Abstract
The purpose of this study is to develop a software for the extraction of the hippocampus and surrounding medial temporal lobe (MTL) regions from T1-weighted magnetic resonance (MR) images with no interactive input from the user, to introduce a novel statistical indicator, computed on the intensities in the automatically extracted MTL regions, which measures atrophy, and to evaluate the accuracy of the newly developed intensity-based measure of MTL atrophy to (a) distinguish between patients with Alzheimer disease (AD), patients with amnestic mild cognitive impairment (aMCI), and elderly controls by using established criteria for patients with AD and aMCI as the reference standard and (b) infer about the clinical outcome of aMCI patients. For the development of the software, the study included 61 patients with mild AD (17 men, 44 women; mean age±standard deviation (SD), 75.8 years±7.8; Mini Mental State Examination (MMSE) score, 24.1±3.1), 42 patients with aMCI (11 men, 31 women; mean age±SD, 75.2 years±4.9; MMSE score, 27.9±1.9), and 30 elderly healthy controls (10 men, 20 women; mean age±SD, 74.7 years±5.2; MMSE score, 29.1±0.8). For the evaluation of the statistical indicator, 150 patients with mild AD (62 men, 88 women; mean age±SD, 76.3 years±5.8; MMSE score, 23.2±4.1), 247 patients with aMCI (143 men, 104 women; mean age±SD, 75.3 years±6.7; MMSE score, 27.0±1.8), and 135 elderly healthy controls (61 men, 74 women; mean age±SD, 76.4 years±6.1). Fifty aMCI patients were evaluated every 6 months over a 3 year period to assess conversion to AD. For each participant, two subimages of the MTL regions were automatically extracted from T1-weighted MR images with high spatial resolution. An intensity-based MTL atrophy measure was found to separate control, MCI, and AD cohorts. Group differences wereassessed by using two-sample t test. Individual classification was analyzed by using receiver operating characteristic (ROC) curves. Compared to controls, significant differences in the intensity-based MTL atrophy measure were detected in both groups of patients (AD vs controls, 0.28±0.03 vs 0.34±0.03, P<0.001; aMCI vs controls, 0.31±0.03 vs 0.34±0.03, P<0.001). Moreover, the subgroup of aMCI converters was significantly different from controls (0.27±0.034 vs 0.34±0.03, P<0.001). Regarding the ROC curve for intergroup discrimination, the area under the curve was 0.863 for AD patients vs controls, 0.746 for all aMCI patients vs controls, and 0.880 for aMCI converters vs controls. With specificity set at 85%, the sensitivity was 74% for AD vs controls, 45% for aMCI vs controls, and 83% for aMCI converters vs controls. The automated analysis of MTL atrophy in the segmented volume is applied to the early assessment of AD, leading to the discrimination of aMCI converters with an average 3 year follow-up. This procedure can provide additional useful information in the early diagnosis of AD.
Keywords: magnetic resonance imaging, image analysis, Alzheimer disease, hippocampus
INTRODUCTION
Early and accurate diagnosis of Alzheimer disease (AD) is challenging. In recent years, the early clinical signs of AD have been extensively investigated, leading to the concept of amnestic mild cognitive impairment (aMCI), an intermediate cognitive state between normal aging and dementia.1, 2, 3, 4, 5 aMCI patients experience memory loss to a greater extent than expected for their age, and they progress to a diagnosis of AD at a faster rate than controls.1, 2
A challenge for modern neuroimaging is to help in the diagnosis of early AD, particularly in aMCI patients. Three-dimensional (3D) magnetic resonance (MR) imaging with high spatial resolution allows visualization of subtle anatomic changes and thus can help detect brain atrophy in the initial stages of the disease.6 For this reason, sensitive neuroimaging measures have been investigated to quantify brain structural changes in early AD which are automated enough to permit large-scale studies of the disease and the factors that affect it. To date, these studies have particularly focused on assessing atrophy of medial temporal lobe (MTL) structures, including the hippocampus (for a review of this topic see Refs. 7, 8, 9, 10, 11).
Methods to assess hippocampal atrophy have largely been based on volumetric measurements,9, 12, 13 on mapping the spatial distribution of atrophy in 3D scans,14, 15, 16, 17, 18 or on visual rating scales.19 Volumetric measurements typically rely on manual outlining of the MTL structures on (serial) MR images, which is time consuming and prone to inter-rater and intrarater variabilities. Thus, large-scale studies of AD-related hippocampal atrophy are often impractical.20 Visual rating scales, although simple to use and suitable for a clinical application, were not designed to detect atrophy progression on serial imaging; their quantized nature makes them insensitive to change over time.21
To accelerate and spread epidemiological studies and clinical trials, some automated systems have been proposed for hippocampal segmentation,22, 23, 24, 25, 26, 27, 28 but none is yet widely used29 due to the high computational burden,30 unsatisfactory results,31 or poor generalization capability.24
In order to overcome these difficulties, in this work we describe the development of a simple, quick, and operator-independent method to extract two fixed size (30×70×30 mm3), parallelepiped-shaped subimages from a MR image. These subimages contain both the hippocampus and the perihippocampal region; in the following they are denoted as hippocampal boxes (HBs).
From the automatically extracted HBs we derive a novel, intensity-based, statistical indicator, which carefully measures the MTL atrophy and is able to distinguish between patients with AD, patients with aMCI, and elderly controls and also between converters and nonconverters to AD within the aMCI population with reasonably good accuracy.
DEVELOPMENT OF THE SOFTWARE
Subjects
At the development stage the study included 61 patients with mild AD (17 men, 44 women; mean age±standard deviation (SD), 75.8 years±7.8; Mini Mental State Examination (MMSE) score, 24.1±3.1), 42 patients with aMCI (11 men, 31 women; mean age±SD, 75.2 years±4.9; MMSE score, 27.9±1.9), and 30 elderly healthy controls (10 men, 20 women; mean age±SD, 74.7 years±5.2; MMSE score, 29.1±0.8) (see Table 1).
Table 1.
Ensemble properties of our “development” subjects. The error is one standard deviation.
| AD | MCI | Controls | |
|---|---|---|---|
| No. of subjects | 61 (17 men, 44 women) | 42 (11 men, 31 women) | 30 (10 men, 20 women) |
| Age (years) | 75.8±7.8 | 75.2±4.9 | 74.7±5.2 |
| MMSE score | 24.1±3.1 | 27.9±1.9 | 29.1±0.8 |
The diagnosis of aMCI was made according to the criteria of Petersen et al.,2 while the diagnosis of AD was made according to the NINCDS-ADRDA (Ref. 32) and the DSM-IV criteria.
