Abstract
Although studies with liver type fatty acid binding protein (L-FABP) gene ablated mice demonstrate a physiological role for L-FABP in hepatic fatty acid metabolism, little is known about the mechanisms whereby L-FABP elicits these effects. Studies indicate that L-FABP may function to shuttle lipids to the nucleus, thereby increasing the availability of ligands of nuclear receptors, such as peroxisome proliferator-activated receptor-α (PPARα). The data herein suggest that such mechanisms involve direct interaction of L-FABP with PPARα. L-FABP was shown to directly interact with PPARα in vitro through co-immunoprecipitation (co-IP) of pure proteins, altered circular dichroic (CD) spectra, and altered fluorescence spectra. In vitro fluorescence resonance energy transfer (FRET) between Cy3-labeled PPARα and Cy5-labeled L-FABP proteins showed that these proteins bound with high affinity (Kd approximately 156 nM) and in close proximity (intermolecular distance of 52Å). This interaction was further substantiated by co-IP of both proteins from liver homogenates of wild-type mice. Moreover, double immunogold electron microscopy and FRET confocal microscopy of cultured primary hepatocytes showed that L-FABP was in close proximity to PPARα (intermolecular distance 40–49Å) in vivo. Taken together, these studies were consistent with L-FABP regulating PPARα transcriptional activity in hepatocytes through direct interaction with PPARα. Our in vitro and imaging experiments demonstrate high affinity, structural molecular interaction of L-FABP with PPARα and suggest a functional role for L-FABP interaction with PPARα in long chain fatty acid (LCFA) metabolism.
Keywords: cytoplasmic lipid binding protein, fluorescence, FRET, liver fatty acid binding protein, nuclei, peroxisome proliferator activated receptor, transcription factor
Peroxisome proliferator-activated receptors (PPAR) are a family of ligand-activated nuclear receptors that induce transcription of multiple genes encoding proteins involved in fatty acid and glucose metabolism, as well as cell differentiation (1, 2). Because abnormal regulation of peroxisome proliferator-activated receptor-α (PPARα), the major PPAR isoform found in liver, is associated with chronic diseases such as diabetes, obesity, and hyperlipidemia, considerable effort extends to understanding how endogenous ligands regulate PPARα transcriptional control (2, 3). Although a broad range of synthetic substances including hypolipidemic agents, plasticizers, and herbicides are known PPARα activators (4), until recently the identity of endogenous, high-affinity PPARα ligands remained elusive.
While both saturated and unsaturated long-chain fatty acids (LCFA) enhance PPARα-activated gene expression (5, 6), only unsaturated LCFA bind to PPARα with high affinity (7). These discrepancies were later explained by studies showing that the activated form of LCFA, long-chain fatty acyl-CoA (LCFA-CoA), could function as high-affinity, endogenous PPARα ligands (6, 8). Binding of both saturated and unsaturated LCFA-CoA induce a conformational change in PPARα, enhance interaction with coactivator steroid receptor coactivator-1 (SRC-1), and enhance PPARα transactivation in cultured cells (6, 8, 9). These effects are not due to hydrolysis of LCFA-CoA, as the nonhydrolyzable S-hexadecyl-CoA is also bound with high affinity, alters PPARα conformation, and alters interaction with coactivators (6, 10). In addition, the high affinity (i.e., nM Kds) binding of LCFA-CoA is in the same range as that of LCFA-CoA concentrations in the nucleoplasm of living cells (11, 12).
As both LCFA and LCFA-CoA have high affinities for membranes, it is unclear how once LCFA are taken up (and/or activated to LCFA-CoA) these ligands are transported to the nuclear envelope and the nucleoplasm to regulate the activity of nuclear receptors such as PPARα (5). Studies with transfected cells overexpressing liver fatty acid binding protein (L-FABP) suggest that this cytoplasmic LCFA and LCFA-CoA binding protein may be a likely candidate for directly enhancing the transfer of these ligands into nuclei (12–15). Further, transactivation assays and colocalization experiments in transfected cells suggest that L-FABP directly interacts with PPARα (12, 13). However, these studies were performed in transformed, tumorigenic cells, and colocalization by confocal imaging (resolution of 2000 Å) is insufficient to demonstrate direct interaction. Because L-FABP protein level is itself regulated by PPARα transcriptional activity, it has been postulated that L-FABP may mediate its own expression by enhancing LCFA and LCFA-CoA transport into nuclei to facilitate transcriptional activity of PPARα (5). Despite these studies, evidence supporting mechanistic details of this hypothesis is lacking. The objective of the present investigation was to use recombinant pure proteins, fluorescently labeled recombinant proteins, and wild-type (L-FABP+/+) and L-FABP gene ablated (L-FABP−/−) mice to begin to resolve some of the mechanistic details of L-FABP–mediated regulation of PPARα in primary hepatocytes; specifically, whether these two proteins directly interacted within nuclei of primary hepatocytes.
MATERIALS AND METHODS
Materials
Palmitic acid (C16:0) was from Sigma-Aldrich (St. Louis, MO). For co-IP, antibodies to the glucocorticoid receptor (GR) and to PPARα were purchased from Affinity BioReagents (Golden, CO), while antibodies to sterol regulatory element binding protein-1 (SREBP-1) and to L-FABP were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). Mammalian co-IP kit, chemiluminescent substrate, and film were from Pierce Biotechnology (Rockford, IL). For western blotting following co-IP, polyclonal antibodies prepared in rabbit to PPARα (Affinity BioReagents) and rabbit polyclonal antiserum to L-FABP (16) were used in combination with HRP-conjugated anti-rabbit IgG secondary antibodies (Sigma-Aldrich). Cy3 and Cy5 protein labeling kits were from Amersham Biosciences (Piscataway, NJ). For double immunogold EM colocalization experiments, LR White resin, donkey anti-rabbit IgG conjugated to 6nm gold, and donkey anti-goat IgG conjugated to 15nm gold were from Electron Microscopy Sciences (Fort Washington, PA); goat anti-human albumin was from Miles-Yeda (Rehovot, Israel); and affinity purified fractions of goat polyclonal antisera to rat L-FABP and rabbit polyclonal antisera to PPARα and SREBP-1 were obtained from Santa Cruz Biotechnology. For double immunofluorescence FRET confocal microscopy, Lab-Tek chambered cover glass slides were from Nunc (Naperville, IL); affinity purified anti-PPARα and anti-SREBP-1 were from Santa Cruz Biotechnology, and polyclonal anti-L-FABP obtained as described earlier (16) was affinity purified on a protein A column according to the manufacturer’s instructions (Bio-Rad Laboratories, Hercules, CA). All reagents and solvents used were of the highest grade available and were cell culture tested.
Recombinant proteins
Mouse recombinant L-FABP protein was produced, purified, and delipidated as previously described (17). The bacterial expression plasmid for mouse recombinant PPARα protein (pET-PPARαΔAB, encoding amino acids 101–468) was a generous gift from Dr. Noa Noy (Case Western University). Mouse PPARα protein was expressed and purified as previously described (6). This truncated version was used for the pure protein studies due to solubility issues with the full-length protein and was expected to show ligand-binding properties identical to those of the full-length receptor based upon similar experiments with PPARγ (18, 19). The bacterial expression plasmid for human recombinant SREBP-1a protein (pGEX4T-SREBP-1a, encoding amino acids 1–460) was generously provided by Dr. Hitoshi Shimano (University of Tsukuba), and the protein was purified as described (20). The mature protein was utilized, rather than the full-length protein, as this is the portion of the protein which is tranlocated to the nucleus for transcriptional control (20, 21). Protein concentrations were determined by Bradford assay.
Recombinant protein co-immunoprecipitation (co-IP)
To determine whether L-FABP could directly interact with PPARα in vitro, the ability of the two proteins to co-immunoprecipitate was examined, and recombinant SREBP-1a was used as a negative control. Purified recombinant proteins were combined as follows: L-FABP and PPARα; L-FABP and SREBP-1a; and PPARα and SREBP-1a. Each protein in the combination was examined for the ability to pull down the other protein. For each sample, 20μg of each protein was mixed and allowed to incubate on ice for 10 min prior to co-IP with the antibody-linked resin from the ProFoundTM co-IP kit (Pierce Biotechnology, Rockford, IL). Proteins unable to bind to the antibody and proteins eluted from the antibody-linked resin were examined by standard SDS-PAGE and Coomassie blue staining for the presence of each protein.
