SUMMARY
Crystallographic studies of the C-terminal, DNA polymerase β-like domain of human DNA polymerase lambda (fPolλ) suggested that the catalytic cycle might not involve a large protein domain rearrangement as observed with several replicative DNA polymerases and DNA polymerase β. To examine solution-phase protein conformation changes in fPolλ, which also contains a breast cancer susceptibility gene 1 C-terminal domain and a Proline-rich domain at its N-terminus, we used a mass spectrometry - based protein footprinting approach. In parallel experiments, surface accessibility maps for Arg residues were compared for the free fPolλ versus the binary complex of enzyme•gapped DNA and the ternary complex of enzyme•gapped DNA•dNTP. These experiments suggested that fPolλ does not undergo major conformational changes during the catalysis in the solution phase. Furthermore, the mass spectrometry-based protein footprinting experiments revealed that active site residue R386 was shielded from the surface only in the presence of both a gapped DNA substrate and an incoming nucleotide dNTP. Site-directed mutagenesis and pre-steady state kinetic studies confirmed the importance of R386 for the enzyme activity, and indicated the key role for its guanidino group in stabilizing the negative charges of an incoming nucleotide and the leaving pyrophosphate product. We suggest that such interactions could be shared by and important for catalytic functions of other DNA polymerases.
Keywords: Human DNA polymerase lambda, pre-steady-state kinetics, protein conformation changes, mass spectrometry-based protein footprinting, nucleotide incorporation
INTRODUCTION
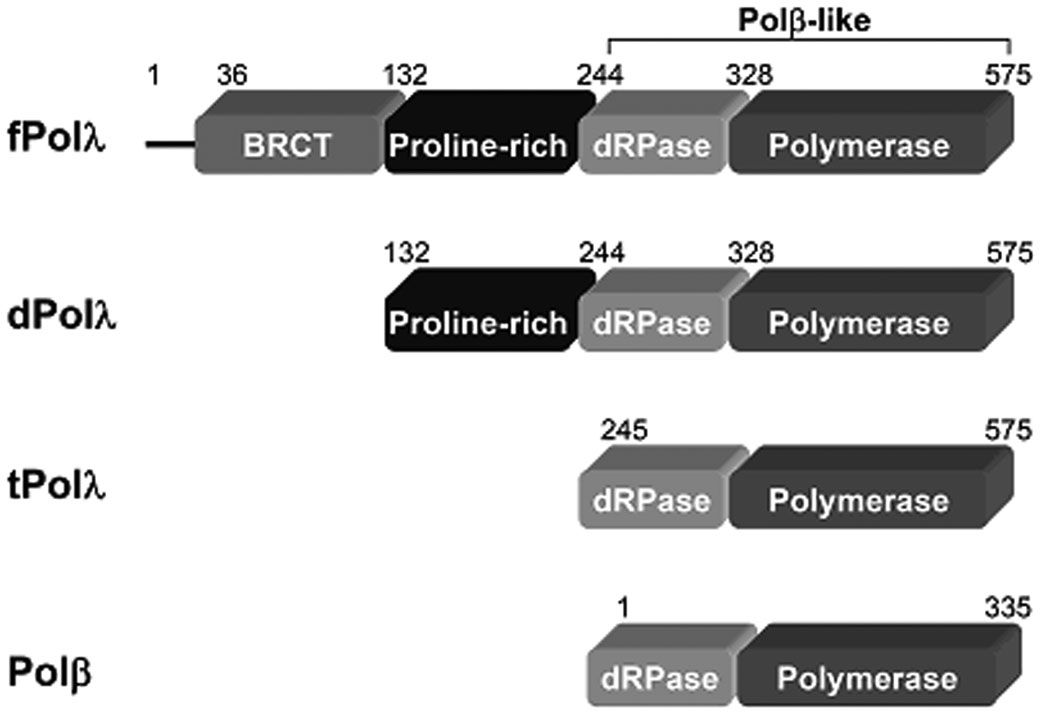
Based upon phylogenetic analyses, DNA polymerases have been arranged into six families which are designated as A, B, C, D, X and Y. Members of the novel X-family of DNA polymerases belong to a larger superfamily of nucleotidyl transferases and are found in all three domains of life 1; 2. Thus far, at least five X-family members have been identified in humans: DNA polymerases lambda (Polλ), beta (Polβ), Mu, sigma, and terminal deoxynucleotidyl transferase (TdT). Polβ is known to function in base excision repair (BER) in vivo 3;4. Physiologically, Polλ has been proposed to function in BER, non-homologous end joining, and V(D)J recombination 5; 6; 7; 8; 9; 10; 11; 12. Polβ and Polλ share 54% sequence homology and 32% sequence identity 13. In addition, both possess gap-filling DNA polymerase activity at their C-terminal polymerase domain, 5'-deoxyribose-5-phosphate lyase (dRPase) activity at their 8-kD dRPase domain, and lack 3'→5' exonuclease activity (Figure 1) 5; 14; 15. However, the full-length Polλ (fPolλ) possesses a breast cancer susceptibility gene 1 C-terminal (BRCT) domain, a Proline-rich domain, and a nuclear localization signal motif on its N-terminus which are absent in the full-length Polβ (Figure 1). Interestingly, the Proline-rich domain has been shown to functionally suppress the polymerase activity of fPolλ 16 and to increase its fidelity by up to 100-fold 17.
Figure 1.
Schematic domain structure of human fPolλ, dPolλ, tPolλ, and Polβ. Each domain, with amino acid residue numbers indicated above, is shown as a rectangle. The N-terminal 35 residues of fPolλ contain a nuclear localization signal motif as represented by the line.
In general, DNA polymerases are structurally and functionally quite diverse, although, commonalities can be found. Firstly, crystal structures for all known template-dependent DNA polymerases reveal that they share a common three-dimensional shape which resembles a human right hand. This feature of E. coli DNA polymerase I 18 led to the “fingers, palm, and thumb” domain nomenclature system. Secondly, all DNA polymerases perform the chemistry of nucleotidyl transfer using the same two divalent metal ion mechanism as first proposed by T. A. Steitz 19. This mechanism, believed to be one of the earliest enzymatic activities to evolve 20 , involves the coordination of two divalent metal ions with three conserved carboxylate residues within a polymerase active site, the primer 3'-OH moiety, and the triphosphate moiety of an incoming nucleotide (dNTP). Finally, a dramatic protein conformational change is observed in the conversion from the binary complex of enzyme•DNA to the ternary complex of enzyme•DNA•dNTP. During this change, the fingers domain moves toward the palm domain forming the hydrophobic polymerase active site. This swing of the fingers domain changes the polymerase from the catalytically inactive “open” to the active “closed” conformation 21. To catalyze nucleotidyl transfer, formation of the closed conformation aligns the polymerase active site by properly orienting the conserved catalytic carboxylates, divalent metal ions, DNA, and dNTP22
Recent crystallographic studies with human truncated Polλ (tPolλ, Figure 1) revealed that unlike other DNA polymerases 23, including Polβ 24; 25 for which atomic structures are available, the catalytic cycle of Polλ might not involve a large, protein domain rearrangement. Rather, when comparing three crystal structures of human tPolλ (tPolλ•gapped DNA, tPolλ•gapped DNA•dNTP, and tPolλ•nicked DNA•pyrophosphate), dNTP binding induces a repositioning of only four side chains (i.e. Y505, F506, R514, and R517) within the active site and a minor shift in the position of two β-strands 23. Together, these movements shift the DNA template strand. However, the crystallographic studies that demonstrate this unique mechanism were performed with tPolλ, rather than fPolλ. The Proline-rich domain absent in tPolλ has recently been shown to increase the fidelity of Polλ by up to 100-fold, although, the reason for this is not yet known 17. It is conceivable that upon dNTP binding, the Proline-rich domain induces either the large swing of the finger domain 26 or relatively modest active site rearrangements in the solution phase. Either of these protein conformational transitions may provide a thermodynamic basis for selecting matched over mismatched incoming dNTPs 27. In the present study, we investigated the extent of protein conformational changes in the solution phase using mass spectrometry (MS)-based protein footprinting methods.
RESULTS
In order to investigate whether or not human fPolλ undergoes a large protein conformational change during catalysis in the solution phase, we employed a mass spectrometry-based protein footprinting method 28; 29; 30. Briefly, a gentle, covalent modification of a protein at solvent accessible arginine residues by p-hydroxyphenylglyoxal (HPG) is conducted under near physiological conditions. After modification, the protein is purified and digested by trypsin, a serine protease, which predominantly cleaves peptide chains at the carboxyl side of the amino acids lysine and arginine, except when either residue is followed by proline or is chemically modified. Thus, trypsin cannot cleave after HPG-modified Arg residues in peptide chains. Following the trypsin digestion, the pool of low molecular weight peptides is analyzed by Matrix Assisted Laser Desorption Ionization Time of Flight (MALDI-ToF) instrument to obtain MS data. Notably, each HPG modification on the side chain of an Arg residue leads to the addition of 131 Daltons to the molecular weight of a peptide 31; 32; 33. Analysis of a peptide MS spectrum will reveal the precise location of the chemical modifications and the extent to which they are modified (Materials and Methods). The solvent accessibility of Arg residues is sensitive to protein conformation. Therefore, comparative analysis of modification patterns in free protein versus its complexes with cognate nucleic acids could reveal protein conformational change(s). A side chain, which becomes less amenable to modification (i.e. a decrease in peak intensity within the MS spectrum compared to control peaks) is deemed “protected” and is less solvent accessible. Conversely, if chemical modification becomes more facile (i.e. an increase in peak intensity within the MS spectrum compared to control peaks), the side chain is said to be “hyper-reactive” and more solvent accessible 29; 30. Here, this MS foot-printing method was employed to analyze the nature of conformational changes that may occur in fPolλ through a single turnover of the catalytic cycle. In addition, the above set of conditions was repeated for dPolλ and tPolλ (Figure 1) to assess any inherent differences between the solution-phase structures of these proteins in the presence or absence of DNA and dNTP.