The presence of dementia was evaluated on the basis of clinical interview with the patient and caregiver, questionnaires for the activities of daily living (ADLs),33 instrumental ADL (IADL),34 and clinical dementia rating (CDR) scale (the result was 0.5 in all aMCI patients and 1.0 in all AD patients). General cognition was assessed by means of MMSE.35
All patients underwent a standard battery of blood count, blood chemical examinations, and urinalysis, according to the commonly followed rules in order to exclude secondary causes of cognitive impairment. The presence of analphabetism, major vision disturbances, psychiatric illnesses, epilepsy, major head trauma, Parkinsonism, previous stroke or TIA, and brain masses was another exclusion criterion. A mild depressive trait, as ascertained by the 15-item geriatric depression scale (GDS), was not an exclusion criterion. Neuropsychiatric symptoms were assessed by interviewing the informant with the neuropsychiatric inventory (NPI).36 Patients scoring higher than 0 on the delusion and the hallucination NPI items were excluded.
MR imaging was performed in all patients by means of a 1.5 T equipment. Only patients with evidence of major stroke were excluded, while white matter (WM) hyperintensities, leukoaraiosis, and lacunae were not exclusion criteria.
The follow-up of the patients began with a clinical examination, also comprehensive of MMSE, ADL, and IADL questionnaires and CDR, and this was repeated every 6 months. A follow-up time of at least 1 year was available for all patients.
The 30 control subjects were recruited among a group of healthy volunteers attending university courses dedicated to elderly people. They all gave their informed consent. Their healthy condition was carefully checked by means of general medical history and clinical examination, and the same exclusion criteria as for patients were applied, with the exception of cognitive complaints. MMSE was performed, and only subjects with a normal score (i.e., ≥26) were considered for this study. Moreover, only subjects with a CDR of 0 were included. The enrolled controls underwent brain MR imaging and the same neuropsychological battery as the patients. The protocol received the approval of the local ethics committee.
Image acquisition
T1-TFE volumetric MR imaging was performed in all patients using 1.5 T equipment (Gyroscan Intera, Philips Medical Systems, Best, The Netherlands) to acquire a sagittal sequence with the following parameters: TRs of 8.7 ms, TEs of 4.1 ms, flip of 8, FOVs of 256 mm, matrix of 256×256, 150 sagittal slices, and voxel size of 0.98×0.98×1.6 mm3. All images were resampled to make them isotropic with voxel size of 1×1×1 mm3.
Extraction of the hippocampal boxes
The extraction method relies on the fact that the gray level contrast displayed by the hippocampal formation and adjacent structures is unique all over the brain. Therefore, a procedure can be developed to identify the hippocampal region unambiguously. Neuroanatomical considerations suggested the size of a HB as a 30×70×30 mm3 parallelepiped-shaped box (sizes of right-to-left, posterior-to-anterior, and inferior-to-superior directions, respectively). The extraction of the 266 HBs (133 right and 133 left) was performed with an automatic procedure, that required minimal interactive intervention. Here we illustrate only the process for the extraction of the right HBs, the procedure for the extraction of the left ones being the same.
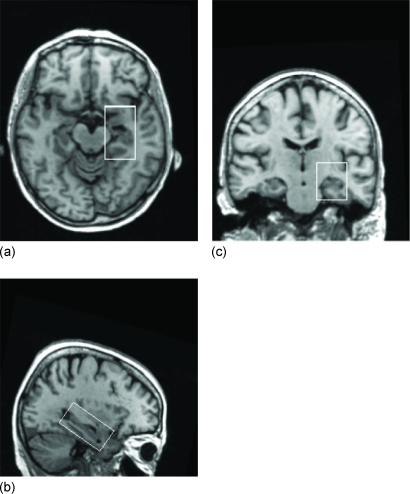
First, all MR scans were labeled and denoted as MR(i) (i=1,…,133). Then all images were spatially normalized to stereotactic space (ICBM152) via a 12-degrees-of-freedom affine transformation,37 which normalizes the brains in terms of dimensions, position, and spatial orientation. Consequently, all hippocampi share similar positions and orientations. Three slices cut from a 3D MR image after spatial normalization and the outline of the hippocampal ROI are shown in Fig. 1.
Figure 1.
(a) Axial, (b) sagittal, (c) and coronal views of an image after alignment with the ICBM152 template. On the three slices an outline of the right hippocampal box is also shown.
After spatial normalization, the first HB was manually extracted by an expert reader from MR(1), the scan of a healthy control displaying minimal atrophy. Particular care was applied in the positioning of the box boundaries in order to place the hippocampal formation in the inner portion of the box. The next step of the procedure consists in extracting the second HB from the remaining 132 MR images.
The extraction procedure is based on the registration of the first HB (the fixed image) onto the remaining 132 images (the moving images) via a six-degrees-of-freedom rigid transformation (three translational and three rotational degrees of freedom).
A definition of distance between two HBs drives the registration procedure in that it provides a measure of how well the transformed moving image matches the fixed image. In this way a quantitative criterion is assigned for finding the optimal values of the transformation parameters. We adopted a definition of distance based on the normalized correlation coefficient C. Assigning the HBs A and B, each one consisting of N voxels (N=63 000 in our case), the corresponding C is given by
| (1) |
where voxel intensities Aα and Bα of HBs A and B, respectively, are labeled by a single index (Greek letter) following lexicographic ordering, and the average intensity is given by
| (2) |
where I=A,B. From this quantification of similarity, one derives the following definition of distance between A and B:
| (3) |
This distance is scale and shift invariant and it produces a cost function with sharp peaks and well defined minima. It has a relatively small capture radius,38 but this fact does not represent a severe limitation in our case because all images are aligned in the same stereotactic space and thus the search for the hippocampal formations requires the exploration of a small parameter space.
The success of the registration of each moving image onto the fixed image is quantified by the minimum reached in distance values, δ. With a moderate computational effort, one could extract all the 132 remaining right HBs by using the first manually defined HB alone, but the quality of the results is not homogeneously good. In fact the fixed image is successful in extracting the HBs which are not too dissimilar from it. However, due to the ample morphological variability contained in the population of MR images, some HBs exist which are unsatisfactorily extracted or not found at all.
Therefore, a more complete approach is required. The population of the remaining 132 MR images is registered onto the fixed image, i.e., the HB extracted from MR(1). Thus, for each given value of index j (2≤j≤133) this operation produces the value of the score, δ1,j, and of the six geometrical parameters, three translations and three rotation angles. Such values are stored in the first row of seven strictly upper triangular matrices. We emphasize that no actual HB extraction is performed at this stage.
On the basis of the presently available score list (the first row of matrix δ), the second box is extracted from MR(j*) where j* is the index of the minimum (not vanishing) value of δ1,j. In our application we obtain j*=13. Now the extraction of the new HB is really performed by using the corresponding values of the geometrical parameters (x1,13,y1,13,z1,13,…).
