Abstract
Studies in the developing spinal cord suggest that different motoneuron (MN) cell types express very different genetic programs, but the degree to which adult programs differ is unknown. To compare genetic programs between adult MN columnar cell types, we used laser capture micro-dissection (LCM) and Affymetrix microarrays to create expression profiles for three columnar cell types: lateral and medial MNs from lumbar segments and sympathetic preganglionic motoneurons located in the thoracic intermediolateral nucleus. A comparison of the three expression profiles indicated that ~7% (813/11,552) of the genes showed significant differences in their expression levels. The largest differences were observed between sympathetic preganglionic MNs and the lateral motor column, with 6% (706/11,552) of the genes being differentially expressed. Significant differences in expression were observed for 1.8% (207/11,552) of the genes when comparing sympathetic preganglionic MNs with the medial motor column. Lateral and medial MNs showed the least divergence, with 1.3% (150/11,552) of the genes being differentially expressed. These data indicate that the amount of divergence in expression profiles between identified columnar MNs does not strictly correlate with divergence of function as defined by innervation patterns (somatic/muscle vs. autonomic/viscera). Classification of the differentially expressed genes with regard to function showed that they underpin all fundamental cell systems and processes, although most differentially expressed genes encode proteins involved in signal transduction. Mining the expression profiles to examine transcription factors essential for MN development suggested that many of the same transcription factors participatein combinatorial codes in embryonic and adult neurons, but patterns of expression change significantly.
Keywords: transcriptome, identified motor neuron, combinatorial code, ion channel, transcription factor, fluorescent laser capture microscopy, spinal cord, lipid raft signaling complex
Contemporary advances in molecular techniques and our understanding of neuronal development in the spinal cord have recently made it possible to identify and manipulate unique populations of mammalian spinal neurons (24, 30). At the same time, transcript levels for thousands of genes expressed in a single sample can now be measured with microarray expression profiling (15). By the combination of transgenic technology, laser capture microdissection (LCM) (6, 9), and microarray expression profiling, the genetic programs operating in identified populations of neurons can be defined (17, 22, 33, 35).
With regard to identified spinal neuronal populations, motoneuron (MN) subtype specification and diversification have been studied for many years at several levels (51). All spinal MNs arise from common progenitor cells that postmitotically differentiate into five columnar subtypes, three of which are considered here. Medial MNs (MMNs) are the first cells to differentiate (50, 62), forming a column along the entire rostrocaudal axis of the spinal cord and innervating axial muscles. Subsequently, lateral MNs (LMNs) form motor columns in the lumbar and brachial spinal segments and innervate limb muscles (52). Visceral MNs located in the intermediolateral column (IML) (also known as sympathetic preganglionic neurons) are among the last subtypes to differentiate (58, 62). They are found only in the thoracic segments through L1. The IML influences the viscera by innervating components of the endocrine system and neurons in the sympathetic chain ganglia, which in turn innervate the viscera. Columnar cell types are not homogeneous and can be further divided into MN pools based on the targets they innervate.
Despite their common developmental lineage, each of the three aforementioned columnar cell types is clearly unique with respect to gene expression during embryonic development (23, 51, 52, 58, 62). These differences help establish appropriate connectivity, leading to the distinct somatic locations and target innervations observed in the adult. Whether differences in MN genetic programs persist into adulthood is a matter of debate. It may be that once spinal circuits are wired, most molecular differences between adult MNs disappear. In this paper we ask the following question: How similar are adult MN columnar subtypes at the molecular level? To answer this question, we develop and critically evaluate a protocol for expression profiling-identified adult spinal MNs and examine differences across three columnar cell types: MMNs, LMNs, and IMLs.
MATERIALS AND METHODS
Animals, surgery, spinal cord dissections, and slide preparation
All animals were cared for in accordance with institutional, United States Department of Agriculture, and National Institutes of Health (NIH) guidelines for the care and use of laboratory animals. Animal protocols were approved by Institutional Animal Care and Use Committee. In total, 8 rats and 11 mice were used in these experiments. Animals were anesthetized with ketamine (100 mg/kg), xylazine (10 mg/kg), and atropine (0.05 mg/kg) for survival surgery. Urethane (2 mg/kg ip) was used for spinal cord isolation.
For some experiments, the complete spinal cord was isolated from rats and mice. Complete rat spinal cords (Sprague Dawley) were dissected out on postnatal day 8 (P8) without any prior manipulations, and the whole cord was stored in RNAlater (Ambion, Austin, TX) at −80°C. These cords were only used in preliminary experiments to develop the amplification protocol. Complete mouse spinal cords [FVB-TgN(GadGFP)45704Swn; Jackson Laboratories] were dissected out on P45, and the whole cord was stored in RNAlater (Ambion) at −80°C until use. These cords were used in preliminary experiments to develop the amplification protocol and to produce targets that were used in various comparisons with identified motoneurons.
In addition to isolating complete spinal cords, we prepared slides containing spinal cord sections that would later be used for isolating identified motoneuron populations with LCM. The five animals used to isolate identified columnar MN populations represent the sham-operated controls for a larger study (data not shown). As such, these animals underwent mock surgery on P24. This surgery exposed the spinal cord at spinal segment T8. There is no reason to suspect that these minor surgical procedures affected the differences between MN populations. In the immunocytochemical (ICC) experiments described in results, animals did not undergo a sham operation. Nevertheless, the data were consistent between sham-operated and unoperated animals. Three days before the mice were killed for LCM, 50 µl of Fluoro-Gold (Fluorochrome, Denver, CO; 1% in saline) were injected intraperitoneally to identify motor efferents. On P45, animals were anesthetized and decapitated, and the spinal cords were dissected out. A roughly 2.5-mm section containing either L2–L4 (for LMN and MMN isolations) or T11–L1 (for IML isolations) was cut, placed on a cryostat chuck covered with tissue-embedding medium (OCT; Electron Microscopy Sciences, Hatfield, PA), and frozen. Fresh frozen 8-µm sections were cut using a cryostat and stored at −80°C with desiccant until use. All sections were mounted on slides that had been coated with a Teflon-like spray, polytetrafluoroethylene (PTEF; Electron Microscopy Sciences), and autoclaved before use. For a given animal, we typically produced 20 slides, each containing 20 8-µm sections representing an ordered array of T11–L4. All tissue manipulation steps were performed using RNase-free conditions. Slides were always transported on dry ice in a sealed storage box with desiccant.
RNA isolation from total spinal cord
For complete spinal cords from mice and rats, the cord was thawed and removed from RNAlater (Ambion), and total RNA was extracted using an RNeasy Mini Kit (Qiagen, Valencia, CA). As stated above, rat total RNA was only used in preliminary experiments to develop the amplification protocol. Mouse total RNA was used in preliminary experiments to develop the amplification protocol and to produce targets used in various comparisons with identified motoneurons.