Investigating the Stability of Human Polλ during HPG Modification
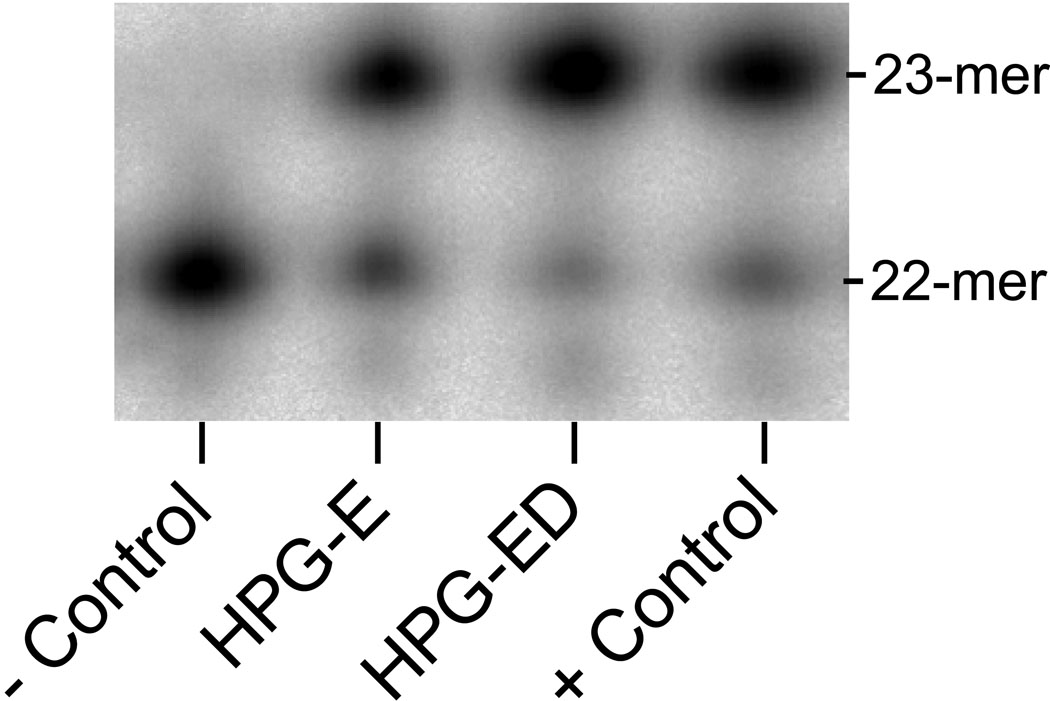
For meaningful protein footprinting experiments, it is essential to establish mild modification conditions under which the integrity of the functional nucleoprotein complexes is preserved. Previous studies indicated that the treatment with 10 mM HPG to be optimal 29; 30. To make sure that these conditions were applicable to fPolλ, we conducted an analysis of the gap-filling DNA polymerase activities following HPG treatments (see Figure 2). The solutions of apo-fPolλ and the binary complex of fPolλ and 22-18/41G-mer (Table 1) were first reacted with 10 mM HPG and then quenched by a molar excess of arginine. The modified fPolλ in these two reaction solutions (lanes “HPG-E” and “HPG-ED” in Figure 2) and unmodified fPolλ (lane “+ Control”) were examined for their ability to incorporate correct dCTP into single-nucleotide gapped 22-18/41G–mer (Materials and Methods). Similar intensities of both product 23-mer and remaining primer 22-mer on the three right lanes in Figure 2 suggested that the gap-filling DNA polymerase activity of fPolλ, in both the apo form and the binary complex with DNA, was almost preserved following HPG modification. Thus, HPG modification conditions were sufficiently mild so that the solution-phase structure of fPolλ remained intact for both its apo form and binary complex with DNA. We did not examine if the HPG modification step altered the integrity of the ternary complex of fPolλ, 22ddC-18/41G-mer (Table 1), and dCTP due to the difficulty to completely replace the fPolλ-bound, dideoxy-terminated DNA substrate with 22- 18/41G–mer in the gap-filling DNA polymerase activity assay (Materials and Methods). However, such an alteration was unlikely to occur considering that the binary complex was not affected by HPG modification. Similar gap-filling DNA polymerase activity assays were performed with dPolλ and tPolλ and demonstrated that HPG modification did not disturb the conformations of these truncated mutants of fPolλ (data not shown).
Figure 2.
Gap-filling DNA polymerase activity of fPolλ following HPG modification. Reactions of fPolλ (10 µM) and 22-18/4 1G–mer (60 µM) were initiated by the addition of 100 µM dCTP at 37 ˚C and terminated after 1 minute by the addition of 0.37 M EDTA. The negative control reaction (− Control) did not have dCTP. “HPG-E” and “HPG-ED” denote HPG-modified apo-fPolλ and the HPG-modified binary complex (fPolλ•22-18/41G–mer), respectively. The positive control reaction (+ Control) contained unmodified fPolλ.
Table 1.
DNA substrates
| 21-19/41A-mer | 5’-CGCAGCCGTCCAACCAACTCA CGTCGATCCAATGCCGTCC-3’ 3’-GCGTCGGCAGGTTGGTTGAGTAGCAGCTAGGTTACGGCAGG-5’ |
| 22-18/41G–mer | 5’-CGCAGCCGTCCAACCAACTCAC GTCGATCCAATGCCGTCC-3’ 3’-GCGTCGGCAGGTTGGTTGAGTGGCAGCTAGGTTACGGCAGG-5’ |
| 22ddC-18/41G–mer | 5’-CGCAGCCGTCCAACCAACTCAC GTCGATCCAATGCCGTCC-3’ 3’-GCGTCGGCAGGTTGGTTGAGTGGCAGCTAGGTTACGGCAGG-5’ |
The downstream 18-mer and 19-mer strands were 5′-phosphorylated. “C” denotes ddCMP.
MS-Based Footprinting of fPolλ, dPol λ, and tPolλ
Representative MS results comparing surface accessible Arg residues of tPolλ in its apo form with the binary and ternary complexes are depicted in Figure 3. Figure 3D is a portion of the mass spectrum for unmodified tPolλ and shows a peak corresponding to tryptic peptide 524–538 which is present in all spectra at the same intensity and is used as an internal control. Figure 3A, 3B, and 3C show the same portion of the MS spectrum of HPG-modified apo-tPolλ, the binary complex of tPolλ•22-18/41G-mer, and the ternary complex of tPolλ•22ddC-18/41G–mer•dCTP, respectively. In the ternary complex, dCTP was not incorporated because 22ddC-18/41G–mer contains a dideoxy-terminated primer. In Figure 3A–3C, peaks resulting from tryptic peptides 562–573, 313–324, and 379–389 correspond to HPG modification of R568, R323, and R386, respectively, thereby shifting their m/z ratios. The spectra in Figure 3A and 3B are almost superimposable, suggesting that the structure of tPolλ is relatively unchanged from apo-tPolλ to the binary complex tPolλ•22-18/41G–mer. In contrast, the intensity of the modified 379–389 (R386+HPG) peak was significantly diminished in the tPolλ•22ddC-18/41G–mer•dCTP complex (Figure 3C) indicating that R386 was shielded from the surface by the bound nucleotide.
Figure 3.
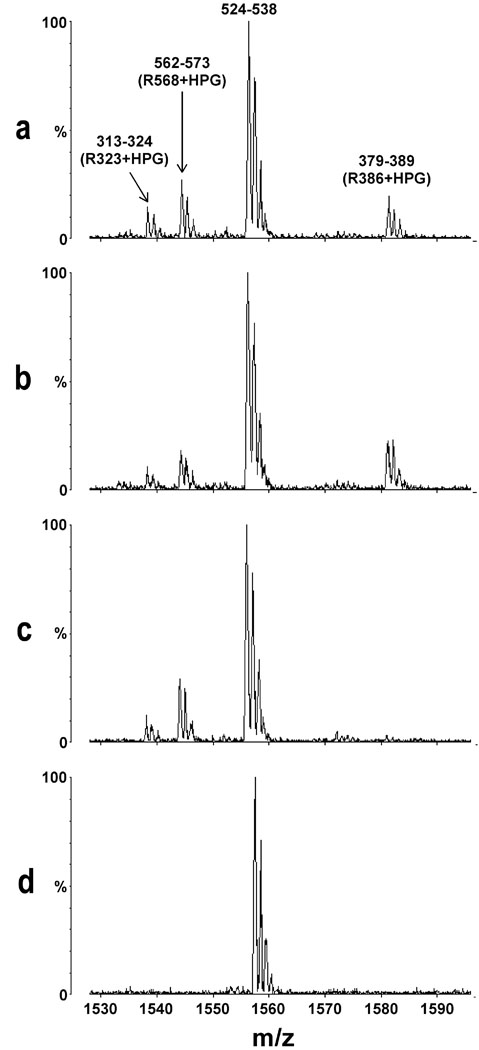
Representative segments of the MALDI-ToF MS spectra. (A) apo-tPolλ was treated with HPG, (B) the Binary complex of tPolλ•22-18/41G–mer DNA was pre-formed and then subjected to HPG modification, (C) the ternary complex of tPolλ•22ddC-18/41G–mer DNA•dCTP was pre-formed and then treated with HPG, (D) unmodified apo-tPolλ.