Once the second HB is available, the remaining 131 moving images are registered onto this new fixed image, and a new set of scores and geometrical parameters are obtained and stored in the second row of the seven matrices. The third extracted HB is selected from MR(j**) where now j** is given by the index of the minimum (not vanishing) value of δ1,j and δ2,j. In this search for the minimum score, the entries corresponding to the already extracted HBs must be skipped.
The procedure for the progressive extraction of all HBs follows this scheme and the extension to an increasing number of HB examples is obvious. The procedure stops once the whole sample of 133 images has been processed and the 133 right HBs have been obtained.
Except for the extraction of the first HB, the whole process runs automatically, without any manual intervention, and no appreciable drift affecting hippocampus orientation or positioning in the HBs is noticeable during the extraction process. Visual inspection of the whole set of 133 HBs shows that the level of spatial registration of similar anatomical structures is very high. Such stability is not surprising if one considers the way the whole procedure works. At the beginning, the early extractions exhaust the set of the HBs which are very similar to the manually defined HB. Then, the procedure starts extracting HBs which are progressively different from the first ones, but diversity creeps into the growing HB database very slowly, thanks to the relevant size of the population of the available MR images. Thus, the orientation and position of the essential geometrical features of the hippocampal formation are preserved during the whole process of HB extraction.
The same procedure was applied to the left hippocampi. A first example of a left HB is manually generated and, then, the whole process just described is run on the left side of the brain. The result consists in the generation of the matrices of scores and of geometrical parameters and in the extraction of the 133 left HBs.
Selection of templates
The procedure described in Sec. 2C to generate the 266 HBs is rather demanding and it is unreasonable to run over it again for extracting the two HBs of any new MR image. In this section we show that the extraction can be successfully performed by a smaller number of properly chosen HBs, in the following denoted as HB templates (HBTs).
The basic idea of the HBT selection process is to create groups of HBs, or clusters, in such a way that the HBs in the same cluster are near and the HBs belonging to different clusters are far. In general, let n be the number of the available HBs (n=133 in our case). Here we denote by di,j the distance between HB(i) and HB(j) as defined in Eq. 3. All di,j are known and can be considered as the entries of a symmetric n×n matrix, with vanishing diagonal elements. Thus, each HB can be considered as a point belonging to an N-dimensional space (N=63 000) and whose distances from all other HBs are known.
The classification of the n HBs in homogeneous clusters is performed by means of the k-means algorithm.39 This algorithm tries to find a partition of the whole set of data into k clusters that are as compact and well separated as possible. Each cluster is defined by its members and by its centroid, i.e., the HB having a minimal sum of distances from all members of the cluster.
For a given value of k, the algorithm needs the initial centroid position as input. These are fixed selecting k randomly chosen HBs among the whole population of n HBs (k⪡n). Since the final result depends on the initial choice, the algorithm is repeated ten times with different initial centroid selections, and the best result is kept. The required set of k HBTs is represented by the centroids of the optimal partition.
With the purpose of finding the appropriate value for the number k of HBTs, some tests were performed by increasing the number of clusters from k=3 up to k=10. The quality of the eight solutions can be evaluated in a quantitative way by means of the corresponding average silhouette values. For a k-clusters partition, the silhouette value for point i is defined as
| (4) |
where ai is the average distance of point i from the other points in its cluster and bi,l is the average distance of point i from the points belonging to cluster l. This quantity is a measure of how similar that point is to points in its own cluster compared to the points in other clusters. Its value ranges from −1 to +1 and is given by
| (5) |
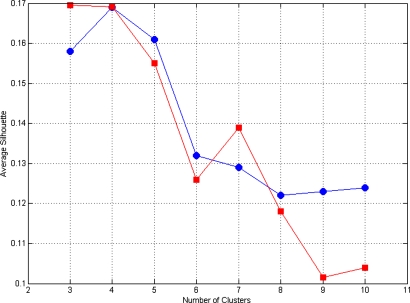
where the sum is over the whole population of n points. The plot of for k=3 to k=10 is shown in Fig. 2 and indicates that average silhouette values begin to decrease for k≥6.
Figure 2.
Average silhouette for k=3 to k=10 for right (circles) and left hippocampi (squares).
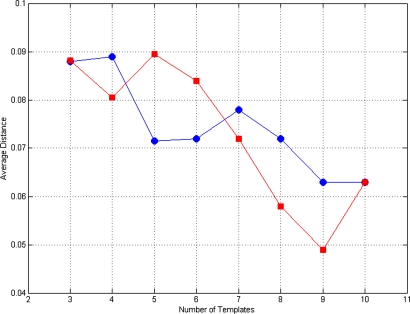
The main objective of the above illustrated clustering procedure consists in selecting a small number of k HBTs as representative of the ample morphological variability contained in the whole MR data set and, finally, in finding an appropriate value for k. Thus, the performance of such small sets of HBTs in extracting all the 133 right HBs was evaluated. The test consisted of extracting all HBs from all MR images by means of every set of k HBTs. Each MR image was registered to all the k HBTs and the actual HB extraction was performed on the basis of the best score obtained (among the k available scores). The test was repeated for k=3–10. The procedure generated eight sets of 133 HBs to be compared to the original set of HBs extracted by means of the general procedure described in Sec. 2C. In order to evaluate the performance of the eight HBT sets in extracting all HBs, we calculated, according to Eq. 3, the average distance between the newly extracted HBs and the original ones. The result is shown in Fig. 3 and displays average distance values less than 0.1 for all values of k=3–10. Obviously such values decrease when the number k of HBTs increases.
Figure 3.
Average distance for k=3 to k=10 for right (circles) and left hippocampi (squares).
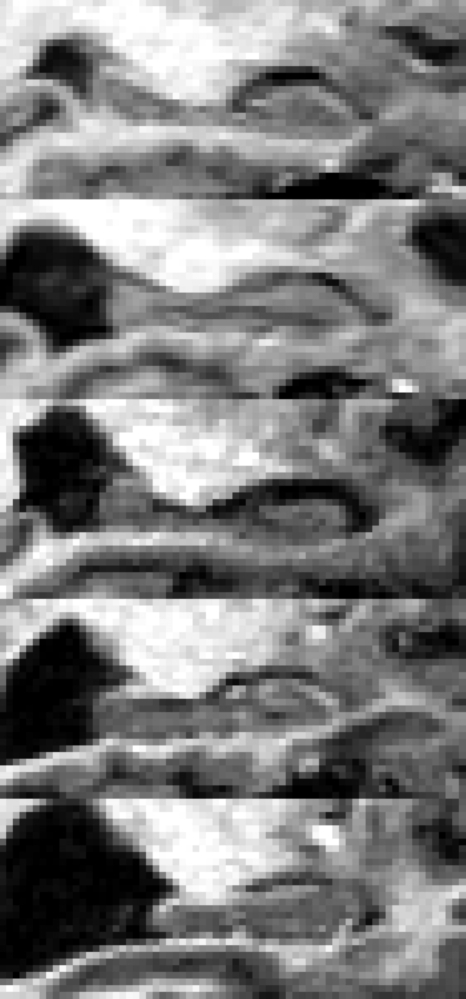
On the basis of the results obtained by the previous tests and shown in Figs. 23, k=5 was chosen for the number of HBTs giving the best trade-off between representative capability and dimension of the template dictionary. A sagittal slice of the five right templates is shown in Fig. 4. The same procedure and the same tests were applied to the left hippocampi (see Figs. 23), and also in this case the choice k=5 represents an excellent trade-off.