LCM and RNA isolation
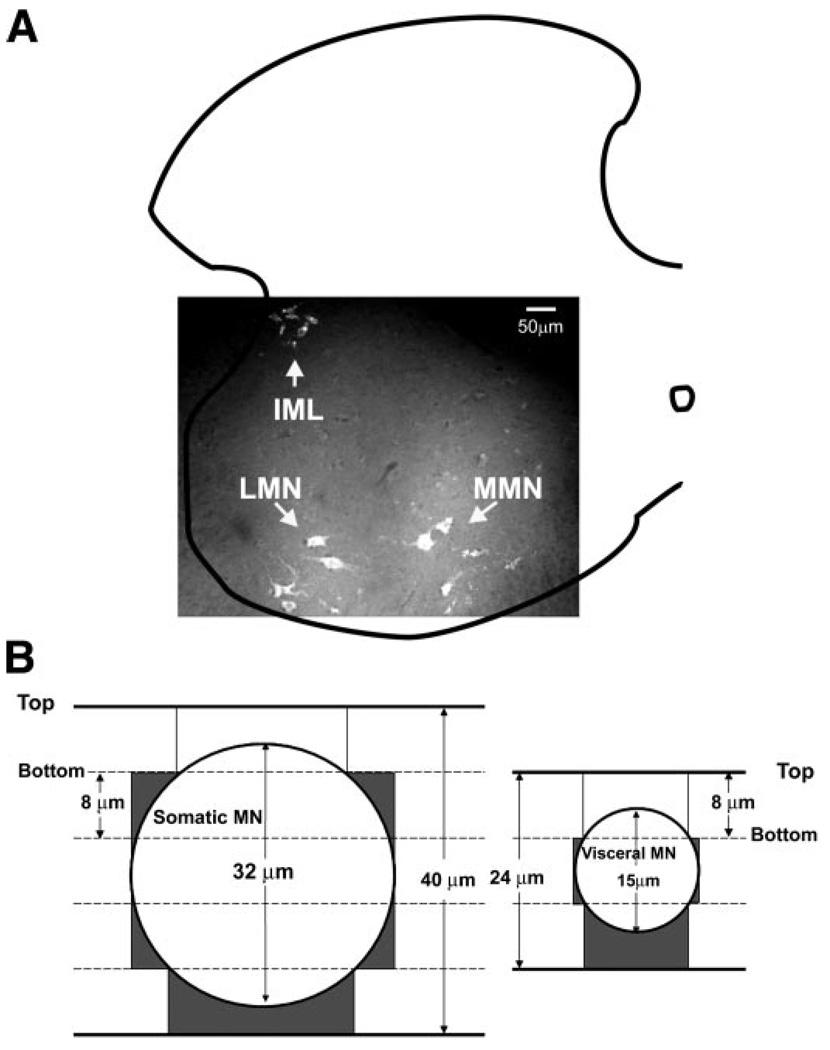
We isolated total RNA from identified populations of MNs using LCM and the previously described slides containing 8-µm spinal cord sections. Immediately before LCM, a single slide was removed from the storage box (which was incubated on dry ice) and fixed in ice-cold acetone for 4 min and dehydrated as follows: 75% ethanol for 30 s, 95% ethanol for 30 s, 100% ethanol for 30 s. The tissue was cleared in xylene for 5 min and vacuum dried in a desiccator for 15 min. The PixCell II LCM system (Arcturus, Mountain View, CA) was used to visualize and capture Fluoro-Gold-labeled neurons (Fig. 1 and Fig. 2). By adjustment of the laser beam diameter, sections of neuronal somata could be isolated (captured) without remarkable contamination of surrounding tissue (Fig. 2, A and B). Visceral MNs (IML) were obtained from spinal segments T11–L1, while populations of somatic MNs (MMN and LMN) were obtained from segments L2–L4, where the LMN innervates the musculature of the hindlimb.
Fig. 1.
Visualizing and sectioning the 3 populations of motoneurons (MNs). A: intraperitoneal injections of Fluoro-Gold and subsequent fluorescent microscopy of cryostat sections were used to visualize individual MNs in the spinal cord. The 3 MN subpopulations discussed in the text are as indicated. A hemisection of the spinal cord is outlined, and ventral is downward. This section is from the 1st lumbar segment (L1), the only segment where the 3 populations overlap. B: somatic and visceral MNs are drawn as circles according to scale (see materials and methods). The no. of 8-µm sections usually required to encompass a single neuron is as indicated, although in some instances this number could be reduced by 1. Top indicates the top surface of a section, while bottom faces the slide. Dark gray shading indicates the nonneuronal area that will be captured in the laser capture microdissection (LCM) experiments. The first section of MNs will most likely not be captured, as the Fluoro-Gold signal will be weakened as it penetrates the nonneuronal covering. IML, intermediolateral column; LMN, lateral MN; MMN, medial MN.
Fig. 2.
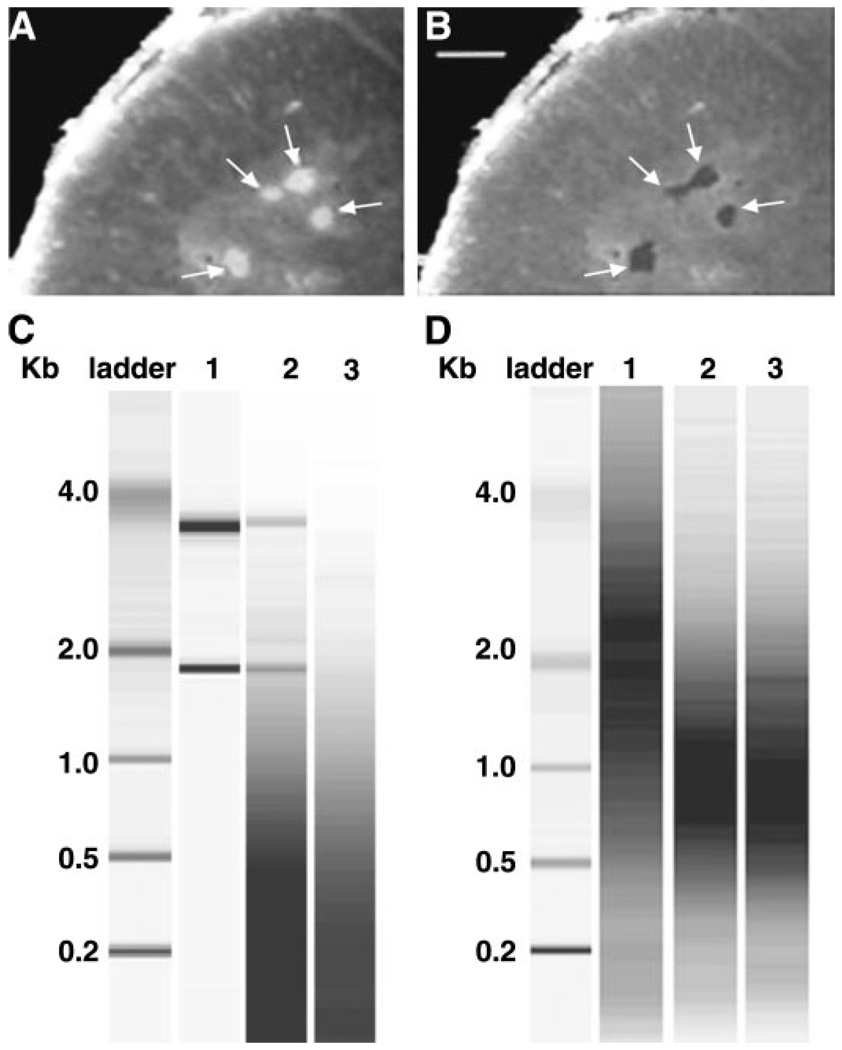
LCM isolation of fluorescently labeled LMNs, RNA extraction, and aRNA target production. A: fresh frozen section before laser capture. Arrows point to Fluoro-Gold-labeled MNs. Ventral is upward. B: 4 captures (fluorescent MN sections) have been physically isolated using LCM, leaving behind surrounding tissues. Scale bar is 100 µm for both A and B. C: bioanalyzer image of total RNA preparations. Lane 1, 100 ng of whole spinal cord total RNA isolated with a Qiagen RNeasy kit; lane 2, total RNA extracted from 100 LCM captures with the PicoPure kit; lane 3, no template negative control using the Picopure kit. The low-molecular-weight smear appearing in lanes 2 and 3 represents the nucleic acid carrier from the PicoPure extraction buffer. D: bioanalyzer image of aRNA targets. Lanes 1–3, aRNA targets produced from 5 µg (lane 1) or 10 ng (lane 2) of the same whole spinal cord total RNA preparation, or total RNA extracted from 100 LCM captures (lane 3).
