]
- 20.Hegde R. S., Bernstein H. D. 2006. The surprising complexity of signal sequences. Trends Biochem. Sci. 31: 563–571 [DOI] [PubMed] [Google Scholar]
- 21.Ghosh D., Sawicki M., Pletnev V., Erman M., Ohno S., Nakajin S., Duax W. L. 2001. Porcine carbonyl reductase. structural basis for a functional monomer in short chain dehydrogenases/reductases. J. Biol. Chem. 276: 18457–18463 [DOI] [PubMed] [Google Scholar]
- 22.Oppermann U., Filling C., Hult M., Shafqat N., Wu X., Lindh M., Shafqat J., Nordling E., Kallberg Y., Persson B., et al. 2003. Short-chain dehydrogenases/reductases (SDR): the 2002 update. Chem. Biol. Interact. 143–144: 247–253 [DOI] [PubMed] [Google Scholar]
- 23.Tusnady G. E., Simon I. 2001. The HMMTOP transmembrane topology prediction server. Bioinformatics. 17: 849–850 [DOI] [PubMed] [Google Scholar]
- 24.Hofmann K., Stoffel W. 1993. TMbase - a database of membrane spanning proteins segments. Biol. Chem. Hoppe Seyler. 374: 166 [Google Scholar]
- 25.von Heijne G.1992. Membrane protein structure prediction: hydrophobicity analysis and the positive-inside rule. J. Mol. Biol. 225: 487–494 [DOI] [PubMed] [Google Scholar]
- 26.Jones D. T., Taylor W. R., Thornton J. M. 1994. A model recognition approach to the prediction of all-helical membrane protein structure and topology. Biochemistry. 33: 3038–3049 [DOI] [PubMed] [Google Scholar]
- 27.Krogh A., Larsson B., von Heijne G., Sonnhammer E. L. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305: 567–580 [DOI] [PubMed] [Google Scholar]
- 28.Rost B., Yachdav G., Liu J. 2004. The PredictProtein server. Nucleic Acids Res. 32: W321–W326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arai M., Mitsuke H., Ikeda M., Xia J. X., Kikuchi T., Satake M., Shimizu T. 2004. ConPred II: a consensus prediction method for obtaining transmembrane topology models with high reliability. Nucleic Acids Res. 32: W390–W393. [DOI] [PMC free article] [PubMed] [Google Scholar]