Figure 4.
Sagittal sections of the five right HBTs.
APPLICATION OF THE SOFTWARE
Subjects
In order to test the HBT extraction efficiency on images acquired with different scanners and∕or with different system settings we processed n=532 images, partly acquired by the National Institute of Cancer (IST) on patients clinically treated at the Clinical Neurophysiology unit in Genoa, Italy, and partly downloaded from the Alzheimer Disease Neuroimaging Initiative (ADNI) database (www.loni.ucla.edu∕ADNI).40 The ADNI was launched in 2003 by the National Institute on Aging (NIA), the National Institute of Biomedical Imaging and Bioengineering (NIBIB), the Food and Drug Administration (FDA), private pharmaceutical companies, and nonprofit organizations as a $60 million, 5 year public-private partnership. ADNI is the result of efforts of many coinvestigators from a broad range of academic institutions and private corporations, and subjects have been recruited from over 50 sites across the US and Canada. The initial goal of ADNI was to recruit 800 adults, ages 55–90, to participate in the research—approximately 200 cognitively normal older individuals to be followed for 3 years, 400 people with MCI to be followed for 3 years, and 200 people with early AD to be followed for 2 years.
The study included 150 patients with mild AD (62 men, 88 women; mean age±SD, 76.3 years±5.8; MMSE score, 23.2±4.1), 247 patients with aMCI (143 men, 104 women; mean age±SD, 75.3 years±6.7; MMSE score, 27.0±1.8), and 135 elderly healthy controls (61 men, 74 women; mean age±SD, 76.4 years±6.1; MMSE score, 29.0±0.8) (see Table 2). For a subset of 50 aMCI patients clinical follow-up data were available (3 year average follow-up time). Among them 25 converted to AD and 5 reverted to normalcy or developed pathologies different from AD, whereas the remaining 20 were still affected by memory deficit but without dementia.
Table 2.
Ensemble properties of our “test” subjects. The error is one standard deviation.
| AD | MCI | Controls | |
|---|---|---|---|
| No. of subjects | 150 (62 M, 88 W) | 247 (143 M, 104 W) | 135 (61 M, 74 W) |
| Age (years) | 76.3±5.8 | 75.3±6.7 | 76.4±6.1 |
| MMSE score | 23.2±4.1 | 27.0±1.8 | 29.0±0.8 |
| Δ-box score | 0.28±0.03 | 0.31±0.03 | 0.34±0.03 |
The images are 1.5 T screening scans, acquired on scanners manufactured by Philips, Siemens, and GE, consisting of a straight sagittal 3D MPRAGE sequence. The images were not corrected for gradient nonlinearity, B1 nonuniformity, and intensity nonuniformity to better represent average scans obtained in “real world” clinical practice. Each image was registered onto the five HBTs and the extraction was successfully carried out as confirmed by visual inspection.
The Δ-box score
In this section we introduce a novel statistical indicator, which measures MTL atrophy and is computed on the intensities in the automatically extracted HBs. Because the absolute intensity of a T1-weighted image is meaningless on its own, after the extraction of the HBs a preliminary equalization of the gray levels was necessary in order to map the mean cerebrospinal fluid (CSF), gray matter (GM), and WM intensities of each HB to common references. This preprocessing, which required a rough segmentation in CSF, GM, and WM, was necessary for obtaining reliable results from voxel based operations and comparisons among HBs.
The next step consisted in preparing the average HBs representative for controls and for AD cohorts. In order to maximize intragroup similarities and intergroup differences, outliers in each group were preliminarily discarded. For each group a distribution of distances was built after defining a metric among HBs in terms of the usual scalar product. Outliers for each group were selected and removed from the least significant 5% of the corresponding distance distributions just obtained. After the outlier removal, the average HBs representative for controls and AD cohorts were prepared by taking the voxel-by-voxel median for each group. Subsequently, the voxel-by-voxel difference between these two medians was performed and denoted as the Δ box. The projection of every HB onto the Δ box, evaluated by means of the scalar product, produces the required intensity-based measure, named hereafter Δ-box score. Age detrending was applied to our variable to ensure that the analysis was not biased by age confounding effects.
Results
Group differences were assessed by using Student’s two-sample t test. Significant differences of MTL atrophy, measured by the Δ-box method, were detected both in AD and in aMCI cohorts (AD vs controls, 0.28±0.03 vs 0.34±0.03, P<0.001; aMCI vs controls, 0.31±0.03 vs 0.34±0.03, P<0.001). MTL atrophy in the subgroup of 25 aMCI converters was similar to the one from the AD group and was significantly different from the one of the controls (0.27±0.03 vs 0.34±0.03, P<0.001).
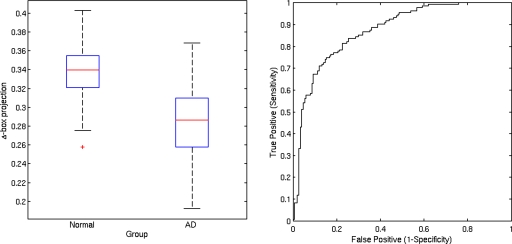
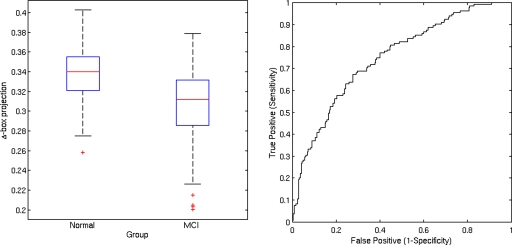
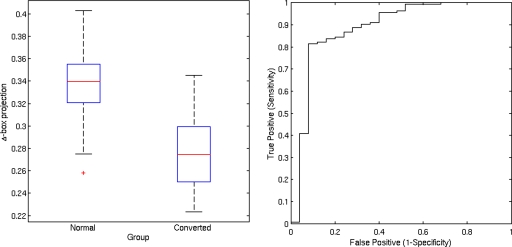
Individual classification on the basis of the Δ-box score was also analyzed by using the receiver operating characteristic (ROC) curves, which indicate the relationship between sensitivity and 1-specificity for each intergroup discrimination. The area under the curve was 0.863 for AD patients vs controls (Fig. 5), 0.746 for aMCI patients vs controls (Fig. 6), and 0.880 for aMCI converters vs controls (Fig. 7). With specificity set at 85%, the sensitivity was 74% for AD vs controls, 45% for aMCI vs controls, and 83% for aMCI converters vs controls.
