Abstract
For many agronomically important plant genes, only their position on a genetic map is known. In the absence of an efficient transposon tagging system, such genes have to be isolated by map-based cloning. In bread wheat Triticum aestivum, the genome is hexaploid, has a size of 1.6 × 1010 bp, and contains more than 80% of repetitive sequences. So far, this genome complexity has not allowed chromosome walking and positional cloning. Here, we demonstrate that chromosome walking using bacterial artificial chromosome (BAC) clones is possible in the diploid wheat Triticum monococcum (Am genome). BAC end sequences were mostly repetitive and could not be used for the first walking step. New probes corresponding to rare low-copy sequences were efficiently identified by low-pass DNA sequencing of the BACs. Two walking steps resulted in a physical contig of 450 kb on chromosome 1AmS. Genetic mapping of the probes derived from the BAC contig demonstrated perfect colinearity between the physical map of T. monococcum and the genetic map of bread wheat on chromosome 1AS. The contig genetically spans the Lr10 leaf rust disease resistance locus in bread wheat, with 0.13 centimorgans corresponding to 300 kb between the closest flanking markers. Comparison of the genetic to physical distances has shown large variations within 350 kb of the contig. The physical contig can now be used for the isolation of the orthologous regions in bread wheat. Thus, subgenome chromosome walking in wheat can produce large physical contigs and saturate genomic regions to support positional cloning.
The isolation of agronomically important genes from crop plants such as rice, maize, and wheat is the basis for molecular improvement of these cereals. Gene isolation in rice is greatly facilitated by a small genome size, high-density genetic maps, a number of large insert libraries, and more recently the development of a retrotransposon tagging system (1). In maize, very active transposon systems can be used to isolate genes and to overcome the difficulty of working with a large and mostly repetitive genome. For barley and wheat, map-based cloning is still the best option to isolate a gene for which only the position on a genetic map is known. Some years ago, comparative genetics in grasses led to the expectation that gene isolation in large grass genomes could be done through cross-genome map-based cloning using rice as a model genome (2). However, comparative analysis at the microlevel revealed many rearrangements between the grass genomes (3–5), suggesting that this approach will not always be successful and that chromosome walking should also be developed in species such as wheat and barley.
In wheat, many genes of agronomic importance such as disease resistance genes are only characterized by their location on the genetic map. A considerable number of markers have been developed for marker-assisted selection (6), but so far no resistance gene has been isolated. Map-based cloning requires the development of a high resolution map and the possibility to do chromosome walks over large distances up to 1 megabase (Mb). Wheat has a very large genome (1.6 × 1010 bp for bread wheat), a high content of repetitive sequences (80%) (7), and different wheat species have different levels of ploidy. For example, the hexaploid genome of bread wheat consists of three homeologous genomes (called A, B, and D). So far, chromosome walking has not been possible in wheat because of the high amount of repetitive sequences. In addition, the presence of three highly similar genomes makes it extremely difficult and tedious to restrict the chromosome walk to a specific genome in hexaploid wheat.
Chromosome walking in large genomes requires large insert libraries. No such library has yet been made from the hexaploid genome of Triticum aestivum. Instead, work has recently focused on the exploitation of ancestral or closely related diploid genomes of the A and D genomes of hexaploid wheat. Comparative genetics has indicated a very high colinearity between chromosome 1Am of Triticum monococcum and chromosome 1A of T. aestivum (8). Similarly, colinearity was found between the D genome of Triticum tauschii and the D genome of bread wheat (9). Thus, the complexity of the analysis in hexaploid wheat could be reduced by using the diploid Am and D genomes of T. monococcum and T. tauschii, respectively, as subgenomes of T. aestivum. Bacterial artificial chromosome (BAC) libraries have been constructed from T. monococcum (10) and from T. tauschii (11), providing access to large genomic fragments orthologous to the A and D genomes of T. aestivum. The efficiency of chromosome walking for gene discovery and isolation also depends on the gene distribution in the genome. Cytogenetic (12, 13) and molecular (4) analysis indicated the existence of gene-rich regions in the wheat genome. These data suggested that chromosome walking should be feasible in the large and repetitive wheat genome at least in regions where the ratio of physical to genetic distance is compatible with the establishment of physical contigs.
In this study, we demonstrate for the first time that chromosome walking can be performed in the diploid wheat T. monococcum. We have established a BAC contig of 450 kb on T. monococcum chromosome 1AmS, which corresponds to the region spanning the Lr10 resistance gene locus on chromosome 1AS of T. aestivum. A number of markers derived from the BAC clones have shown a perfect colinearity between the genetic map of T. aestivum and the physical map of T. monococcum. These markers have also revealed a number of polymorphisms in wheat germplasm, suggesting their usefulness for mapping in other crosses. In addition, we have observed uneven distribution of the recombination frequency in this region associated with the presence of putative coding regions. We conclude that subgenome chromosome walking provides an efficient and generally applicable method to establish physical contigs and to saturate high-density genetic maps in targeted regions of the wheat genome.