Circular dichroism (CD)
Circular dichroism was used to examine changes in conformation upon L-FABP and PPARα interaction using recombinant SREBP-1a as a negative control. A J-710 spectropolarimeter (JASCO Inc., Easton, MD) was used to record circular dichroic spectra of PPARα (0.8 μM), SREBP-1a (0.7 μM), L-FABP (2.4 μM), [0.4 μM PPARα + 1.2 μM L-FABP], and [0.35 μM SREBP-1a + 1.2 μM L-FABP] (final amino acid molarity in each sample was equal to 0.0003 M) in 125 μM HEPES, pH 8.0, 12.5 μM DTT, 5 mM KCl, 0.3% glycerol at 23°C in a 1 mm cuvette as described previously (6, 22). Replicate spectra were recorded ten times over the far-UV region from 186 to 260nm with a 2 nm bandwidth, 10 millidegree sensitivity, 50 nm/min scan rate, and 1 s time constant. The spectral result obtained by averaging the ten scans was used to determine percent composition of α-helices, β-strands, turns, and unordered structures with CDPro software (http://lamar.colostate.edu/∼sreeram/CDPro) by the following methods: SELCON3, CDSSTR, and CONTIN/LL (23). The CD spectrum of the mixed proteins was compared with a theoretical spectrum of combined but noninteracting proteins. This spectrum was calculated by averaging the spectra of each protein analyzed separately at a concentration equal to that in the mixture (22).
Fluorescence resonance energy transfer (FRET)
Recombinant PPARα, SREBP-1a, and L-FABP proteins were fluorescently labeled with Cy3 or Cy5 dye using Fluorolink-antibody Cy3 and Cy5 labeling kits (Amersham Biosciences). Absorbance measurements were used to determine protein concentrations and dye-to-protein ratios. Emission spectra (560–700 nm) were obtained of 25 nM donor (Cy3-labeled PPARα, Cy-3-labeled SREBP-1a) in PBS upon excitation at 550 nm with increasing concentration of acceptor (Cy5-labeled L-FABP) in a PC1 photon counting spectrofluorometer (ISS Inc., Champaign, IL) at 24°C. The spectra were corrected for background (buffer only and acceptor only) and the maximal intensities measured using Vinci 1.5 software (ISS Inc., Champaign, IL). The energy transfer efficiency was calculated using the amount of sensitized acceptor fluorescence, and the intermolecular distance was calculated according to the Förster equation as described earlier (6, 24).
Animals
All animal protocols were approved by the Institutional Animal Care and Use Committee (IACUC) at Texas A&M University. L-FABP null mice (L-FABP−/−), generated by targeted disruption of the L-FABP gene through homologous recombination, were obtained as described earlier (25, 26). All experiments were performed with livers or hepatocytes derived from N6 backcross generation male mice ranging in age from 2 to 4 months (25–35 g). Hepatocytes from male age-matched, wild-type littermates of the same backcross generation were used as controls. Animals were kept under constant light-dark cycles and had access to food and water ad libitum.
Co-immunoprecipitation
The co-IP of native proteins from mouse liver homogenates was performed as previously described (6). Following homogenization of livers, PPARα and L-FABP were coimmunoprecipitated from liver homogenate (2 mg protein) with antibodies to PPARα or L-FABP (100 μg of antibody) using the ProFoundTM mammalian co-IP kit (Pierce Biotechnology, Rockford, IL) according to the manufacturer’s instructions. Immunoprecipitated proteins were eluted in 100 μl of buffer, and 1/4 of the eluted proteins were loaded onto SDS-PAGE gels and visualized by Western blot for PPARα and L-FABP. Specificity of L-FABP co-IP with PPARα, as well as specificity of PPARα for L-FABP, was determined by examination of the eluted proteins by Western blot for other transcription factors (GR; SREBP-1). Specificity was further examined by immunoprecipitation with antibodies to GR and SREBP-1 followed by Western blotting of eluted proteins for L-FABP as previously described for PPARα and cofactors (6).
Immunoelectron microscopy
Liver tissue segments from wild-type L-FABP+/+ and L-FABP−/− mice were fixed by immersion in 4% formaldehyde, 0.1% glut in 0.1 M sodium phosphate buffer (pH 7.4) for 20 h at 4°C. The tissue segments were washed with 0.1 M sodium phosphate, dehydrated in an ethanol series, and then embedded in LR White resin at 48°C for two days. Ultrathin sections (60–80 nm) were placed on Formvar-coated nickel grids and immunogold-stained with rabbit anti-L-FABP antiserum (diluted 1:600) alone or in a mixture with goat anti-PPARα (diluted 1:125). For comparison, other sections from the same tissue segments were immunostained with a mixture of goat anti-LFABP antiserum (diluted 1:50) and rabbit anti-SREBP-1 (diluted 1:50). These sections were washed and incubated with a mixture of donkey anti-rabbit IgG conjugated to 6 nm gold and donkey anti-goat IgG conjugated to 15 nm gold. Controls included (a) anti-L-FABP incubation with sections of liver from an L-FABP−/− mouse and (b) incubations without primary antibodies. All sections were post stained very briefly with aqueous uranyl acetate and Reynold’s lead citrate and examined with a Zeiss 10c TEM (Carl Zeiss Microimaging Inc., Thornwood, NY).
To evaluate the significance of clusters with two sizes of gold particles in immunogold stains of anti-L-FABP and anti-PPARα, images of nuclear colocalization patterns were statistically analyzed by the method of Philimonenko, Janacek, and Hozak (27). For this, 13 random images of hepatocyte nuclei from immunogold-stained sections were filmed. The sites of gold particles in the nucleoplasm (200 µm2 per group) were manually marked with Adobe Photoshop on layers of digitized versions of these images, and the spatial locations were identified with ImageJ (http://rsb.info.nih.gov/ij/) available from the National Institutes of Health (Bethesda, MD). These data were loaded into the Gold program (27) for calculating the pair cross-correlation function and the cross-K function to evaluate the level of significance of particle size distributions at various selected distances. Thus the extent of colocalization can be analyzed. For comparison with these measurements of anti-L-FABP and anti-PPARα labeling, an identical analysis was performed on a similar set of nucleoplasm images from sections that were immunogold-stained with anti–L-FABP and anti–SREBP-1.
Hepatocyte isolation, fixation, and staining
Hepatocytes from wild-type (L-FABP+/+) mice were isolated as described earlier (26, 28), seeded (125,000 cells/well) onto collagen-coated 4-well Lab-Tek chambered slides, and incubated overnight at 37°C in a humidified incubator with 5% carbon dioxide. After 24 h, cultured primary hepatocytes were rinsed twice in PBS and fixed in 4% formaldehyde (methanol-free) with 0.1% glutaraldehyde in 0.1 M sodium phosphate buffer (pH 7.4) for 20 h at 4°C. The fixed cultures were washed with PBS containing 0.05 M glycine to quench remaining glutaraldehyde autofluorescence, treated with 5% BSA to block nonspecific protein binding, and then incubated for 2 h at room temperature with a mixture of the antibody:dye conjugates in PBS containing 0.5% BSA, washed in PBS, and coverslipped with Gel Mount (Polysciences). Controls included (a) hepatocytes individually stained with each antibody and (b) incubations in the absence of antibodies.
Immunofluorescence confocal microscopy
For FRET determined by laser scanning confocal microscopy, affinity purified anti-L-FABP was labeled with Cy3 while anti-PPARα and anti–SREBP-1 were labeled with Cy5 using Fluorolink-antibody Cy3 and Cy5 labeling kits (Amersham Biosciences). The stained hepatocytes were imaged with a laser scanning confocal microscope (LSCM) consisting of an MRC-1024 fluorescence imaging system (Bio-Rad) with an Axiovert 135 microscope (Zeiss, NY). Excitation light (488, 568, and 647 nm) from a 15-milliwatt krypton-argon laser was delivered to the sample through 63× Zeiss Plan-Fluor oil immersion objective, numerical aperture 1.4.
To estimate the intermolecular distance between L-FABP and PPARα in hepatocyte nuclei, FRET from Cy3 to Cy5 was detected as sensitized emission of Cy5 (through the 680/32 bandpass filter) upon excitation of Cy3 at 488 nm after correction of some bleed through of Cy3 emission through the same filter as described earlier (22). Quantitative measurements for FRET efficiency estimation were carried out by the acceptor photobleaching method as described (22). Cellular images produced by fluorescence emission of Cy3 (488 nm excitation, 598/40 filter) and Cy5 (647 nm excitation, 680/32 filter) were sequentially acquired. The cells were then photobleached for 3 min with the 647 nm laser (the conditions of photobleaching were optimized such that a good decrease in Cy5 fluorescence was obtained without affecting the emission intensity of Cy3), and then the Cy5 and Cy3 post-bleaching images were recorded at their original instrument settings. The image sets were analyzed with ImageJ using the FRETcalculation plugin written by David Stepensky (29). FRET efficiency was calculated from measurements of the increased Cy3 fluorescence emission after Cy5 photobleaching in 71 different 1 µm2 regions of 3 hepatocyte nuclei and used to estimate the intermolecular distance between Cy3 and Cy5 (distance ultimately dictated in this experiment by the proximity between the selected protein pair) according to the Förster equation (22, 30). To determine the specificity of the interaction of L-FABP with PPARα, these methods were repeated with hepatocytes stained with Cy3-anti-L-FABP and Cy5-anti-SREBP-1, and an identical analysis was performed on images with similar staining.
Statistics
Values represented the mean ± SE with n and P indicated as described. Statistical analyses were performed using Student’s t-test or two-way ANOVA (ANOVA) (GraphPad Prism, San Diego, CA). Values with P < 0.05 were considered statistically significant.