Figure 5.
Box plot of Δ-box score for controls and AD. Lines represent the median, boxes the interquartile range, and whiskers the range; stars=outliers. The area under the ROC curve is 86.3%.
Figure 6.
Box plot of Δ-box score for controls and aMCI. The area under the ROC curve is 74.6%.
Figure 7.
Box plot of Δ-box score for controls and aMCI converters. The area under the ROC curve is 88%.
DISCUSSION
While several automated hippocampal segmentation methods have been proposed,25, 41, 42, 43, 44, 45 most of them rely either on the manual identification of several hippocampal landmarks on each scan16, 25, 46, 47 or on algorithms based on the intensities and spatial anatomical relationship of different brain structures to guide hippocampal outlining.24, 48 Webb et al.49 devised an automated method to detect hippocampal atrophy in patients with temporal lobe epilepsy based on the analysis of the image intensity differences between patients and controls within a volume of interest centered on the hippocampus. Thompson et al.18 generated color-coded maps to visualize the hippocampal atrophy rate using 3D parametric mesh models of manually segmented hippocampal regions on serial scans. In addition, an automatic measure of hippocampal atrophy rates has been derived using regional fluid registration23 or by calculating the regional boundary shift integral.22 However, both methods require manual segmentation of the baseline hippocampal region. Rusinek et al.50 used the boundary shift integral analysis applied to a volume of interest centered on the hippocampus to calculate the rate of MTL atrophy. Compared to these methods our procedure requires no prerequisites for automation.
Moreover, few of these methods have been applied to patients with AD and∕or MCI and rarely did researchers report the accuracy of their techniques in the differentiation among MCI, AD, and controls. Carmichael et al.20 assessed the performance of automated atlas-based segmentation by using several freely available registration methods [automated image registration (University of California at Los Angeles, Los Angeles, CA), statistical parametric mapping (Wellcome Department of Imaging Neuroscience), functional MR imaging linear image registration tool (University of Oxford, Oxford, England), and a fully deformable approach] in AD and MCI patients. They came to the conclusion that these approaches are less precise when applied to AD patients than controls but this should be tempered by the fact that these techniques were not specifically designed for this task. Fischl et al.24 proposed a general method, derived from a probabilistic atlas, to automatically label different noncortical structures, including the hippocampus, and applied this technique to patients with mild and questionable AD. The method helped identify significant group differences in terms of hippocampal volume but the authors did not investigate the classification of individual participants. Csernansky et al.51 used the high-dimensional brain mapping approach, on the basis of fluid registration with a template, to obtain hippocampal volumes and hippocampal shape differences between patients with very mild AD and controls. By using a classification on the basis of volume and shape features, they achieved a sensitivity of 83% and a specificity of 78%. By using a similar high-dimensional brain mapping approach, Hsu et al.46 compared automated and manual segmentations in AD and cognitively impaired patients. They reported good correlations between manual and automated measurements of the hippocampal volume. However, they did not investigate the accuracy of this technique for the classification of individual patients. Colliot et al.52 evaluated the accuracy of automated hippocampal volumetry to help distinguish between patients with AD, patients with MCI, and elderly controls. Individual classification on the basis of hippocampal volume resulted in 84% correct classification (sensitivity, 84%; specificity, 84%) between AD patients and controls and 73% correct classification (sensitivity, 75%; specificity, 70%) between MCI patients and controls. Ridha et al.21 compared an automated intensity-based measure of medial temporal lobe atrophy (ATLAS) with volumetric and visually based methods. Their measure differentiates patients with AD from controls at cross-sectional and longitudinal levels. At baseline, for a specificity of 85%, the sensitivity of hippocampal volume measurement and visual rating scale19 were similar (84% vs 86%), whereas the sensitivity of the ATLAS measure was lower at 73%.
In this work we introduced a novel approach, based on the measure of a new statistical indicator, the Δ-box score, able to separate the AD, aMCI, and controls cohorts. Since it is well known that MTL atrophy is associated with declining cognitive function,13 we showed that our method is able to capture differences between subgroups of interest with different stages of cognitive impairment, with comparable discriminating capability between aMCI converters and controls and between AD patients and controls. This result should be considered with caution owing to the relatively small number of converters. Anyway, this is in agreement with several studies regarding manual segmentation of hippocampus, which have reported that baseline hippocampal volume is an indicator of future progression to AD.13, 53, 54, 55, 56 This is also in agreement with studies based on visual rating, which clearly found MTL atrophy in patients who subsequently converted to AD.57, 58, 59
Compared to other methods of hippocampal or MTL atrophy measurement, our method, which does not directly tackle the objective of hippocampus segmentation, is fully automated, allowing the analysis of large sets of data, and requires relatively moderate image postprocessing and prerequisites for automation. Therefore, it could be a good candidate for being more widely used than other automatic methods.
In conclusion, we report a novel procedure for assessing MTL atrophy based on intensity measurement in a standardized perihippocampal volume using established T1-weighted volumetric scans. This measure significantly differentiates patients with AD from controls and MCI converters from controls. The technique is simple to use and may be of value in clinical practice for an early diagnosis of AD, without the need for expert assessment or labor intensive manual measures.