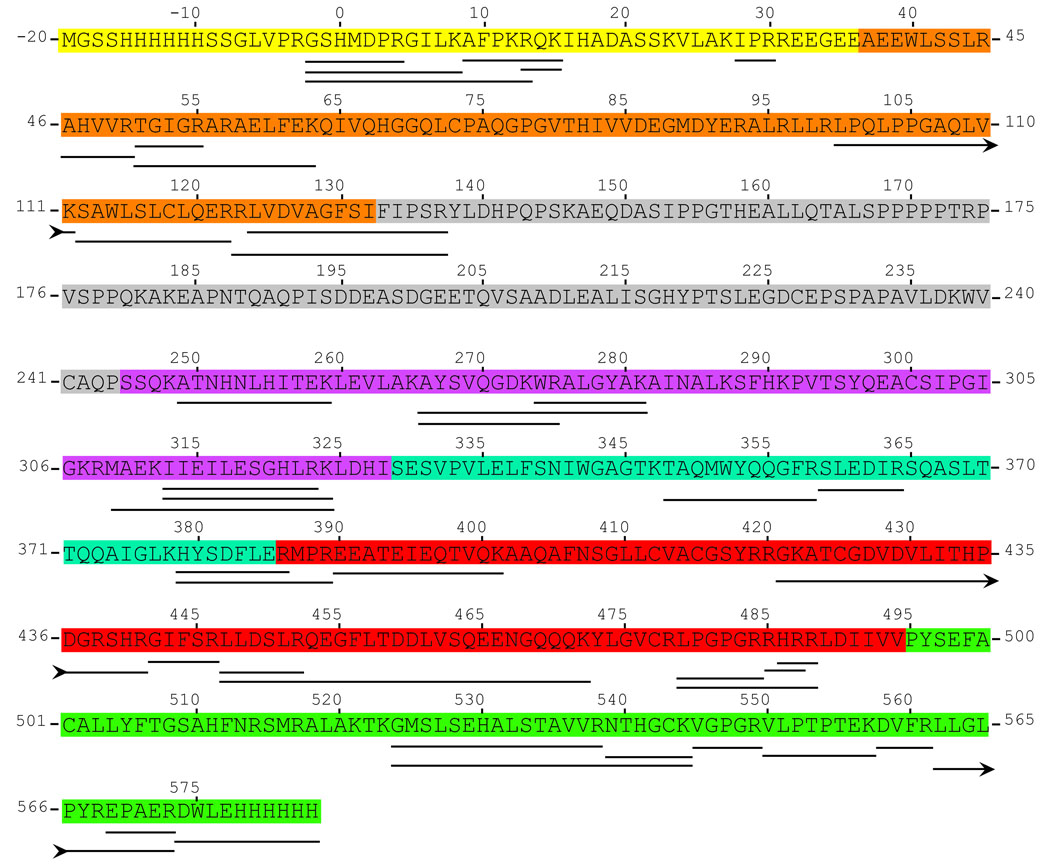
The purified, recombinant fPolλ, dPolλ, and tPolλ possess 41, 28, and 25 Arg residues, respectively (Figure 4). In the absence of DNA and dNTP, HPG modified 13, 10, and 10 Arg residues of the apo forms of fPolλ, dPolλ, and tPolλ, respectively (Table 2). Similar or lower Arg coverages as probed by HPG have been reported in other protein systems 30; 34; 35. The HPG-modified Arg residues of apo-dPolλ and apo-tPolλ are identical, indicating that the Proline-rich domain did not shield any of the exposed Arg residues in the Polβ–like domain (Figure1). At the same time it is important to note that not all the surface exposed Arg residues could be detected by our approach as certain large tryptic peptide fragments may not be readily amenable for MALDI-ToF analysis. For example, the single Arg residue (R174) in the Proline-rich domain (Figure 4) is followed immediately by a Pro residue thereby making trypsin cleavage at R174 impossible regardless of HPG modification. Therefore, the shortest peptide resulting from trypsin digestion to contain R174 is 147–181, which likely escaped detection due to its large size. Thus, modification at R174 is beyond the limits of detection for this assay and may or may not be modified. Finally, R50, R55, and R57 which are located in the BRCT domain (Figure 4), were modified by HPG in apo-fPolλ, but not in apo-dPolλ or apo-tPolλ, because these two Polλ fragments do not contain the BRCT domain (Figure 1).
Figure 4.
The tryptic digestion map of human fPolλ. Residues 0-575 are encoded by the fPolλ gene. The protein contained N-terminal and C-terminal hexahistidine tags. The tryptic peptide peaks that were detected by our MALDI-ToF instrument are underlined. Arrow heads indicate that the peptide sequence continues on the next lower line at the arrow tail. Protein domains are colored as follows: Yellow, N-terminal nuclear localization sequence and hexahistidine tag; Orange, BRCT domain; Grey, Proline-rich domain; Purple, dRPase domain; Blue, Fingers subdomain; Red, Palm subdomain; Green, Thumb subdomain and C-terminal hexahistidine tag.
Table 2.
Summary of modified arginine residues.
| tPolλ | dPolλ | fPolλ | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Apo | Binary | Ternary | Apo | Binary | Ternary | Apo | Binary | Ternary | |
| R50 | NP | NP | NP | NP | NP | NP | + | + | + |
| R55 | NP | NP | NP | NP | NP | NP | + | + | + |
| R57 | NP | NP | NP | NP | NP | NP | + | + | + |
| R275 | + | + | − | + | + | − | + | + | − |
| R323 | + | + | + | + | + | + | + | + | + |
| R386 | + | + | − | + | + | − | + | + | − |
| R441 | + | + | + | + | + | + | + | + | + |
| R446 | + | + | + | + | + | + | + | + | + |
| R484 | + | + | + | + | + | + | + | + | + |
| R485 | + | + | + | + | + | + | + | + | + |
| R549 | + | + | + | + | + | + | + | + | + |
| R561 | + | + | + | + | + | + | + | + | + |
| R568 | + | + | + | + | + | + | + | + | + |
“+” Arg residues susceptible to modification by HPG;
“−” Arg residues shielded from HPG in the nucleoprotein complexes.
“NP” means that indicated Arg are not present in the protein constructs.
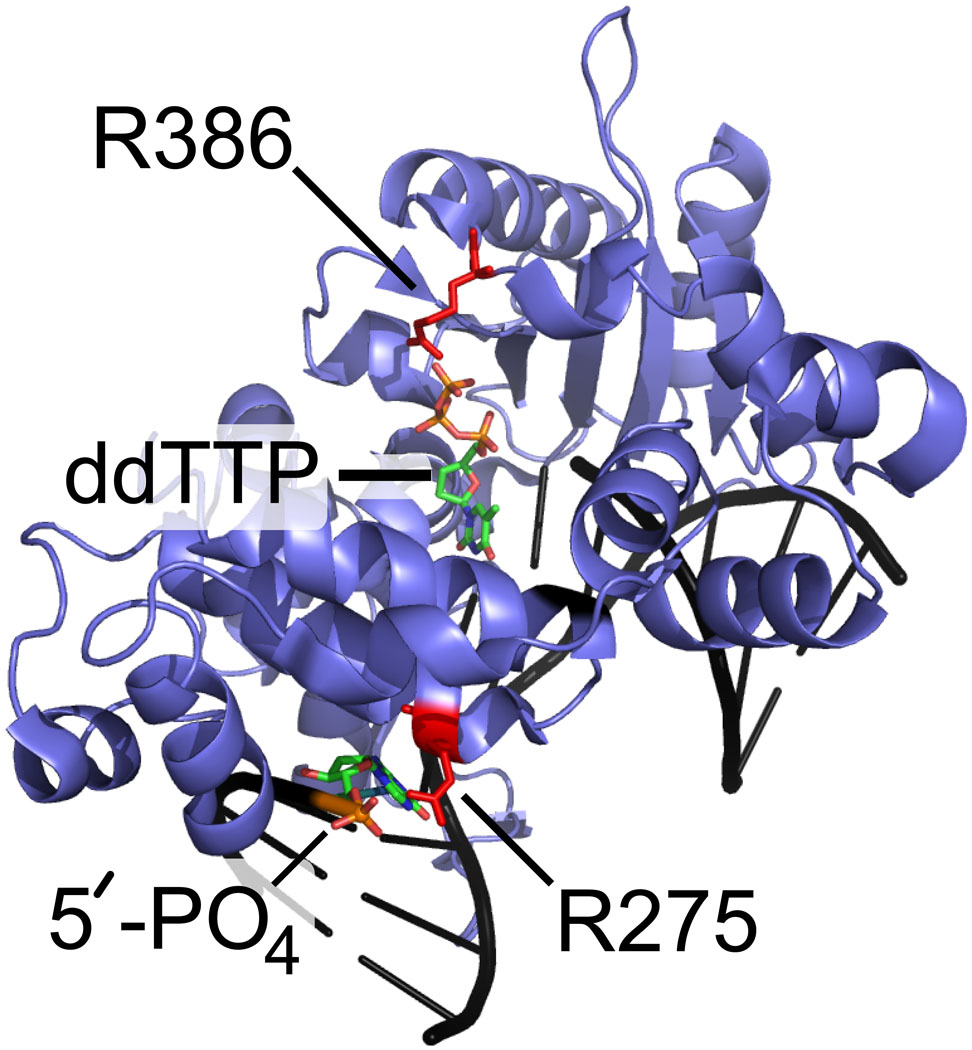
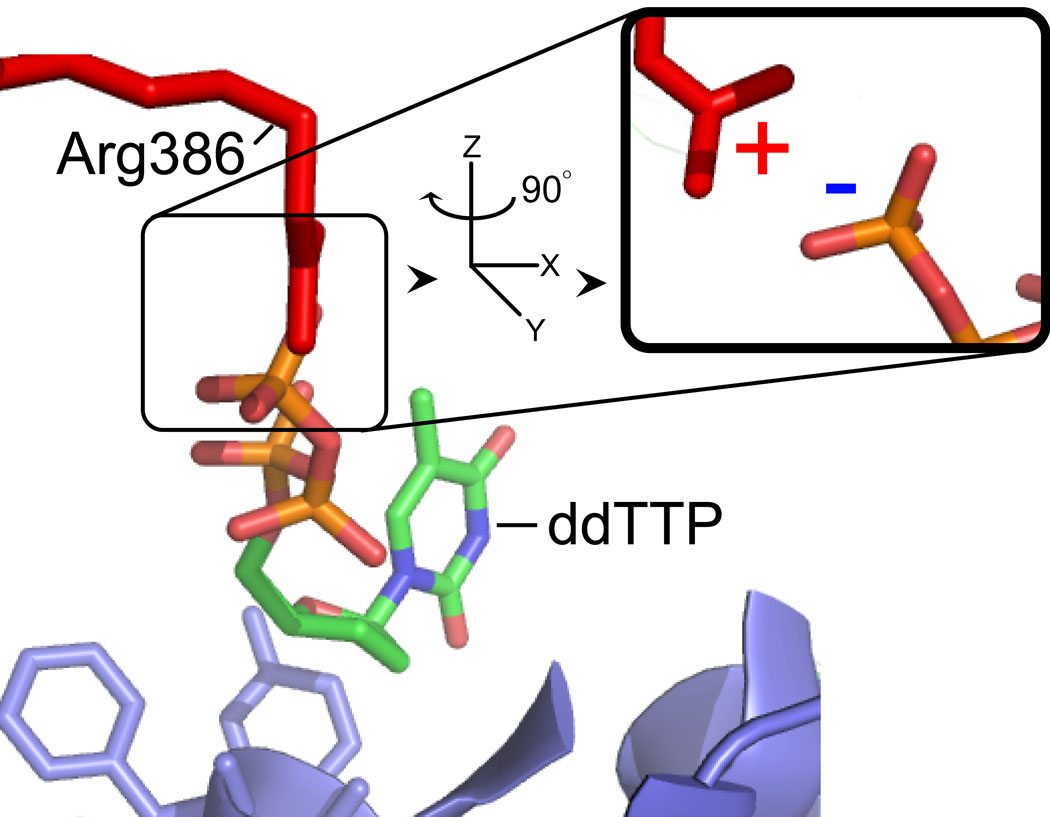
