Abstract
Omi/HtrA2 is a mitochondrial serine protease that has a dual function: while confined in the mitochondria, it promotes cell survival, but when released into the cytoplasm, it participates in caspase-dependent as well as caspase-independent cell death. To investigate the mechanism of Omi/HtrA2's function, we set out to isolate and characterize novel substrates for this protease. We have identified Thanatos-associated protein 5 (THAP5) as a specific interactor and substrate of Omi/HtrA2 in cells undergoing apoptosis. This protein is an uncharacterized member of the THAP family of proteins. THAP5 has a unique pattern of expression and is found predominantly in the human heart, although a very low expression is also seen in the human brain and muscle. THAP5 protein is localized in the nucleus and, when ectopically expressed, induces cell cycle arrest. During apoptosis, THAP5 protein is degraded, and this process can be blocked using a specific Omi/HtrA2 inhibitor, leading to reduced cell death. In patients with coronary artery disease, THAP5 protein levels substantially decrease in the myocardial infarction area, suggesting a potential role of this protein in human heart disease. This work identifies human THAP5 as a cardiac-specific nuclear protein that controls cell cycle progression. Furthermore, during apoptosis, THAP5 is cleaved and removed by the proapoptotic Omi/HtrA2 protease. Taken together, we provide evidence to support that THAP5 and its regulation by Omi/HtrA2 provide a new link between cell cycle control and apoptosis in cardiomyocytes.
Keywords: Omi/HtrA2, Thanatos-associated protein 5, coronary artery disease, apoptosis
omi/htra2 is a mitochondrial serine protease that has an essential role in both mitochondrial homeostasis, as well as cell death (49). Most of the studies on Omi/HtrA2 have substantially contributed to our understanding of the mechanism of its proapoptotic function. On the contrary, very little is known about its normal function within the mitochondria. Upon induction of apoptosis, Omi/HtrA2 is released to the cytoplasm, where it participates in caspase-dependent and caspase-independent cell death (21, 34, 45, 48, 50). The mechanism of its proapoptotic activity involves degradation of specific substrates that include mitochondrial protein HS1-associated protein X-1, cytoplasmic proteins X-chromosome-linked inhibitor, PEA/PED, and Apollon/Bruce (10, 41, 44, 47, 51), as well as nuclear factors Grim-19, WARTS, and p73 (28, 29, 31, 33). The protease activity of Omi/HtrA2 is always necessary and essential for its proapoptotic function. We have used the yeast two-hybrid system to isolate and characterize new Omi/HtrA2 interacting proteins (10). These interacting proteins could be new substrates of Omi/HtrA2 or modulators of its proteolytic activity. Previous studies have shown that the proteolytic activity of Omi/HtrA2 can be regulated through specific protein-protein interactions mediated via its PDZ domain (36). In this report, we describe one such new Omi/HtrA2 interactor, the Thanatos-associated protein 5 (THAP5) protein. The THAP family of proteins comprises a group of nuclear factors defined by the presence of an ∼90-residue protein motif (the THAP domain) (40). THAP domains are atypical zinc fingers with specific zinc-dependent DNA binding activity and show similarity to the site-specific DNA binding domain of the P element transposase from Drosophila (38, 40). THAP proteins are transcription factors, and the limited information that exists on their function suggests that they might be involved in gene regulation, cell cycle control, and/or apoptosis (6, 12, 39). THAP5 is the fifth member in the 12-member family of human THAP proteins and is unique since, outside its THAP domain, it shares no sequence homology with any other reported protein. THAP5 interacted with Omi/HtrA2 both in yeast and mammalian cells under proapoptotic conditions, where Omi/HtrA2 is known to be released from mitochondria to the cytoplasm. Furthermore, THAP5 could be cleaved very efficiently in vitro by Omi/HtrA2 protease. Since very little is known about the function of THAP5, we performed a detailed study to characterize its normal function and the significance of its interaction and degradation by Omi/HtrA2. We found THAP5 to be a tissue-specific nuclear factor that is predominantly expressed in the human heart. Interestingly, there is no mouse or rat ortholog of THAP5; this is a characteristic of some THAP family members, since it has also been reported for four other proteins, namely THAP6, THAP8, THAP9, and THAP10 (12, 38). The normal function of THAP5 is the regulation of cell cycle, and ectopic expression of the protein caused cell cycle arrest. During cell death, THAP5 was cleaved and removed by Omi/HtrA2 in cells treated with cisplatin and H2O2, but it was not affected in cells treated with etoposide or camptothecin. Using the ucf-101 inhibitor of Omi/HtrA2, we could very effectively block THAP5 degradation and protect cells from undergoing apoptosis. The degradation of THAP5 seen during experimentally induced cell death or cell injury is a physiological event that follows cellular damage and was observed in the myocardial infarction (MI) area of the heart tissues from patients with coronary artery disease (CAD).
MATERIALS AND METHODS
Yeast two-hybrid screen.
We used the yeast two-hybrid system to screen a HeLa, as well as a melanocyte cDNA library, as previously described (10). The bait used was the mature, proteolytically active form of the Omi/HtrA2 protein (aa 134-458) cloned in the pGilda (Clontech) bait vector. Several interacting proteins were identified in this screen. One of these Omi/HtrA2 interactors isolated from the melanocyte cDNA library was a partial clone of a previously uncharacterized protein called THAP5. The full-length cDNA for THAP5 encodes 395 amino acids and was isolated from a Marathon Ready human heart cDNA library (Clontech). The specificity of THAP5 interaction with Omi/HtrA2 in yeast was tested using HtrA1, a mammalian homolog of Omi/HtrA2 that has 68% amino acid sequence similarity. The presence and stability of the recombinant proteins in yeast cells was monitored by Western blot analysis using LexA antibodies (for baits) or HA antibodies (for preys).
Interaction between Omi/HtrA2 and THAP5 in mammalian cells.
Human embryonic kidney (HEK)-293 cells were transfected in duplicates with either pEGFP-C1 empty vector (Clontech) or enhanced green fluorescent protein (EGFP)-THAP5 plasmid using Lipofectamine 2000 reagent (Invitrogen). EGFP-THAP5 encodes the full-length THAP5 protein fused in frame to EGFP-C1 vector. Fourteen hours later, one-half of the cells were treated with cisplatin (50 μM) for 10 h. Cell lysates were prepared using RIPA buffer (150 mM NaCl, 50 mM Tris·HCl, pH 7.5, 1% Nonidet P-40, 0.25% deoxycholic acid sodium salt) containing the protease-inhibitor cocktail (Roche). Approximately 200 μg of total protein cell lysates were precleared by mixing with protein G-agarose beads (Roche) for 1 h, followed by incubation with the Omi/HtrA2 polyclonal antibody (10) for 2 h at 4°C. Protein G-agarose beads were then added and allowed to bind overnight at 4°C. Immunoprecipitates were collected by brief centrifugation, washed extensively with RIPA buffer, and resolved by SDS-PAGE. They were then electro-transferred onto a polyvinylidene difluoride (PVDF) membrane and probed with a mouse monoclonal green fluorescent protein (GFP) antibody (Santa Cruz Biotechnology), followed by a secondary goat anti-mouse horseradish peroxidase-conjugated antibody, and the immunocomplex was visualized by enhanced chemiluminescence (Pierce). We also performed the reverse of this experiment by transfecting HEK-293 cells with pEGFP-N1-Omi (1-458). Approximately 200 μg of total protein cell lysates were precleared by mixing with protein G-agarose beads (Roche) for 1 h and then incubated with THAP5 polyclonal antibody, followed by Western blot using the GFP antibody, as described above.
Northern blot analysis of THAP5 mRNA expression in human tissues.
Human mRNA tissue blot (Clontech), representing 12 human tissues, was probed with a radiolabeled DNA probe corresponding to THAP5 protein sequence residues 163-395. This DNA sequence is unique and has no homology to any other known gene in the GenBank. The blot was hybridized with the radiolabeled probe at 42°C, washed at 65°C, and subjected to autoradiography (17). To verify that an equal amount of mRNA was present on each lane, the blot was stripped and reprobed for β-actin mRNA expression.
Degradation assay.