With the use of a single slide, ~50 captures for 1 cell type were performed with 1 Capsure HS LCM cap in a 30-min period of time. Arcturus denaturing buffer was then added to the cap to lyse cells and denature proteins, and the cap was incubated on the bench at room temperature while a new cap was inserted into the instrument; more cells were collected in additional rounds of capture as necessary. As a precaution against RNA degradation, cells never remained in a cap for longer than 30 min before exposure to denaturing buffer. After collecting 200 captures in 4–5 caps, the caps containing cells and buffer were heated to 42°C for 30 min, and cell lysates were spun into a microfuge tube. The lysates were frozen at −80°C for up to 1 wk while remaining cells were captured. After completion of all captures for a given animal, the lysates for that animal were pooled according to cell type, and RNA was isolated by adding the total lysate to a column followed by washing and elution of the column-bound RNA using an Arcturus Picopure kit (Arcturus) and the instructions provided by the manufacturer. Figure 2C indicates that high-quality RNA was obtained with this method. The 2 ribosomal RNA (rRNA) bands from as few as 100 captures were easily visualized with a Bioanalyzer (Agilent). Typically, the ratio of rRNA bands (28S/18S) is used as a measure of RNA quality, with acceptable ratios being >2. Unfortunately, the ratio of 28S to 18S rRNA could not be determined for LCM samples because the distribution of the rRNA bands overlapped with the distribution of carrier DNA, which is included in the process to minimize sample loss due to nonspecific sticking of nucleic acids (see Fig. 2C, lane 3, for intensity and distribution of carrier). Despite this, we judged the integrity of the RNA to be excellent, since two strong bands representing 28S and 18S were always observed. This technical artifact also prevented the exact quantification of RNA; however, on the basis of signal intensity, we estimate that 100 LMN captures yields 10–20 ng of total RNA. Arcturus has changed their kit since these studies were completed, and carrier DNA no longer obscures RNA concentration determinations.
We considered the likelihood of isolating unidentified cells along with MNs by examining the relationship between MN diameter and the thickness of the sections (8 µm). The average size of each of the three classes of MNs was determined with a few of the slides used in the LCM procedure. Slides were removed from the freezer, quickly dried with a blow dryer, and examined on a Nikon E-800 epifluorescent microscope. Regions of interest were identified and cells traced at 40× using a neurolucida program (Microbrightfield, Williston, VT). Cell diameters were determined by averaging the minimum and maximum distances from the cell perimeter. The average diameters are 33 ± 1.0, 31 ± 1.5, and 15 ± 0.9 µm for LMN, MMN, and IML, respectively. The somatic cell types are significantly larger than the visceral (P < 10−12). Figure 1B illustrates the average number of 8-µm sections that comprise a single neuron for somatic (LMN or MMN) vs. visceral (IML) MN cell types. Note, however, that the number of sections could be reduced by one in the rare event that the beginning of the neuron coincided with the beginning of the section. We expect that either the first or last section will not be captured, as in one case nonneuronal cells will cover and weaken the Fluoro-Gold signal. If we assume that nonneuronal cells fill the space that is not occupied by a neuron, then nonneuronal contamination in our captured samples can be approximated by the dark gray regions in Fig. 1B. The dark gray regions represent ~24 and 42% of the total captured area for somatic and visceral MNs, respectively, assuming that we capture four slices for every somatic MN and two slices for every visceral MN and that we capture the entire 8-µm section in every case. Thus, theoretically, different levels of nonneuronal contamination could lead to some of the observed differences between somatic (LMN and MMN) vs. visceral (IML) samples. This should be considered when interpreting the data. However, in practice, this problem appears to be minimal. For example, glial cells are expected to be a major nonneuronal contaminant, but transcripts for glial-specific proteins, such as glial fibrillary acidic protein (GFAP) and myelin-associated oligodendrocyte basic protein (Mobp), do not vary significantly between the three cell types (see results and Supplemental Table S1; Supplemental Material is available at the Physiological Genomics web site).1 Furthermore, if differences in cell size alone were responsible for divergent expression profiles, then the smaller IML should be equally different from MMN and LMN; this is not the case. Similarly, the difference between IML and both somatic MNs should be much larger than the difference between LMN and MMN, which is not true. These results might indicate that the adhesive forces between the neuronal and nonneuronal cells are much weaker than the adhesive forces between the slide and the nonneuronal cells. In this case, when the neuron is pulled from the slide, the bonds to other cells break, and the nonneuronal cells remain on the slide rather than being captured along with the neuron. Alternatively, the researcher may be biased toward capturing only the largest, brightest sections of a neuron, which represent mostly neuronal tissue.
aRNA amplification and labeling
For experiments involving samples of the complete spinal cord RNA, total RNA was extracted from a cord, and 5 µg were used to make an aRNA target, as described in a protocol supplied by Affymetrix (Santa Clara, CA; http://www. affymetrix.com/support/technical/manual/expression_manual.affx). A high-quality aRNA target was represent by an OD260/280 of 1.9~2.1, a yield of 40 µg, and a smear centered at ~2 kb.
For samples starting with 10 ng of RNA or 200 laser captures, we used a slight modification of Baugh et al. (4). The two changes to the original protocol were the following. 1) All of the first-round cDNA was used as the template in a 20 µl in vitro transcription reaction using an Enzo kit. 2) In the second-round RT, the first-round aRNA was primed at 70°C for 4 min with 20 pmol of random hexamer and 10 pmol of TC primers (5′-TATCAACGCAGAGTCCC-3′, 5′-TATCAACGCAGAGTCGG-3′) (13). The resulting biotinylated aRNA was only used as a target if it was of high quality (OD260/280 of 1.9~2.1, yield of 20 µg, and a smear centered at ~1 kb).
Microarray hybridization and scanning
The high-quality aRNA target was express mailed on dry ice to the Genomics Core Laboratory at the Medical College of Georgia, (Augusta, GA), whereupon it was fragmented and hybridized to Affymetrix oligonucleotide microarrays (Murine Genome Chip U74Av2 or Rat Neurobiology Chip U34) according to the manufacturer’s instructions. Mouse aRNA targets were always hybridized to the Murine Genome Chip, while rat aRNA targets were always hybridized to the Rat Neurobiology Chip; there was never any mixing of species targets and chips. The arrays were then washed and scanned. Excellent and reproducible results were always obtained with this Genomics Core Laboratory.
Microarray data analysis
U74Av2 contains oligonucleotides representing roughly 6,000 mouse probe sets with known functions and another 6,000 probe sets representing expressed sequence tags (ESTs). These chips were used to establish expression profiles for each of the three columnar cell types and for control experiments with spinal cord. U34 contains 1,200 rat neurobiology-related probe sets. These chips were used in preliminary studies to develop the protocol. Raw data (dat and cel files) were downloaded from the Genomics Core Laboratory web page. Cel files representing the complete data set were normalized with robust multiarray average (RMA) (5, 21). For each cell type from a single animal, there were two technical replicates: A and B. Thus 30 chips were used to determine the expression profiles for these 3 columnar cell types (5 animals × 3 cell types/animal × 2 technical replicates/cell type). The inverse log of the RMA-normalized data for each of the 30 mouse U74Av2 chips used in this study has been submitted to the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/projects/geo) under accession number GSE2595.
Before averaging and analysis of the normalized technical replicates, the normalized data were prefiltered as follows. For each cell type, averaged signals of the five technical replicates (replicates A from the 5 animals) were compared with the averaged signals for the other five technical replicates for that cell type (replicates B from the same 5 animals). We prefiltered our data such that, if a gene showed >20% variability between averaged technical replicates (A vs. B) for any cell type, that gene was removed from all further analyses for all cell types. We assumed that technical artifacts, such as an undetectable air bubble during the hybridization process or a scratch on the chip, were responsible for irreproducible data so that removal of these genes from the analyses was valid. This reduced the number of probe sets we employed in the final analyses from 12,488 to 11,552. However, we also analyzed the data with these genes included, and they did not produce large changes in any of the results or figures.