Materials and Methods
Plant Material.
Genetic mapping of the Lr10 gene in T. aestivum was performed in three steps with F2 populations of increasing size (128, 385, and 3,120 plants) derived from a cross between the resistant near isogenic line ThatcherLr10 (ThLr10, syn. Exchange/6*Thatcher) (14) and the susceptible Swiss variety Frisal. Segregation for the Lr10 resistance gene in the F2 population was evaluated by artificial infection as previously described (15). In the high-resolution mapping population, the phenotype was determined after artificial infection of 20 F3 seedlings to distinguish between homozygous and heterozygous resistant individuals for each of the 96 recombinants. Seeds of the T. monococcum accession DV92 were kindly provided by J. Dubcovsky (University of California, Davis).
Restriction Fragment Length Polymorphism (RFLP) and Sequence-Tagged-Site (STS) Analysis.
Isolation of genomic DNA, Southern hybridization, and labeling experiments were performed as described by Graner et al. (16). For high-resolution mapping, the amount of fresh plant material used for DNA extraction was scaled down to 200 mg per sample, and the DNA extraction procedure was optimized for high-throughput of samples (N.S. and B.K., unpublished results). Linkage analysis was performed with RFLP probes except for Lrk10–6 (15), which was converted to a codominant STS marker (primers for the ThLr10 allele, Lrk10D1/Lrk10D2 (15); primers for the Frisal allele, Lrk10D3 5′-GCTCGTCATCTCCACAGG-3′/Lrk10D4 5′-ACCTCATGCGGATGTAG-3′, for 30 cycles: 94°C, 45 s; 55°C, 45 s; 72°C, 30 s). PCR amplification was performed in a PTC-200 thermocycler (MJ Research, Cambridge, MA).
BAC Library Screening and Manipulation of BAC DNA.
Screening of the T. monococcum BAC library was carried out under the same conditions as RFLP hybridization. DNA for fingerprint analysis and BAC end cloning was prepared by standard alkaline lysis procedures (17). End sequences of BAC insert DNA were subcloned by plasmid-rescue and inverse PCR (18). Five micrograms of pure BAC DNA (QIAGEN plasmid midi kit) were used for direct cycle sequencing of BAC ends (Thermo sequenase premixed cycle sequencing kit, Amersham Pharmacia) using an automatic DNA Sequencer 4200 (Li-Cor).
Shotgun Cloning and Low-Pass Sequencing of BAC Clones.
Ten micrograms of BAC DNA free of bacterial genomic DNA (QIAGEN large-construct kit) were mechanically sheared (HydroShear, Genemachines), and end repair of DNA fragments was performed by mung bean nuclease treatment. A DNA size fraction between 1 and 3 kb was gel purified, dephosphorylated, extracted once with phenol/chloroform, ethanol precipitated, and cloned using the Zero Blunt TOPO PCR cloning kit (Invitrogen) according to the manufacturer's instructions. Plasmid DNA was automatically purified (BioRobot 9600, QIAGEN) with the Wizard SV96 plasmid DNA purification system (Promega) and used for cycle sequencing on a ABI PRISM 377 automatic sequencer (PE Applied Biosystems). Sequence comparison was performed by using the BLASTx and n algorithms (19).
Results
High-Resolution Genetic Mapping of the Lr10 Locus in Hexaploid Wheat.
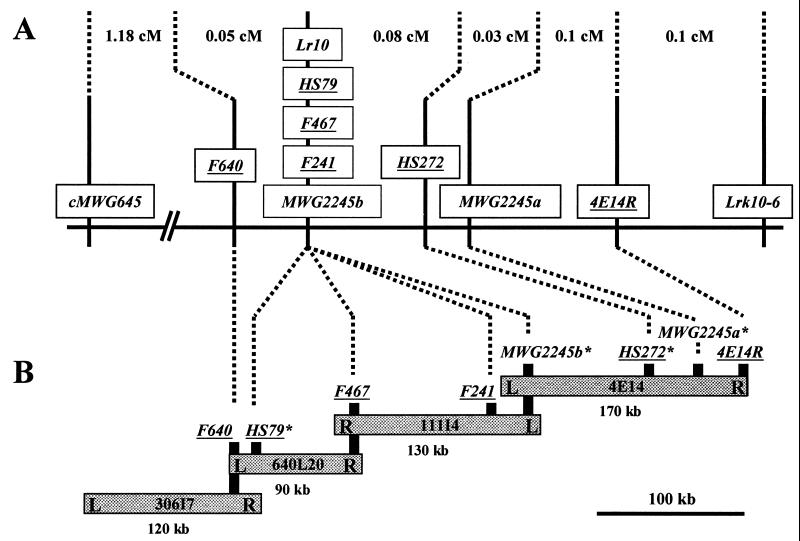
Lr10 was mapped on chromosome 1AS and cosegregated with the RFLP marker Lrk10 in a small segregating population of the cross ThatcherLr10 × Frisal (20). In total, 385 susceptible F2 plants from the same segregating population were subsequently used for fine mapping. The Lr10 gene was mapped in a 2-centimorgan (cM) interval between the RFLP markers cMWG645 (1.2 cM proximal) and Lrk10–6 (0.8 cM distal), whereas the RFLP marker MWG2245 showed cosegregation with the gene. A high-resolution mapping population of 3,120 F2 plants was then screened with the two flanking probes. Ninety-six plants with a recombination event between cMWG645 and Lrk10–6 were selected and represented recombinants in a genetic interval of 1.54 cM around the Lr10 gene (Fig. 1A). After determining the resistance phenotype of the recombinant plants, cMWG645 and Lrk10–6 were mapped at 1.23 cM proximal and 0.31 cM distal to Lr10, respectively. In addition, segregation of two different loci detected by MWG2245 (designated as a and b) was observed. The b locus showed complete linkage with the Lr10 gene, whereas the a locus mapped 0.11 cM distal to Lr10 (Fig. 1A). MWG2245 was then used to start subgenome chromosome walking.