RESULTS
Co-immunoprecipitation: direct interaction of L-FABP and PPARα recombinant proteins
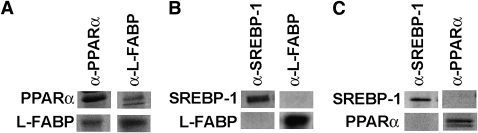
One possible mechanism whereby L-FABP expression may influence PPARα-mediated regulation of fatty acid metabolism is through direct interaction of L-FABP with PPARα. To determine whether L-FABP and PPARα proteins interact in vitro, recombinant proteins were mixed, precipitated with antibodies to L-FABP or PPARα, and examined by SDS-PAGE for coprecipitation of both proteins. Whether the antibody to PPARα or the antibody to L-FABP was used, both proteins were pulled down by the antibody (Fig. 1A), suggesting a direct interaction in vitro. To examine the specificity of L-FABP for PPARα versus other transcription factors, the ability of anti–SREBP-1 and anti–L-FABP to pull down SREBP-1a and L-FABP was examined. Neither antibody was capable of co-immunoprecipitating both L-FABP and SREBP-1a (Fig. 1B), suggesting that L-FABP and SREBP-1a do not interact and that the L-FABP interaction with PPARα is specific. To further confirm the specificity of this technique, the ability of anti–SREBP-1 and anti–PPARα to pull down SREBP-1a and PPARα was examined. Again, neither antibody was capable of co-immunoprecipitating both SREBP-1a and PPARα (Fig. 1C), suggesting that the L-FABP and PPARα interaction is specific.
Fig. 1.
Co-IP of L-FABP and PPARα recombinant proteins. A: L-FABP and PPARα proteins (20 μg each) were mixed, immunoprecipitated with anti-PPARα (α-PPARα) or anti-L-FABP (α-L-FABP), and examined by SDS-PAGE and Coomassie blue staining for each protein. B: L-FABP and SREBP-1a proteins (20 μg each) were mixed, immunoprecipitated with anti-SREBP-1 (α-SREBP-1) or anti-L-FABP (α-L-FABP), and examined by SDS-PAGE and Coomassie blue staining for the presence of each protein. C: SREBP-1a and PPARα proteins (20 μg each) were mixed, immunoprecipitated with anti-SREBP-1 (α-SREBP-1) or anti-PPARα (α-PPARα), and examined by SDS-PAGE and Coomassie blue staining for each protein.
Circular dichroism: effect of L-FABP interaction with PPARα on conformation
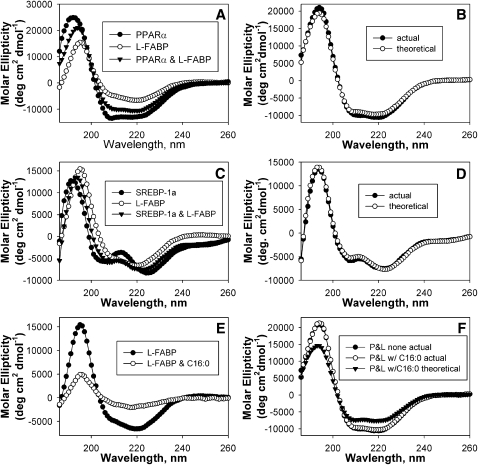
Different proteins such as L-FABP and PPARα may interact with or without undergoing conformational changes. This possibility was examined by circular dichroism, a method that determines the secondary structure of proteins. The shapes of the circular dichroic spectra of L-FABP and PPARα were markedly different, consistent with PPARα alone having a high content of α-helical structure (Fig. 2A, closed circles) and L-FABP alone having a high content of β-sheet (Fig. 2A, open circles). For the mixture containing both proteins, the theoretically expected circular dichroic spectrum based upon the assumption of zero interaction between L-FABP and PPARα (Fig. 2B, open circles) was not superimposable upon the experimentally measured spectrum of the combination of L-FABP and PPARα (Fig. 2B, closed circles), although only small changes in spectra were observed. Results from the compositional analysis of the α-helices, β-strands, turns, and unordered structures confirmed small conformational changes in the mixture of these proteins, with a small increase in α-helical structure concomitant with a decrease in unordered structure (Table 1). The presence of small conformational changes upon L-FABP interaction with PPARα suggests a direct interaction between these proteins. However, the magnitude of these protein–protein conformational changes was 2- to 3-fold smaller than those exhibited by PPARα in response to LCFA or LCFA-CoA binding (6, 8).
Fig. 2.
Circular dichroism of PPARα and L-FABP. A: Far-UV CD spectra of PPARα (closed circles), L-FABP (open circles), and a mixture of equal amino acid molarities of PPARα and L-FABP (closed inverted triangles). B: Comparison of the far-UV CD spectra of PPARα and L-FABP obtained experimentally (closed circles) and the theoretically expected spectrum (open circles). The theoretically expected value of the combined proteins assuming no interaction was determined by averaging the spectra of each protein analyzed separately at a concentration equal to that in the mixture. C: Far-UV CD spectra of SREBP-1a (closed circles), L-FABP (open circles), and a mixture of equal amino acid molarities of SREBP-1a and L-FABP (closed inverted triangles). D: Comparison of the far-UV CD spectra of SREBP-1a and L-FABP obtained experimentally (closed circles) and the theoretically expected spectrum (open circles). E: CD spectra of L-FABP in the absence (closed circles) and presence of 10 μM palmitic acid (open circles). F: Comparison of the experimentally obtained spectrum PPARα and L-FABP in the absence (filled circles, P and L none actual) and presence of 10 μM palmitic acid (open circles, P and L w/C16:0 actual) with the theoretically obtained spectrum for PPARα and L-FABP in the presence of 10 μM palmitic acid if no interaction occurred between the proteins (closed inverted triangles, P and L w/C16:0 theoretical). Each spectrum represents an average of ten scans for a given representative spectrum from four replicates.
TABLE 1.
Secondary structures of PPARα, L-FABP, SREBP-1a, and complexes of these proteins in the absence and presence of palmitic acid (C16:0)
| Proteins | α-helix regular H(r) % | α-helix distort H(d) % | β-sheet regular S(r) % | β-sheet distort S(d) % | Turns T % | Unrd U % |
|---|---|---|---|---|---|---|
| PPARα | 24.1 ± 0.5 | 15.8 ± 0.1 | 8.3 ± 0.1 | 7.1 ± 0.1 | 18.9 ± 0.4 | 26.1 ± 0.2 |
| L-FABP | 10.1 ± 0.4 | 9.9 ± 0.2 | 18.7 ± 0.5 | 10.3 ± 0.1 | 22.1 ± 0.2 | 27.5 ± 0.4 |
| SREBP-1a | 16.1 ± 3.8 | 11.1 ± 1.9 | 9.3 ± 2.1 | 11.1 ± 0.8 | 20.8 ± 3.0 | 31.4 ± 3.1 |
| PPARα + L-FABP | 18.6 ± 0.4* | 13.9 ± 0.3 | 11.4 ± 0.3 | 8.6 ± 0.1 | 21.8 ± 0.5 | 26.1 ± 0.4* |
| Theoretical | 17.0 ± 0.4 | 13.2 ± 0.2 | 12.0 ± 0.2 | 8.8 ± 0.07 | 21.9 ± 0.5 | 27.4 ± 0.4 |
| SREBP-1a + L-FABP | 18.0 ± 2.3 | 12.9 ± 1.1 | 11.8 ± 1.1 | 10.3 ± 0.8 | 19.7 ± 1.3 | 26.9 ± 2.0 |
| Theoretical | 18.5 ± 3.1 | 13.8 ± 1.8 | 11.6 ± 0.5 | 10.8 ± 0.7 | 19.6 ± 1.4 | 25.4 ± 2.8 |
| PPARα + C16:0 | 22.6 ± 3.0 | 15.2 ± 0.7 | 8.7 ± 2.3 | 7.3 ± 0.7 | 19.5 ± 3.1 | 26.6 ± 0.9 |
| L-FABP + C16:0 | 1.1 ± 0.8 | 4.4 ± 0.6 | 28.1 ± 2.3 | 14.5 ± 0.8 | 22.1 ± 2.9 | 29.8 ± 2.7 |
| PPARα+L-FABP+C16:0 | 18.0 ± 0.5* | 13.5 ± 0.4* | 11.5 ± 0.3* | 8.9 ± 0.2* | 22.1 ± 0.6 | 29.1 ± 2.5* |
| Theoretical | 12.6 ± 0.7 | 10.7 ± 1.3 | 15.7 ± 4.4 | 9.5 ± 0.7 | 21.4 ± 1.6 | 29.1 ± 2.5 |
L-FABP, liver fatty acid binding protein; PPARα, peroxisome proliferator activated receptor-α; SREBP-1, sterol regulatory element-binding protein-1; Unrd, unordered.
Significant differences between actual and theoretical values for protein mixtures determined by student’s t-test; * = P < 0.05; n = 4–6.