After prefiltering, the normalized technical replicates were averaged. GeneSpring 6.0 (Silicon Genetics, Redwood City, CA) was then used to analyze and manipulate the data. Log-log plots and linear regressions were used to compare global signal intensities between two samples. To compare the expression profiles from the three neuronal populations, a one-way ANOVA followed by a Tukey’s post hoc test was performed using averaged signals for all three cell types from all five animals. In some cases, we only compared two sets of data (e.g., whole cord vs. LMN; see results), and, in these instances, Student’s t-tests were performed with GeneSpring 6.0 on the same normalized data. In addition to log-log plots, ANOVAs, and t-tests, we obtained a present/absent call using Affymetrix Microarray Suite (MAS)5 software. Here we emphasize that this detection call was not used as a prefilter. Rather, it was used for the sake of comparison in Supplemental Table S1. When this software was used, the data were not normalized with RMA but with MAS5.
Because results are known to vary with the normalization procedure (5), we also performed a complete set of analyses on data that were normalized with MAS5. As expected, the R2 values for the log-log plots were significantly reduced. In addition, there were differences in the results of the ANOVA such that many more genes showed significant differences in expression when normalized with MAS5. Furthermore, a small fraction of genes that showed significant differences in expression when normalized with RMA were not significant when normalized with MAS5. Here we present only the results of RMA-normalized data, but the 30 cel files used in these studies are available upon request for investigators wishing to examine the data with other normalization procedures.
Quantitative real-time RT-PCR
Gene-specific probes and primers were designed, and quantitative real-time RT-PCR (qRT-PCR) was performed using an ABI PRISM 7700 Sequence Detection System (Applied Biosystems, Foster City, CA). Sequences of primer sets and probes for adenylate cyclase-activating polypeptide1 (Adcyap1), pre-proenkephalin1, and classic myelin basic protein (MBP) were custom designed and are listed in Table 1. On the other hand, primer sets and probes for GFAP, Mobp, apolipoprotein D, Peripherin1, vesicular acetylcholine transporter (VAchT), dopamine receptor 2 (D2R), D3R, and D4R were Assay-on-Demand gene expression products (Applied Biosystems). PCRs were performed in a 30-µl volume containing 800 nM primers and 100 nM probe in Taqman Universal PCR Master Mix. Cycling conditions were as follows: AmpliTaq Gold DNA Polymerase activation at 95°C for 10 min followed by 40 cycles of amplification (denaturation at 95°C for 15 s, anneal-extension at 60°C for 1 min). No template controls were always performed.
Table 1.
Real-time PCR primers and probes
| Gene | Oligonucleotides (5′ to 3′) |
|---|---|
| Adenylate cyclase-activating polypeptide 1 (accession no. AB010149) | |
| Forward primer | CTTTTGGCTGTCCCGCAG |
| Reverse primer | AGTCTTGCAGCGGGTTTCC |
| Probe | CCAGAAGACGAGGCTTACGACCAGGA |
| Preproenkephalin 1 (accession no. M55181) | |
| Forward primer | TGGGTCCTGCCTCCTGG |
| Reverse primer | TACGTGCATTTAGCGCAGTCC |
| Probe | TACAGTGCAGGCGGAATGCAGCC |
| Myelin basic protein (accession no. M11532) | |
| Forward primer | GATTTGGCTACGGAGGCAGA |
| Reverse primer | CGTCGTAGGCCCCCTTG |
| Probe | CTTCCGACTATAAATCGGCTCACAAGGGAT |
Expression levels of D2R, D3R, D4R, Adcyap1, preproenkephalin1, MBP, Mobp, apolipoprotein D, Peripherin1, and VAchT were compared between isolated LMN and whole spinal cord using the relative standard curve method as previously described (ABI Sequence Detection System User Bulletin). Serial dilutions of spinal cord cDNA were used as the template to construct standard curves, and gene-specific signals obtained from spinal cord and LMN templates were normalized to the endogenous control 18S RNA signals.
Fold change analyses
A fold change for a given gene was obtained by dividing one signal by another (e.g., whole spinal cord signal/LMN signal). For fold changes obtained with microarrays (as opposed to qRT-PCR), the production of targets for LMN captures was as previously described. Spinal cord targets were produced from 10 ng of input RNA. We chose to produce spinal cord targets from 10 ng of input RNA, rather than 5 µg, to minimize error caused by differences in amounts of input RNA (see results). We estimated that 200 captures yield 10–40 ng of input RNA (Fig. 2C), and the intensity and size distribution for a 10-ng target were similar to 100 captures (Fig. 2D); thus we used 10 ng of RNA to produce the spinal cord targets.
Immunohistochemistry
Adult male GAD-EGFP mice were injected intraperitoneally with 50 µl of 1% Fluoro-Gold diluted in 0.9% NaCl 24 h before transcardial perfusion with ice-cold 1:3 (vol/wt) 0.9% NaCl-0.1% Na-nitrite-0.01% heparin followed by equal volume/weight Lana’s fixative (4% paraformaldehyde, 0.16 M phosphate buffer, 0.2% picric acid, pH 6.9). Spinal cords were removed and postfixed in Lana’s fixative for 1 h and then cryoprotected for at least 24 h in 10% sucrose, 0.1 M phosphate buffer, pH 7.4, at 4°C. Spinal segments T11–L1 and L2–L4 were sectioned at 8 µm on a Leica 1720 cryostat. Sections were thaw mounted onto microscope slides and stored at −80°C until use. Sections were rehydrated in 0.1 M PBS, pH 7.4, for 4 h at room temperature. Slides were then incubated in rabbit anti-reticulocalbin 1 (Bethyl Laboratories) diluted 1:500 in PBS containing 0.3% Triton X-100 (PBS-T) and 1% donkey serum (Jackson Immunoresearch) for 48–72 h at 4°C. Slides were then washed 3 × 30 min in PBS-T followed by incubation in biotinylated donkey anti-rabbit IgG (Jackson Immunoresearch) diluted 1:250 in PBS-T containing 1% donkey serum for 1.5 h at room temperature. Slides were then washed 3 × 20 min in PBS-T followed by incubation in Cy3-conjugated extravidin (Sigma Laboratories) diluted 1:1,000 in PBS-T for 1.5 h at room temperature. Slides were then washed 20 min in PBS-T followed by 2 × 20 min in 50 mM Tris·HCl, pH 7.4. Slides were then coverslipped with Fluoromount mounting medium (Vector Laboratories). Individual cells were identified on a Nikon E-800 microscope with epifluorescence and digitally photographed, and densitometry was performed with Neurolucida software (Microbright-field). A one-way ANOVA followed by Tukey’s post hoc test was performed on the data.
RESULTS
Isolating identified spinal MN populations
We physically isolated identified MNs to determine their expression profiles. All MNs can be labeled by intraperitoneal injection of Fluoro-Gold, as their terminal fields access this retrograde tracer via fenestrated capillaries (39). With the use of fluorescent microscopy, three different Fluoro-Gold labeled populations are recognized in spinal cord sections based on location: IML, MMN, and LMN (Fig. 1). These represent three distinct populations that have been well characterized at anatomic, physiological, and developmental levels (see introduction). Cells from each of these three columnar cell types were physically isolated, or captured, using LCM, as described in detail in materials and methods (Fig. 2). Here we define a capture to mean a single, physically isolated 8-µm section of an identified, fluorescently labeled MN.