Figure 1.
Subgenome chromosome walking on chromosome 1AmS. (A) High-resolution genetic map of the Lr10 locus on chromosome 1AS of hexaploid wheat derived from 3120 F2 plants of a cross between ThLr10 and Frisal. Six of the 12 probes derived from T. monococcum BACs are shown and underlined. The remaining 6 probes were derived from BAC 111I4 and 640L20, and all of them cosegregated with the Lr10 gene. Probe names beginning with an F represent PCR products derived from specific regions of either BAC ends (F640) or shotgun clones (F241, F467). Probe names beginning with HS represent inserts from shotgun clones, which were directly used as RFLP probes. 4E14R is the right BAC end of 4E14 derived by plasmid rescue. Genetic distances are in centimorgans (cM). (B) Physical contig at the MWG2245 locus in diploid T. monococcum (DV92). The total contig length is about 450 kb. Left and right ends of BAC clones are indicated as L and R, respectively. Probes for which the physical location is only approximately known are marked with an asterisk.
Subgenome Chromosome Walking in Diploid Wheat T. monococcum.
The RFLP probe MWG2245 was used to screen the BAC library of T. monococcum accession DV92 (10). In agreement with the finding of two genetic loci in the Lr10 region in hexaploid wheat, two fragments were detected by Southern hybridization with MWG2245 on BglII-digested DV92 genomic DNA (data not shown). Seven BAC clones were isolated and assigned to the same genomic region based on DNA fingerprinting with the restriction endonucleases NotI, SalI, NotI/SalI, and HindIII and the hybridization pattern with the MWG2245 probe. The BAC clone 4E14 contained both MWG2245 hybridizing BglII fragments (data not shown), whereas the remaining six BACs only contained one fragment. The maximum contig length of about 270 kb was represented by two BAC clones (4E14 and 111I4, Fig. 1B) overlapping in a region of about 30 kb.
The right end of BAC 4E14 (4E14R) mapped at 0.1 cM in between the two markers MWG2245a and Lrk10–6 (Fig. 1A), allowing the orientation of the physical contig with BAC 111I4 in the direction of Lr10 (Fig. 1B). Only one additional probe (4E14L) derived by BAC end cloning from the seven BAC clones of the contig was a low copy sequence. However, polymorphisms revealed by this probe did not map to chromosome 1AS. Screening the BAC library with 4E14L did not identify additional BACs belonging to the initial contig. The end probes from BAC111I4 could neither be mapped in the recombinant population nor used to screen the BAC library to identify additional BACs for extension of the contig. As an alternative strategy to develop new probes, low-pass sequencing of a shotgun library of the BAC clone 111I4 was performed. After BLAST searches, 14 primer pairs were designed to PCR-amplify putatively nonrepetitive regions of independent shotgun sequences. The PCR products were tested by Southern hybridization to digested genomic DNA of the parents of the mapping population. Two probes, F241 and F467, were genetically mapped in the recombinant population. They showed complete linkage with the Lr10 gene and with the marker MWG2245b (Fig. 1). Physical restriction mapping on the BAC clones showed that F241 was located at about 8 kb proximal to the left end of 4E14, whereas F467 was located 9 kb from the right end of 111I4 (Fig. 1B). Thus, within a physical distance of about 120 kb (between markers F467 and MWG2245b), no recombination occurred between Lr10 and the markers present in this region (Fig. 1). The first step of chromosome walking was then performed using F467 as a probe. This probe detected at least ten loci by Southern hybridization to HindIII-digested DV92 DNA (data not shown) and identified 136 BAC clones in the library. Based on HindIII fingerprints of all 136 clones, Southern hybridization with F467 and sequencing of the BAC DNA corresponding to F467, three BAC clones were identified to belong to the existing contig. Thus, using a probe obtained by low-pass sequencing, it was possible to identify a new BAC clone (640L20) that extended the contig length by about 80 kb in a first step of chromosome walking (Fig. 1B).