The ability of His-Omi134-458 to cleave recombinant His-THAP5 protein in vitro was investigated. The full-length cDNA for the THAP5 protein was cloned in frame in pET-28 vector (Novagen). Bacterially expressed recombinant His-THAP5 was purified on nickel-nitrilotriacetic acid (NTA-agarose) affinity resin, as described (10). The synthesis and properties of the His-Omi134-458 have been previously reported (10). His-Omi134-458 was incubated with His-THAP5 in 15 μl of reaction volume in assay buffer (20 mM Na2HPO4, pH 8, 200 mM NaCl, and 5% glycerol). In some of the reactions, the ucf-101 inhibitor was used. After various incubation times at 37°C, the reactions were stopped through the addition of SDS sample buffer. Reaction products were analyzed by SDS-PAGE followed by Coomassie blue staining.
Cell culture.
HEK-293 cells were grown in Dulbecco's modified Eagle's medium, supplemented with 10% fetal calf serum (Hyclone), 2 mM l-glutamine, 1.5 g/l sodium bicarbonate, 1 mM sodium pyruvate, 50 U/ml penicillin, and 50 μg/ml streptomycin (Invitrogen). HeLa cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum, 2 mM l-glutamine, 50 U/ml penicillin, and 50 μg/ml streptomycin.
Subcellular localization of the THAP5 protein.
To investigate the subcellular localization of the THAP5 protein, the full-length cDNA (aa 1-395) was cloned in frame into EGFP-C1 vector (Clontech). Furthermore, DNA sequence corresponding to amino acids 1-162 or 163-395 of THAP5 protein were also amplified by PCR and cloned into EGFP-C1 vector. HeLa cells were grown on glass coverslips in 12-well plates. Approximately 60% confluent cells were transiently transfected with 1 μg of the various GFP constructs using Lipofectamine 2000 Transfection Reagent (Invitrogen). Twenty-four hours after transfection, cells were washed and fixed in 4% paraformaldehyde, permeabilized with 0.2% Triton X-100, and incubated with Texas red-phalloidin (Molecular Probes) to stain cytoplasm. The coverslips were then washed and placed on microscope slides using Fluoromount-G as the mounting solution. Slides were observed using a LSM510 confocal laser-scanning microscope (Zeiss). The expression and stability of the various GFP fusion proteins were verified by Western blot analysis using GFP-specific antibodies (Santa Cruz Biotechnology).
Expression, purification of His-tagged THAP5163-395 protein, and antibody production.
PCR was used to amplify DNA sequence corresponding to a partial carboxyl terminus THAP5 (aa 163-395) polypeptide. The PCR product was cloned in-frame in the bacterial expression vector pET-28 (Novagen). For protein expression, BL21 (DE3) (Novagen) bacteria were transformed with pET-THAP5 and single colonies were grown overnight in Luria Bertani medium containing kanamycin. The overnight culture (1 ml) was used to inoculate 1 liter of Luria Bertani medium, and growth was continued at 37°C until the 600-nm optical density was ∼0.8. At this time, 2 mM isopropylthiogalactoside was added, and the culture was placed in a shaking incubator for 4 h at 25°C. Bacteria were harvested by centrifugation and lysed in a buffer containing 6 M urea, 0.1 M NaH2PO4, 20 mM Tris·HCl, pH 8, and a protease inhibitor cocktail (Sigma). The bacterial suspension was then sonicated, and recombinant His-THAP5163-395 protein was purified using Ni-NTA His-Bind Resin (Novagen). The quality of the His-THAP5163-395 protein was monitored by SDS-PAGE, followed by Coomassie blue staining. To make the polyclonal antibody, 15 mg of purified human His-THAP5163-395 were used to immunize two rabbits. Immunization, production, and purification of the polyclonal THAP5 antibody were performed by New England Peptide.
Western blot analysis of THAP5 protein expression in various human tissues.
Because THAP5 is expressed only in human cells, we used a commercially available Western blot that represents five human tissues (Calbiochem). Each lane of the blot contains 10 μg of crude protein extract from the corresponding tissue. We used our THAP5 polyclonal antibody at a 1:5,000 dilution. We also tested THAP5 expression in HEK-293 and HeLa cell lines. The method and conditions for the Western blot analysis have been previously described (8, 10).
Cell cycle analysis.
HEK-293 cells were seeded onto six-well plates, synchronized with serum starvation (13, 24), and then transiently transfected with EGFP-C1 empty vector (Clontech) or EGFP-THAP5 using Lipofectamine 2000 Transfection Reagent (Invitrogen). Forty or fifty hours after transfection, cells were washed and fixed in 4% paraformaldehyde. The cells were then washed, permeabilized with 0.1% saponin, and stained with 7-amino-actinomycin D (5 μl per 100 μl of PBS) and incubated for 30 min at 37°C. GFP-positive cells in the transfected population were measured using a FACSCalibur Flow Cytometer (BD Biosciences) (7) and analyzed using ModFitLT software.
Cell death assays.
HeLa cells were treated for 12 h with different chemicals: camptothecin (100 μM), cisplatin (5 μM), etoposide (20 μM), H2O2 (0.2 mM), and staurosporine (100 μM). HeLa cells were grown in six-well plates in the appropriate medium until they reached 70–80% confluency; they were then treated with ucf-101 (20 or 30 μM) for 20 min followed by cisplatin (5 μM) or H2O2 (0.2 mM) treatment for 12 h. Cells were detached with 1× Trypsin-EDTA (Gibco) and washed twice with ice-cold PBS. One-half of them were used for Western blot analysis, and the rest for apoptotic assay. Western Blot analysis was performed as described (8, 10). The apoptotic assay was performed according to BD Biosciences protocol. Briefly, cells were suspended in 1× binding buffer and stained with annexin V (apoptotic cell) and 7-amino-actinomycin D (necrotic cells) (15, 16, 22). Samples were analyzed on a FACSCalibur Flow Cytometer (BD Biosciences).
Expression of THAP5 in the heart of patients with CAD.
Human cardiac tissues were prepared as described (42). Briefly, human cardiac tissue samples were taken from the left ventricles of failing human hearts that were explanted in the course of heart transplantation. The study's protocol was approved by the local ethics committee, and written, informed consent was given by patients, according to the National Disease Research Interchange. Hearts from patients with end-stage heart failure who were undergoing cardiac transplantation because of either dilated cardiomyopathy (DCM) or CAD were investigated. Furthermore, healthy donor (HD) hearts that were ultimately rejected for transplantation because of technical reasons were also included in this study as healthy tissue control. For Western blot analysis, 100 mg of tissue were ground in liquid nitrogen and homogenized with an Ultra-Turrax T8 (IKA-werke) in ice-cold lysis buffer containing 150 mM NaCl, 20 mM Tris·HCl, pH 7.6, 1 mM CaCl2, 1 mM MgCl2, 10% glycerol, 1% Nonidet P-40, and protease and phosphatase inhibitors cocktail (Roche). Homogenate was cleared by centrifugation for 10 min at 14,000 g. Protein concentrations were determined using the Bio-Rad protein assay. Approximately 20 μg of whole cell extracts were resuspended in SDS sample buffer and boiled for 3 min. Samples were resolved by SDS-PAGE and electrotransferred onto PVDF membranes (Pall Life Sciences) using a semi-dry cell transfer blot (Bio-Rad). Nonfat dry milk (4%) in TBST buffer was used to block any nonspecific binding. The membrane was incubated with our THAP5 polyclonal antibody (1:5,000), followed by a secondary horseradish peroxidase-conjugated goat anti-rabbit (Jackson ImmunoResearch) (1:15,000) and visualization by enhanced chemiluminescence (Pierce).
Statistical analysis.
All quantitative data are expressed as means ± SD. Differences among groups were analyzed by one-way ANOVA, followed by Tukey's post hoc test. A value of P < 0.05 was considered significant.
RESULTS
Isolation of THAP5 as an Omi/HtrA2 interactor.