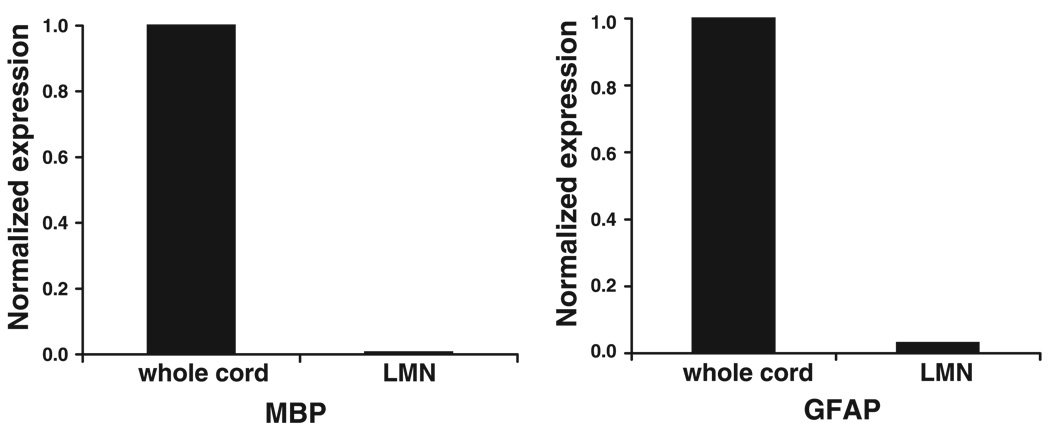
Although individual neurons are clearly visible and well separated, they are surrounded by glia, micro-blood vessels, and neurites. To address the possibility that glial cells were captured along with MNs, we performed qRT-PCRs on cDNA from isolated neuronal populations and whole spinal cord, which contains a variety of neuronal and glial cell types. If LCM successfully excludes glia, then the number of glial-specific transcripts should be greatly reduced in an LCM population relative to a whole spinal cord sample. Classic MBP is expressed only in oligodendrocytes (14); therefore, classic MBP transcript number will be proportional to the number of oligodendrocytes present in a sample. Importantly, transcripts from the MBP gene can be alternately spliced to yield multiple products, including a second protein called golli, which is found in both neurons and glia (29, 43). The exons found in golli transcripts cannot be used to discriminate between neurons and glia. We carefully designed primers and a probe across an exon that is found in classic MBP, but not golli transcripts, to quantify classic MBP expression. Figure 3 illustrates that the classic MBP signal is reduced by over two orders of magnitude in our captured neuronal populations relative to whole spinal cord cDNA. Thus oligodendrocyte contamination is minimal in our captured sample. Similarly, qRT-PCRs performed with primers and a probe designed against the glial astrocyte-specific protein GFAP (10) suggest that astrocytes are reduced in number by 97% in isolated LMN populations relative to whole spinal cord samples. Unfortunately, to our knowledge there are no transcript markers specific for microglia, and so we have no measure of the microglial contamination in our LCM samples.
Fig. 3.
LCM successfully excludes oligodendrocytes and astrocytes from captured neuronal populations. Quantitative real-time RT-PCR (qRT-PCR) was performed on spinal cord and LMN cDNA templates with primers and probes that specifically recognize classic myelin basic protein (MBP; oligodendrocytes) or glial fibrillary acidic protein (GFAP; astrocytes), and relative signal intensities were obtained. Spinal cord and LMN signals were normalized by the spinal cord signals. Normalized qRT-PCR signals reveal a 99.4 and 97% reduction in classic MBP and GFAP expression, respectively, in the LMN relative to the spinal cord template.
Expression profiling identified MN populations
The ultimate goal of this work was to produce an RNA expression profile for each of the three cell types using Affymetrix oligonucleotide arrays. In this method, the RNA isolated from a given population of cells is converted into a biotinylated target. That target is then hybridized to an Affymetrix oligonucleotide array containing >12,000 probe sets, where each probe set corresponds to a uniquely identified gene. After hybridization, the biotin is reacted with a fluorescent tag, and the intensity of the fluorescent signal associated with each probe set is used as a quantitative measure of transcript levels for the gene associated with that probe set.
Typically, 5 µg of RNA are required to produce a target for an Affymetrix chip; however, it is impossible to obtain 5 µg of RNA from an identified population of spinal MNs from a single animal. Thus we first designed a protocol for producing MN targets from small amounts of input RNA, based on two previously published methods (4, 13). When producing a biotinylated target from the standard 5 µg of RNA, a reverse transcription reaction (RT) is followed by a second-strand synthesis reaction (SSS), which is followed by an in vitro transcription reaction (IVT) containing biotinylated UTP. On the other hand, when starting with smaller, nonstandard amounts of RNA, such as 10 ng or 100 captures, the first round of RT-SSS-IVT must be followed by a second round to produce enough target for hybridization to the Affymetrix array. Figure 2D illustrates that the size distribution of the biotinylated target varied with the amount of input RNA.
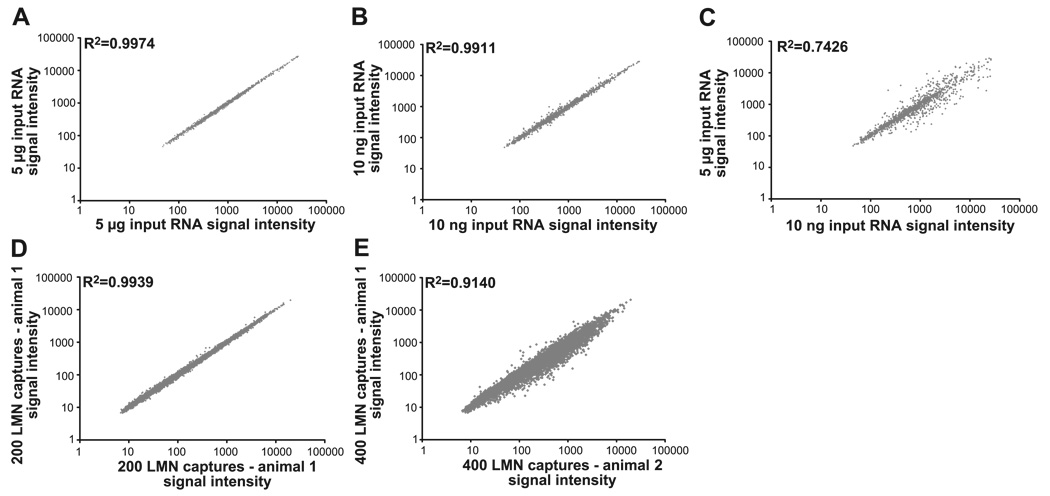
We asked whether the difference in the size of the biotinylated targets reflected a difference in the representation of genes in each target, as has been previously demonstrated by Baugh et al. (4). First, we prepared four biotinylated targets from the same RNA, but we varied the amounts of input RNA such that two of the targets were produced from 5 µg of the RNA and the other two from 10 ng. Next, we hybridized each of the four targets to Affymetrix oligonucleotide arrays, normalized the data with RMA, and compared the resulting expression profiles with GeneSpring software. When targets are produced starting with the same amounts of the same RNA, the expression profiles are reproducible so that R2 > 0.99 when comparing the two chips. This is true no matter whether the targets are prepared from large amounts of RNA (5 µg; Fig. 4A) or small amounts of RNA (10 ng; Fig. 4B). However, when comparing expression profiles for targets made from different amounts of the same input RNA (5 µg vs. 10 ng), the R value was drastically reduced to 0.74 (Fig. 4C). This experiment was repeated twice more, and the findings were reproducible for targets made from the three different animals. There are several possible reasons for this technical artifact, as detailed in the discussion.
Fig. 4.
Comparisons of expression profiles reveal that biological variability is greater than technical variability, as long as the targets are produced from similar amounts of input RNA. All panels represent the signal intensities for the same probe sets (genes) on 2 microarrays. Each of the data points represents 1 probe set. The y-axis represents the signal intensities for the probe sets on microarray 1, and the x-axis represents the signal intensities on microarray 2. Linear regressions were used to calculate R2. A–C: varying the amount of input RNA alters gene representation in the target. Each panel represents a comparison of the expression profiles obtained from 2 microarrays hybridized to different targets. In all cases, the targets were prepared from the same RNA extracted from the spinal cord of a single rat; however, the amount of input RNA used in target production varied, as indicated on the x-and y-axes (see text). The same ~1,200 genes are represented in all 3 panels. A: each of the 2 targets was produced with 5 µg of same total rat RNA. B: each of the 2 targets was produced with 10n g of same total rat RNA. C: each target was produced with either 5 µg or 10 ng of the same total rat RNA. D and E: examining technical and biological variability within an LCM population. The no. of probe sets represented in the 2 panels is the same (12,488). D: 400 LMNs were captured from a single mouse, and RNA was extracted (materials and methods). The RNA was split in half, and each one-half was served as the template in target production. The resulting expression profiles of these technical replicates are compared. E: the expression profiles shown in D were averaged and compared with the averaged technical replicates for the LMN population isolated from a second mouse.