The interaction between L-FABP and SREBP-1a was examined to determine specificity of L-FABP for PPARα versus other transcription factors. The circular dichroic spectrum of these proteins varied, with SREBP-1a (Fig. 2C, closed circles) having more unordered structures and L-FABP (Fig. 2C, open circles) having more β-sheets. However, the spectrum of the experimentally obtained combination of these proteins (Fig. 2D, closed circles) was superimposable upon the theoretical spectrum for no interaction (Fig. 2D, open circles); suggesting that these proteins do not undergo conformational changes or do not directly interact. Results from the compositional analysis supported this suggestion, as no significant differences were noted (Table 1).
To determine the effect of LCFA on this interaction, the CD experiment between L-FABP and PPARα was repeated in the presence of palmitic acid, a strong L-FABP ligand (31) which is not bound by PPARα (6). Although the presence of palmitic acid does not affect PPARα secondary structure (6), the spectrum of L-FABP was strongly altered (Fig. 2E), resulting in strongly decreased α-helical content and a concomitant increase in β-sheets (Table 1). Upon comparison of these spectra, the experimentally obtained spectrum for L-FABP and PPARα in the presence of palmitic acid (Fig. 2F, open circles, P and L w/C16:0 actual) was superimposable upon the experimentally obtained spectrum for L-FABP and PPARα in the absence of ligand (Fig. 2F, closed circles, P and L none actual); suggesting that the presence of palmitic acid did not affect the L-FABP-PPARα interaction. This was further confirmed by the analysis of the percent composition (Table 1), which showed no significant difference between the experimentally obtained values in the presence or absence of palmitic acid.
Protein binding assay: effect of L-FABP interaction with PPARα on conformation
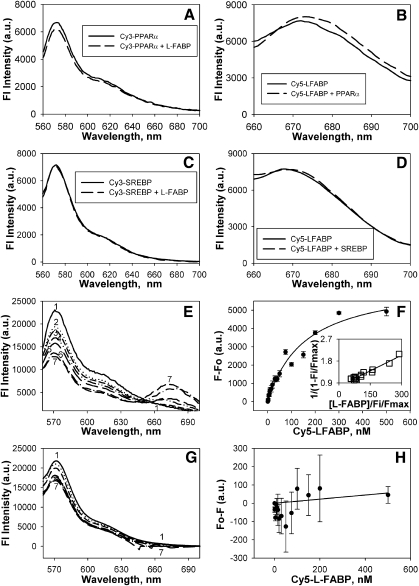
To further examine the effect of protein-protein binding on conformation, these respective recombinant proteins were fluorescently labeled with Cy3 for the nuclear receptor (PPARα or SREBP-1a) and Cy5 for the LCFA/LCFA-CoA binding protein (L-FABP). Labeling ratios of fluorescent dye/protein were maintained low (1.1 for Cy3-labeled PPARα; 0.8 for Cy5-labeled L-FABP; 1.0 for Cy3-labeled SREBP-1a) to assure essentially 1 tag/protein on average. First, to determine if L-FABP binding altered the conformation of PPARα, the fluorescence emission of Cy3-labeled PPARα was determined in the absence (Fig. 3A, solid line) and presence (Fig. 3A, dashed line) of an equal molar amount of L-FABP. The addition of L-FABP slightly decreased the emission of Cy3-PPARα. Second, in the converse experiment to determine if PPARα binding altered the conformation of L-FABP, the fluorescence emission of Cy5-labeled L-FABP was determined in the absence (Fig. 3B, solid line) and presence (Fig. 3B, dashed line) of PPARα. PPARα increased and red-shifted the emission of Cy5-L-FABP, further confirming that the interaction of these two proteins result in altered conformational structures. Third, to ensure that this effect was due to binding and not an effect of the dye, the effect of L-FABP on Cy3-labeled SREBP-1a (Fig. 3C) and the effect of SREBP-1a on Cy5-labeled L-FABP (Fig. 3D) were examined. In both instances, no changes were noted. These data further supported the conformational change seen by CD and suggested that the addition of the small dye molecules (at approximately 1:1 dye to protein ratio) does not interfere with such conformational changes. This is similar to previous data with another nuclear receptor and lipid binding protein showing that the presence of Cy3 or Cy5 dye molecules does not alter protein–protein interactions or ligand binding (24).
Fig. 3.
Fluorescence detection of L-FABP interaction with PPARα: FRET. Recombinant PPARα, SREBP-1a, and L-FABP proteins were chemically labeled with Cy3 and Cy5, respectively as described in Materials and Methods. FRET from donor Cy3-PPARα or Cy3-SREBP-1a to acceptor Cy5-L-FABP was detected as quenching of Cy3 fluorescence emission (near 575 nm) and as appearance of sensitized emission from Cy5 (near 670 nm). A: To determine if L-FABP binding altered the conformation of PPARα, the emission spectrum of Cy3-PPARα (25 nM) was determined in the absence (solid line) and presence (dashed line) of L-FABP (25 nM). B: To determine if PPARα binding altered the conformation of L-FABP, the emission spectrum of Cy5-L-FABP (25 nM) was determined in the absence (solid line) and presence (dashed line) of PPARα (25 nM). C: To determine if L-FABP binding altered the conformation of SREBP-1a, the emission spectrum of Cy3-SREBP-1a (25 nM) was determined in the absence (solid line) and presence (dashed line) of L-FABP (25 nM). D: To determine if SREBP-1a binding altered the conformation of L-FABP, the emission spectrum of Cy5-L-FABP (25 nM) was determined in the absence (solid line) and presence (dashed line) of SREBP-1a (25 nM). E: Emission spectra of Cy3-PPARα and Cy5-L-FABP upon excitation of Cy3 at 550 nm. Spectrum 1: Cy3-labeled PPARα with no acceptor; Spectra 2 7: Cy3-labeled PPARα with the addition of 4, 10, 50, 100, 200, and 300 nM Cy5-labeled L-FABP, respectively. F: Plot of the average change in maximal fluorescence emission intensity at 670 nm (F0–F) of Cy5- L-FABP upon excitation at 550 nm as a function of Cy5-labeled L-FABP concentration. Inset: Linear plot of the binding curve. Values represent the mean ± SE; n = 4. G: Emission spectra of Cy3-SREBP-1a and Cy5-L-FABP upon excitation of Cy3 at 550 nm. Spectrum 1: Cy3-labeled SREBP-1a with no acceptor; Spectra 2 7: Cy3-labeled SREBP-1a with the addition of 4, 10, 50, 100, 200, and 300 nM Cy5-labeled L-FABP, respectively. H: Plot of the average change in Cy5-L-FABP fluorescence emission (F0–F) at 670 nm upon excitation at 550n m as a function of Cy5-labeled L-FABP concentration. Values represent the mean ± SE; n = 4.
FRET: Direct interaction between pure recombinant PPARα and L-FABP proteins
To determine the affinity of PPARα for L-FABP and the distance between these proteins, an in vitro FRET experiment was performed with the fluorescently labeled proteins mentioned above. Cy3 and Cy5 dyes are small (less than 1 kDa) fluorescent tags that form an efficient FRET donor/acceptor pair (22, 32). The donor (Cy3-PPARα) was excited at 550 nm, and an emission spectrum was recorded over the emission range for Cy3- and Cy5-labeled proteins (Fig. 3E, spectrum 1). Next, spectra from 560–700 nm were recorded after addition of increasing concentrations of Cy5-L-FABP acceptor (Fig. 3E, spectra 2–7). With increasing concentration of the Cy5-L-FABP, decreased Cy3 emission intensity was observed near 575 nm concomitant with the appearance of increasing sensitized emission of Cy5 near 670 nm (Fig. 3E), consistent with FRET between the two fluorophores. Transformation of the sensitized acceptor emission data into a binding curve revealed that Cy5-L-FABP binding to Cy3-PPARα was saturable (Fig. 3F) with 1:1 stoichiometry (Fig. 3F, inset). Quantitative analysis of multiple replicates yielded a Kd = 156.5 ± 18.1 nM. Furthermore, the efficiency of energy transfer was calculated to be E = 44 ± 1% with an interaction distance of r = 52 ± 1Å using the sensitized emission of Cy5, consistent with direct interaction between Cy3-L-FABP and Cy5-PPARα. Although a slight decrease in Cy3-SREBP-1a fluorescence intensity was noted at approximately 575 nm with the addition of Cy5-L-FABP, no concomitant increase in sensitized acceptor emission was noted (Fig. 3G). Transformation of the sensitized acceptor emission data into a binding curve revealed that Cy5-L-FABP binding to Cy3-SREBP-1a was not saturable (Fig. 3H), and no binding curve or energy transfer efficiency could be calculated. Thus, only the direct interaction of PPARα with L-FABP resulted in FRET, demonstrating high affinity binding of these two proteins.