ACKNOWLEDGMENTS
The authors thank Mr. E. Deseri for image acquisition and Dr. C. E. Neumaier for MR image reporting. Data collection and sharing for this project was funded by the ADNI (Principal Investigator: Michael Weiner; NIH Grant No. U01 AG024904). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering (NIBIB), and through generous contributions from the following: Pfizer Inc., Wyeth Research, Bristol-Myers Squibb, Eli Lilly and Co., GlaxoSmithKline, Merck & Co. Inc., AstraZeneca AB, Novartis Pharmaceuticals Corporation, Alzheimer Association, Eisai Global Clinical Development, Elan Corporation plc, Forest Laboratories, and the Institute for the Study of Aging, with participation from the US Food and Drug Administration. Industry partnerships are coordinated through the Foundation for the National Institutes of Health. The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer Disease Cooperative Study at the University of California, San Diego. ADNI data are disseminated by the Laboratory of Neuro Imaging at the University of California, Los Angeles. This work benefited from the use of the Insight Segmentation and Registration Toolkit (ITK), an open source software developed as an initiative of the US National Library of Medicine and available at http://www.itk.org.60
References
- Petersen R. C., Smith G. E., Waring S. C., Ivnik R. J., Tangalos E. G., and Kokmen E., “Mild cognitive impairment: Clinical characterization and outcome,” Arch. Neurol. 56(3), 303–308 (1999). 10.1001/archneur.56.3.303 [DOI] [PubMed] [Google Scholar]
- Petersen R. C., Doody R., Kurz A., Mohs R. C., Morris J. C., Rabins P. V., Ritchie K., Rossor M., Thal L., and Winblad B., “Current concepts in mild cognitive impairment,” Arch. Neurol. 58(12), 1985–1992 (2001). 10.1001/archneur.58.12.1985 [DOI] [PubMed] [Google Scholar]
- Dubois B. and Albert M. L., “Amnestic MCI or prodromal Alzheimer’s disease?,” Lancet Neurol. 3(4), 246–248 (2004). 10.1016/S1474-4422(04)00710-0 [DOI] [PubMed] [Google Scholar]
- Petersen R. C., “Mild cognitive impairment as a diagnostic entity,” J. Intern. Med. 256(3), 183–194 (2004). 10.1111/j.1365-2796.2004.01388.x [DOI] [PubMed] [Google Scholar]
- Winblad B., Palmer K., Kivipelto M., Jelic V., Fratiglioni L., Wahlund L. -O., Nordberg A., Bäckman L., Albert M., Almkvist O., Arai H., Basun H., Blennow K., de Leon M., DeCarli C., Erkinjuntti T., Giacobini E., Graff C., Hardy J., Jack C., Jorm A., Ritchie K., van Duijn C., Visser P., and Petersen R. C., “Mild cognitive impairment–beyond controversies, towards a consensus: Report of the International Working Group on Mild Cognitive Impairment,” J. Intern. Med. 256(3), 240–246 (2004). 10.1111/j.1365-2796.2004.01380.x [DOI] [PubMed] [Google Scholar]
- Kantarci K. and Jack C. R., “Quantitative magnetic resonance techniques as surrogate markers of Alzheimer’s disease,” NeuroRx: The Journal of the American Society for Experimental NeuroTherapeutics 1(12), 196–205 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atiya M., Hyman B. T., Albert M. S., and Killiany R., “Structural magnetic resonance imaging in established and prodromal Alzheimer disease: A review,” Alzheimer Dis. Assoc. Disord. 17(3), 177–195 (2003). 10.1097/00002093-200307000-00010 [DOI] [PubMed] [Google Scholar]
- Chetelat G. and Baron J. -C., “Early diagnosis of Alzheimer’s disease: Contribution of structural neuroimaging,” Neuroimage 18(2), 525–541 (2003). 10.1016/S1053-8119(02)00026-5 [DOI] [PubMed] [Google Scholar]
- Kantarci K. and Jack C. R., “Neuroimaging in Alzheimer disease: An evidence-based review,” Neuroimaging Clin. N. Am. 13(2), 197–209 (2003). 10.1016/S1052-5149(03)00025-X [DOI] [PubMed] [Google Scholar]
- Anderson V. C., Litvack Z. N., and Kaye J. A., “Magnetic resonance approaches to brain aging and Alzheimer disease-associated neuropathology,” Top. Magn. Reson Imaging 16(6), 439–452 (2005). 10.1097/01.rmr.0000245458.05654.d0 [DOI] [PubMed] [Google Scholar]
- MICCAI 2008 Workshop on the Computational Anatomy and Physiology of the Hippocampus (unpublished),; http://picsl.upenn.edu/caph08.
- Jack C. R., Petersen R. C., O’Brien P. C., and Tangalos E. G., “MR-based hippocampal volumetry in the diagnosis of Alzheimer’s disease,” Neurology 42(1), 183–188 (1992). [DOI] [PubMed] [Google Scholar]
- Jack C. R., Petersen R. C., Xu Y. C., O’Brien P. C., Smith G. E., Ivnik R. J., Boeve B. F., Waring S. C., Tangalos E. G., and Kokmen E., “Prediction of AD with MRI-based hippocampal volume in mild cognitive impairment,” Neurology 52(7), 1397–1403 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apostolova L. G., Lu P. H., Rogers S., Dutton R. A., Hayashi K. M., Toga A. W., Cummings J. L., and Thompson P. M., “3D mapping of mini-mental state examination performance in clinical and preclinical Alzheimer disease,” Alzheimer Dis. Assoc. Disord. 20(4), 224–231 (2006). 10.1097/01.wad.0000213857.89613.10 [DOI] [PubMed] [Google Scholar]
- Apostolova L. G., Dinov I. D., Dutton R. A., Hayashi K. M., Toga A. W., Cummings J. L., and Thompson P. M., “3D comparison of hippocampal atrophy in amnestic mild cognitive impairment and Alzheimer’s disease,” Brain: A Journal of Neurology 129, 2867–2873 (2006). [DOI] [PubMed] [Google Scholar]
- Csernansky J. G., Joshi S., Wang L., Haller J. W., Gado M., Miller J. P., Grenander U., and Miller M. I., “Hippocampal morphometry in schizophrenia by high dimensional brain mapping,” Proc. Natl. Acad. Sci. U.S.A. 95(19), 11406–11411 (1998). 10.1073/pnas.95.19.11406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisoni G. B., Sabattoli F., Lee A. D., Dutton R. A., Toga A. W., and Thompson P. M., “In vivo neuropathology of the hippocampal formation in AD: A radial mapping MR-based study,” Neuroimage 32(1), 104–110 (2006). 10.1016/j.neuroimage.2006.03.015 [DOI] [PubMed] [Google Scholar]
- Thompson P. M., Hayashi K. M., De Zubicaray G. I., Janke A. L., Rose S. E., Semple J., Hong M. S., Herman D. H., Gravano D., Doddrell D. M., and Toga A. W., “Mapping hippocampal and ventricular change in Alzheimer disease,” Neuroimage 22(4), 1754–1766 (2004). 10.1016/j.neuroimage.2004.03.040 [DOI] [PubMed] [Google Scholar]