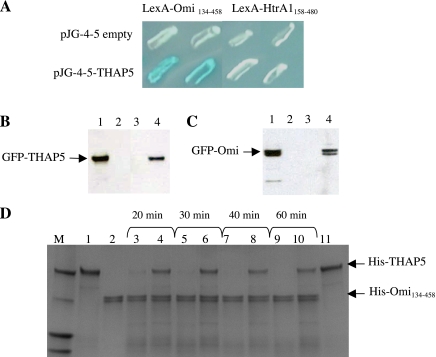
We used the yeast two-hybrid system to isolate Omi/HtrA2 interactors. We screened two different cDNA libraries derived from HeLa cells and primary human melanocytes. We used two different cDNA libraries to screen as many diverse proteins as possible, including any potential tissue-specific interactors of Omi/HtrA2. Furthermore, one of the cDNA libraries was prepared from primary cells to avoid a potential problem often seen in cDNA libraries prepared from transformed cell lines. Cells lines such as HeLa used here often have deregulated expression of genes involved in cell growth as well as underrepresentation of proapoptotic cDNAs. The bait in this screen was the LexA-Omi134-458, which represents the mature active form of the Omi/HtrA2 enzyme (10). The screen was performed as described, and several novel Omi/HtrA2 interacting proteins were identified (10). One of the specific interactors from the human melanocyte cDNA library was a partial polypeptide of THAP5. THAP5 is the fifth member of a recently described 12-member family of proteins characterized by the presence of a THAP (Thanatos, Greek for death) motif at the amino terminus of the protein (40). Using specific primers and rapid amplification of cDNA ends, we were able to isolate the full-length cDNA, which was then cloned back into the pJG4-5 vector, and its interaction with Omi/HtrA2 was monitored. Figure 1A shows that full-length THAP5 interacts strongly with Omi/HtrA2. Furthermore, the specificity of the interaction was tested using HtrA1, a human homolog of Omi/HtrA2 that has 68% amino acid sequence similarity. No interaction was observed in this yeast two-hybrid assay between these two proteins (Fig. 1A). The expression and stability of the different baits and preys were monitored and verified by Western blot analysis (results not shown).
Fig. 1.
Thanatos-associated protein (THAP) 5 is an interactor and substrate of Omi/HtrA2 protease. A: interaction of Omi/HtrA2 with THAP5 protein in yeast. Yeast colonies were transformed with plasmids encoding the indicated baits and full-length THAP5 cloned into the prey vector. The specificity of THAP5 and Omi/HtrA2 interaction was verified using a closely related homolog, the HtrA1 protein. Blue color results are from a positive protein-protein interaction. B: interaction of Omi/HtrA2 and THAP5 in mammalian cells during apoptosis. Human embryonic kidney (HEK)-293 cells were plated in duplicates in 60-mm dishes and then transfected with either enhanced green fluorescent protein (EGFP), EGFP-THAP5, or green fluorescent protein (GFP)-Omi. Twelve hours after transfection, one plate was treated with 50 μM cisplatin to induce apoptosis, and one plate was used as control. Cell lysates were prepared as described in materials and methods. A polyclonal Omi/HtrA2 antibody was used to immunoprecipitate the endogenous Omi/HtrA2 and any associated proteins. The immunoprecipitated complex was resolved on SDS-PAGE and transferred to a polyvinylidene difluoride (PVDF) membrane, and the presence of GFP-THAP5 fusion protein was detected using a specific anti-GFP antibody. Lane 1 shows crude lysates of GFP-THAP5 transfected cells. Lane 2 shows coimmunoprecipitation (co-IP) lysates obtained from cells transfected with GFP empty vector. Lane 3 shows co-IP lysates obtained from cells transfected with GFP-THAP5 control cells. Lane 4 shows co-IP lysates obtained from cells transfected with GFP-THAP5 and then treated with cisplatin. C: the reverse experiment described in B. HEK-293 cells were now transfected with GFP-Omi and processed exactly as in B, except THAP5 antiserum was used in the co-IP and anti-GFP on the Western blot to detect the GFP-Omi. Lane 1 shows total cell lysates of GFP-Omi transfected cells. Lane 2 shows co-IP lysates obtained from cells transfected with GFP empty vector. Lane 3 shows co-IP lysates obtained from cells transfected with GFP-Omi control cells. Lane 4 shows co-IP lysates obtained from cells transfected with GFP-Omi and then treated with cisplatin. In both B and C, THAP5 was coprecipitated with Omi/HtrA2, but only in cells in which apoptosis was induced. D: THAP5 is cleaved by Omi/HtrA2 protease in vitro. Purified His-THAP5 was incubated with His-Omi134-458 at 37°C for the indicated time periods. For some assays, Omi/HtrA2 was preincubated with ucf-101 inhibitor 10 min before the addition of His-THAP5. The reactions were resolved on SDS-PAGE, and the gel stained with Coomassie blue. Lane 1: His-THAP5 control (400 ng); lane 2: His-Omi134-458 control (400 ng); lanes 3, 5, 7, and 9: His-Omi+His-THAP5 at different time points; lane 4, 6, 8, and 10: His-Omi + His-THAP5 + ucf-101 (50 μM); lane 11: His-THAP5 + ucf-101 control.
Interaction of Omi/HtrA2 with THAP5 in mammalian cells during apoptosis.
To investigate whether Omi/HtrA2 can interact with THAP5 in vivo, HEK-293 cells were transfected with a construct encoding GFP-THAP5. Cells were treated with cisplatin for 12 h to induce apoptosis. After this time, Omi/HtrA2 antibodies were used to precipitate endogenous Omi/HtrA2 protein, and the presence of any GFP-THAP5 protein in the complex was monitored by Western blot analysis using a GFP-specific monoclonal antibody (Santa Cruz Biotechnology). Figure 1B shows that endogenous Omi/HtrA2 interacts with GFP-THAP5 in HEK-293 only in cells treated with cisplatin and undergoing apoptosis. No interaction between Omi/HtrA2 and GFP-THAP5 was observed under normal nonapoptotic conditions. We also performed the reverse experiment by transfecting HEK-293 cells with GFP-Omi and using the THAP5 antibody to precipitate endogenous protein and any GFP-Omi associated with it. This experiment also clearly shows that THAP5 and GFP-Omi can associate in mammalian cells, but only during apoptotic conditions (Fig. 1C).
THAP5 is cleaved by Omi/HtrA2 protease in vitro.
To test the ability of Omi/HtrA2 protease to cleave the THAP5 protein in vitro, bacterially made His-THAP5 was incubated with His-Omi/HtrA2134-458 protease for different time periods. Figure 1D shows that His-Omi/HtrA2134-458 was able to degrade the THAP5 protein in this assay. To verify that cleavage of THAP5 was due to Omi/HtrA2 activity and not by some other contaminated bacterial protease, we used its specific inhibitor ucf-101 (9). When used in this assay, ucf-101 was able to prevent the degradation of THAP5 by Omi/HtrA2 (Fig. 1D).
Expression of THAP5: mRNA and protein.
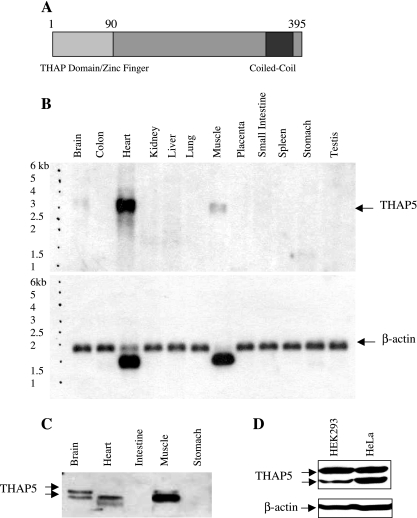
Figure 2A is a schematic diagram of the THAP5 protein. The nucleotide sequence predicts a protein consisting of 395 amino acids. The THAP domain is located at its amino terminus and shows high homology to the other 11 members of the human THAP family of proteins. Outside the THAP domain, THAP5 protein has no similarity to any other known protein sequence that is uploaded in the GenBank. A coiled-coil region is also predicted at its carboxy terminus. Since there is no mouse or rat ortholog for THAP5, a human Northern blot representing various tissues was used to investigate THAP5 mRNA expression. THAP5 expression is represented by a specific mRNA band of ∼3.2 kb (Fig. 2B). THAP5 mRNA shows very high expression in the human heart, and, when the blot is overexposed, some expression is also seen in the human brain and skeletal muscle (Fig. 2B). No expression of THAP5 is detected in any of the other human tissues represented on this blot. To verify that there is a bona fide THAP5 protein present in these human tissues, we raised a polyclonal antibody against a recombinant His-THAP5163-395. This antibody was used on a Western blot containing extracts from various human tissues (Calbiochem). Figure 2C shows that a 50-kDa polypeptide is readily detectable in human heart, brain, and skeletal muscle. A lower molecular weight band is also detected with the same THAP5 antibody. The relative levels of the THAP5 protein vs. mRNA in the three human tissues do not correspond, and this is probably due to the poor quality (protein degradation) of the commercially available human Western blot. Figure 2D shows the relative expression of THAP5 protein in HeLa and HEK-293 cells. The lower band recognized by this antibody probably corresponds to a proteolytic product of THAP5. Its intensity increases after several cycles of freeze-thawing the lysates. Both HEK-293 and HeLa cell lines express similar levels of THAP5 protein, but THAP5 protein is 30 times more abundant in the human heart (results not shown).