It is important to note that energy transfer between donors and acceptors that are randomly distributed in solutions would occur at a concentration much higher (mM) than used in this experiment (nanomolar to micromolar). The critical concentration (C0) at which the acceptor concentration would result in 76% energy transfer can be calculated from the expression C0 = 447/R03. Using the Förster distance R0 = 50Å for the Cy3/Cy5 pair, the critical concentration would be C0 = 3.6 mM. Also, diffusion-enhanced energy transfer would not be considered a factor as the lifetimes of Cy3 and Cy5 are in the ns range (33). Since FRET can only occur at an optimal distance (1–100Å) (33), molecules farther apart than 100Å will not undergo energy transfer. So although the exact location of the dye on each molecule of protein is unknown, these data are an average of the interactions from all of the labeled molecules and suggest that the overall average is in close molecular proximity.
Co-immunoprecipitation: Direct interaction of L-FABP and PPARα in liver homogenates
To determine whether the direct interaction of L-FABP and PPARα seen in vitro could occur in vivo, native proteins were immunoprecipitated from liver homogenates of L-FABP+/+ (Fig. 4A and B, lanes 1–3) and L-FABP−/− (Fig. 4A and B, lanes 4–6) mice. Whether anti–L-FABP (Fig. 4A) or anti–PPARα (Fig. 4B) was used for co-immunoprecipitation, L-FABP and PPARα proteins co-immunoprecipitated in the absence (−) (lane 1) and presence (+) (lane 2) of palmitic acid. As a negative control, these experiments were repeated with liver homogenates from L-FABP gene ablated mice. No coimmunoprecipitation was noted in the absence (−) (lane 4) or presence (+) (lane 5) of palmitic acid. As a Western blot control, the proportional amounts of each protein per 10 μg of liver homogenate (lane 3, WT, “L”; lane 6, L-FABP−/−, “L”) were determined by Western blotting. To examine the specificity of the co-IP experiment, both anti–L-FABP and anti-PPARα immunoprecipitated samples from wild-type (L-FABP+/+) liver homogenates were analyzed by Western blot for the presence of other transcription factors. Although both GR and SREBP-1 proteins were detected in liver homogenate (Fig. 4C, lane 3), neither protein was co-immunoprecipitated with PPARα (Fig. 4C, lane 1) or L-FABP (Fig. 4C, lane 2). To further confirm the L-FABP specificity for PPARα versus other transcription factors, wild-type (L-FABP+/+) liver homogenates were also immunoprecipitated with antibodies to GR and SREBP-1, followed by Western blotting to probe for co-immunoprecipitated L-FABP (Fig. 4D). As a positive control, L-FABP was detected prominently in liver homogenates (Fig. 4D, lane 3). L-FABP did not co-immunoprecipitate with antibodies to either of these other transcription factors (GR, Fig. 4D, lane 1; SREBP-1 and Fig. 4D, lane 2), suggesting that even though co-IP experiments tend to have some artifacts, the L-FABP interaction with PPARα seems specific. These data also suggest that the presence of palmitic acid does not inhibit or alter L-FABP interaction with PPARα.
Fig. 4.
Co-IP of native PPARα and L-FABP proteins from liver homogenates. A: Immunoprecipitates of proteins from wild-type (L-FABP+/+, lanes 1–3) and L-FABP gene ablated (−/−, lanes 4–6) mice liver homogenates were obtained with anti-L-FABP in the absence (lanes 1 and 4) or presence (lanes 2 and 5) of palmitic acid. Lanes 3 and 6 are of 10 μg of liver homogenate. Western blots are shown for PPARα and L-FABP. B: Immunoprecipitates of proteins from wild-type (L-FABP+/+, lanes 1–3) and L-FABP gene ablated (−/−, lanes 4–6) mice liver homogenates were obtained with anti-PPARα in the absence (lanes 1 and 4) or presence (lanes 2 and 5) of palmitic acid. Lanes 3 and 6 are of 10 μg of liver homogenate. Western blots are shown for PPARα and L-FABP. C: To determine specificity of the co-IP, immunoprecipitates of proteins from wild-type (L-FABP+/+) mice livers obtained with anti-PPARα (lane 1) or anti L-FABP (lane 2) were examined by Western blot for the presence of GR and SREBP-1. As a positive control for the Western blot, GR and SREBP-1 proteins were detected prominently in liver homogenates (lane 3). D: Wild-type (L-FABP+/+) liver homogenates were immunoprecipitated with antibodies to GR (lane 1) and SREBP-1 (lane 2), followed by Western blotting to probe for coimmunoprecipitated L-FABP. Again, Western blotting of L-FABP (lane 3) in the liver homogenate was used as a positive control.
Immunoelectron microscopy of fixed hepatocytes: direct L-FABP and PPARα interaction
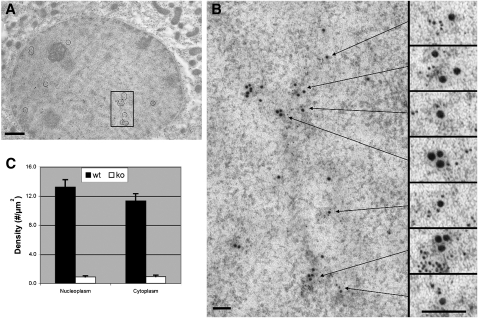
For L-FABP expression to influence PPARα transcriptional activity in vivo, L-FABP would need to directly interact with PPARα in the nucleus. To determine if L-FABP and PPARα are in sufficiently close physical proximity for direct interaction within hepatic nuclei, hepatocytes from L-FABP+/+ and L-FABP−/− mice were examined by double immunolabeling electron microscopy. These studies revealed two important observations: antigenic sites of L-FABP (6 nm gold particle size) were observed in nuclei of cultured mouse primary hepatocytes from L-FABP+/+ (Fig. 5A, small gold particles, shown as small black dots located within the circled regions of the nucleus) but not in L-FABP−/− (not shown) mice. Likewise, L-FABP (6 nm gold particles) was also detected outside the nucleus in the cytoplasm (Fig. 5A, small gold particles, shown as small black dots located outside of the nucleus). Examination of multiple hepatocytes (n = 21) revealed that the anti–L-FABP 6 nm gold particle density in the nucleoplasm was not significantly different from that in the cytoplasm (Fig. 5C). Control experiments for antibody specificity revealed a near absence of immunogold anti–L-FABP staining on hepatocytes derived from L-FABP−/− mice (not shown). Nonspecific staining of L-FABP, represented by the immunogold labeled hepatocytes from the L-FABP−/− mice, accounted for 7% and 8% of the particle density in nucleoplasm and cytoplasm, respectively (Fig. 5C). Importantly, double immunogold electron microscopy of antigenic sites of L-FABP (6 nm gold particle size) and PPARα (15 nm gold particle size) showed that the L-FABP in nuclei of hepatocytes from L-FABP+/+ was significantly colocalized with PPARα as shown in the boxed area (Fig. 5A), which was further examined under high magnification (Fig. 5B). Seven regions of colocalizing L-FABP (6 nm gold particle size) with PPARα (15 nm gold particle size) were subsequently magnified 2.5× to more effectively visualize the two sizes of gold particles (Fig. 5B, boxes on right). The statistical significance of the colocalization of L-FABP with PPARα and SREBP-1 (control) was further examined and revealed that the greatest probability of finding L-FABP was within 0–10 nm from PPARα, in contrast to a null probability of finding L-FABP in close proximity of SREBP-1 (Table 2). Using closest pairs, a mean separation distance from edge to edge between the colocalizing gold particles was within 40 ± 5 Å (n = 21). Small amounts of PPARα also appeared in the cytoplasm colocalized with L-FABP (Fig. 5A, areas outside of the nucleus where large and small gold particles are in close proximity, shown as large and small black dots, respectively). Subsequently, control experiments for antibody specificity revealed a near absence of immunogold anti–L-FABP staining on hepatocytes derived from L-FABP−/− mice (not shown). These immunoelectron microscopic results revealed significant nuclear localization of L-FABP and the clustering of L-FABP with PPARα in sufficiently close proximity (0–100 Å) for direct interaction.
Fig. 5.
Double immunogold labeling and electron microscopy of L-FABP and PPARα binding sites in mouse liver. Antigenic sites of L-FABP were labeled with 6 nm gold particles and antigenic sites of PPARα were labeled with 15 nm gold. A: Multiple sites of colocalization in a control mouse hepatocyte were marked by circles in this low magnification image. This image focuses on the hepatic nucleus (darker circle which comprises most of the image) and some surrounding cytoplasm. B: The boxed region of the nucleus in (A) was enlarged and seven individual sites of colocalization were subsequently enlarged 2.5× to better visualize the localization of the 6 nm gold particles of L-FABP in proximity with the 15 nm gold particles of PPARα. C: Bar graph of the antibody-L-FABP labeling particle density in nucleoplasmic and cytoplasmic regions of hepatocytes obtained from L-FABP+/+ and L-FABP−/− mice. Bars = 1.0 μm in (A); 100 nm in (B). The images (A) and (B) were modified in Adobe Photoshop to adjust the brightness and contrast ( Curves ), remove random noise (“Despeckle”), and clarify positions of gold particles (“Unsharp Mask”).
TABLE 2.