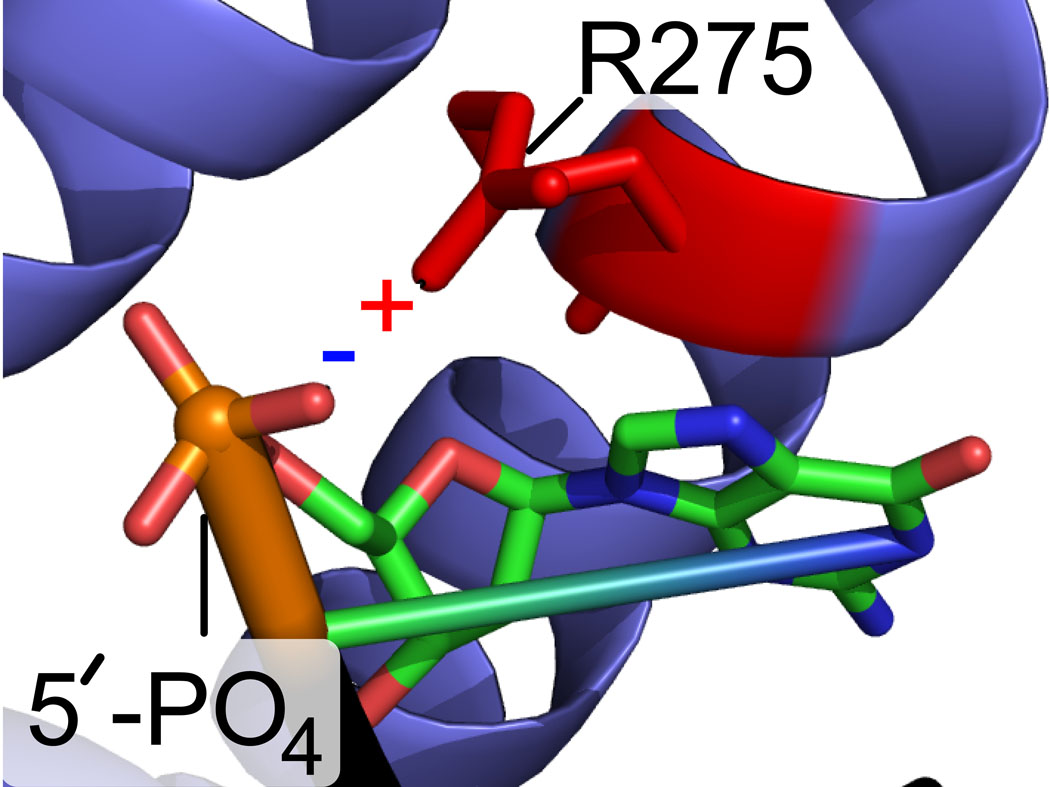
In the presence of 22-18/41G–mer (60 µM, Table 1), the HPG-modified Arg residues in fPolλ, dPolλ, and tPolλ were identical to those detected in the corresponding apo forms of these Polλ constructs (Table 2). This suggested that the solution phase structures of fPolλ, dPolλ, and tPolλ were not significantly altered in the presence of DNA. In contrast, upon formation of the ternary complex of Polλ•gapped DNA•dNTP, two residues R275 and R386 were shielded from HPG modification in all three Polλ constructs (Table 2). Interestingly, the X-ray crystal structure of the ternary complex tPolλ•gapped DNA•ddTTP shows that R386 forms a salt bridge with the γ-phosphate moiety of the incoming ddTTP (Figure 5). Thus, the binding of a dNTP likely shielded R386 from HPG modification. In addition, the X-ray crystal structure23 reveals that R275 forms a salt bridge with the 5’ terminal phosphate moiety of the downstream DNA primer (Figure 5C).
Figure 5.
The crystal structure of the ternary complex of tPolλ (blue), gapped DNA substrate (black), and ddTTP (multi-colored) 23. (A) overall structure; (B) a close-up view detailing the interaction between R386 (red) and ddTTP; (C) a close-up view detailing the interaction between R275 (red) and the 5’-phosphate of the downstream primer terminus.
Pre-steady state kinetic analysis of two R386 mutants of fPolλ
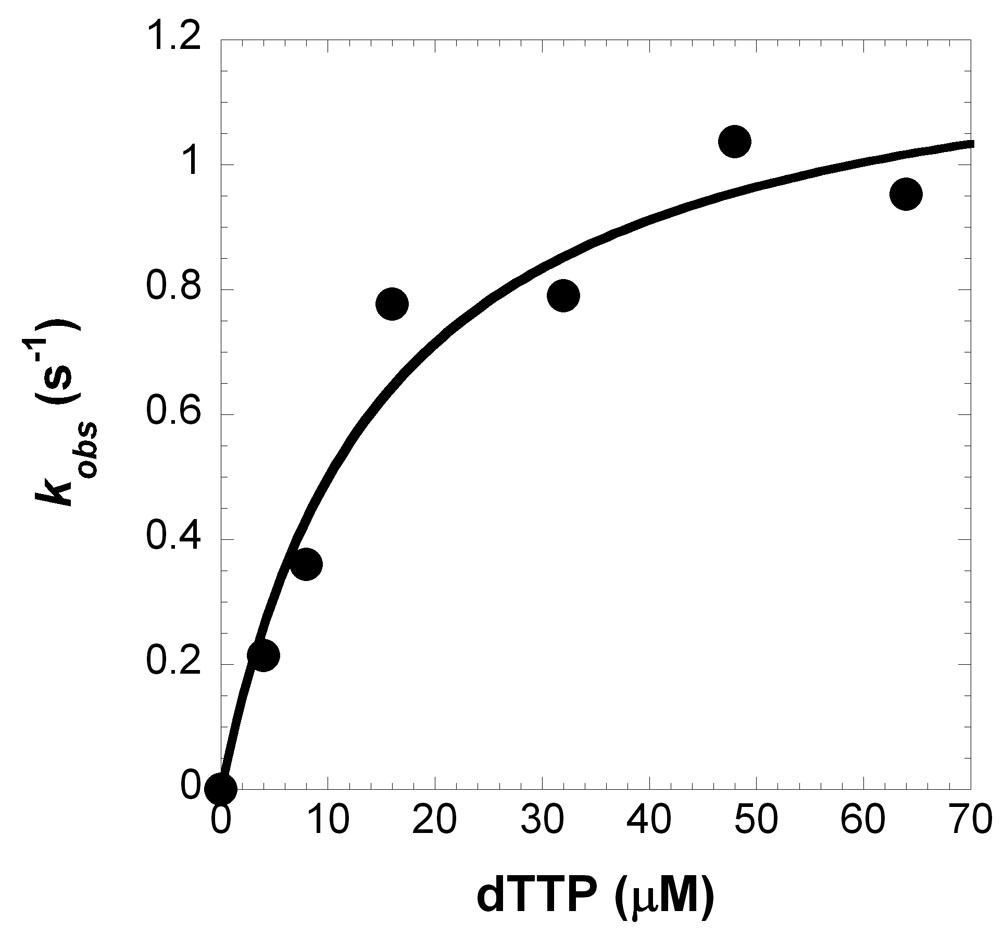
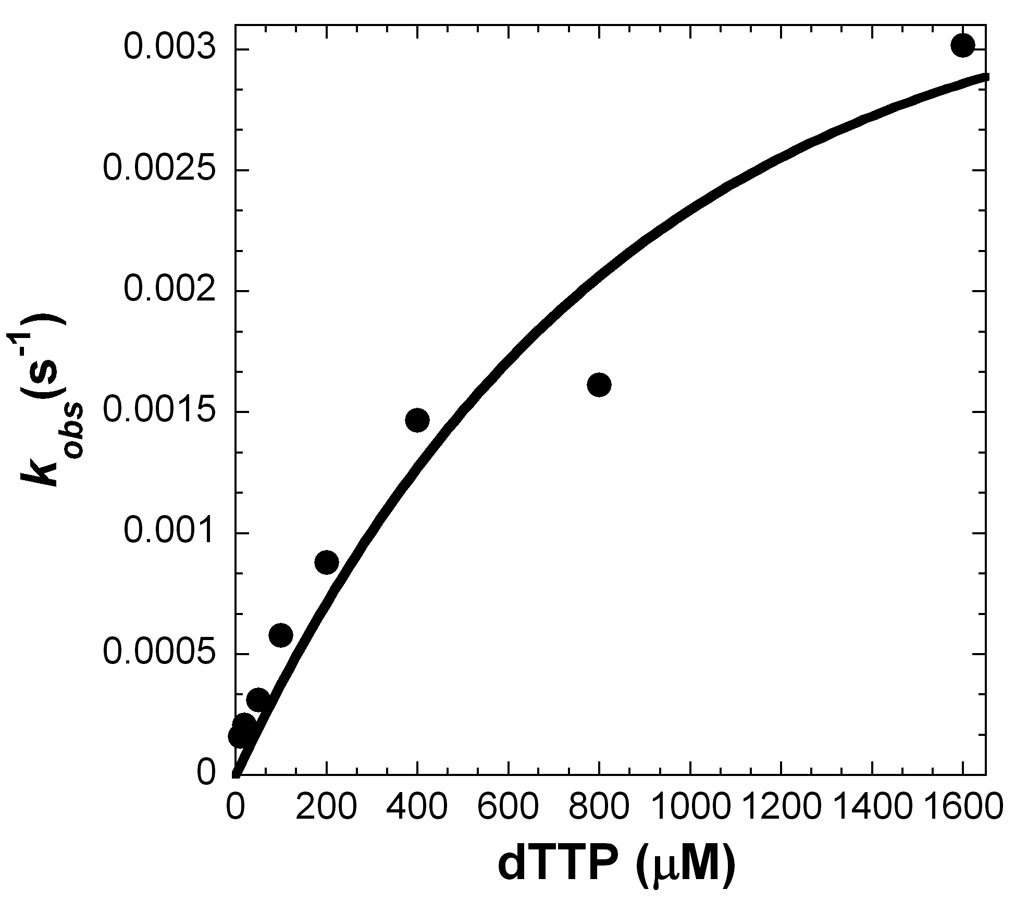
To investigate the significance of the salt bridge formed between R386 and a dNTP, we created two point mutants of fPolλ, R386A and R386E. The purpose of the alanine substitution was to generate a small, neutral side chain at residue 386, while the glutamate substitution was to create a nearly isosteric side chain with a negative charge which could repel a dNTP through charge-charge interactions. To kinetically characterize these two mutants, we determined their gap-filling DNA polymerase activity separately under single-turnover reaction conditions (Materials and Methods). For example, a pre-incubated solution of R386A (300 nM) and 5'-[32P]-labeled 21-19/41A-mer (30 nM, Table 1) was mixed with increasing concentrations of dTTP (4–64 µM) for varying times before being quenched by 0.37 M EDTA. Each time course of product formation was fit to Equation 1 (Materials and Methods) to yield an observed rate constant (kobs) (data not shown). The kobs values were then plotted against the corresponding dTTP concentration (Figure 6A), and the data were fit to Equation 2 (Materials and Methods) to generate a maximum rate of nucleotide incorporation (kp) of 1.3 ± 0.1 s−1 and an equilibrium dissociation constant (Kd) of 15 ± 5 µM (Table 3). Under the same reaction conditions, R386E catalyzed the incorporation of dTTP into 21-19/41A-mer with a kp of 0.005 ± 0.001 s−1 and a Kd of 1000 ± 410 µM (Table 3 and Figure 6B). The substrate specificity, kp/Kd, was calculated to be 0.087 and 5×10−6 µM−1s−1 for R386A and R386E, respectively (Table 3). These values are much lower than 1.5 µM−1s−1 observed with the wild-type fPolλ 17. Thus, both the charge and size of the side chain of R386 are important to the catalytic activity of fPolλ.