To determine an acceptable range of variability for amounts of input RNA and whether targets made from two different protocols (i.e., 1 vs. 2 rounds of amplification) could be compared, we expanded these studies with varying amounts of the same input RNA. One or two rounds of amplification were used to produce targets beginning with > or < 1.0 µg of input RNA, respectively. Table 2 illustrates that, when using the same protocol, the amount of input RNA can vary by fivefold without producing a considerable change in the representation of genes in the target (e.g., R2 = 0.96 when comparing 0.5 with 0.1 µg, both 2 rounds of amplification). On the other hand, 10-and 50-fold differences in starting amounts of RNA produce substantial alterations in the target (e.g., R2 = 0.9 and 0.83 when comparing 0.1 vs. 0.01 and 0.5 vs. 0.01 µg, respectively, all double rounds). The data also suggest that results obtained with two different target production protocols cannot be compared. A roughly threefold change in the amount of input RNA did not result in a significant difference when using the same protocol (e.g., R2 = 0.97 when comparing 5.0 vs. 1.5 µg, respectively, both single rounds); however, when using two different protocols, a threefold difference in input RNA results in significant differences (e.g., R2 = 0.82 when comparing 1.5 vs. 0.5 µg, single round vs. double round).
Table 2.
R2 values for linear regressions of expression profiles
| Nanograms of input RNA |
5,000 (sr) | 2,500 (sr) | 1,500 (sr) | 500 (dr) | 100 (dr) | 10 (dr) |
|---|---|---|---|---|---|---|
| 5,000 (sr) | 1.0 | 0.97 | 0.97 | 0.82 | 0.82 | 0.74 |
| 2,500 (sr) | 0.99 | 0.82 | ||||
| 1,500 (sr) | 0.82 | |||||
| 500 (dr) | 0.96 | 0.83 | ||||
| 100 (dr) | 0.9 | |||||
| 10 (dr) | 0.99 |
R2 values for linear regressions of expression profiles generated with the indicated amounts of the same mouse total RNA. sr, Single-round amplification; dr, double-round amplification.
The previous experiments suggested that, to accurately compare different cell types, the amounts of input RNA should not vary by more than a factor of five (approximately) for each cell type, and the same protocol should be used to produce the target. Typically, somatic MNs are larger than visceral MNs (26), and, as discussed in materials and methods, the diameter of the somatic and visceral MNs used in these studies differed by a factor of two. If we assume that the nucleus occupies a similar fraction of the cell volume in all cells, and that cytosolic RNA concentrations are comparable, then because section thickness is constant, the amount of input RNA/capture should vary with the area of the cell according to the equation πr2. In this case, we can compare results across the three columnar cell types if we start with the same number of captures, because the differences in the amount of input RNA between somatic and visceral MNs should be approximately fourfold.
To obtain expression profiles that could be compared between cell types, we isolated LMN, MMN, and IML from adult male mice at P45. Five animals were used in these experiments, and all three populations were isolated from each animal. For a given cell type, we always isolated RNA from 400 captures. We then split this RNA in half, and each half was treated in an identical fashion to produce technical replicates. This was true for all three cell types from all five animals; therefore, our data are derived from 30 Affymetrix arrays (10 LMN, 10 MMN, 10 IML), which have been submitted to the NCBI GEO (http://www.ncbi.nlm.nih.gov/projects/geo) under accession number GSE2595. Figure 4D illustrates that technical replicates were highly reproducible such that, when compared, the R2 >0.99. As has been previously demonstrated in Churchill et al. (8), we found that the observed technical variability was much lower than the biological variability. When the two technical replicates representing the LMN expression profile from one animal are averaged and compared with the averaged LMN technical replicates from a second animal, the R2 value drops to 0.91 (Fig. 4E). Expression profiles (inverse log2 of the RMA-normalized, averaged technical replicates) were prepared for each cell type in each of the five animals. Mean expression profiles for a given cell type were obtained by averaging the data for the five individuals.
Comparing expression profiles between cell types
Next, we sought to compare the mean expression profile for each of the three different MN subtypes to establish how these columnar cell types differed from one another. No genes were excluded from our analyses on the basis of signal intensity or the present/absent call; however, data were prefiltered according to technical reproducibility [see materials and methods; 7.5% (936/12,488) of probe sets were removed]. After determining that the data for all cell types displayed equal variance (Bartlett’s test of homogeneity of variance; Prism), we performed a one-way ANOVA using Tukey’s post hoc test (Gene-Spring).
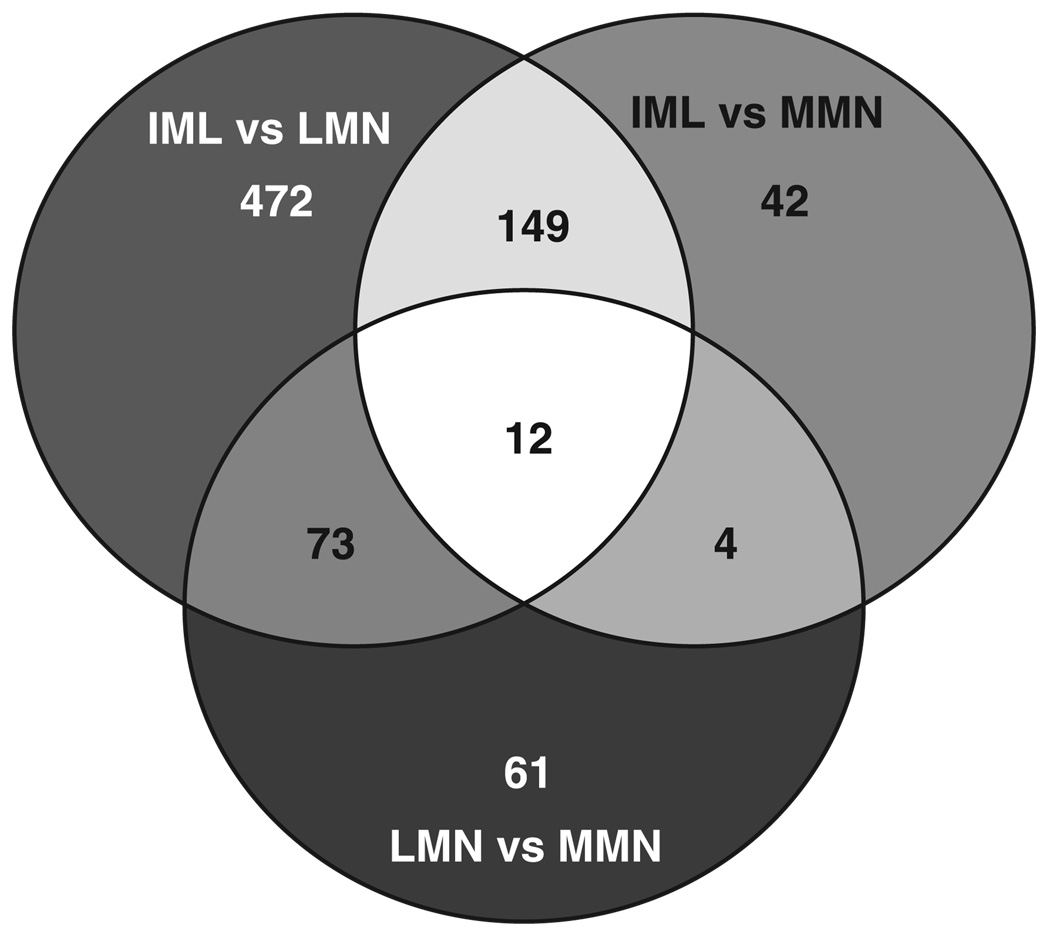
Genes that showed significant differences in expression levels across cell types are shown in Venn diagram format in Fig. 5. Interestingly, 7.0% (813/11,552) of the genes were expressed at significantly different levels across the three cell types, and only 0.6% (74/11,552) showed a greater than twofold change. The 813 differentially expressed genes fall into 7 classes, each represented by a sliver of color. Class I consists of 12 genes that are uniquely expressed in each cell type, class II contains 149 genes that are differentially expressed in IML (expression levels are significantly different between IML vs. MMN and between IML vs. LMN, but not between LMN vs. MMN), and class III contains 73 genes that are uniquely expressed in LMN. Class IV contains four genes that are uniquely expressed in MMN. Class V contains 472 genes that are differentially expressed between IML and LMN (expression levels are significantly different between IML and LMN but not between IML and MMN or LMN and MMN). Class VI contains 42 genes that are differentially expressed between IML and MMN. Class VII contains 61 genes that are differentially expressed between LMN and MMN. The differentially expressed genes, along with their subcellular location, function, detection call, and associated signal intensities for each cell type in every animal, are presented according to class in Excel spreadsheet format in Supplemental Table S1.