Statistical significance of colocalization
| Label | 0–10 nm | 10–20 nm | 20–30 nm | 30–40 nm | 40–50 nm | 50–100 nm | 100–200 nm |
|---|---|---|---|---|---|---|---|
| α-L-FABP, α-PPARα | P ≤ 0.05 | P ≤ 0.01 | P ≤ 0.01 | P ≤ 0.01 | P ≤ 0.01 | NS | NS |
| α-L-FABP, α-SREBP-1 | NS | NS | NS | NS | NS | NS | NS |
Statistical analysis demonstrated significant numbers of L-FABP and PPARα within a distance of 10 nm of each other but found no significant colocalization of LFABP and SREBP-1.
L-FABP, liver fatty acid binding protein; NS, not significant; PPARα, peroxisome proliferator-activated receptor-α; SREBP-1, sterol regulatory element-binding protein-1.
Immunofluorescence confocal microscopy and FRET in fixed hepatocytes: direct interaction between L-FABP and PPARα
Although confocal imaging of transfected cells overexpressing L-FABP detected L-FABP in nuclei where it colocalized with PPARα (12), the resolution of confocal microscopy alone is insufficient to determine if L-FABP and PPARα are in sufficiently close physical proximity for direct interaction. Therefore to further confirm the interaction in nuclei between L-FABP and PPARα indicated by electron microscopy, hepatocytes were examined by double immunolabeling fluorescence confocal microscopy and FRET. Since FRET efficiency varies inversely as the sixth root of intermolecular distance, L-FABP and PPARα must be in close proximity (i.e., 0–100Å) for efficient FRET to occur.
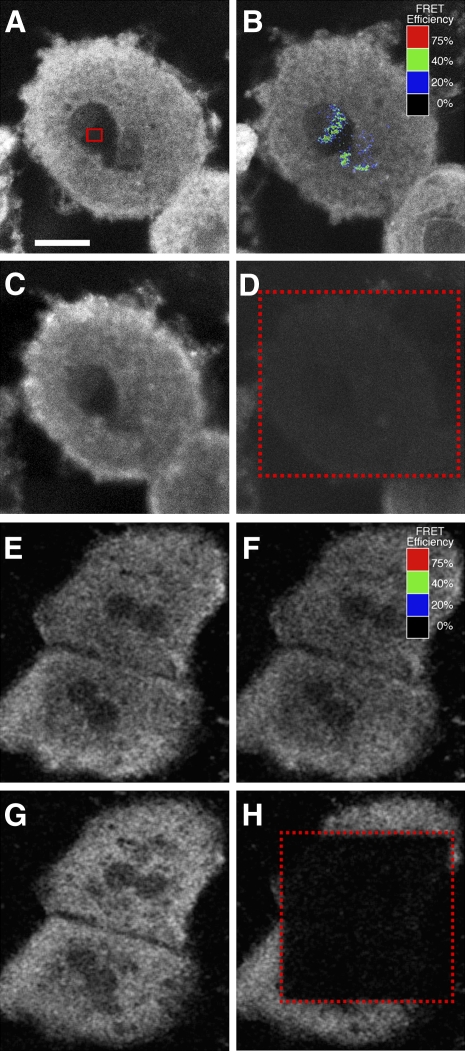
The data revealed two important observations. First, Cy3-anti-L-FABP detected L-FABP in both the nuclei and cytoplasm as shown by a representative image of Cy3-anti-L-FABP in cultured mouse primary hepatocytes (Fig. 6A, white pixels) upon excitation of the Cy3 dye. Quantitative analysis of multiple images showed that the ratio of Cy3-anti-L-FABP intensity in the cytoplasm/nucleoplasm was 1.64 ± 0.14 (n = 5). Second, double immunolabeling FRET confocal microscopy showed significant FRET between Cy3-anti-L-FABP and Cy5-anti-PPARα in the nucleus (Fig. 6B, blue/green pixels); with the area of highest energy transfer shown within the red box in Fig. 6A. In this experiment, FRET was performed by measuring the increase in intensity of Cy3-anti-L-FABP in the nucleus after photobleaching the acceptor Cy5-anti-PPARα (Fig. 6B) compared with the Cy3-anti-L-FABP intensity before photobleaching the acceptor Cy5-anti-PPARα (Fig. 6A). Cy5-anti-PPARα emission upon excitation of the Cy5 dye at 647 nm is shown as a control to ensure that the bleach pulse was sufficiently powerful to completely bleach the Cy5-anti-PPARα emission (Fig. 6D, area inside the dotted box) compared with the Cy5-anti-PPARα intensity before photobleaching the Cy5-anti-PPARα acceptor (Fig. 6C). Quantitative analysis of multiple regions of highest FRET in nuclei showed that the closest intermolecular distance between Cy3-anti-L-FABP and Cy5-anti-PPARα was 49.2 ± 2.8 Å (n = 26).
Fig. 6.
Double immunofluorescence labeling and FRET confocal microscopy of L-FABP and PPARα interaction in cultured primary hepatocytes. Cultured primary hepatocytes were fixed and double immunostained with Cy3-anti-L-FABP and either Cy5-anti-PPARα (A–D) or Cy5-anti-SREBP-1 (E–H) for FRET confocal microscopy as described in Materials and Methods. A: Cy3-anti-L-FABP fluorescence emission upon excitation at 488 nm before photobleaching of Cy5-anti-PPARα. The red box indicates the region within the nucleus where the greatest amount of energy transfer was detected. B: Cy3-anti-L-FABP fluorescence emission upon excitation at 488 nm after photobleaching of Cy5-anti-PPARα at 647 nm. The colored pixels overlaying the cell nucleus indicate the level of FRET efficiency. The scale correlates overlay colors to the level of FRET efficiency. C: Cy5-anti-PPARα fluorescence emission upon excitation at 647 nm before photobleaching at 647 nm. D: Cy5-anti-PPARα fluorescence emission upon excitation at 647 nm after photobleaching at 647 nm. The red rectangle indicates limits of the region bleached by 647 nm laser irradiation. E: Cy3-anti-L-FABP fluorescence emission upon excitation at 488 nm before photobleaching of Cy5-anti-SREBP-1. F: Cy3-anti-L-FABP fluorescence emission upon excitation at 488 nm after photobleaching of Cy5-anti-SREBP-1 at 647 nm. The colored pixels (black) overlaying the cell nucleus indicate the level of FRET efficiency. The scale correlates overlay colors to the level of FRET efficiency. G: Cy5-anti-SREBP-1 fluorescence emission upon excitation at 647 nm before photobleaching at 647 nm. H: Cy5-anti-SREBP-1 fluorescence emission upon excitation at 647 nm after photobleaching at 647 nm. The red rectangle indicates limits of the region bleached by 647 nm laser irradiation. Bar = 10 μm.
Although hepatocytes stained with Cy3-anti-L-FABP and Cy5-anti-SREBP-1 show similar distributions of L-FABP between the cytoplasm and nucleoplasm (Fig. 6E), very few pixels colocalize in the nucleus (data not shown), and no significant FRET is noted within nuclei (Fig. 6F). Images of the Cy5-anti-SREBP-1 show that the bleach pulse was sufficiently powerful to completely bleach the Cy5-anti-SREBP-1 emission (Fig. 6H, area inside the dotted box) compared with the intensity before photobleaching the Cy5-anti-SREBP-1 (Fig. 6G). Quantitative analysis of multiple nuclear regions showing the highest FRET had a maximal energy transfer of less than 1%, and no intermolecular distance between Cy3-anti-L-FABP and Cy5-anti-SREBP-1 could be calculated. In summary, the immunofluorescence FRET confocal imaging studies were overall consistent with the results above involving electron microscopy; i.e., L-FABP was significantly present in nuclei where it clustered in sufficiently close proximity (within a few Å) for direct interaction between L-FABP and PPARα.
DISCUSSION
It has been postulated that L-FABP may play a longer-acting role in regulating hepatic fatty acid oxidation/metabolism by participating in an intricate interplay between both cytoplasmic (e.g., L-FABP) and nuclear receptor (e.g., PPARα) proteins that bind and are activated by LCFA and LCFA-CoA (5, 6, 8, 13). In this scheme, it is hypothesized that L-FABP binds and cotransports the bound ligands into the nucleus for direct interaction with PPARα to induce PPARα transcription of the gene encoding L-FABP itself (i.e., coordinated regulation) and numerous genes encoding enzymes/proteins involved in LCFA oxidation and gluconeogenesis. While appealing, this hypothesis is based largely on assays performed in vitro with transfected cells overexpressing L-FABP in culture. The present work provides studies with purified recombinant L-FABP and PPARα, fluorescently-labeled L-FABP and PPARα proteins, and double immunolabeling imaging experiments in hepatocytes from wild-type L-FABP (+/+) and gene ablated L-FABP (−/−) mice that yield significant new mechanistic insights into this evolving hypothesis.