Fig. 2.
Expression of THAP5: mRNA and protein in human tissues. A: schematic representation of the THAP5 protein encoded by 395 amino acids. The light gray box represents the 90aa THAP domain, which is characteristic of all THAP proteins. B: Northern blot analysis of THAP5 expression in human tissues. A commercially available Northern blot (Clontech) containing 2 μg/lane poly (A) mRNA from various adult human tissues was probed with 32P-THAP5 (539-1342) cDNA. A single transcript was detected of ∼3.2 kb. The blot clearly shows high expression of THAP5 in the human heart; some expression was also seen in skeletal muscle and brain. β-Actin probe was used to verify that equal amounts of mRNA were present in each lane. C: Western blot analysis of THAP5 protein in multiple human tissues. A commercial Western blot (INSTA-blot Calbiochem) containing 15 μg/lane of total protein from adult human tissues was incubated with rabbit-polyclonal anti-THAP5 antibody followed by a secondary antibody, horseradish peroxidase-conjugated goat anti-rabbit and chemiluminescence detection. D: Western blot analysis showing the relative expression of THAP5 in HEK-293 and HeLa cells. Thirty micrograms of whole cell lysates were resolved on SDS-PAGE, and the membrane was incubated with THAP5 polyclonal antibody. β-Actin antibody was used to verify that an equal amount of protein was loaded in each lane.
Subcellular localization of the THAP5 protein.
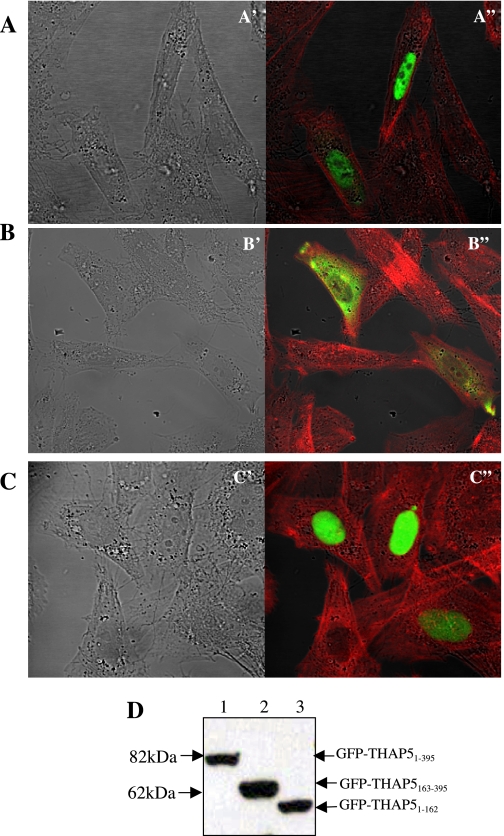
The cDNA sequence corresponding to the full-length THAP5 protein was cloned into the GFPC1 vector. The GFP-THAP5 vector was transfected into HeLa cells, and 24 h later the subcellular localization of the protein was observed using a confocal microscope, as previously described (8). Figure 3A shows that the full-length GFP-THAP5 protein is localized exclusively in the cell nucleus, but is excluded from the nucleoli. There are two discernible domains in the THAP5 protein: 1) the THAP domain (90aa) present at the amino terminus that shares homology with the corresponding domain in the other 11 members of the THAP family of proteins; and 2) the rest of the THAP5 protein (91-395aa) that shows no homology to any other known protein sequence in the data banks. Therefore, we cloned DNA sequence corresponding to the amino terminus of the protein, including the THAP domain or the unique carboxy terminus of THAP5 into the GFP vector to investigate their potential role in targeting the protein to the nucleus. Figure 3B shows that the THAP5 protein that lacks the amino terminus is now predominantly cytoplasmic, whereas the THAP domain alone localizes exclusively in the cell nucleus (Fig. 3C). Figure 3D is a Western blot to verify the expression and stability of the three different GFP fusion proteins used in this experiment.
Fig. 3.
Subcellular localization of the THAP5 protein in mammalian cells. A: confocal image of HeLa cells transfected with GFP-THAP51-395 shows exclusive nuclear localization (green in A"). B: HeLa cells transfected with GFP-THAP5163-395 show cytoplasmic localization (green in B"). C: HeLa cells transfected with GFP-THAP51-162 show exclusive nuclear localization (green in C") of this protein. The cells were also stained with Texas red-phalloidin (Molecular Probes) that stain actin filaments (red). Nomarski/DIC images of the same cells are shown in A′, B′, and C′, respectively. D: the stability of EGFP-fusion proteins was monitored by Western blot analysis using an anti-GFP antibody. Equal amounts of whole cell lysates, obtained 24 h posttransfection, were subjected to SDS-PAGE followed by Western blot analysis using anti-GFP monoclonal antibody, as described in materials and methods. Lanes 1, 2, and 3 represent lysates obtained after transfection of HeLa cells with GFP-THAP51-395, GFP-THAP5163-395, and GFP-THAP51-162, respectively.
THAP5 is a potential regulator of cell cycle.
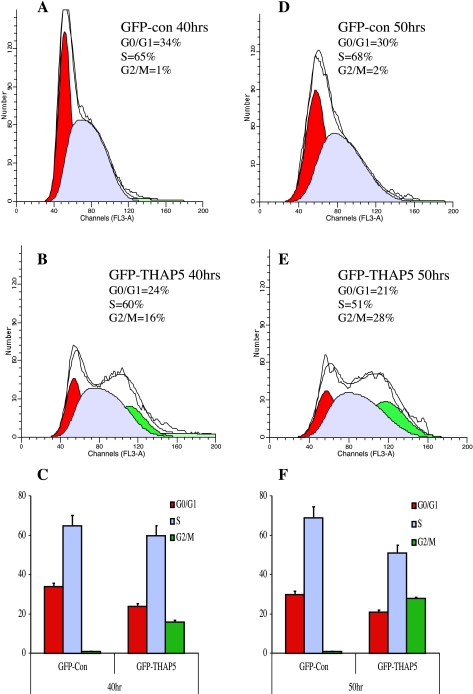
Previous reports suggested that some members of the THAP family of proteins could inhibit cell cycle progression (6). Therefore, we investigated whether ectopic expression of a GFP-THAP5 protein in HEK-293 will interfere with the cell cycle progression. Cells were synchronized and transfected with GFP-THAP5 or the empty GFP vector, and 40 or 50 h later the percentage of transfected cells at various points of the cell cycle were estimated. Figure 4 shows an accumulation of GFP-THAP5 expressing cells at the G2/M phase compared with cells expressing GFP alone 40 h posttransfection. This difference becomes more pronounced after 50 h, where over 28% of the GFP-THAP5 expressing cells were found in the G2/M phase vs. 2% of cells transfected with GFP vector. The same experiment was also performed using HeLa cells transfected with GFP vector or GFP-THAP5. Here too, overexpression of THAP5 causes accumulation of cells at G2/M phase (results not shown).
Fig. 4.
Overexpression of GFP-THAP5 causes accumulation of cells in G2/M phase. HEK-293 cells were transiently transfected with GFP vector alone or GFP-THAP5. Forty and fifty hours posttransfection, cells were stained with 7-amino-actinomycin D (7-AAD), and the DNA content was analyzed using ModFitLT software. The percentages of GFP positive cells at different phases of cell cycle 40 h (C) and 50 h (F) after transfection are shown. Histogram results are shown from representative experiments in cells transfected with GFP vector at 40 h (A) and 50 h (D). Cells transfected with GFP-THAP5 for 40 h (B) and 50 h (E) show significant increase in the percentage of cells in G2/M phase. Data are means ± SD of four independent experiments.
THAP5 is degraded in cells treated with cisplatin or H2O2.