Fig. 5.
Seven classes of differentially expressed genes among three cell types. The 3 circles represent 3 groups of genes that are significantly different between IML/LMN (706 probe sets in total), IML/MMN (207 probe sets in total), and LMN/MMN (150 probe sets in total), as determined with a 1-way ANOVA. The Venn diagram suggests that the probe sets can be divided into 7 classes: genes that are uniquely expressed in all 3 cell types (12), genes that are uniquely differentially expressed in IML (149), genes that are uniquely differentially expressed in LMN (73), genes that are uniquely differentially expressed in MMN (4), genes that are differentially expressed between LMN and IML (472), genes that are differentially expressed between MMN and IML (42), and genes that are differentially expressed between LMN and MMN (61). The identities of these genes are listed in Supplemental Table S1.
Approximately 76% (618/813) of the differentially expressed genes have known functions. The differentially expressed genes were categorized according to biological processes using the Gene Ontology Functions provided by Gene-Spring and Affymetrix software as well as database searches with NCBI software, such as Entrez Gene. A summary of our classification of genes is shown in Table 3. The differentially expressed genes can be divided into 12 classes, where 1 gene can fall into multiple classes. There appeared to be no major differences in the classes of differentially expressed genes across cell types.
Table 3.
Categories of differentially expressed genes represented in Fig. 5
| Category | Percentage of Differentially Expressed Genes with Known Functions |
|---|---|
| Signal transduction | 22% (135/618) |
| Secreted | 20% (121/618) |
| Transcription factors | 10% (66/618) |
| Mitochondrial | 7% (42/618) |
| Cell growth and maintenance | 7% (41/618) |
| Cell adhesion | 6% (34/618) |
| Cytoskeletal | 6% (36/618) |
| Ribosomal | 3% (16/618) |
| Ion channels | 2% (12/618) |
| Putative non-MN transcripts | 2% (12/618) |
| Endomembrane | 3% (17/618) |
| Neuroptide and neurotransmitters | 1% (6/618) |
MN, motoneuron.
Proteins involved in signal transduction made up the largest class of differentially expressed genes (22% or 135/618). Interestingly, in some cases, several components of a multiprotein signaling complex show similar expression patterns across cell types. For example, NCam1 is involved in cell adhesion, synaptic plasticity, and release (41, 42, 46). Many components of the NCam1 signaling pathway exist together in lipid rafts and appear to be upregulated to similar extents in visceral MNs, including NCam1 (2.5-fold higher in IML; Supplemental Table S1, class I), GAP43 (2.64-fold higher in IML; Supplemental Table S1, class I), MARCKS (2.59-fold higher in IML; Supplemental Table S1, class I), Basp1 (also known as CAP23, 1.6-fold higher in IML; Supplemental Table S1, class I), and gelsolin (2.85-fold higher in IML; Supplemental Table S1, class I). Thus expression of multiple components of this lipid raft signaling complex appear to be co-regulated at the level of the transcript.
Secreted proteins comprise the next largest class of differentially expressed genes. These include several neuropeptides and neurohormones that show greater than twofold differences between cell types: cerebellin 1, preproenkephalin 1, tachykinin 1, and pituitary adenylate cyclase-activating peptide. Components of the release machinery, such as synaptotagmins, vary as well.
Members of several classes of cell adhesion molecules show cell type-specific expression, including members of the immunoglobulin superfamily (NCam1 and Igsf8), cadherins (Cdh5 and PCdha12), integrins (Itga3), and ephrins (Epha7). Many proteins involved in cell growth and maintenance vary, as do a significant number of mitochondrial and ribosomal proteins. Approximately 2% of the differentially expressed probe sets represent transcripts that are currently thought to be specific for non-MN cell types. For example, transcripts for hemoglobin, a putatively hematopoietic-specific gene, are readily detected with microarray expression profiling (Supplemental Table S1, class V).
A surprisingly small number of ion channels and receptors are significantly different across columnar cell types. We previously showed that MN transcript levels linearly correlate with conductance for a shal (Kv4) K+ channel (3). Furthermore, while transcript and conductance levels varied within an identified MN cell type, we could detect significant differences between MN cell types using qRT-PCR. This current unexpected finding that expression of few ion channels is significantly different across MN cell types may reflect the fact that transcripts for many of these genes are low abundance, so small signal-to-noise ratios might obscure important changes in their expression when using microarrays (see below). Alternatively, it has been shown that ionic conductances can vary several fold for a given MN cell type, depending on the state of the cell (16, 44, 45). Such variability in ion channel gene expression within a cell type could diminish significant differences between related cell types.
Adult expression patterns for transcription factors that generate embryonic combinatorial codes
Dynamic, combinatorial transcription factor codes operate throughout development to establish unique columnar cell types (51). However, the transcription factors operating during prenatal development have not been examined in adult MNs, and it is not known whether and how they participate in combinatorial codes that maintain differences between adult columnar cell types. We mined the data set submitted to the NCBI Omnibus under accession number GSE2595 to examine differences in transcription factors that are known to be involved in establishing MN identity. Supplemental Table S2 lists the expression of genes on the U74Av2 microarray that belong to families of transcription factors that play important roles in the development and differentiation of MNs. Significant differences across cell types is as indicated. This is by no means a complete list of transcription factors on the array.
During embryogenesis, at least 10 known homeodomain and basic helix-loop-helix transcription factors are differentially expressed along the dorsal-ventral axis of the ventral neural tube, producing a combinatorial code that divides the developing ventral spinal cord into 5 progenitor domains: p0–p3 and pMN (51). Each progenitor domain gives rise to a different class of postmitotic neurons. The pMN progenitor domain gives rise to all MNs. Five of the ten genes generating the code are not on the chip: Pax 7, Dbx1, Dbx2, Nkx6.1, and Olig2. Of the remaining five, two transcription factors expressed within the pMN domain, NKx6.2 and Pax6, appear to be expressed in all three adult columnar cell types at similar levels (Supplemental Table S2). Nkx2.9, which is exclusively expressed in the p3 progenitor domain, exhibits nearly identical average signal intensities in all three adult MN cell types (Supplemental Table S2). NKx2.2 and Irx3 are expressed in the progenitor domains that abut the pMN domain and are essential for establishing the ventral and dorsal boundaries of pMN, respectively. Both of these genes are differentially expressed in adult MNs. Irx3 expression is approximately twofold higher in somatic vs. visceral MNs (class 2, Supplemental Table S1), and NKx2.2 is differentially expressed between the LMN and MMN somatic motor columns (class VII, Supplemental Table S1). Thus each adult columnar cell type shows a unique expression pattern, or code, with regard to these five transcription factors, and it is different from that observed during embryonic development.