Sequence analysis of the new BAC ends revealed that the left end of clone 640L20 shared 88% sequence identity in a stretch of 199 bp to an exon of a maize actin gene (ZMU60509). A probe, F640, amplified from this region detected a single copy locus in DV92 and one locus for each of the three homeologous chromosomes of group 1 in hexaploid wheat (data not shown). Genetic mapping placed F640 at 0.05 cM proximal to Lr10. Thus, with subgenome chromosome walking, it was possible to develop markers flanking the Lr10 resistance gene.
To identify the region between F467 and F640 where recombination took place, low-pass sequencing was done on BAC 640L20. A single copy probe (HS79) located on the same 30-kb SalI fragment as F640 cosegregated with the Lr10 gene. This demonstrated that the three recombination events (0.05 cM) between F640 and Lr10 occurred in a 30-kb region (Fig. 1). Low-pass sequencing for marker development was also performed on BAC 4E14. A new probe (HS272) derived from this BAC mapped 0.08 cM distal to Lr10 (Fig. 1). Thus, the Lr10 resistance gene was located in a 0.13-cM interval in hexaploid wheat, which represents 300 kb of physical distance in T. monococcum. F640 was identified to be part of a putative actin gene. Interestingly, the barley RFLP probe cMWG645 corresponds also to a cDNA sequence of an actin gene. To study if the contig would extend into a cluster of actin genes, we performed a second chromosome walking step by screening the library with the single copy probe F640. Thirty-three new BACs were identified. Fingerprinting and hybridization analysis demonstrated that a single clone 306I7 belonged to the established contig, expanding its total length to 450 kb (Fig. 1B). Hybridization with cMWG645 detected no additional actin gene on BAC 306I7.
Thus, in two steps of subgenome chromosome walking, we have established a 450-kb physical contig from the T. monococcum Am genome that genetically spans the Lr10 locus in hexaploid wheat and contains flanking markers at less than 0.1 cM from the gene. All of the six probes that were derived from a 350-kb region of this contig and that were mapped in the high-resolution population confirmed complete colinearity between the physical map on chromosome 1AmS of T. monococcum and the genetic map of this locus on chromosome 1AS of hexaploid wheat.
Low-Pass Sequencing as a Key Strategy for Successful Chromosome Walking in Wheat.
Relying only on the cloning and sequencing of BAC ends was not sufficient to extend the contig in the direction of the Lr10 gene after the first BAC clones were isolated. In total, 18 BAC ends (corresponding to 9 BACs covering the whole 450-kb contig) were analyzed during this study (Table 1). Based on sequence comparison (BLAST) against public databases, 15 BAC ends (83%) corresponded to known repetitive sequences. Only two end probes (4E14R, F640) detected a polymorphism between ThLr10 and Frisal that mapped to chromosome 1AS. In contrast, low-pass sequencing of a limited number of shotgun clones of the BACs 111I4, 640L20, and 4E14 allowed the development of 12 new probes, which detected polymorphisms between ThLr10 and Frisal that mapped to chromosome 1AS (Table 1). Sequences from shotgun clones were first analyzed using the BLASTx and n algorithms. Sequences showing similarity to known repetitive plant DNA or to noncoding plant DNA were classified as being putatively repetitive. All sequences with similarity to plant open reading frames (ORF) or with no similarity to any plant DNA were selected as being putatively nonrepetitive. Out of 234 sequences from BAC 111I4 that were analyzed using BLAST search, 61 (26%) were classified as nonrepetitive. Nine of them shared sequence similarity to the same plant ORF (Table 1). Fourteen putatively nonrepetitive sequences were explored for probe development. Eight of these probes showed polymorphisms between ThLr10 and Frisal on chromosome 1AS (Table 1), one was monomorphic, and five probes revealed hybridization signals typical for repetitive DNA (data not shown). This result indicates that the systematic comparison of the sequences against the databases greatly helped to identify nonrepetitive sequences. By low-pass sequencing of the BAC clones 640L20 and 4E14, a smaller portion of repetitive sequences (52% and 46%, respectively) was found compared with 111I4 (Table 1). Interestingly, 15 and 17 individual sequences on each of these two BACs showed similarity to sequences of three different putative ORFs in the databases (data not shown). Thus, the lower amount of repetitive sequences found on BACs 640L20 and 4E14 is likely attributable to a higher gene content. We conclude that low-pass sequencing is a reliable and successful strategy for marker development from wheat BAC clones when no markers can be derived from the BAC ends.
Table 1.