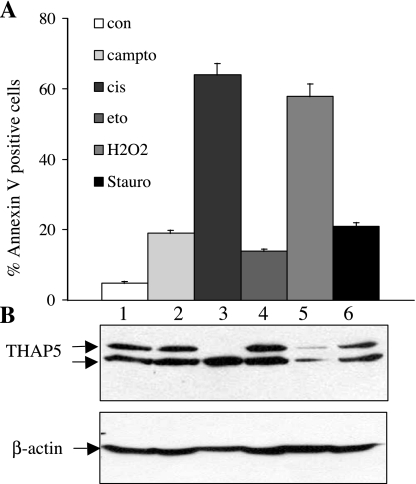
To investigate whether THAP5 protein is regulated during apoptosis, HeLa cells were treated with various chemicals known to induce apoptosis, including camptothecin, cisplatin, etoposide, H2O2, and staurosporine; subsequently, cell death was measured using annexin V staining and flow cytometry (10). THAP5 protein level was monitored by Western blot analysis of cell extracts, and it was significantly reduced in cells treated with cisplatin or H2O2 (Fig. 5). These chemicals also induced the maximum cell death (64 and 58%, respectively) in this assay. In cells treated with cisplatin, the upper band of THAP5 was preferentially cleaved, whereas, in cells treated with H2O2, both bands of the protein were equally degraded.
Fig. 5.
THAP5 protein level is regulated during apoptosis. Total cell lysates were prepared from HeLa cells after induction of apoptosis using different chemicals (camptothecin, cisplatin, etoposide, H2O2, and staurosporine). A: cell death was monitored in the treated cell populations by annexin V and 7-AAD staining and analyzed by flow cytometry. B: cell extracts were prepared followed by Western blot analysis of the same samples. Lane 1 shows lysates from control, untreated cells; lanes 2, 3, and 4 show cell extracts from HeLa treated with the chemical indicated above. β-Actin antibody was used to verify that equal amounts of protein were present in each lane. Data are means ± SD of four independent experiments.
Ucf-101 inhibitor prevents THAP5 degradation.
We investigated whether the Omi/HtrA2 protease is responsible for the degradation of THAP5 observed in HeLa cells during apoptosis. For this, HeLa cells were treated with cisplatin, the percentage of cell death was monitored by annexin V staining and flow cytometry, and the levels of THAP5 protein were monitored by Western blot analysis. HeLa cells treated with 5 μM cisplatin for 12 h resulted in 42% cell death of the population (Fig. 6). The cell death in these cells coincided with a dramatic reduction in the level of THAP5 protein. We also performed the same experiment in the presence of two different concentrations of ucf-101, a specific inhibitor of the proteolytic activity of Omi/HtrA2 (9). In this case, the percentage of apoptotic cells in the population was reduced to 21%. The ucf-101 inhibitor was also able to protect the degradation of the THAP5 protein, implicating Omi/HtrA2 protease in this process.
Fig. 6.
A specific inhibitor of Omi/HtrA2 blocks THAP5 cleavage and protects HeLa cells from apoptosis. A: HeLa cells were treated with 20 or 30 μM of ucf-101, and apoptosis was induced with 5 μM cisplatin for 12 h. Apoptosis was monitored using annexin V and 7-AAD staining and analyzed by flow cytometry. B: extracts were prepared from the same cell population and analyzed by SDS-PAGE and Western blot analysis using THAP5 antibody. Cisplatin treatment caused a dramatic reduction in THAP5 protein level (lane 2). This corresponds with increased apoptosis in the cell population. When HeLa cells were treated with ucf-101 followed by cisplatin, the inhibitor substantially protected THAP5 proteins, and the percentage of apoptotic cells was significantly reduced (lanes 3 and 4). β-Actin antibody was used to verify that equal amounts of protein were present in each lane. Data are means ± SD of three independent experiments.
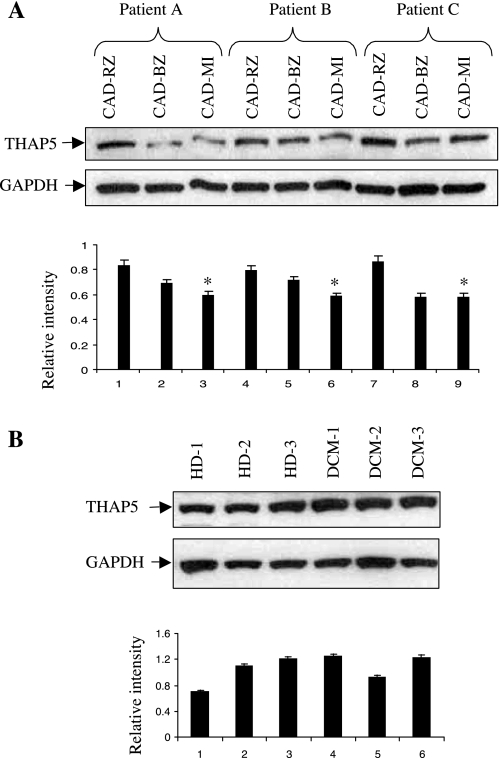
Expression of THAP5 protein in heart tissues derived from patients with CAD.
THAP5 expression is highest in the human heart, suggesting a potential role of this protein in the normal function of the cardiomyocytes. We monitored the protein level of THAP5 in the heart of patients with various heart diseases, including CAD and DCM. Figure 7A shows the expression of THAP5 protein in the remote zone (CAD-RZ), border zone (CAD-BZ), and MI area (CAD-MI) in the heart of three CAD patients. The relative THAP5-to-GAPDH ratio for each sample was quantified by densitometry. In all three patients, there is clear reduction in the level of THAP5 in the CAD-MI area compared with the CAD-RZ and CAD-BZ. The MI area is the part of the heart that sustains maximum injury in CAD. Therefore, this area is expected to contain the most damaged and apoptotic cells. Figure 7B shows the expression of THAP5 protein in the heart tissue of three patients with DCM and three HDs. No significant difference is seen in the level of THAP5 protein in the hearts of DCM patients or HDs. The blot was also probed with GAPDH-specific antibody to verify that an equal amount of protein extract was used in each lane.
Fig. 7.
THAP5 protein levels in heart tissues from patients with coronary artery disease (CAD). Heart tissues were obtained from patients with end-stage heart failure due to CAD or dilated cardiomyopathy (DCM) and were undergoing cardiac transplantation. A: heart tissue extracts from three patients with CAD were used in a Western blot to monitor the expression of THAP5 protein. Top: extracts were prepared from three areas on each heart corresponding to the remote zone (CAD-RZ), border zone (CAD-BZ), and the infarction (CAD-MI) area. Bottom: THAP5-to-GAPDH ratios were calculated after densitometry. *P < 0.05 vs. CAD-RZ. B, top: human heart tissue lysates from three healthy donors (HD-1, -2, and -3) and three DCM patients (DCM-1, -2, and -3) were probed for THAP5 expression. Data are means ± SD (n = 6 patients/group). Bottom: THAP5-to-GAPDH ratios were calculated after densitometry.