First, L-FABP and PPARα were shown to directly interact in vitro. These two proteins were each able to pull down the other protein in pure protein co-immunoprecipitation studies. CD and fluorescence spectroscopy of recombinant proteins showed that L-FABP interaction with PPARα elicited small but significant alterations in the conformation of both proteins. Moreover, in vitro FRET studies showed that PPARα bound L-FAPB with high affinity (Kd = 156.5 ± 18.1 nM) and in close molecular proximity (average intermolecular distance of 52 ± 1Å). Furthermore, CD and co-IP results were unaltered by the presence of palmitic acid, a LCFA known to interact with L-FABP (31) but not PPARα (6, 7). This suggests that structural studies regarding the interaction of L-FABP with PPARα are physiologically relevant even in the absence of endogenous ligands. However, the effect of these interactions in the presence of ligands bound with high affinity by both proteins remains to be elucidated.
Second, double immunogold electron microscopy and double immunofluorescence confocal FRET microscopy detected L-FABP in nuclei at relatively high amounts as shown by cytoplasm-to-nucleus ratios between 1 and 1.6. While the mechanism in which L-FABP distributes into the nucleus has not yet been resolved, either passive diffusion or bidirectional active transport may contribute. Since the hydrodynamic diameter of L-FABP (14 kDa) is only about 36 Å (31, 34) and other small proteins (e.g., cytochrome C, 13 kDa) diffuse freely through the nuclear pores (90 Å diameter), L-FABP is sufficiently small to passively diffuse through nuclear pores. Passive distribution of L-FABP between the nucleus and cytoplasm is also supported by findings with transfected L-cell fibroblasts overexpressing L-FABP (12). Although L-FABP levels were from 5- to 10-fold lower in transfected L-cells overexpressing L-FABP than in liver hepatocytes, the proportion of nuclear-to-cytoplasmic L-FABP levels were again nearly the same as in hepatocytes.
Third, L-FABP and PPARα interact in vivo and are in close molecular proximity in hepatocyte nuclei. Both L-FABP and PPARα antibodies were able to co-immunoprecipitate the respective protein from mouse liver homogenates, in both the presence and absence of palmitic acid. Double immunogold labeling electron microscopy detected significant clustering of L-FABP with PPARα in hepatocyte nuclei, with a mean separation distance of 40 ± 5 Å. This finding was further confirmed by double immunofluorescence FRET confocal microscopy, which estimated the intermolecular distance to be 49.2 ± 2.8 Å, similar to that obtained for the recombinant proteins in solution. The interaction of L-FABP with PPARα was specific, as no interaction between L-FABP and SREBP-1a was observed by any of these methods.
The results of these studies may contribute to our understanding of lipid disorders in humans. For example, human variants in the L-FABP gene exhibit elevated fasting LDL-cholesterol and triglyceride levels—traits associated with increased risk of CVD, type 2 diabetes, and metabolic syndrome (35, 36). This phenotype was exacerbated by treatment with fenofibrate (36). The genetic mutations PPARα L162V and L-FABP T94A together show a synergistic effect on the basal metabolic index in humans, suggesting that the L-FABP T94A missense mutation might influence obesity indices and increase the risk of residual hypertriglyceridemia following a lipid lowering therapy with fenofibrate (36). Finally, increased L-FABP expression is associated with insulin-dependent diabetes and gestational diabetes in humans, streptozotocin-induced diabetes or obesity in rats, and type 1 diabetes in mice (37–40).
In summary, these new in vitro and imaging experiments demonstrate a high affinity, structural molecular interaction of L-FABP with PPARα and suggest a functional role for L-FABP interaction with PPARα in LCFA metabolism. The potential importance of this interaction is underscored by studies with the closely related cellular retinoic acid binding protein-1 and -2 (CRABP-1 and CRABP-2) and retinoid X receptor (5, 41, 42). The latter studies showed that CRABP-2 (but not CRABP-1) binds to retinoid X receptor (RXR) and distributes to the nucleus. Thus, direct interaction of L-FABP with PPARα may determine L-FABP distribution to the nucleus, facilitate delivery of L-FABP bound ligand (LCFA, LCFA-CoA) into the nucleus, and potentially directly channel L-FABP bound ligand to PPARα. In support of this possibility, the intermolecular distance observed between L-FABP and PPARα (i.e., 40–50 Å) was in the same range as reported for PPARα intrinsic aromatic amino acids and bound fluorescent ligands such as trans-parinaric acid and cis-parinaroyl-CoA located deep within the binding pocket (6). Thus, L-FABP and PPARα were in sufficiently close proximity for direct interaction and transfer of L-FABP bound ligand to PPARα.
Acknowledgments
The authors appreciate the technical assistance of Ms. Aude Vespa in purifying recombinant SREBP-1a. Facilities of the Microscopy and Imaging Center at Texas A&M University were used in one of the steps of specimen preparation for electron microscopy.
Footnotes
Abbreviations:
- CD
- circular dichroism
- co-IP
- co-immunoprecipitation
- FRET
- fluorescence resonance energy transfer
- GR
- glucocorticoid receptor
- LCFA
- long chain fatty acid
- LCFA-CoA
- long chain fatty acyl CoA
- L-FABP
- liver fatty acid binding protein
- LSCM
- laser scanning confocal microscopy
- PPARα
- peroxisome proliferator activated receptor-α
- SRC-1
- steroid receptor coactivator-1
- SREBP-1
- sterol regulatory element-binding protein-1
This work was supported in part by the United States Public Health Service (USPHS), and National Institutes of Health (NIH) Grant DK-41402 (F.S. and A.K), National Research Service Award DK-066732 (H.A.H.), and K99 Award DK-77573 (H.A.H).
REFERENCES
- 1.Francis G. A., Fayard E., Picard F., Auwerx J. 2003. Nuclear receptors and the control of metabolism. Annu. Rev. Physiol. 65: 261–311 [DOI] [PubMed] [Google Scholar]
- 2.Desvergne B., Wahli W. 1999. Peroxisome proliferator activated receptors: nuclear control of metabolism. Endocr. Rev. 20: 649–688 [DOI] [PubMed] [Google Scholar]
- 3.Kersten S., Desvergne B., Wahli W. 2000. Roles of PPARs in health and disease. Nature. 405: 421–424 [DOI] [PubMed] [Google Scholar]
- 4.Green S., Wahli W. 1994. Peroxisome proliferator-activated receptors: finding the orphan a home. [Review] Mol. Cell. Endocrinol. 100: 149–153 [DOI] [PubMed] [Google Scholar]
- 5.Schroeder F., Petrescu A. D., Huang H., Atshaves B. P., McIntosh A. L., Martin G. G., Hostetler H. A., Vespa A., Landrock K., Landrock D., et al. 2008. Role of fatty acid binding proteins and long chain fatty acids in modulating nuclear receptors and gene transcription. Lipids. 43: 1–17 [DOI] [PubMed] [Google Scholar]
- 6.Hostetler H. A., Petrescu A. D., Kier A. B., Schroeder F. 2005. Peroxisome proliferator activated receptor alpha (PPARalpha) interacts with high affinity and is conformationally responsive to endogenous ligands. J. Biol. Chem. 280: 18667–18682 [DOI] [PubMed] [Google Scholar]
- 7.Lin Q., Ruuska S. E., Shaw N. S., Dong D., Noy N. 1999. Ligand selectivity of the peroxisome proliferator-activated receptor α. Biochemistry. 38: 185–190 [DOI] [PubMed] [Google Scholar]
- 8.Hostetler H. A., Kier A. B., Schroeder F. 2006. Very-long-chain and branched-chain fatty acyl CoAs are high affinity ligands for the peroxisome proliferator-activated receptor alpha (PPARalpha). Biochemistry. 45: 7669–7681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hostetler H. A., Huang H., Kier A. B., Schroeder F. 2008. Glucose directly links to lipid metabolism through high-affinity interactionwith peroxisome proliferator activated receptor-alpha. J. Biol. Chem. 283: 2246–2254 [DOI] [PubMed] [Google Scholar]
- 10.Elholm M., Dam I., Jorgenesen C., Krogsdam A-M., Holst D., Kratchamarova I., Gottlicher M., Gustafsson J. A., Berge R. K., Flatmark T., et al. 2001. Acyl CoA esters antagonize the effects of ligands on peroxisome proliferator activated receptor α conformation, DNA binding, and interaction with cofactors. J. Biol. Chem. 276: 21410–21416 [DOI] [PubMed] [Google Scholar]