Strategies for probe development from BAC clones
| BAC end cloning/sequencing | Low-pass sequencing of BAC
|
|||
|---|---|---|---|---|
| 111I4* | 640L20* | 4E14* | ||
| Number of sequences | 18 | 234 | 183 | 236 |
| DNA sequence (kb) | 15.8 | 156.3 | 119.3 | 159.2 |
| Repetitive sequences | 15 (83%) | 173 (74%) | 95 (52%) | 108 (46%) |
| Putative nonrepetitive sequences | 3 (17%) | 61 (26%) | 88 (48%) | 128 (54%) |
| No homology to ORF | 2 | 52 | 73 | 111 |
| Homology to ORF | 1 | 9 | 15 | 17 |
| Polymorphic probes† | 2/3‡ | 8/14‡ | 3/4‡ | 1/6‡ |
BAC length is 130, 90, and 170 kb, respectively.
Polymorphism between ThatcherLr10 and Frisal linked to the Lr10 locus.
Number of polymorphic probes/number of tested nonrepetitive sequences.
The Ratio of Physical to Genetic Distance Is Highly Variable.
Extensive genetic and physical mapping at the Lr10 locus has allowed us to analyze the ratio of physical to genetic distance over a physical contig of about 350 kb. With a genetic distance of 0.26 cM between the two probes F640 and 4E14R (Fig. 1), the overall ratio of physical to genetic distance was about 1.4 Mb/cM. In the region between MWG2245b and HS272, a similar ratio of 950 kb/cM was observed. Lower ratios of 400 kb/cM between 4E14R and MWG2245a and 600 kb/cM between F640 and HS79 were found in the adjacent regions. Interestingly, three putative ORFs have been identified in the 30-kb interval between F640 and HS79 (data not shown). No recombination event was found in 200 kb between HS79 and MWG2245b, resulting in a minimal ratio of 12 Mb/cM, which is ten times more than the overall average ratio. These data showed that the ratio of physical to genetic distance is very variable in this region of chromosome 1AS.
BAC Clones of T. monococcum Are a Valuable Source of Markers for Wheat.
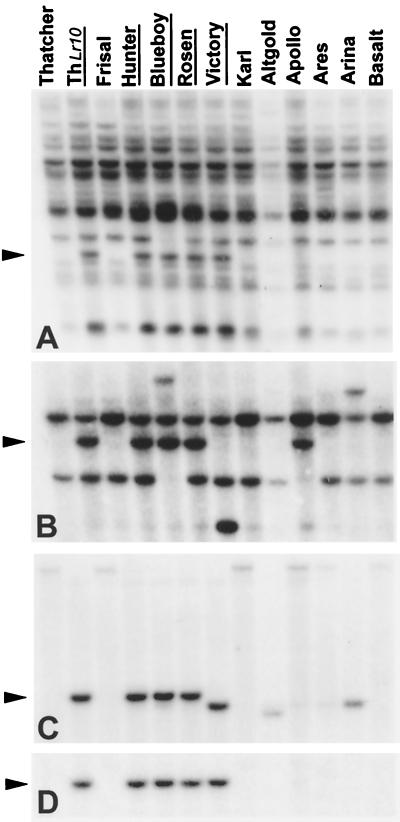
We have derived 12 RFLP probes from T. monococcum BAC clones that detected polymorphisms between the hexaploid wheat lines Frisal and ThLr10 and mapped to chromosome 1AS. These probes fell into different categories: four probes gave a high copy number hybridization signal (more than 12 bands) on a Southern blot. Five single copy number probes detected loci in all three homoeologous group 1 chromosomes, and three single copy number probes were A genome-specific. Four probes (F467, F640, HS272, and F241) representing the different categories were tested for polymorphism on a collection of 58 wheat and spelt breeding lines (15). The high copy number probe F467 (Fig. 2A) detected the same polymorphic fragment as in ThLr10 in five lines containing the Lr10 gene (14, 15) but also in Can3842 (data not shown), which is not known to carry Lr10. For the group 1 chromosome specific probe F640 (Fig. 2B), 14 lines showed the same fragment as ThLr10 at the Lr10 locus including Blueboy, Greif (data not shown), Hunter, and Rosen. In the other lines, F640 detected a number of additional polymorphisms corresponding to three different alleles. The four lines Blueboy, Greif (data not shown), Hunter, and Rosen also showed the same allele as ThLr10 when hybridized with the A genome-specific single copy probe HS272 (Fig. 2C). Interestingly, the A genome-specific single copy probe F241 (Fig. 2D) detected the same fragment as in ThLr10, only in lines known to contain Lr10. These data showed that clones from the DV92 BAC library are a valuable source of polymorphic markers for the analysis of wheat germplasm and can be generally applied for mapping in other crosses.
Figure 2.