DISCUSSION
Omi/HtrA2 is a nuclear-encoded mitochondrial protease that has homology to bacterial HtrA chaperones (17, 27, 43). Recent evidence suggests that Omi/HtrA2 has two distinct functions, both requiring its protease activity. Omi/HtrA2 can be a proapoptotic protein, as well as a prosurvival factor, depending on its subcellular location. The prosurvival function of Omi/HtrA2 was based on studies on motor neuron degeneration 2 (mnd2) mice that carry a single mutation affecting the proteolytic activity of Omi/HtrA2 (25). Mnd2 homozygous animals suffer muscle wasting, neurodegeneration, and die by 6 wk of age. Based on these studies, it is assumed that the primary function (physiological function) of Omi/HtrA2 in mammalian cells is to somehow maintain mitochondrial homeostasis necessary for cell survival (25, 35). The other function of Omi/HtrA2 (pathological function) becomes operational only under conditions leading to cell death, where Omi/HtrA2 is released to the cytoplasm as a proapoptotic protein and participates in caspase-dependent as well as caspase-independent apoptosis (21, 45, 50). Most of the studies reported so far have provided significant new information on the mechanism of Omi/HtrA2's proapoptotic function, but little if anything on its prosurvival function is known. Omi/HtrA2, when released to the cytoplasm, binds and degrades several substrates, including inhibitors of apoptosis protein, PEA/PED, and Apollon/Bruce (41, 44, 47, 51). Some of these proteins are clearly antiapoptotic factors, and their removal by Omi/HtrA2 can undoubtedly accelerate cell death. Others have a more obscure function, and their association and cleavage by Omi/HtrA2, in the context of inducing cell death, is more difficult to justify. In the present study, we report the characterization of THAP5, the fifth member of the THAP family of proteins, as a specific interactor and substrate of Omi/HtrA2 protease. THAP proteins comprise a recently described family of cellular factors with unique structural and functional characteristics. They are defined by the presence of the THAP domain, an atypical zinc-dependent DNA-binding domain of ∼90 amino acids. There are 113 THAP proteins listed in the databases, and in humans there are 12 distinct proteins (THAP0-THAP11) (40). All 12 human THAP proteins have a single THAP domain at their amino terminus. In Drosophila, there are 7 THAP proteins, 2 of which contain more than 1 THAP domain, CG14860 has 2 THAP domains, and the CG10631 is predicted to have as many as 27 THAP domains (40). THAP domains are unique as DNA binding motifs due to a significant similarity with the DNA binding domain of Drosophila P element transposase. This similarity includes the size of the THAP domain, its location in the protein, and the conservation in the number and residues that define the domain (40). THAP0, also known as the death-associated protein DAP4/p52rIPK, was identified as one of the genes induced by IFN-γ-mediated apoptosis and in an independent study, as an interactor of protein kinase R (14, 18). THAP1 was originally defined as a proapoptotic nuclear factor localized at the promyelocytic leukemia nuclear bodies and interacting with prostate apoptosis response-4 protein (39). Since very little is known about THAP5, we embarked on a detailed study, beginning with its expression. In human tissues, THAP5 mRNA is predominantly expressed in the human heart, although low expression is also detected in skeletal muscle and brain. More significantly, we found no orthologs for THAP5 in mouse (Mus musculus) or rat (Rattus norvegicus). The closest orthologs were detected in Macaca fascicularis, Bos taurus, and Gallus gallus. The absence of orthologs in mouse and rat has also been reported for four other members of the THAP family, the THAP6, THAP8, THAP9, and THAP10 proteins (37). It was suggested that some THAP genes might have originated from a domestication event of a single copy of P element transposase that took place in the human lineage after it had diverged from the rodent lineage (38). Alternatively, THAP gene domestication might have occurred earlier in the evolution, but was later lost in the lineage leading to rodents (38).
To verify that THAP5 is a bona fide human protein, we raised a polyclonal antibody against the unique part of the protein (aa163-395) that does not include the THAP domain. This polyclonal antibody clearly recognized a specific band of 50 kDa, corresponding to THAP5 in human heart, skeletal muscle, and brain. The THAP5 antibody did not detect any specific protein bands when tested against various mouse or rat tissue extracts (results not shown).
To investigate the subcellular localization of THAP5, we expressed the full-length protein fused to GFP in HeLa cells. The fusion protein, GFP-THAP5, shows exclusive nuclear localization but is excluded from the nucleoli. The amino terminus of the protein that includes the THAP5 domain is responsible for its nuclear localization. This is the part of the protein implicated in DNA binding, as well as protein-protein interactions (32). We also stained HeLa cells for endogenous THAP5 protein using our THAP5 antibody. In this experiment, the THAP5 protein was found to be exclusively located in the nucleus (results not shown). The interaction with Omi/HtrA2 is mediated by the carboxy terminus of THAP5 that binds the PDZ domain of Omi/HtrA2. The carboxy terminus of THAP5 has the amino acid sequence EVTMI* that conforms to the consensus sequence for PDZ (Omi) binding proteins (26, 36).
Previous studies have shown two members of the same family, THAP1 and THAP11, can regulate cell proliferation (6, 52). Furthermore, genetic studies using C. elegans support a role of THAP proteins in cell cycle regulation. These studies demonstrated a genetic interaction between LIN-35/Rb, which is the C. elegans retinoblastoma homolog, and four C. elegans THAP proteins, LIN-36, LIN-15B, LIN-15A, and HIM-17. LIN-36 and LIN-15B are known inhibitors of G1/S cell cycle transition (4, 5, 11, 12, 23, 46). Also, in zebrafish the ortholog of the cell cycle transcription factor E2F6 has a THAP domain and functions as a repressor of E2-F-dependent transcription during S phase (20). Based on these reports, we decided to investigate if THAP5 plays any role in the regulation of cell cycle.
For these studies, we ectopically expressed GFP-THAP5 in HEK-293. This cell line expresses endogenous THAP5 protein and can be transfected with high efficiency. The results from these experiments clearly show that GFP-THAP5 had a dramatic effect, blocking the progression of transfected cells through the cell cycle and causing high accumulation of cells at the G2/M phase. The effect of THAP5 overexpression is not unique to HEK-293 cells, and similar experiments performed in HeLa cells also caused arrest at G2/M phase. The mechanism by which THAP5 inhibits cell cycle is not known, and it most likely will involve the direct regulation of cell cycle genes. THAP5 has an atypical zinc finger domain at its amino terminus that could bind to specific DNA sequences on different promoters. A recent study showed that another member of the same family, THAP11, could inhibit cell cycle by binding to the promoter of c-Myc and downregulating its expression (52).
To investigate the role of the THAP5-Omi/HtrA2 interaction during apoptosis, HeLa cells that normally express THAP5 were treated with various chemicals to induce cell death. Under these conditions, Omi/HtrA2 is released from mitochondria and becomes a proapoptotic protein. In these experiments, the THAP5 protein was cleaved in cells treated with cisplatin or H2O2, and the degree of THAP5 degradation increased proportionally with the rate of apoptosis in the cell population. No significant cleavage of THAP5 was seen in cells treated with etoposide and camptothecin, but these chemicals also caused minimal cell death in this protocol and cell line used for these experiments.
We used a specific inhibitor to show that Omi/HtrA2 is responsible for THAP5 degradation. This inhibitor, ucf-101, was developed in our laboratory and has been extensively studied in various systems of cell death (8, 9, 30). When the ucf-101 inhibitor was used, it clearly blocked THAP5 cleavage and protected cells from apoptosis.
THAP5 is hyperexpressed in the human heart, but there are no rodent orthologs, therefore limiting the type of studies that can be done to investigate its potential involvement in the development or progression of heart disease. We obtained small tissue samples from patients with several human heart diseases, including CAD and DCM. In CAD patients, we were able to detect a decrease in the THAP5 protein level in the MI area compared with RZ or BZ areas of the same heart. These results, although preliminary, suggest that, during pathological conditions resulting in myocardial cell death, such as MI, the THAP5 protein decreases significantly in areas of the heart that sustain maximum cell damage. There was no difference in THAP5 protein levels in DCM patients compared with normal (donor) heart. Previous studies have shown that Omi/HtrA2 is a key player in myocardial ischemia-reperfusion injury (2). Furthermore, inhibition of the proteolytic activity of Omi/HtrA2 using the ucf-101 inhibitor improved postischemic myocardial function and reduction of the myocardial infarct size (2, 3, 30). THAP5 is the first human, heart-specific substrate of Omi/HtrA2 identified thus far. Under normal, physiological conditions, THAP5 functions as an inhibitor of cell cycle. This dual function of THAP5 supports previous studies that have shown the existence of a cross talk between cell cycle control and apoptosis in cardiomyocytes. For example, overexpression of the cell cycle inhibitor p57kip2 in cardiomyocytes was shown to attenuate ischemia-reperfusion injury in the mouse heart (19). In another study, cyclin A/cdk2 activation was involved in hypoxia-induced apoptosis in cardiomyocytes (1). The hyperexpression of THAP5 in human heart and its regulation during cell death by Omi/HtrA2 suggest that this pathway plays a significant role in the normal function of the heart. Furthermore, THAP5 might be involved in the development and progression of heart disease, especially in CAD.
GRANTS
This work was supported by National Institute of Diabetes and Digestive and Kidney Diseases Grant 5R01 DK55734 (to A. S. Zervos) and National Institute of General Medicine Grant GM076176 (to E. S. Alnemri).
Acknowledgments
We are grateful to Drs Federica del Monte, Judyth K. Gwathmey, and Thomas Macgillivray for providing the human heart tissues. We also thank Dr. Debabrata Chakravarti for comments, suggestions, and critical reading of the manuscript.