- 11.Elholm M., Garras A., Neve S., Tarnehave D., Lund T. B., Skorve J., Flatmark T., Kristiansen K., Berge R. K. 2000. Long chain acyl CoA esters and acyl CoA binding protein are present in the nucleus of rat liver cells. J. Lipid Res. 41: 538–545 [PubMed] [Google Scholar]
- 12.Huang H., Starodub O., McIntosh A., Atshaves B. P., Woldegiorgis G., Kier A. B., Schroeder F. 2004. Liver fatty acid binding protein colocalizes with peroxisome proliferator receptor alpha and enhances ligand distribution to nuclei of living cells. Biochemistry. 43: 2484–2500 [DOI] [PubMed] [Google Scholar]
- 13.Wolfrum C., Borrmann C. M., Borchers T., Spener F. 2001. Fatty acids and hypolipidemic drugs regulate PPARalpha and PPARgamma gene expresion via L-FABP: a signaling path to the nucleus. Proc. Natl. Acad. Sci. USA. 98: 2323–2328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lawrence J. W., Kroll D. J., Eacho P. I. 2000. Ligand dependent interaction of hepatic fatty acid binding protein with the nucleus. J. Lipid Res. 41: 1390–1401 [PubMed] [Google Scholar]
- 15.Huang H., Starodub O., McIntosh A., Kier A. B., Schroeder F. 2002. Liver fatty acid binding protein targets fatty acids to the nucleus: real-time confocal and multiphoton fluorescence imaging in living cells. J. Biol. Chem. 277: 29139–29151 [DOI] [PubMed] [Google Scholar]
- 16.Atshaves B. P., Petrescu A., Starodub O., Roths J., Kier A. B., Schroeder F. 1999. Expression and intracellular processing of the 58 kDa sterol carrier protein 2/3-oxoacyl-CoA thiolase in transfected mouse L-cell fibroblasts. J. Lipid Res. 40: 610–622 [PubMed] [Google Scholar]
- 17.Murphy E. J., Edmondson R. D., Russell D. H., Schroeder F. 1999. Isolation and characterization of two distinct forms of liver fatty acid binding protein from the rat. Biochim. Biophys. Acta. 1436: 413–425 [DOI] [PubMed] [Google Scholar]
- 18.Forman B. M., Tontonoz P., Chen J., Brun R. P., Spiegelman B. M., Evans R. M. 1995. 15-Deoxy-delta(12,14)-prostaglandin J(2) is a ligand for the adipocyte determination factor PPARgamma. Cell. 83: 803–812 [DOI] [PubMed] [Google Scholar]
- 19.Kliewer S. A., Lenhard J. M., Willson T. M., Patel I., Morris D. C., Lehmann J. M. 1995. A prostaglandin J(2) metabolite binds peroxisome proliferator activated receptor gamma and promotes adipocyte differentiation. Cell. 83: 813–819 [DOI] [PubMed] [Google Scholar]
- 20.Yamamoto T., Shimano H., Nakagawa Y., Ide T., Yagagi N., Matsuzaka T., Nakakuki M., Takahashi A., Suzuki H., Sone H., et al. 2004. SREBP-1 interacts with HNF4alpha and interferes with PGC-1 recruitment to suppress hepatic gluconeogenic genes. J. Biol. Chem. 279: 12027–12035 [DOI] [PubMed] [Google Scholar]
- 21.Wang X., Sato R., Brown M. S., Hua X., Goldstein J. L. 1994. SREBP-1, a membrane-bound transcription factor released by sterol-regulated proteolysis. Cell. 77: 53–62 [DOI] [PubMed] [Google Scholar]
- 22.Petrescu A. D., Payne H. R., Boedeker A. L., Chao H., Hertz R., Bar-Tana J., Schroeder F., Kier A. B. 2003. Physical and functional interaction of acyl CoA binding protein (ACBP) with hepatocyte nuclear factor-4alpha (HNF4alpha). J. Biol. Chem. 278: 51813–51824 [DOI] [PubMed] [Google Scholar]
- 23.Sreerama N., Woody R. 2000. Estimation of protein secondary structure from circular dichroism spectra: comparison of CONTIN, SELCON, and DCSSTR methods with an expanded reference set. Anal. Biochem. 287: 252–260 [DOI] [PubMed] [Google Scholar]
- 24.Petrescu A. D., Huang H., Hostetler H. A., Schroeder F., Kier A. B. 2008. Structural and functional characterization of a new recombinant histidine-tagged acyl CoA binding protein (ACBP) from mouse. Protein Expr. Purif. 58: 184–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martin G. G., Danneberg H., Kumar L. S., Atshaves B. P., Erol E., Bader M., Schroeder F., Binas B. 2003. Decreased liver fatty acid binding capacity and altered liver lipid distribution in mice lacking the liver fatty acid binding protein (L-FABP) gene. J. Biol. Chem. 278: 21429–21438 [DOI] [PubMed] [Google Scholar]
- 26.Atshaves B. P., McIntosh A. L., Lyuksyutova O. I., Zipfel W. R., Webb W. W., Schroeder F. 2004. Liver fatty acid binding protein gene ablation inhibits branched-chain fatty acid metabolism in cultured primary hepatocytes. J. Biol. Chem. 279: 30954–30965 [DOI] [PubMed] [Google Scholar]
- 27.Philimonenko A. A., Janacek J., Hozak P. 2000. Statistical evaluation of colocalization patterns in immunogold labeling experiments. J. Struct. Biol. 132: 201–210 [DOI] [PubMed] [Google Scholar]
- 28.Atshaves B. P., McIntosh A. L., Payne H. R., Gallegos A. M., Landrock K., Maeda N., Kier A. B., Schroeder F. 2007. Sterol carrier protein-2/sterol carrier protein-x gene ablation alters lipid raft domains in primary cultured mouse hepatocytes. J. Lipid Res. 48: 2193–2211 [DOI] [PubMed] [Google Scholar]
- 29.Stepensky D.2007. FRETcalc plugin for calculation of FRET in non-continuous intracellular compartments. Biochem. Biophys. Res. Commun. 359: 752–758 [DOI] [PubMed] [Google Scholar]
- 30.Wouters F. S., Bastiaens P. I., Wirtz K. W., Jovin T. M. 1998. FRET microscopy demonstrates molecular association of non-specific lipid transfer protein (nsL-TP) with fatty acid oxidation enzymes. EMBO J. 17: 7179–7189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Frolov A., Cho T. H., Murphy E. J., Schroeder F. 1997. Isoforms of rat liver fatty acid binding protein differ in structure and affinity for fatty acids and fatty acyl CoAs. Biochemistry. 36: 6545–6555 [DOI] [PubMed] [Google Scholar]
- 32.Martin G. G., Hostetler H. A., Tichy S. E., Russell D. H., Berg J. M., Woldegiorgis G., Spencer T. A., Ball J. A., Kier A. B., Schroeder F. 2008. Structure and function of the sterol carrier protein-2 (SCP-2) N-terminal pre-sequence. Biochemistry 47: 5915–5934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lakowicz J. R.2006. Energy transfer. Lakowicz J. R., editor Springer Science, New York: 443–475 [Google Scholar]
- 34.Thompson J., Winter N., Terwey D., Bratt J., Banaszak L. 1997. The crystal structure of the liver fatty acid-binding protein. J. Biol. Chem. 272: 7140–7150 [DOI] [PubMed] [Google Scholar]
- 35.Fisher E., Weikert C., Klapper M., Lindner I., Mohlig M., Spranger J., Boeing H., Schrezenmeir J., Doring F. 2007. L-FABP T94A is associated with fasting triglycerides and LDL-cholesterol in women. Mol. Genet. Metab. 91: 278–284 [DOI] [PubMed] [Google Scholar]
- 36.Brouillette C., Bose Y., Perusse L., Gaudet D., Vohl M-C. 2004. Effect of liver fatty acid binding protein (FABP) T94A missense mutation on plasma lipoprotein responsiveness to treatment with fenofibrate. J. Hum. Genet. 49: 424–432 [DOI] [PubMed] [Google Scholar]
- 37.Magnusson A. L., Waterman I. J., Jansson T., Powell T. L. 2004. Triglyceride hydrolase activities and expression of fatty acid binding proteins in the human placenta in pregnancies complicated by intrauterine growth restriction and diabetes. J. Clin. Endocrinol. Metab. 89: 4607–4614 [DOI] [PubMed] [Google Scholar]
- 38.Engels W., van Bilsen M., Wolffenbuttel B. H. R., Van der Vusse G. J., Glatz J. F. 1999. Cytochrome P450, peroxisome proliferation, and cytoplasmic fatty acid binding protein content in liver, heart, and kidney of the diabetic rat. Mol. Cell. Biochem. 192: 53–61 [PubMed] [Google Scholar]
- 39.Paulussen R. J. A., Verkamp J. H. 1990. Intracellular fatty acid binding proteins: characteristics and function. In Subcellular Biochemistry: Intracellular Transfer of Lipid Molecules. Vol. 16 Hilderson H. J., editor Plenum Press, New York: [DOI] [PubMed] [Google Scholar]
- 40.Kamijo-Ikemori A., Sugaya T., Sekizuka A., Hirata K., Kimura K. 2009. Amelioration of diabetic tubulointerstitial damage in liver type fatty acid binding protein transgenic mice. Nephrol. Dial. Transplant. 24: 788–800 [DOI] [PubMed] [Google Scholar]
- 41.Budhu A. S., Noy N. 2002. Direct channeling of retinoic acid between cellular retinoic acid binding protein II and retinoic acid receptor sensitizes mammary carcinoma cells to retinoic acid induced growth arrest. Mol. Cell. Biol. 22: 2632–2641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tan N-S., Shaw N. S., Vinckenbosch N., Liu P., Yasmin R., Desvergne B., Wahli W., Noy N. 2002. Selective cooperation between fatty acid binding proteins and peroxisome proliferator activated receptors in regulating transcription. Mol. Cell. Biol. 22: 5114–5127 [DOI] [PMC free article] [PubMed] [Google Scholar]