Figure 6.
Concentration dependence on the rate of dTTP incorporation into 21-19/41A-mer (Table 1). A pre-incubated solution of enzyme (300 nM) and 5′-[32P]-labeled 21-19/41A-mer DNA (30 nM) was mixed with increasing concentrations of dTTP for various times prior to be quenched by 0.37 M EDTA. The observed rate constants (kobs) were plotted against the concentrations of dTTP and the data were fit to Equation 2 (Materials and Methods). (A) For fPolλ R386A, a kp of 1.3 ± 0.1 s−1 and a Kd of 15 ± 5 µM were determined, and (B) for fPolλ R386E, a kp of 0.005 ± 0.001 s−1 and a Kd of 1000 ± 410 µM were determined.
Table 3.
Kinetic parameters of dTTP incorporation into single-nucleotide gapped 21-19/41A-mer catalyzed by fPolλ variants at 37 °C
| fPolλ Mutant | kp (s−1) | Kd (µM) | kp/Kd (µM−1s−1) | Efficiency Ratioa |
|---|---|---|---|---|
| WTb | 3.9 ± 0.2 | 2.6 ± 0.4 | 1.5 | 1 |
| R386A | 1.3 ± 0.1 | 15 ± 5 | 0.087 | 17 |
| R386E | 0.005 ± 0.001 | 1000 ± 410 | 0.000005 | 300,000 |
Calculated as (kp/Kd)WT/(kp/Kd)Mutant
Values for the WT enzyme are from Fiala et. al.17
DISCUSSION
Structural Implications of Our MS-Based Protein Footprinting Data
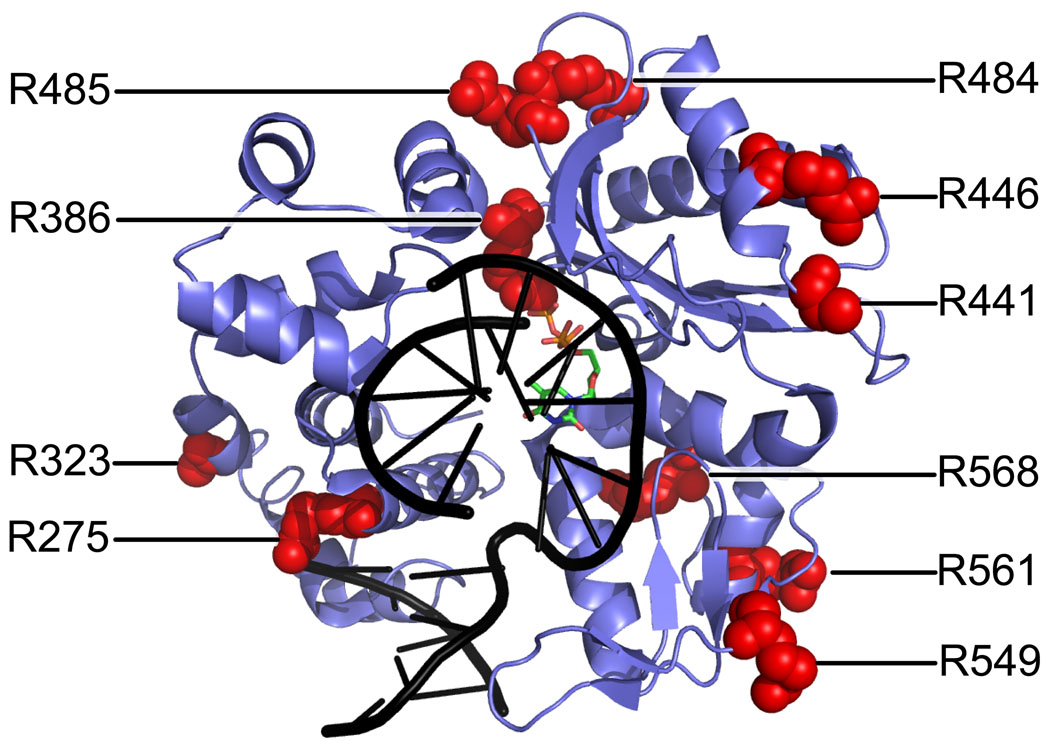
For the MS-based footprinting method, a small molecule like HPG is a proven chemical to readily modify the most solvent accessible Arg residues of a protein in the solution phase 28; 34; 36;37. The reagent modified 13, 10, and 10 Arg residues in the apo forms of fPolλ, dPolλ, and tPolλ, respectively (Table 2). The affected amino acids in the context of the available crystal structure are depicted in Figure 7. Of these only two residues R275 and R386 were selectively protected in the ternary complex (Polλ•DNA•dNTP), not in the binary complex (Polλ•DNA) and apo-Polλ.
Figure 7.
Ternary complex of tPolλ (blue), gapped DNA substrate (black), and incoming ddTTP (multiple colors) 23 showing locations of the arginine residues (red space filling models) modified by HPG in apo-tPolλ.
The shielding of R275 in the ternary complex is consistent with the X-ray crystal structure of tPolλ•gapped DNA•dNTP 23 implicating this residue in charge-charge interactions with the 5′-phosphate of the downstream strand. Since R275 was modified in the tPolλ•gapped DNA complex, it is logical to suggest that these interactions in the context of the binary complex are less stable or highly transient. In contrast, in the presence of an incoming dNTP, R275 was protected from modification. This may have been due to dNTP-induced stabilization of the ternary complex. Such stabilization may have arisen by equalizing unsatisfied positive charges (for example R386, R420) in the polymerase active site. Thus, by introducing the dNTP’s negatively charged triphosphate moiety, positively charged surfaces on the interior of the protein may have experienced less charge-charge repulsion and thus settled into greater proximity. In addition, a resulting reduction of protein dynamics may have stabilized R275 by immobilizing the thumb domain in the tPolλ•DNA•dNTP structure, thus leading to greater interaction of R275 with 5’-phosphate of the downstream strand.
MS-based protein footprinting also revealed selective protection of R386 in the ternary complex (Polλ•DNA•dNTP) and not in the binary complex (Polλ•DNA) and apo-Polλ. These results are in excellent agreement with the X-ray crystal structure of the ternary complex tPolλ•gapped DNA•ddTTP showing that R386 forms a salt bridge with the γ-phosphate moiety of the incoming ddTTP (Figure 5). The fact that we did not observe additional protections or hyper-reactive Arg residues in the ternary complexes suggests that dNTP binding does not induce significant protein conformational changes. Consistently, crystallographic studies of tPolλ 23 have indicated that dNTP binding induces only a repositioning of four active site side chains and a minor shift in the position of two β-strands. Taken together, our MS-based footprinting data indicated that the solution-phase and solid-phase structures of tPolλ were similar but not identical, especially at the local structure surrounding R275.
Comparing the MS-based footprinting spectra of fPolλ, dPolλ and tPolλ (Table 2) in the same substrate binding states reveals remarkable similarity, with the exception of R50, R55, and R57 which reside in the BRCT domain and were expected not to be probed by HPG in dPolλ and tPolλ. Given that nearly all probed Arg residues were modified to similar extents, and that fPolλ, dPolλ, and tPolλ were subjected to conditions designed to mimic discrete states of the catalytic cycle, it is reasonable to assume that this polymerase does not undergo a radical conformational change involving Arg residues as it carries out gap-filling DNA synthesis. However, more experimental evidence is required to substantiate this conclusion. Additionally, few differences between the MS-based footprinting spectra of Polλ•DNA and Polλ•DNA•dNTP (Table 2) do not exclude the possibility of small, local structural rearrangements within the polymerase active site as catalysis occurs.