Southern hybridization of HindIII-digested genomic DNA of wheat and spelt breeding lines (13 of 58 varieties tested are shown) with four different RFLP probes derived from T. monococcum BACs. Filled arrowheads indicate the allele specific for chromosome 1AS found in ThLr10. Lines carrying the Lr10 gene are underlined. (A) Hybridization with the probe F467 resulted in a complex pattern with a single dominant fragment for ThLr10. (B) Different alleles were detected by the single-copy probe F640 (one locus per genome A, B, and D as demonstrated by aneuploid nulli-tetrasomic analysis; data not shown). (C) The probe HS272 was mapped as a codominant, A genome-specific probe. In addition to the two alleles of ThLr10 and Frisal, two further alleles were detected, e.g., in the lines Victory and Altgold. (D) Probe F241 detected a genome-specific fragment in lines containing the Lr10 resistance gene.
Discussion
Identification of Mapping Probes from Wheat BAC Sequences.
Chromosome walking and positional cloning were so far considered as not feasible in bread wheat because of the large genome size, the amount of repetitive sequences, and the presence of three closely related genomes. The strategy of shuttle mapping between diploid progenitor species and polyploid relatives was previously proposed to support positional cloning in wheat (21, 22). In this study, we explored the possibility of chromosome walking on the Am genome of the diploid wheat T. monococcum and its use for high-density mapping of the Lr10 resistance gene locus on chromosome 1AS of T. aestivum. Chromosome walking was initiated by screening the T. monococcum BAC library developed by Lijavetzky et al. (10) with an RFLP marker completely linked to the Lr10 resistance gene. During chromosome walking, new probes have to be derived from isolated BACs for the next walking step. For most of the plant genomes studied so far, this was easily achieved by deriving probes from the BAC ends. In genomes such as wheat, however, more than 80% of the genome consists of repetitive sequences. Consequently, in most of the cases, the BAC ends cannot be mapped because of their repetitive nature. Kleine et al. (23) have shown that only 19% of yeast artificial chromosome (YAC) ends from 54 randomly chosen barley YACs contained low-copy sequences. Similarly, we have found that only 17% of the probes that were generated by BAC end sequencing or cloning were not repetitive. Thus, chromosome walking cannot solely rely on BAC end probes but other approaches have to be used. In barley, chromosome walking at the Mla locus required intensive work combining RFLP and Amplified Fragment Length Polymorphism (AFLP) analysis, the development of sequence-specific AFLP, and STS markers derived from BAC and YAC end sequences (24). Here, we have developed a strategy of low-pass sequencing of shotgun clones isolated from the T. monococcum BACs and comparison of the sequences with known sequences in the databases. Between 46 and 74% of the sequences have been identified as repetitive. Of the remaining sequences, only very few probes proved repetitive, demonstrating that most of the repetitive sequences were identified by BLAST. Among the nonrepetitive sequences identified in the three analyzed BACs, 41 showed significant homology with putative coding regions in the databases, indicating the presence of six to seven different genes in 350 kb of contig (data not shown). This suggests that the gene density at this locus is lower than in a more distal chromosomal region at the Lrk locus (4).
A Perfect Colinearity Between the 1Am and 1A Chromosomes.
A total of 12 probes were derived from a 350-kb BAC contig of T. monococcum. Using six of them, we were able to saturate the chromosomal region of the Lr10 resistance gene in hexaploid wheat with new markers. The order of these markers on the genetic map of hexaploid wheat was the same as the order of their corresponding probes on the physical contig in T. monococcum. This demonstrated perfect colinearity between chromosome 1Am of T. monococcum and chromosome 1A of T. aestivum and confirmed, at the microlevel, data obtained by comparative genetics (8). Apparently, only very minor rearrangements, if any, have occurred in this region between the Am and the A genomes. These data confirmed that the Am genome of T. monococcum is an excellent model for the A genome of hexaploid wheat and that the colinearity between these two genomes can be used to sequentially develop new markers for high-resolution mapping at a specific locus in hexaploid wheat.
The Recombination Frequency Is Unevenly Distributed Over 350 kb of Contig.