REFERENCES
- 1.Adachi S, Ito H, Tamamori-Adachi M, Ono Y, Nozato T, Abe S, Ikeda M, Marumo F, Hiroe M. Cyclin A/cdk2 activation is involved in hypoxia-induced apoptosis in cardiomyocytes. Circ Res 88: 408–414, 2001. [DOI] [PubMed] [Google Scholar]
- 2.Bhuiyan MS, Fukunaga K. Activation of HtrA2, a mitochondrial serine protease mediates apoptosis: current knowledge on HtrA2 mediated myocardial ischemia/reperfusion injury. Cardiovasc Ther 26: 224–232, 2008. [DOI] [PubMed] [Google Scholar]
- 3.Bhuiyan MS, Fukunaga K. Inhibition of HtrA2/Omi ameliorates heart dysfunction following ischemia/reperfusion injury in rat heart in vivo. Eur J Pharmacol 557: 168–177, 2007. [DOI] [PubMed] [Google Scholar]
- 4.Boxem M, van den Heuvel S. C. elegans class B synthetic multivulva genes act in G(1) regulation. Curr Biol 12: 906–911, 2002. [DOI] [PubMed] [Google Scholar]
- 5.Boxem M, van den Heuvel S. lin-35 Rb and cki-1 Cip/Kip cooperate in developmental regulation of G1 progression in C. elegans. Development 128: 4349–4359, 2001. [DOI] [PubMed] [Google Scholar]
- 6.Cayrol C, Lacroix C, Mathe C, Ecochard V, Ceribelli M, Loreau E, Lazar V, Dessen P, Mantovani R, Aguilar L, Girard JP. The THAP-zinc finger protein THAP1 regulates endothelial cell proliferation through modulation of pRB/E2F cell-cycle target genes. Blood 109: 584–594, 2007. [DOI] [PubMed] [Google Scholar]
- 7.Chu YW, Wang R, Schmid I, Sakamoto KM. Analysis with flow cytometry of green fluorescent protein expression in leukemic cells. Cytometry 36: 333–339, 1999. [DOI] [PubMed] [Google Scholar]
- 8.Cilenti L, Kyriazis GA, Soundarapandian MM, Stratico V, Yerkes A, Park KM, Sheridan AM, Alnemri ES, Bonventre JV, Zervos AS. Omi/HtrA2 protease mediates cisplatin-induced cell death in renal cells. Am J Physiol Renal Physiol 288: F371–F379, 2005. [DOI] [PubMed] [Google Scholar]
- 9.Cilenti L, Lee Y, Hess S, Srinivasula S, Park KM, Junqueira D, Davis H, Bonventre JV, Alnemri ES, Zervos AS. Characterization of a novel and specific inhibitor for the pro-apoptotic protease Omi/HtrA2. J Biol Chem 278: 11489–11494, 2003. [DOI] [PubMed] [Google Scholar]
- 10.Cilenti L, Soundarapandian MM, Kyriazis GA, Stratico V, Singh S, Gupta S, Bonventre JV, Alnemri ES, Zervos AS. Regulation of HAX-1 anti-apoptotic protein by Omi/HtrA2 protease during cell death. J Biol Chem 279: 50295–50301, 2004. [DOI] [PubMed] [Google Scholar]
- 11.Clark SG, Lu X, Horvitz HR. The Caenorhabditis elegans locus lin-15, a negative regulator of a tyrosine kinase signaling pathway, encodes two different proteins. Genetics 137: 987–997, 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clouaire T, Roussigne M, Ecochard V, Mathe C, Amalric F, Girard JP. The THAP domain of THAP1 is a large C2CH module with zinc-dependent sequence-specific DNA-binding activity. Proc Natl Acad Sci USA 102: 6907–6912, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davis PK, Ho A, Dowdy SF. Biological methods for cell-cycle synchronization of mammalian cells. Biotechniques 30: 1322–1326, 1328, 1330–1321, 2001. [DOI] [PubMed] [Google Scholar]
- 14.Deiss LP, Feinstein E, Berissi H, Cohen O, Kimchi A. Identification of a novel serine/threonine kinase and a novel 15-kD protein as potential mediators of the gamma interferon-induced cell death. Genes Dev 9: 15–30, 1995. [DOI] [PubMed] [Google Scholar]
- 15.Derby E, Reddy V, Baseler M, Malyguine A. Flow cytometric assay for the simultaneous analysis of cell-mediated cytotoxicity and effector cell phenotype. Biotechniques 31: 660, 664–665, 2001. [DOI] [PubMed] [Google Scholar]
- 16.Derby E, Reddy V, Kopp W, Nelson E, Baseler M, Sayers T, Malyguine A. Three-color flow cytometric assay for the study of the mechanisms of cell-mediated cytotoxicity. Immunol Lett 78: 35–39, 2001. [DOI] [PubMed] [Google Scholar]
- 17.Faccio L, Fusco C, Chen A, Martinotti S, Bonventre JV, Zervos AS. Characterization of a novel human serine protease that has extensive homology to bacterial heat shock endoprotease HtrA and is regulated by kidney ischemia. J Biol Chem 275: 2581–2588, 2000. [DOI] [PubMed] [Google Scholar]
- 18.Gale M, Blakely CM, Hopkins DA, Melville MW, Wambach M, Romano PR, Katze MG. Regulation of interferon-induced protein kinase PKR: modulation of P58IPK inhibitory function by a novel protein, P52rIPK. Mol Cell Biol 18: 859–871, 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haley SA, Zhao T, Zou L, Klysik JE, Padbury JF, Kochilas LK. Forced expression of the cell cycle inhibitor p57Kip2 in cardiomyocytes attenuates ischemia-reperfusion injury in the mouse heart. BMC Physiol 8: 4, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hammer SE, Strehl S, Hagemann S. Homologs of Drosophila P transposons were mobile in zebrafish but have been domesticated in a common ancestor of chicken and human. Mol Biol Evol 22: 833–844, 2005. [DOI] [PubMed] [Google Scholar]
- 21.Hegde R, Srinivasula SM, Zhang Z, Wassell R, Mukattash R, Cilenti L, DuBois G, Lazebnik Y, Zervos AS, Fernandes-Alnemri T, Alnemri ES. Identification of Omi/HtrA2 as a mitochondrial apoptotic serine protease that disrupts inhibitor of apoptosis protein-caspase interaction. J Biol Chem 277: 432–438, 2002. [DOI] [PubMed] [Google Scholar]
- 22.Herault O, Colombat P, Domenech J, Degenne M, Bremond JL, Sensebe L, Bernard MC, Binet C. A rapid single-laser flow cytometric method for discrimination of early apoptotic cells in a heterogenous cell population. Br J Haematol 104: 530–537, 1999. [DOI] [PubMed] [Google Scholar]
- 23.Huang LS, Tzou P, Sternberg PW. The lin-15 locus encodes two negative regulators of Caenorhabditis elegans vulval development. Mol Biol Cell 5: 395–411, 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jeanblanc M, Mousli M, Hopfner R, Bathami K, Martinet N, Abbady AQ, Siffert JC, Mathieu E, Muller CD, Bronner C. The retinoblastoma gene and its product are targeted by ICBP90: a key mechanism in the G1/S transition during the cell cycle. Oncogene 24: 7337–7345, 2005. [DOI] [PubMed] [Google Scholar]
- 25.Jones JM, Datta P, Srinivasula SM, Ji W, Gupta S, Zhang Z, Davies E, Hajnoczky G, Saunders TL, Van Keuren ML, Fernandes-Alnemri T, Meisler MH, Alnemri ES. Loss of Omi mitochondrial protease activity causes the neuromuscular disorder of mnd2 mutant mice. Nature 425: 721–727, 2003. [DOI] [PubMed] [Google Scholar]
- 26.Junqueira D, Cilenti L, Musumeci L, Sedivy JM, Zervos AS. Random mutagenesis of PDZ(Omi) domain and selection of mutants that specifically bind the Myc proto-oncogene and induce apoptosis. Oncogene 22: 2772–2781, 2003. [DOI] [PubMed] [Google Scholar]