Structural and Functional Roles of R386
Table 2 illustrates that nucleotide binding to the binary complexes of all three Polλ constructs protected R386 from chemical modification. In the crystal structure of tPolλ•gapped DNA•ddTTP (Figure 5), a strong salt bridge (2.90 and 3.04 Å) likely forms with the γ-phosphate of the bound ddTTP due to charge-charge attraction. Thus, it is most likely the salt bridge that prevents the guanidinium moiety of R386 to react with HPG. Such a salt bridge perhaps strengthens the ground-state binding affinity of an incoming nucleotide, positions it for catalysis, and stabilizes pyrophosphate, the leaving group. This possibility was strongly supported by the kinetic data of R386A and R386E, two point mutants of fPolλ (Table 3). Relative to wild-type fPolλ, R386A catalyzed correct dTTP incorporation into single-nucleotide gapped DNA with a 3-fold lower kp, 6-fold higher Kd, and 17-fold lower incorporation efficiency (kp/Kd). These kinetic effects from the side chain of R386 could be contributed to the positive charge, size, or a combination of both properties. When R386 was mutated to a glutamic acid residue which is of similar size but opposite charge, the kp was reduced by 780-fold, the Kd was increased by 385-fold, and the nucleotide incorporation efficiency was decreased by 300,000-fold. Thus, the positive charge of R386 has a more important role in nucleotide binding and catalysis than the size of its side chain. The R386E mutation could repel the negatively-charged dNTP and significantly weaken its binding. Since the rate-limiting step of the kinetic mechanism for nucleotide incorporation catalyzed by Polλ has not been established, the charge-charge repulsion could either destabilize the transition state or alter the positioning of dNTP during catalysis.
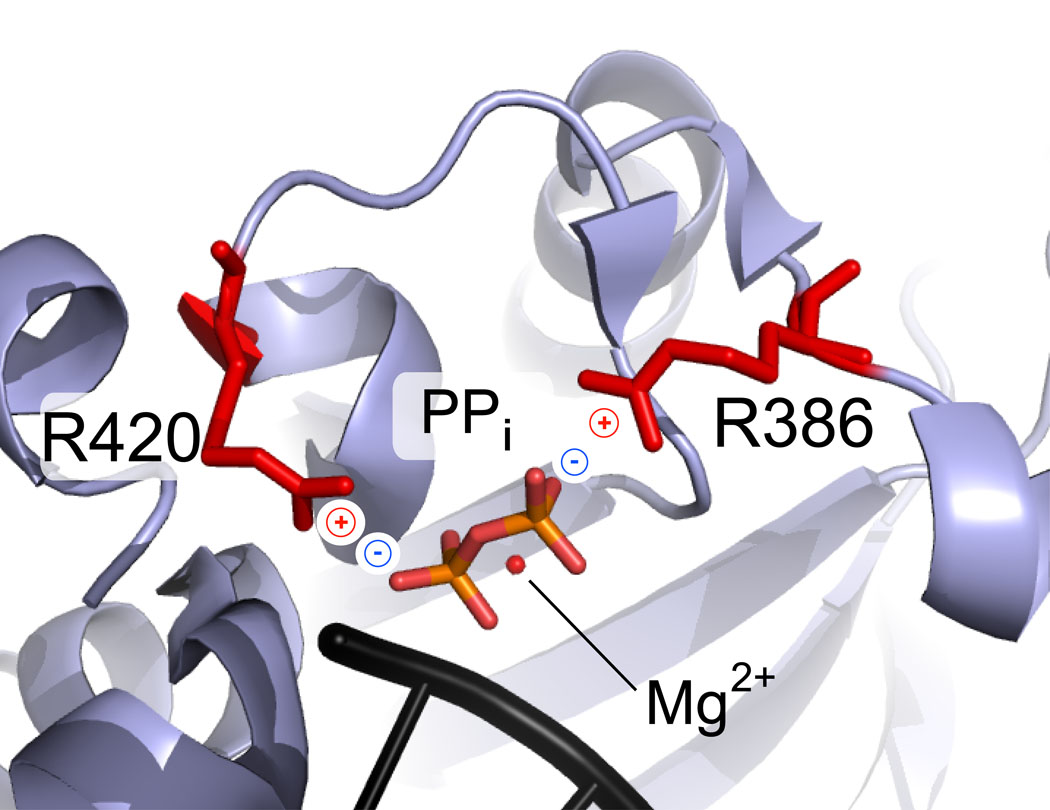
Interestingly, in the crystal structure of the product ternary complex tPolλ•nicked DNA•pyrophosphate 23, R386, R420, and Mg2+ stabilize the negative charges on the pyrophosphate leaving group and facilitate catalysis (Figure 8). The R386E substitution is likely to inhibit the formation of the pyrophosphate product through charge-charge repulsion during catalysis. Thus, the R386E mutation could compromise coordination of both dNTP and pyrophosphate in the enzyme active site, leading to the greatly reduced kp.
Figure 8.
Active site of tPolλ (blue) in the ternary complex with a gapped DNA (black), after the chemistry step but before product release 23. The charge of the leaving group, pyrophosphate (PPi, red-orange), is stabilized by two Arg residues (red) and a divalent metal ion (red sphere).
Conservation of R386 and R420 in Other DNA Polymerases
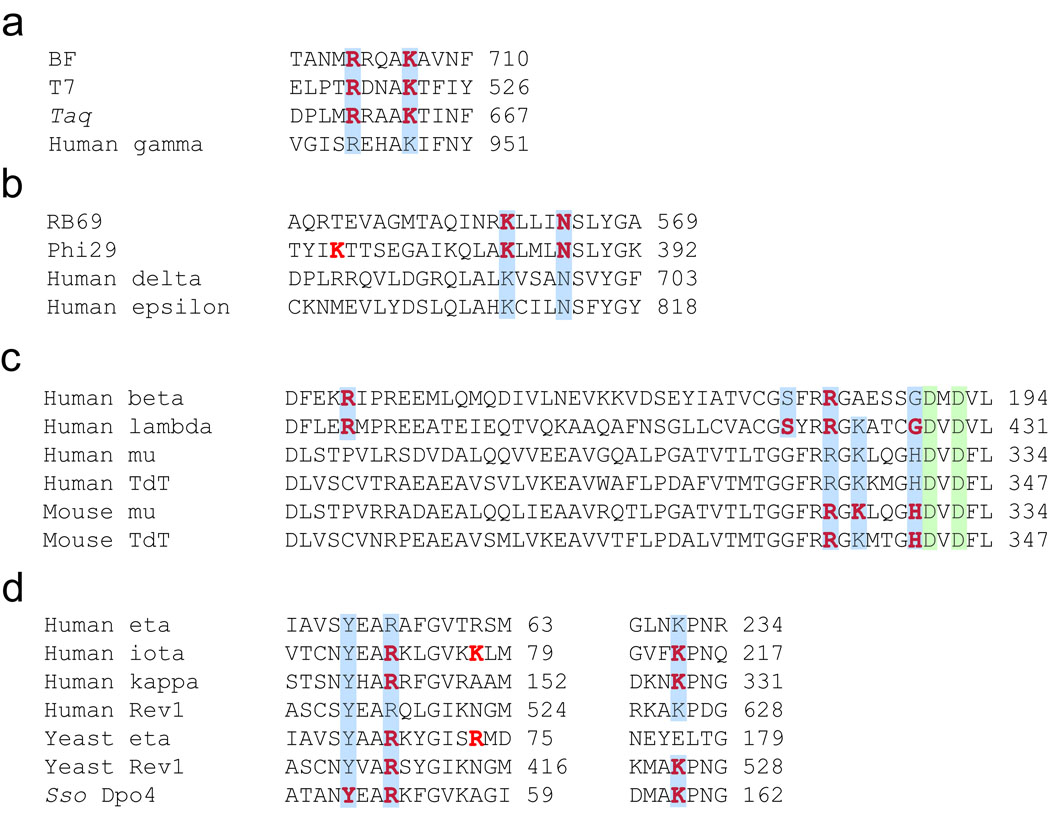
The structural and functional importance of R386 and R420 suggested that other DNA polymerases may use similar positively-charged residues, arginine or lysine, to anchor both the dNTP and pyrophosphate. Thus, we analyzed the available X-ray crystal structures of the other ternary complexes in order to determine whether this feature was conserved among DNA polymerases (Supplementary Table 1). The X-ray crystal structure of Polβ•gapped DNA•ddCTP shows that R149 and R183 are oriented near the triphosphate group of ddCTP in a manner similar to Polλ’s R386 and R420 24. For DNA polymerase Mu and TdT, these two members of the X-family possess the structural homolog of R420 which corresponds to R323 for DNA polymerase Mu and R336 for TdT 38; 39. However, in lieu of a positively-charged residue structurally homologous to R386 and/or R420, the negative charge on the dNTP phosphate can be stabilized by utilizing lysine, histidine, asparagine, or possibly the amide nitrogen of the peptide backbone (Supplementary Table 1) 38; 39. As an example, DNA polymerase IV from Sulfolobus solfataricus employs a network of residues, an arginine (R51), a lysine (K159), a phenylalanine (F11), and two tyrosines (Y10 and Y48), to cooperatively stabilize the negatively-charged phosphates through salt bridge formation and hydrogen-bonding interactions 40; 41.
After examining the published ternary structures of several DNA polymerases listed in Supplementary Table 1, we concluded that the presence of positively-charged residues, either arginine or lysine, at the dNTP and pyrophosphate binding sites of a DNA polymerase active site is almost universal. The alignment of the amino acid sequences of several DNA polymerases, including the A-, B-, X-, and Y-family members for which crystal structures are not available, is shown in Figure 9. Although DNA polymerases are structurally and functionally quite diverse, all DNA polymerases analyzed in this work showed that at least one-positively charged residue was conserved in each DNA polymerase family (Figure 9). In addition to the two metal ion mechanism proposed by Steitz 19, our finding reveals another conserved feature among the DNA polymerases examined herein. However, based upon our mutagenesis results, these positively-charged residues are not an absolute requisite for catalysis, since the catalytic activity of Polλ R386A remained fairly robust (Table 3).
Figure 9.
Conservation of positively charged residues involved in stabilizing the triphosphate moiety and/or the pyrophosphate product. Based upon X-ray crystal structural analysis, amino acid residues within 4.0 Å of the triphosphate moiety (red) were identified. Amino acid sequences of selected (A) A-family, (B) B-family, (C) X-family, and (D) Y-family DNA polymerases were aligned using ClustalW2. Conserved residues are shaded in blue and the conserved catalytic aspartic acid residues are shaded in green.
In summary, our mass spectrometry-based protein footprinting method suggested that tPolλ, dPolλ, and fPolλ do not undergo a dramatic conformational change during catalysis in the solution phase. Moreover, our work identified the importance of stabilizing the negative charges of an incoming nucleotide and the pyrophosphate product, a feature shared by a myriad of DNA polymerases.