A population size representing 6,240 gametes was sufficient to develop markers flanking the Lr10 gene with a genetic distance of less than 0.1 cM. With this population size, we could detect recombination within 0.13 cM, a distance that corresponds to approximately 300 kb on the physical map of T. monococcum. It is known that the recombination frequency is not evenly distributed along the chromosomes. Thus, the number of gametes required to observe the necessary recombination rate can vary depending on the target region. Map-based isolation of the Mla, mlo, and Rar-1 genes from barley was achieved with the development of large populations of 3,600, 4,044, and more than 8,000 gametes, respectively (24–26). We have studied the ratio between the physical and the genetic distances at the Lr10 locus. Interestingly, we have observed that probes that were physically very close such as F640 and HS79 (30 kb) were separated by three recombination events, giving a ratio of physical to genetic distance of <600 kb/cM. In contrast, the probes HS79 and MWG2245b, which are roughly 200 kb apart, cosegregated on the genetic map, indicating a ratio of more than 12 Mb/cM. These data revealed large differences in the recombination frequency within as little as 230 kb. The average relationship between genetic and physical distance in wheat, based on a genome size of 16,000 Mb and a genetic map of 2,900 cM, is about 5.5 Mb/cM. Cytogenetic analysis, however, has clearly shown the existence of hot spots of recombination on wheat chromosomes (12, 13). On chromosome 1, the genetic to physical distance ratio was estimated to be 118 kb/cM in gene-rich regions and 22 Mb/cM in gene-poor regions (12). Mechanisms underlying the discrepancy in the rate of recombination and the occurrence of hot spots of recombination have been extensively studied in yeast (27). In has been observed that within genes, the recombination rate can be one to two orders of magnitude higher than the average (27). In plants, a striking example of a hot spot of recombination was found in the 5′-transcribed region of the a1 gene in maize. There, 20% of the recombination observed in a 140-kb interval occurred within 377 bp (28). In our study, we have observed a higher rate of recombination than the average for the wheat genome in a 30-kb region proximal to the Lr10 gene. Interestingly, three putative ORFs have been identified in this region (data not shown). In T. tauschii, Spielmeyer et al. (29) have identified a highly recombinogenic region containing genes encoding seed storage proteins at the telomeric end of chromosome 1DS. In this region, the ratio of physical to genetic distance has been estimated to be around 20 kb/cM, a similar ratio as the one observed at the mlo resistance gene in barley (25). Together, these data confirm the hypothesis that in large and repetitive genomes, gene-containing regions are likely targets for recombination. In contrast, in the interval spanning the Lr10 resistance gene, we have found an apparent suppression of recombination (no recombination within 200 kb). Interestingly, Wei et al. (24) also observed the same discrepancy at the Mla locus in barley with 5 Mb/cM close to the gene and 176 kb/cM in the flanking interval. A high rate of polymorphism between allelic sequences can be responsible for the suppression of recombination (24). Lr10 originates from the wheat gene pool (14), and the mechanisms underlying suppression of recombination in the Lr10 interval in the cross between ThLr10 and Frisal remain to be studied.
Subgenome Chromosome Walking: An Efficient Strategy for Targeted Genome Characterization in Hexaploid Wheat.
The 12 different RFLP probes that were derived from the 350-kb BAC contig have revealed different numbers of fragments (1 to >12) after Southern hybridization. These probes have revealed polymorphisms in many wheat and spelt breeding lines. In general, two to three different alleles were detected for each low-copy probe. Thus, probes derived from the T. monococcum BAC clones can generally be used for genetic mapping in wheat crosses. In addition to Lr10, 14 agronomically important genes have been located in the distal region of chromosome group 1S of hexaploid wheat (12). The approach described in this paper will support the isolation of other genes, e.g., the powdery mildew resistance gene Pm3 from the same genetic region of chromosome 1AS (14).
In three cases, the derived probes were single copy and specific for the A genome. This proportion (25%) is higher than the 7% of genome-specific probes found among 71 probes analyzed for their location on the homoeologous genomes of wheat (30). Interestingly, two A genome-specific probes belonged to putative ORFs. This finding differs from the observation that cDNA probes usually hybridize to all three genomes (30) and that coding regions are very well conserved between the A, B, and D genomes. The fact that the derived probes revealed polymorphisms in the wheat gene pool suggests that the strategy of developing new probes by chromosome walking on T. monococcum and subsequent genetic mapping in hexaploid wheat can generally be applied to saturate regions of interest on the A genome of hexaploid wheat. The same approach can be used for genes located on the D genome by using the T. tauschii BAC library. Many probes derived from the Am or the D genome will reveal the three alleles on the homeologous genomes of hexaploid wheat. Therefore, the use of a diploid BAC library will not be limited to genes from the corresponding subgenome but will certainly be useful to saturate orthologous regions on the other genomes.
In this study, we have genetically saturated and physically covered the Lr10 locus in wheat. Six to seven putative ORFs have been identified on the 350-kb BAC contig (unpublished results). These putative coding regions can now be used to screen a small insert size library (lambda or cosmid) of the hexaploid wheat variety containing the Lr10 resistance gene to isolate the corresponding candidate genes. Subgenome chromosome walking offers the possibility to isolate bread wheat genes through positional cloning and is likely to have a major impact on wheat improvement in the future.
Acknowledgments
We are very grateful to Geri Herren and Kathi Stein for their excellent technical assistance. We thank Dr. C. Ringli and Dr. N. Yahiaoui for critical reading of the manuscript. This project was supported by Swiss Priority Program Biotechnology Grants 5002-45033 and 5002-57824.
Abbreviations
- BAC
bacterial artificial chromosome
- cM
centimorgan
- Mb
megabase
- RFLP
restriction fragment length polymorphism
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AZ254919–AZ254921).