- Scheltens P., Leys D., Barkhof F., Huglo D., Weinstein H. C., Vermersch P., Kuiper M., Steinling M., Wolters E. C., and Valk J., “Atrophy of medial temporal lobes on MRI in ‘probable’ Alzheimer’s disease and normal ageing: Diagnostic value and neuropsychological correlates,” J. Neurol., Neurosurg. Psychiatry 55(10), 967–972 (1992). 10.1136/jnnp.55.10.967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmichael O. T., Aizenstein H. A., Davis S. W., Becker J. T., Thompson P. M., Meltzer C. C., and Liu Y., “Atlas-based hippocampus segmentation in Alzheimer’s disease and mild cognitive impairment,” Neuroimage 27(4), 979–990 (2005). 10.1016/j.neuroimage.2005.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridha B. H., Barnes J., van de Pol L. A., Schott J. M., Boyes R. G., Siddique M. M., Rossor M. N., Scheltens P., and Fox N. C., “Application of automated medial temporal lobe atrophy scale to Alzheimer disease,” Arch. Neurol. 64(6), 849–854 (2007). 10.1001/archneur.64.6.849 [DOI] [PubMed] [Google Scholar]
- Barnes J., Scahill R. I., Boyes R. G., Frost C., Lewis E. B., Rossor C. L., Rossor M. N., and Fox N. C., “Differentiating AD from aging using semiautomated measurement of hippocampal atrophy rates,” Neuroimage 23(2), 574–581 (2004). 10.1016/j.neuroimage.2004.06.028 [DOI] [PubMed] [Google Scholar]
- Crum W. R., Scahill R. I., and Fox N. C., “Automated hippocampal segmentation by regional fluid registration of serial MRI: Validation and application in Alzheimer’s disease,” Neuroimage 13(5), 847–855 (2001). 10.1006/nimg.2001.0744 [DOI] [PubMed] [Google Scholar]
- Fischl B., Salat D. H., Busa E., Albert M., Dieterich M., Haselgrove C., van der Kouwe A., Killiany R., Kennedy D., Klaveness S., Montillo A., Makris N., Rosen B., and Dale A. M., “Whole brain segmentation: Automated labeling of neuroanatomical structures in the human brain,” Neuron 33(3), 341–355 (2002). 10.1016/S0896-6273(02)00569-X [DOI] [PubMed] [Google Scholar]
- Hogan R. E., Mark K. E., Wang L., Joshi S., Miller M. I., and Bucholz R. D., “Mesial temporal sclerosis and temporal lobe epilepsy: MR imaging deformation-based segmentation of the hippocampus in five patients,” Radiology 216(1), 291–297 (2000). [DOI] [PubMed] [Google Scholar]
- Powell S., Magnotta V. A., Johnson H., Jammalamadaka V. K., Pierson R., and Andreasen N. C., “Registration and machine learning-based automated segmentation of subcortical and cerebellar brain structures,” Neuroimage 39(1), 238–247 (2008). 10.1016/j.neuroimage.2007.05.063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Beg F., Ratnanather T., Ceritoglu C., Younes L., Morris J. C., Csernansky J. G., and Miller M. I., “Large deformation diffeomorphism and momentum based hippocampal shape discrimination in dementia of the Alzheimer type,” IEEE Trans. Med. Imaging 26(4), 462–470 (2007). 10.1109/TMI.2006.887380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yushkevich P. A., Piven J., Hazlett H. C., Smith R. G., Ho S., Gee J. C., and Gerig G., “User-guided 3D active contour segmentation of anatomical structures: significantly improved efficiency and reliability,” Neuroimage 31(3), 1116–1128 (2006). 10.1016/j.neuroimage.2006.01.015 [DOI] [PubMed] [Google Scholar]
- Morra J. H., Tu Z., Apostolova L. G., Green A. E., Avedissian C., Madsen S. K., Parikshak N., Hua X., Toga A. W., Jack C. R., Weiner M. W., and Thompson P. M., “Validation of a fully automated 3D hippocampal segmentation method using subjects with Alzheimer’s disease mild cognitive impairment, and elderly controls,” Neuroimage 43(1), 59–68 (2008). 10.1016/j.neuroimage.2008.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker J. T., Davis S. W., Hayashi K. M., Meltzer C. C., Toga A. W., Lopez O. L., and Thompson P. M., “Three-dimensional patterns of hippocampal atrophy in mild cognitive impairment,” Arch. Neurol. 63(1), 97–101 (2006). 10.1001/archneur.63.1.97 [DOI] [PubMed] [Google Scholar]
- Collins D. L., Neelin P., Peters T. M., and Evans A. C., “Automatic 3D intersubject registration of MR volumetric data in standardized talairach space,” J. Comput. Assist. Tomogr. 18(2), 192–205 (1994). 10.1097/00004728-199403000-00005 [DOI] [PubMed] [Google Scholar]
- McKhann G., Drachman D., Folstein M., Katzman R., Price D., and Stadlan E. M., “Clinical diagnosis of Alzheimer’s disease: Report of the NINCDS-ADRDA work group under the auspices of department of health and human services task force on Alzheimer’s disease,” Neurology 34(7), 939–944 (1984). [DOI] [PubMed] [Google Scholar]
- Katz S., Downs T. D., Cash H. R., and Grotz R. C., “Progress in development of the index of ADL,” Gerontologist 10(1), 20–30 (1970). [DOI] [PubMed] [Google Scholar]
- Lawton M. P. and Brody E. M., “Assessment of older people: Self-maintaining and instrumental activities of daily living,” Gerontologist 9(3), 179–186 (1969). [PubMed] [Google Scholar]
- Folstein M. F., Folstein S. E., and McHugh P. R., “Mini-mental state. A practical method for grading the cognitive state of patients for the clinician,” J. Psychiatr. Res. 12(3), 189–198 (1975). 10.1016/0022-3956(75)90026-6 [DOI] [PubMed] [Google Scholar]
- Cummings J. L., Mega M., Gray K., Rosenberg-Thompson S., Carusi D. A., and Gornbein J., “The neuropsychiatric inventory: Comprehensive assessment of psychopathology in dementia,” Neurology 44(12), 2308–2314 (1994). [DOI] [PubMed] [Google Scholar]
- Evans A. C., Kamber M., Collins D. L., and MacDonald D., “An MRI–based probabilistic atlas of neuroanatomy,” Magnetic Resonance Scanning and Epilepsy, NATO Advance Studies Institute, Series A: Life Sciences Vol. 264, edited by Shorvon S., Fish D., Andermann F., Bydder G., and Stefan H. (Plenum, New York, 1994), pp. 263–274. [Google Scholar]
- Ibanez L., Schroeder W., Ng L., and Cates J., The ITK software guide, 2nd ed., 2005.
- Seber G. A. F., Multivariate Observations (Wiley, New York, 1984). [Google Scholar]
- Alzheimer’s Disease Neuroimaging Initiative (ADNI), http://www.loni.ucla.edu/ADNI/, 2007.