- 27.Krojer T, Garrido-Franco M, Huber R, Ehrmann M, Clausen T. Crystal structure of DegP (HtrA) reveals a new protease-chaperone machine. Nature 416: 455–459, 2002. [DOI] [PubMed] [Google Scholar]
- 28.Kuninaka S, Iida SI, Hara T, Nomura M, Naoe H, Morisaki T, Nitta M, Arima Y, Mimori T, Yonehara S, Saya H. Serine protease Omi/HtrA2 targets WARTS kinase to control cell proliferation. Oncogene 26: 2395–2406, 2007. [DOI] [PubMed] [Google Scholar]
- 29.Kuninaka S, Nomura M, Hirota T, Iida S, Hara T, Honda S, Kunitoku N, Sasayama T, Arima Y, Marumoto T, Koja K, Yonehara S, Saya H. The tumor suppressor WARTS activates the Omi/HtrA2-dependent pathway of cell death. Oncogene 24: 5287–5298, 2005. [DOI] [PubMed] [Google Scholar]
- 30.Liu HR, Gao E, Hu A, Tao L, Qu Y, Most P, Koch WJ, Christopher TA, Lopez BL, Alnemri ES, Zervos AS, Ma XL. Role of Omi/HtrA2 in apoptotic cell death after myocardial ischemia and reperfusion. Circulation 111: 90–96, 2005. [DOI] [PubMed] [Google Scholar]
- 31.Ma X, Kalakonda S, Srinivasula SM, Reddy SP, Platanias LC, Kalvakolanu DV. GRIM-19 associates with the serine protease HtrA2 for promoting cell death. Oncogene 26: 4842–4849, 2007. [DOI] [PubMed] [Google Scholar]
- 32.Macfarlan T, Parker JB, Nagata K, Chakravarti D. Thanatos-associated protein 7 associates with template activating factor-Ibeta and inhibits histone acetylation to repress transcription. Mol Endocrinol 20: 335–347, 2006. [DOI] [PubMed] [Google Scholar]
- 33.Marabese M, Mazzoletti M, Vikhanskaya F, Broggini M. HtrA2 enhances the apoptotic functions of p73 on bax. Cell Death Differ 15: 849–858, 2008. [DOI] [PubMed] [Google Scholar]
- 34.Martins LM, Iaccarino I, Tenev T, Gschmeissner S, Totty NF, Lemoine NR, Savopoulos J, Gray CW, Creasy CL, Dingwall C, Downward J. The serine protease Omi/HtrA2 regulates apoptosis by binding XIAP through a reaper-like motif. J Biol Chem 277: 439–444, 2002. [DOI] [PubMed] [Google Scholar]
- 35.Martins LM, Morrison A, Klupsch K, Fedele V, Moisoi N, Teismann P, Abuin A, Grau E, Geppert M, Livi GP, Creasy CL, Martin A, Hargreaves I, Heales SJ, Okada H, Brandner S, Schulz JB, Mak T, Downward J. Neuroprotective role of the Reaper-related serine protease HtrA2/Omi revealed by targeted deletion in mice. Mol Cell Biol 24: 9848–9862, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martins LM, Turk BE, Cowling V, Borg A, Jarrell ET, Cantley LC, Downward J. Binding specificity and regulation of the serine protease and PDZ domains of HtrA2/Omi. J Biol Chem 278: 49417–49427, 2003. [DOI] [PubMed] [Google Scholar]
- 37.Quesneville H, Nouaud D, Anxolabehere D. Detection of new transposable element families in Drosophila melanogaster and Anopheles gambiae genomes. J Mol Evol 57, Suppl 1: S50–S59, 2003. [DOI] [PubMed] [Google Scholar]
- 38.Quesneville H, Nouaud D, Anxolabehere D. Recurrent recruitment of the THAP DNA-binding domain and molecular domestication of the P-transposable element. Mol Biol Evol 22: 741–746, 2005. [DOI] [PubMed] [Google Scholar]
- 39.Roussigne M, Cayrol C, Clouaire T, Amalric F, Girard JP. THAP1 is a nuclear proapoptotic factor that links prostate-apoptosis-response-4 (Par-4) to PML nuclear bodies. Oncogene 22: 2432–2442, 2003. [DOI] [PubMed] [Google Scholar]
- 40.Roussigne M, Kossida S, Lavigne AC, Clouaire T, Ecochard V, Glories A, Amalric F, Girard JP. The THAP domain: a novel protein motif with similarity to the DNA-binding domain of P element transposase. Trends Biochem Sci 28: 66–69, 2003. [DOI] [PubMed] [Google Scholar]
- 41.Sekine K, Hao Y, Suzuki Y, Takahashi R, Tsuruo T, Naito M. HtrA2 cleaves Apollon and induces cell death by IAP-binding motif in Apollon-deficient cells. Biochem Biophys Res Commun 330: 279–285, 2005. [DOI] [PubMed] [Google Scholar]
- 42.Song G, Campos B, Wagoner LE, Dedman JR, Walsh RA. Altered cardiac annexin mRNA and protein levels in the left ventricle of patients with end-stage heart failure. J Mol Cell Cardiol 30: 443–451, 1998. [DOI] [PubMed] [Google Scholar]
- 43.Spiess C, Beil A, Ehrmann M. A temperature-dependent switch from chaperone to protease in a widely conserved heat shock protein. Cell 97: 339–347, 1999. [DOI] [PubMed] [Google Scholar]
- 44.Srinivasula SM, Gupta S, Datta P, Zhang Z, Hegde R, Cheong N, Fernandes-Alnemri T, Alnemri ES. Inhibitor of apoptosis proteins are substrates for the mitochondrial serine protease Omi/HtrA2. J Biol Chem 278: 31469–31472, 2003. [DOI] [PubMed] [Google Scholar]
- 45.Suzuki Y, Imai Y, Nakayama H, Takahashi K, Takio K, Takahashi R. A serine protease, HtrA2, is released from the mitochondria and interacts with XIAP, inducing cell death. Mol Cell 8: 613–621, 2001. [DOI] [PubMed] [Google Scholar]
- 46.Thomas JH, Horvitz HR. The C. elegans gene lin-36 acts cell autonomously in the lin-35 Rb pathway. Development 126: 3449–3459, 1999. [DOI] [PubMed] [Google Scholar]
- 47.Trencia A, Fiory F, Maitan MA, Vito P, Barbagallo AP, Perfetti A, Miele C, Ungaro P, Oriente F, Cilenti L, Zervos AS, Formisano P, Beguinot F. Omi/HtrA2 promotes cell death by binding and degrading the anti-apoptotic protein ped/pea-15. J Biol Chem 279: 46566–46572, 2004. [DOI] [PubMed] [Google Scholar]
- 48.van Loo G, van Gurp M, Depuydt B, Srinivasula SM, Rodriguez I, Alnemri ES, Gevaert K, Vandekerckhove J, Declercq W, Vandenabeele P. The serine protease Omi/HtrA2 is released from mitochondria during apoptosis. Omi interacts with caspase-inhibitor XIAP and induces enhanced caspase activity. Cell Death Differ 9: 20–26, 2002. [DOI] [PubMed] [Google Scholar]
- 49.Vande Walle L, Lamkanfi M, Vandenabeele P. The mitochondrial serine protease HtrA2/Omi: an overview. Cell Death Differ 15: 453–460, 2008. [DOI] [PubMed] [Google Scholar]
- 50.Verhagen AM, Silke J, Ekert PG, Pakusch M, Kaufmann H, Connolly LM, Day CL, Tikoo A, Burke R, Wrobel C, Moritz RL, Simpson RJ, Vaux DL. HtrA2 promotes cell death through its serine protease activity and its ability to antagonize inhibitor of apoptosis proteins. J Biol Chem 277: 445–454, 2002. [DOI] [PubMed] [Google Scholar]
- 51.Yang QH, Church-Hajduk R, Ren J, Newton ML, Du C. Omi/HtrA2 catalytic cleavage of inhibitor of apoptosis (IAP) irreversibly inactivates IAPs and facilitates caspase activity in apoptosis. Genes Dev 17: 1487–1496, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhu CY, Li CY, Li Y, Zhan YQ, Li YH, Xu CW, Xu WX, Sun HB, Yang XM. Cell growth suppression by thanatos-associated protein 11 (THAP11) is mediated by transcriptional downregulation of c-Myc. Cell Death Differ 16: 395–405, 2009. [DOI] [PubMed] [Google Scholar]