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.230361597.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.230361597
References
- 1.Hiruchika H. Plant Mol Biol. 1997;35:231–240. [PubMed] [Google Scholar]
- 2.Havukkala I J. Curr Opin Genet Dev. 1996;6:711–714. doi: 10.1016/s0959-437x(96)80025-6. [DOI] [PubMed] [Google Scholar]
- 3.Kilian A, Chen J, Han F, Steffenson B, Kleinhofs A. Plant Mol Biol. 1997;35:187–195. [PubMed] [Google Scholar]
- 4.Feuillet C, Keller B. Proc Natl Acad Sci USA. 1999;96:8665–8670. doi: 10.1073/pnas.96.14.8265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tarchini R, Biddle P, Wineland R, Tingey S, Rafalski A. Plant Cell. 2000;12:381–391. doi: 10.1105/tpc.12.3.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feuillet C, Keller B. Proceedings 9. 1998. thInternational Wheat Genetics Symposium 1, 171–177. [Google Scholar]
- 7.Smith D B, Flavell R B. Chromosoma. 1975;50:223–242. [Google Scholar]
- 8.Dubcovsky J, Luo M-C, Dvorak J. Proc Natl Acad Sci USA. 1995;92:6645–6649. doi: 10.1073/pnas.92.14.6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gill K S, Lubbers E L, Gill B S, Raupp W P, Cox T S. Genome. 1991;34:362–374. [Google Scholar]
- 10.Lijavetzky D, Muzzi G, Wicker T, Keller B, Wing R, Dubcovsky J. Genome. 1999;42:1176–1182. [PubMed] [Google Scholar]
- 11.Moullet O, Zhang H-B, Lagudah E S. Theor Appl Genet. 1999;99:305–313. [Google Scholar]
- 12.Gill K S, Gill B S, Endo T R, Taylor T. Genetics. 1996;144:1883–1891. doi: 10.1093/genetics/144.4.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Faris J D, Haen K, Gill B S. Genetics. 2000;154:823–835. doi: 10.1093/genetics/154.2.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McIntosh R A, Wellings C R, Park R F. In: Wheat Rusts: An Atlas of Resistance Genes. Cloud-Guest A, Jeans K, editors. Dordrecht: Kluwer; 1995. [Google Scholar]
- 15.Schachermayr G, Feuillet C, Keller B. Mol Breed. 1997;3:65–74. [Google Scholar]
- 16.Graner A, Siedler H, Jahoor A, Hermann R G, Wenzel G. Theor Appl Genet. 1990;80:826–832. doi: 10.1007/BF00224200. [DOI] [PubMed] [Google Scholar]
- 17.Sambrook J, Fritsch E F, Maniatis T. In: Molecular Cloning: A Laboratory Manual. For d N, Nolan C, Ferguson M., editors. Plainview, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 18.Woo S S, Jiang J, Gill B S, Paterson A H, Wing R A. Nucleic Acids Res. 1994;22:4922–4931. doi: 10.1093/nar/22.23.4922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Altschul S F, Madden T L, Schäffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feuillet C, Schachermayr G, Keller B. Plant J. 1997;11:45–52. doi: 10.1046/j.1365-313x.1997.11010045.x. [DOI] [PubMed] [Google Scholar]
- 21.Kam-Mogan L N W, Gill B S, Muthukrishan S. Genome. 1989;32:724–732. [Google Scholar]
- 22.Gill K S, Gill B S. BioEssays. 1994;16:841–846. [Google Scholar]
- 23.Kleine M W, Michalek W, Diefenthal T, Dargatz H, Jung C. Genome. 1997;40:896–902. doi: 10.1139/g97-116. [DOI] [PubMed] [Google Scholar]
- 24.Wei F, Gobelman-Werner K, Moroll S M, Kurth J, Mao L, Wing R, Leister D, Schulze-Lefert P, Wise R. Genetics. 1999;153:1929–1948. doi: 10.1093/genetics/153.4.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Buschges R, Hollricher K, Panstruga R, Simons G, Wolter M, Frijters G, van Daelen R, van der Lee T, Diergaarde P, Groenendijk J, et al. Cell. 1997;88:695–705. doi: 10.1016/s0092-8674(00)81912-1. [DOI] [PubMed] [Google Scholar]
- 26.Shirasu K, Lahaye T, Tan M- H, Zhou F, Azevedo C, Schulze-Lefert P. Cell. 1999;99:355–366. doi: 10.1016/s0092-8674(00)81522-6. [DOI] [PubMed] [Google Scholar]
- 27.Lichten M, Goldman A S H. Annu Rev Genet. 1995;29:423–444. doi: 10.1146/annurev.ge.29.120195.002231. [DOI] [PubMed] [Google Scholar]
- 28.Xu X, Hsia A-P, Zhang L, Nikolau B J, Schnable P S. Plant Cell. 1995;7:2151–2161. doi: 10.1105/tpc.7.12.2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Spielmeyer W, Moullet O, Laroche A, Lagudah E S. Genetics. 2000;155:361–367. doi: 10.1093/genetics/155.1.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Devey M E, Hart G E. Genome. 1993;36:913–918. doi: 10.1139/g93-120. [DOI] [PubMed] [Google Scholar]