MATERIALS AND METHODS
Preparation of Human fPolλ, dPolλ, and tPolλ
Cloning, expression, and purification of human fPolλ17, dPolλ17, and tPolλ 42 were described previously.
Synthetic Oligodeoxyribonucleotides
The oligodeoxyribonucleotides in Table 1 were purchased from Integrated DNA Technologies (Coralville, IA) and purified by denaturing polyacrylamide gel electrophoresis (17% acrylamide, 8 M urea, Tris-borate-EDTA running buffer). Their concentrations were determined by UV absorbance at 260 nm with calculated extinction coefficients. Each single-nucleotide gapped DNA substrate was prepared by heating a mixture of 21-mer (or 22-mer), 19-mer (or 18-mer), and 41-mer in a 1:1.25:1.15 molar ratio, respectively, for 8 min at 95 °C and then cooling the mixture slowly to room temperature over 3 h as described previously 42. For polymerization assays, a DNA primer was 5'-[32P]-labeled by incubating [γ-32P]ATP (GE Healthcare) and T4 polynucleotide kinase (New England BioLabs, Inc) for 1 hour at 37 °C. The unreacted [γ-32P]ATP was subsequently removed by centrifugation via a Bio-Spin-6 column (Bio-Rad Laboratories). Lastly, the radiolabeled primer was annealed to form the appropriate gapped DNA substrate as described above.
Reaction Buffer
Reaction buffer L contained 50 mM Tris-Cl (pH 8.4 at 37 °C), 5 mM MgCl2, 100 mM NaCl, and 0.1mM EDTA and 10% glycerol. For the kinetic assays, reaction buffer L was supplemented with 0.1 mg/mL BSA and 5 mM DTT. This reaction buffer was optimized previously for transient state kinetic analysis of fPolλ and its deletion constructs 17; 42. All reactions reported herein were carried out in the appropriate reaction buffer at 37 °C, and all concentrations refer to the final concentration of the components after mixing.
Mass Spectrometry-Based Protein Footprinting Assay
In parallel experiments using fPolλ, dPolλ, and tPolλ: enzyme (10 µM), the enzyme (10 µM)•22-18/41G–mer (60 µM) binary complex, and the enzyme (10 µM)•22ddC-18/41G–mer (60 µM)•dCTP (100 µM) ternary complex were subjected to chemical modification by HPG. HPG reacts specifically with the guanidine group in an arginine residue resulting in 131 Da mass increase 31; 32; 33;. Previous optimization experiments indicated the Arg/HPG ratio in the range of 1:40 to 1:20 was optimal for achieving very mild modification conditions, under which the integrity of the functional complexes were preserved 29; 35. These conditions were adopted for footprinting purified free Polλ and its binary and ternary complexes. The HPG treatments were carried out at 37 ˚C in the dark for 60 minutes and terminated by addition of 160 mM final concentration of arginine in its free form. Polλ was then separated from DNA and dNTP by SDS-PAGE. The protein bands were excised, destained, dehydrated, and digested with 1 µg of trypsin in 50 mM NH4HCO3 at 25 °C overnight.
Small molecular weight peptides were analyzed by MALDI-ToF MS using AXIMA-CFR instrument (Shimadzu Scientific Instruments). The samples were analyzed with an α-cyano-4-hydroxycinnamic acid matrix as described previously28. Sequence data and Protein Prospector v4.0.6 (http://prospector.ucsf.edu) were used to identify Polλ peptide peaks. Modified arginine residues were assigned by identifying mass peaks that appear only in the spectra of HPG-modified Polλ and that have a molecular weight corresponding to the sum of the predicted peptide fragment plus the 131 Da HPG adduct. For accurate qualitative analysis of the modified peptide peaks, at least two unmodified proteolytic peptide peaks were used as internal controls. A protection was considered to be significant when the intensity of the given modified peptide peak derived from HPG treated free protein was reduced at least 10-fold in the context of the nucleoprotein complexes. A modified peptide peak was considered unprotected when the intensities of the given peptide obtained from free protein and nucleoprotein complexes were within ± 20% of each other. The data were reproducibly compiled and analyzed from at least three independent experimental groups.
Gap-Filling DNA Polymerase Activity Assay for HPG-Modified Enzymes
Following chemical modification with 10 mM HPG at 37 ˚C for 60 minutes (see above) and quenching of the reaction with arginine (160 mM), the gap-filling DNA polymerase activity of HPG-modified Polλ was tested by pre-incubating the modified enzyme (10 µM) with 5’-[32P]-labeled 22-18/41G–mer (60 nM) for 5 min at 37 ˚C. The polymerization reaction was then initiated by the addition of dCTP (100 µM), and the reaction was terminated with EDTA (0.37 M) after 1 minute. Reaction products were separated using denaturing polyacrylamide gel electrophoresis and visualized using autoradiography.
Determination of kp and Kd Values
Both the maximum rate of nucleotide incorporation (kp) and the equilibrium dissociation constant (Kd) of an incoming nucleotide were determined under single-turnover conditions: a pre- incubated solution of Polλ (300 nM) and 5′-[32P]-labeled 21-19/41A-mer (30 nM) was mixed with increasing concentrations of dTTP (GE Healthcare) in reaction buffer L at 37 °C. Reactions were terminated by adding 0.37 M EDTA at various times. For rapid nucleotide incorporations, experiments were performed using a rapid chemical-quench flow apparatus (KinTek). Reaction aliquots, sampled at discrete time points, were analyzed by sequencing gel analysis and quantitated using a Typhoon TRIO PhosphorImager (GE Healthcare). Each time course of product formation was fit to a single-exponential equation (Equation 1) to yield an observed rate constant of nucleotide incorporation (kobs) and reaction amplitude (A). The kobs values were subsequently plotted against the corresponding dTTP concentration, and these data were fit to a hyperbolic equation (Equation 2) using non-linear regression to yield the kp and Kd values.
| 1 |
| 2 |
Supplementary Material
Acknowledgments
Funding
This work was supported by the National Institutes of Health (GM079403 to Z.S., AI062520 and AI077341 to M.K.). J.D.F. and J.A.B. were respectively supported by American Heart Association Predoctoral Fellowships (Grants 0615091B and 0815382D).
LIST OF ABBREVIATIONS
- BER
Base Excision Repair
- BRCT
breast cancer susceptibility gene 1 C-terminal
- dNTP
2′-deoxynucleotide triphosphate
- dRPase
5′-deoxyribose-5-phosphate lyase
- dPolλ
DNA polymerase lambda deletion construct (AA 132-575)
- fPolλ
full-length DNA polymerase lambda
- HPG
p-Hydroxyphenylglyoxal
- MALDI-ToF MS
Matrix Assisted Laser Desorption Ionization Time of Flight Mass Spectrometry
- Polβ
DNA polymerase beta
- Polλ
DNA polymerase lambda
- TdT
terminal deoxynucleotidyltransferase
- tPolλ
truncated DNA polymerase lambda (AA 245-575)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Aravind L, Koonin EV. DNA polymerase beta-like nucleotidyltransferase superfamily: identification of three new families, classification and evolutionary history. Nucleic Acids Res. 1999;27:1609–1618. doi: 10.1093/nar/27.7.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Filee J, Forterre P, Sen-Lin T, Laurent J. Evolution of DNA polymerase families: evidences for multiple gene exchange between cellular and viral proteins. J Mol Evol. 2002;54:763–773. doi: 10.1007/s00239-001-0078-x. [DOI] [PubMed] [Google Scholar]
- 3.Singhal RK, Prasad R, Wilson SH. DNA polymerase beta conducts the gap-filling step in uracil-initiated base excision repair in a bovine testis nuclear extract. J Biol Chem. 1995;270:949–957. doi: 10.1074/jbc.270.2.949. [DOI] [PubMed] [Google Scholar]
- 4.Sobol RW, Horton JK, Kuhn R, Gu H, Singhal RK, Prasad R, Rajewsky K, Wilson SH. Requirement of mammalian DNA polymerase-beta in base-excision repair. Nature. 1996;379:183–186. doi: 10.1038/379183a0. [DOI] [PubMed] [Google Scholar]
- 5.Garcia-Diaz M, Bebenek K, Kunkel TA, Blanco L. Identification of an intrinsic 5’-deoxyribose-5-phosphate lyase activity in human DNA polymerase lambda: a possible role in base excision repair. J Biol Chem. 2001;276:34659–34663. doi: 10.1074/jbc.M106336200. [DOI] [PubMed] [Google Scholar]
- 6.Braithwaite EK, Prasad R, Shock DD, Hou EW, Beard WA, Wilson SH. DNA Polymerase lambda mediates a back-up base excision repair activity in extracts of mouse embryonic fibroblasts. J Biol Chem. 2005 doi: 10.1074/jbc.M411864200. [DOI] [PubMed] [Google Scholar]
- 7.Braithwaite EK, Kedar PS, Lan L, Polosina YY, Asagoshi K, Poltoratsky VP, Horton JK, Miller H, Teebor GW, Yasui A, Wilson SH. DNA polymerase lambda protects mouse fibroblasts against oxidative DNA damage and is recruited to sites of DNA damage/repair. J Biol Chem. 2005;280:31641–31647. doi: 10.1074/jbc.C500256200. [DOI] [PubMed] [Google Scholar]