- Kelemen A., Székely G., and Gerig G., “Elastic model-based segmentation of 3-D neuroradiological data sets,” IEEE Trans. Med. Imaging 18(10), 828–839 (1999). 10.1109/42.811260 [DOI] [PubMed] [Google Scholar]
- Yang J. and Duncan J. S., “3D image segmentation of deformable objects with joint shape-intensity prior models using level sets,” Med. Image Anal. 8(3), 285–294 (2004). 10.1016/j.media.2004.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitiot A., Delingette H., Thompson P. M., and Ayache N., “Expert knowledge-guided segmentation system for brain MRI,” Neuroimage 23, S85–S96 (2004). 10.1016/j.neuroimage.2004.07.040 [DOI] [PubMed] [Google Scholar]
- Shen D., Moffat S., Resnick S. M., and Davatzikos C., “Measuring size and shape of the hippocampus in MR images using a deformable shape model,” Neuroimage 15(2), 422–434 (2002). 10.1006/nimg.2001.0987 [DOI] [PubMed] [Google Scholar]
- Duchesne S., Pruessner J., and Collins D. L., “Appearance-based segmentation of medial temporal lobe structures,” Neuroimage 17(2), 515–531 (2002). 10.1016/S1053-8119(02)91188-2 [DOI] [PubMed] [Google Scholar]
- Hsu Y. -Y., Schuff N., Du A. -T., Mark K., Zhu X., Hardin D., and Weiner M. W., “Comparison of automated and manual MRI volumetry of hippocampus in normal aging and dementia,” J. Magn. Reson Imaging 16(3), 305–310 (2002). 10.1002/jmri.10163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haller J. W., Christensen G. E., Joshi S. C., Newcomer J. W., Miller M. I., Csernansky J. G., and Vannier M. W., “Hippocampal MR imaging morphometry by means of general pattern matching,” Radiology 199(3), 787–791 (1996). [DOI] [PubMed] [Google Scholar]
- Gosche K. M., Mortimer J. A., Smith C. D., Markesbery W. R., and Snowdon D. A., “An automated technique for measuring hippocampal volumes from MR imaging studies,” AJNR Am. J. Neuroradiol. 22(9), 1686–1689 (2001). [PMC free article] [PubMed] [Google Scholar]
- Webb J., Guimond A., Eldridge P., Chadwick D., Meunier J., Thirion J. P., and Roberts N., “Automatic detection of hippocampal atrophy on magnetic resonance images,” Magn. Reson. Imaging 17(8), 1149–1161 (1999). 10.1016/S0730-725X(99)00044-2 [DOI] [PubMed] [Google Scholar]
- Rusinek H., De Santi S., Frid D., Tsui W. -H., Tarshish C. Y., Convit A., and de Leon M. J.,“Regional brain atrophy rate predicts future cognitive decline: 6-year longitudinal MR imaging study of normal aging,” Radiology 229(3), 691–696 (2003). 10.1148/radiol.2293021299 [DOI] [PubMed] [Google Scholar]
- Csernansky J. G., Wang L., Joshi S., Miller J. P., Gado M., Kido D., McKeel D., Morris J. C., and Miller M. I., “Early DAT is distinguished from aging by high-dimensional mapping of the hippocampus. Dementia of the Alzheimer type,” Neurology 55(11), 1636–1643 (2000). [DOI] [PubMed] [Google Scholar]
- Colliot O., Chételat G., Chupin M., Desgranges B., Magnin B., Benali H., Dubois B., Garnero L., Eustache F., and Lehéricy S., “Discrimination between Alzheimer disease, mild cognitive impairment, and normal aging by using automated segmentation of the hippocampus,” Radiology 248(1), 194–201 (2008). 10.1148/radiol.2481070876 [DOI] [PubMed] [Google Scholar]
- Tapiola T., Pennanen C., Tapiola M., Tervo S., Kivipelto M., Hänninen T., Pihlajamäki M., Laakso M. P., Hallikainen M., Hämäläinen A., Vanhanen M., Helkala E. -L., Vanninen R., Nissinen A., Rossi R., Frisoni G. B., and Soininen H., “MRI of hippocampus and entorhinal cortex in mild cognitive impairment: A follow-up study,” Neurobiol. Aging 29(1), 31–38 (2008). 10.1016/j.neurobiolaging.2006.09.007 [DOI] [PubMed] [Google Scholar]
- Devanand D. P., Pradhaban G., Liu X., Khandji A., De Santi S., Segal S., Rusinek H., Pelton G. H., Honig L. S., Mayeux R., Stern Y., Tabert M. H., and de Leon M. J., “Hippocampal and entorhinal atrophy in mild cognitive impairment: Prediction of Alzheimer disease,” Neurology 68(11), 828–836 (2007) 10.1212/01.wnl.0000256697.20968.d7 [DOI] [PubMed] [Google Scholar]
- Convit A., de Asis J., de Leon M. J., Tarshish C. Y., De Santi S., and Rusinek H., “Atrophy of the medial occipitotemporal, inferior, and middle temporal gyri in non-demented elderly predict decline to Alzheimer’s disease,” Neurobiol. Aging 21(1), 19–26 (2000). 10.1016/S0197-4580(99)00107-4 [DOI] [PubMed] [Google Scholar]
- Jack C. R., Shiung M. M., Weigand S. D., O’Brien P. C., Gunter J. L., Boeve B. F., Knopman D. S., Smith G. E., Ivnik R. J., Tangalos E. G., and Petersen R. C., “Brain atrophy rates predict subsequent clinical conversion in normal elderly and amnestic MCI,” Neurology 65(8), 1227–1231 (2005). 10.1212/01.wnl.0000180958.22678.91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeCarli C., Frisoni G. B., Clark C. M., Harvey D., Grundman M., Petersen R. C., Thal L. J., Jin S., Jack C. R., and Scheltens P., “Qualitative estimates of medial temporal atrophy as a predictor of progression from mild cognitive impairment to dementia,” Arch. Neurol. 64(1), 108–115 (2007). 10.1001/archneur.64.1.108 [DOI] [PubMed] [Google Scholar]
- Geroldi C., Rossi R., Calvagna C., Testa C., Bresciani L., Binetti G., Zanetti O., and Frisoni G. B., “Medial temporal atrophy but not memory deficit predicts progression to dementia in patients with mild cognitive impairment,” J. Neurol., Neurosurg. Psychiatry 77(11), 1219–1222 (2006). 10.1136/jnnp.2005.082651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Leon M. J., Golomb J., George A. E., Convit A., Tarshish C. Y., McRae T., De Santi S., Smith G., Ferris S. H., and Noz M.., “The radiologic prediction of Alzheimer disease: The atrophic hippocampal formation,” AJNR Am. J. Neuroradiol. 14(4), 897–906 (1993). [PMC free article] [PubMed] [Google Scholar]
- Yoo T. S., Ackerman M. J., Lorensen W. E., Schroeder W., Chalana V., Aylward S., Metaxes D., and Whitaker R., in Proceedings of Medicine Meets Virtual Reality Conference, edited by Westwood J. (IOS, Amsterdam, 2002), pp. 586–592. [Google Scholar]