- 8.Fan W, Wu X. DNA polymerase lambda can elongate on DNA substrates mimicking non-homologous end joining and interact with XRCC4-ligase IV complex. Biochem Biophys Res Commun. 2004;323:1328–1333. doi: 10.1016/j.bbrc.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 9.Lee JW, Blanco L, Zhou T, Garcia-Diaz M, Bebenek K, Kunkel TA, Wang Z, Povirk LF. Implication of DNA polymerase lambda in alignment-based gap filling for nonhomologous DNA end joining in human nuclear extracts. J Biol Chem. 2004;279:805–811. doi: 10.1074/jbc.M307913200. [DOI] [PubMed] [Google Scholar]
- 10.Ma Y, Lu H, Tippin B, Goodman MF, Shimazaki N, Koiwai O, Hsieh CL, Schwarz K, Lieber MR. A biochemically defined system for mammalian nonhomologous DNA end joining. Mol Cell. 2004;16:701–713. doi: 10.1016/j.molcel.2004.11.017. [DOI] [PubMed] [Google Scholar]
- 11.Capp JP, Boudsocq F, Bertrand P, Laroche-Clary A, Pourquier P, Lopez BS, Cazaux C, Hoffmann JS, Canitrot Y. The DNA polymerase lambda is required for the repair of non-compatible DNA double strand breaks by NHEJ in mammalian cells. Nucleic Acids Res. 2006;34:2998–3007. doi: 10.1093/nar/gkl380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bertocci B, De Smet A, Weill JC, Reynaud CA. Nonoverlapping functions of DNA polymerases mu, lambda, and terminal deoxynucleotidyltransferase during immunoglobulin V(D)J recombination in vivo. Immunity. 2006;25:31–41. doi: 10.1016/j.immuni.2006.04.013. [DOI] [PubMed] [Google Scholar]
- 13.Garcia-Diaz M, Dominguez O, Lopez-Fernandez LA, de Lera LT, Saniger ML, Ruiz JF, Parraga M, Garcia-Ortiz MJ, Kirchhoff T, del Mazo J, Bernad A, Blanco L. DNA polymerase lambda (Pol lambda), a novel eukaryotic DNA polymerase with a potential role in meiosis. J Mol Biol. 2000;301:851–867. doi: 10.1006/jmbi.2000.4005. [DOI] [PubMed] [Google Scholar]
- 14.Nagasawa K, Kitamura K, Yasui A, Nimura Y, Ikeda K, Hirai M, Matsukage A, Nakanishi M. Identification and characterization of human DNA polymerase beta 2, a DNA polymerase beta -related enzyme. J Biol Chem. 2000;275:31233–31238. doi: 10.1074/jbc.M004263200. [DOI] [PubMed] [Google Scholar]
- 15.Aoufouchi S, Flatter E, Dahan A, Faili A, Bertocci B, Storck S, Delbos F, Cocea L, Gupta N, Weill JC, Reynaud CA. Two novel human and mouse DNA polymerases of the polX family. Nucleic Acids Res. 2000;28:3684–3693. doi: 10.1093/nar/28.18.3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shimazaki N, Yoshida K, Kobayashi T, Toji S, Tamai K, Koiwai O. Over-expression of human DNA polymerase lambda in E. coli and characterization of the recombinant enzyme. Genes Cells. 2002;7:639–651. doi: 10.1046/j.1365-2443.2002.00547.x. [DOI] [PubMed] [Google Scholar]
- 17.Fiala KA, Duym WW, Zhang J, Suo Z. Upregulation of the fidelity of human DNA polymerase lambda by its non-enzymatic proline-rich domain. J Biol Chem. 2006 doi: 10.1074/jbc.M601178200. [DOI] [PubMed] [Google Scholar]
- 18.Ollis DL, Brick P, Hamlin R, Xuong NG, Steitz TA. Structure of large fragment of Escherichia coli DNA polymerase I complexed with dTMP. Nature. 1985;313:762–766. doi: 10.1038/313762a0. [DOI] [PubMed] [Google Scholar]
- 19.Steitz TA. DNA- and RNA-dependent DNA polymerases. Curr Opin Struct Biol. 1993;3:31–38. [Google Scholar]
- 20.Steitz TA. A mechanism for all polymerases. Nature. 1998;391:231–232. doi: 10.1038/34542. [DOI] [PubMed] [Google Scholar]
- 21.Hubscher U, Maga G, Spadari S. Eukaryotic DNA polymerases. Annu Rev Biochem. 2002;71:133–163. doi: 10.1146/annurev.biochem.71.090501.150041. [DOI] [PubMed] [Google Scholar]
- 22.Johnson KA. Conformational coupling in DNA polymerase fidelity. Annu Rev Biochem. 1993;62:685–713. doi: 10.1146/annurev.bi.62.070193.003345. [DOI] [PubMed] [Google Scholar]
- 23.Garcia-Diaz M, Bebenek K, Krahn JM, Kunkel TA, Pedersen LC. A closed conformation for the Pol lambda catalytic cycle. Nat Struct Mol Biol. 2005;12:97–98. doi: 10.1038/nsmb876. [DOI] [PubMed] [Google Scholar]
- 24.Sawaya MR, Prasad R, Wilson SH, Kraut J, Pelletier H. Crystal structures of human DNA polymerase beta complexed with gapped and nicked DNA: evidence for an induced fit mechanism. Biochemistry. 1997;36:11205–11215. doi: 10.1021/bi9703812. [DOI] [PubMed] [Google Scholar]
- 25.Pelletier H, Sawaya MR, Kumar A, Wilson SH, Kraut J. Structures of ternary complexes of rat DNA polymerase beta, a DNA template-primer, and ddCTP. Science. 1994;264:1891–1903. [PubMed] [Google Scholar]
- 26.Kunkel TA, Bebenek K. DNA replication fidelity. Annu Rev Biochem. 2000;69:497–529. doi: 10.1146/annurev.biochem.69.1.497. [DOI] [PubMed] [Google Scholar]
- 27.Petruska J, Sowers LC, Goodman MF. Comparison of nucleotide interactions in water, proteins, and vacuum: model for DNA polymerase fidelity. Proc Natl Acad Sci U S A. 1986;83:1559–1562. doi: 10.1073/pnas.83.6.1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kvaratskhelia M, Miller JT, Budihas SR, Pannell LK, Le Grice, F S. Identification of specific HIV-1 reverse transcriptase contacts to the viral RNA:tRNA complex by mass spectrometry and a primary amine selective reagent. Proc Natl Acad Sci U S A. 2002;99:15988–15993. doi: 10.1073/pnas.252550199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao Z, McKee CJ, Kessl JJ, Santos WL, Daigle JE, Engelman A, Verdine G, Kvaratskhelia M. Subunit-specific protein footprinting reveals significant structural rearrangements and a role for N-terminal Lys-14 of HIV-1 Integrase during viral DNA binding. J Biol Chem. 2008;283:5632–5641. doi: 10.1074/jbc.M705241200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McKee CJ, Kessl JJ, Shkriabai N, Dar MJ, Engelman A, Kvaratskhelia M. Dynamic Modulation of HIV-1 Integrase Structure and Function by Cellular Lens Epithelium-derived Growth Factor (LEDGF) Protein. J Biol Chem. 2008;283:31802–31812. doi: 10.1074/jbc.M805843200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wood TD, Guan Z, Borders CL, Jr, Chen LH, Kenyon GL, McLafferty FW. Creatine kinase: essential arginine residues at the nucleotide binding site identified by chemical modification and high-resolution tandem mass spectrometry. Proc Natl Acad Sci U S A. 1998;95:3362–3365. doi: 10.1073/pnas.95.7.3362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McLafferty FW, Kelleher NL, Begley TP, Fridriksson EK, Zubarev RA, Horn DM. Two-dimensional mass spectrometry of biomolecules at the subfemtomole level. Curr Opin Chem Biol. 1998;2:571–578. doi: 10.1016/s1367-5931(98)80085-9. [DOI] [PubMed] [Google Scholar]
- 33.Hager-Braun C, Tomer KB. Characterization of the tertiary structure of soluble CD4 bound to glycosylated full-length HIVgp120 by chemical modification of arginine residues and mass spectrometric analysis. Biochemistry. 2002;41:1759–1766. doi: 10.1021/bi011626k. [DOI] [PubMed] [Google Scholar]
- 34.Liu Y, Kvaratskhelia M, Hess S, Qu Y, Zou Y. Modulation of replication protein A function by its hyperphosphorylation-induced conformational change involving DNA binding domain B. J Biol Chem. 2005;280:32775–32783. doi: 10.1074/jbc.M505705200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Deval J, D'Abramo CM, Zhao Z, McCormick S, Coutsinos D, Hess S, Kvaratskhelia M, Gotte M. High resolution footprinting of the hepatitis C virus polymerase NS5B in complex with RNA. J Biol Chem. 2007;282:16907–16916. doi: 10.1074/jbc.M701973200. [DOI] [PubMed] [Google Scholar]
- 36.Williams KL, Zhang Y, Shkriabai N, Karki RG, Nicklaus MC, Kotrikadze N, Hess S, Le Grice SF, Craigie R, Pathak VK, Kvaratskhelia M. Mass spectrometric analysis of the HIV-1 integrase-pyridoxal 5'-phosphate complex reveals a new binding site for a nucleotide inhibitor. J Biol Chem. 2005;280:7949–7955. doi: 10.1074/jbc.M413579200. [DOI] [PubMed] [Google Scholar]
- 37.Shell SM, Hess S, Kvaratskhelia M, Zou Y. Mass spectrometric identification of lysines involved in the interaction of human replication protein a with single-stranded DNA. Biochemistry. 2005;44:971–978. doi: 10.1021/bi048208a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wilson RC, Pata JD. Structural insights into the generation of single-base deletions by the Y family DNA polymerase dbh. Mol Cell. 2008;29:767–779. doi: 10.1016/j.molcel.2008.01.014. [DOI] [PubMed] [Google Scholar]
- 39.Moon AF, Garcia-Diaz M, Bebenek K, Davis BJ, Zhong X, Ramsden DA, Kunkel TA, Pedersen LC. Structural insight into the substrate specificity of DNA Polymerase mu. Nat Struct Mol Biol. 2007;14:45–53. doi: 10.1038/nsmb1180. [DOI] [PubMed] [Google Scholar]
- 40.Ling H, Boudsocq F, Woodgate R, Yang W. Crystal structure of a Y-family DNA polymerase in action: a mechanism for error-prone and lesion-bypass replication. Cell. 2001;107:91–102. doi: 10.1016/s0092-8674(01)00515-3. [DOI] [PubMed] [Google Scholar]
- 41.Vaisman A, Ling H, Woodgate R, Yang W. Fidelity of Dpo4: effect of metal ions, nucleotide selection and pyrophosphorolysis. Embo J. 2005;24:2957–2967. doi: 10.1038/sj.emboj.7600786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fiala KA, Abdel-Gawad W, Suo Z. Pre-Steady-State Kinetic Studies of the Fidelity and Mechanism of Polymerization Catalyzed by Truncated Human DNA Polymerase lambda. Biochemistry. 2004;43:6751–6762. doi: 10.1021/bi049975c. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.