Once pMN cells exit the cell cycle, postmitotic neurons migrate and extend axons to innervate targets. The family of LIM homeodomain (LIM-HD) transcription factors plays an important role in regulating these series of events (51). The mouse LIM-HD subfamily contains 12 genes that can be subdivided into 6 subgroups based on conserved sequences (20): APTEROUS (Lhx2 and Lhx9), LHX6/7 (Lhx6 and Lhx7, also known as Lhx8), ISLET (Isl1 and Isl2), LMX (Lmx1a and Lmx1b), LIM-3 (Lhx4 and Lhx3), and LIN-11 (Lim1 and Lhx5). Supplemental Table S2 shows that the eight LIM-HD genes represented on the chip are not differentially expressed across cell types. However, many other transcription factors contain LIM but not HD domains. Three of these transcription factors may be differentially expressed across cell types (Lasp1, Pdlim 1, and Limk1).
Several other classes of transcription factors play a role in the differentiation of MNs and the formation of motor pools, including homeobox (Hox), ETS, and forkhead box (Fox) families (18, 27, 28, 49, 56, 60). The Hox and ETS genes on the chip do not appear to be differentially expressed across columnar cell types (Supplemental Table S2). Supplemental Table S1 suggests that Hoxa4 gene expression is significantly different between LMN and IML, but the signal intensity for this probe set is extremely low (<15), and additional probe sets for this gene do not suggest differential expression (Supplemental Table S2). On the other hand, four fox genes appear to be differentially expressed between columnar cell types. In summary, the data suggest that transcription factors that participate in establishing MN identity early in development are also expressed in the adult, but their expression patterns have changed dramatically.
Verification of chip data
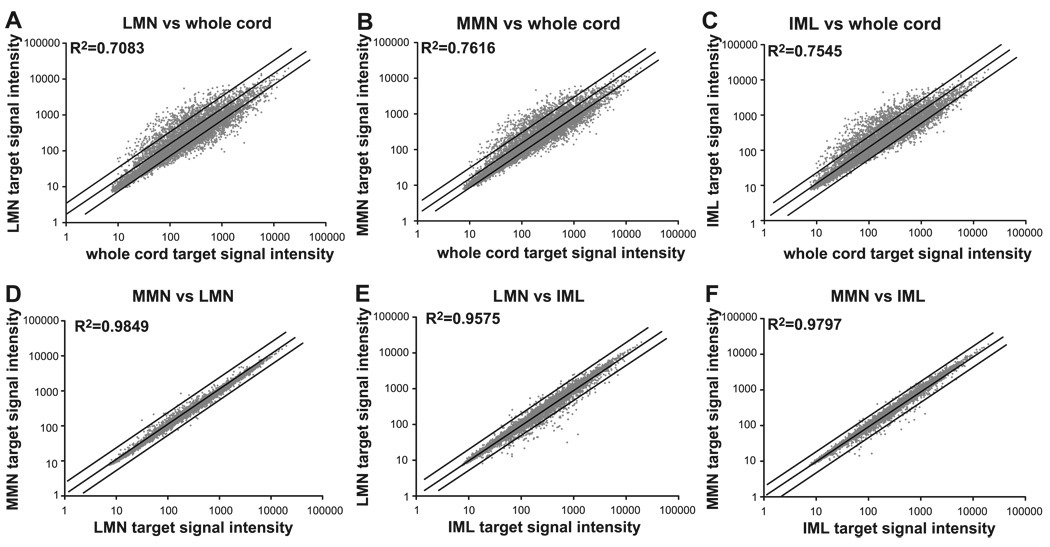
Expression profiles must be verified with a variety of methods. As part of the verification process, we first compared expression profiles between cell types and spinal cord. A global view of the changes between spinal cord and the three cell types is shown in Fig. 6, A–C. As expected, the data suggest that MN expression profiles are much more similar to each other than any one is to whole spinal cord. The data show that the profiles for individual cell types and spinal cord are highly divergent. The R2 values for targets made from similar amounts of input RNA are reduced to as little as 0.71 when comparing total spinal cord with MN expression. On the other hand, there is little divergence between MN populations. The results shown in Fig. 6, D–F, are consistent with the results of the ANOVA (Fig. 5). The greatest divergence is between IML and LMN, while the MMN vs. LMN comparison is the least divergent.
Fig. 6.
MN expression profiles are more similar to each other than to whole cord expression profiles. Averaged expression profiles for each cell type (n = 5 animals, 10 chips) and whole spinal cord (n = 3 animals, 3 chips) are compared. Each of the 12,488 diamonds represents the average signal intensity for 1 probe set in the 2 different samples. Lines on either side of the midline represent a 2-fold change between samples. Same number of probe sets are examined in all panels (12,488). Linear regressions were used to calculate R2.
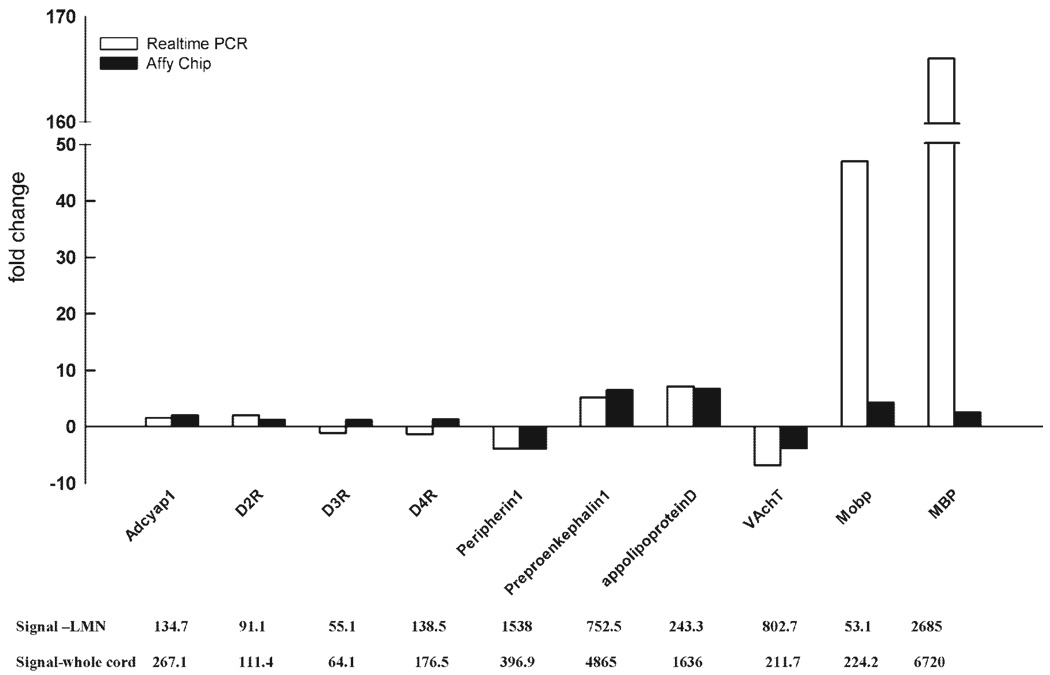
Next, data were verified by comparing the results obtained from gene chips with the results obtained from qRT-PCR. Using these two independent methods, we measured the expression level of a given gene in whole spinal cord cDNA and in LMN cDNA (Fig. 7). A comparison of the fold changes acquired with the two different protocols is an indication of the robustness of the expression profile data. For example, using either procedure, we would expect that the signal for the oligodendrocyte-specific transcript, classic MBP, would be much higher in whole spinal cord vs. LMN cDNA, and thus we should obtain similar fold changes with both protocols.
Fig. 7.
Two independent measures of differential gene expression. A: for each gene, expression levels were measured with qRT-PCR (open bars) and microarrays (solid bars). Expression levels were measured in 2 different samples: whole spinal cord cDNA (10 ng of starting RNA used in the double-amplification protocol, as described in materials and methods) or LMN cDNA (400 LMN captures used in a double-amplification protocol, as described in materials and methods). For each gene, the larger signal is normalized by the smaller signal, and the resulting fold change is plotted. Positive fold changes indicate that the spinal cord signal > LMN signal. Negative fold changes indicate that the LMN signal > spinal cord signal. Note, the hatches on the y-axis and in the bars representing MBP indicate that large portions of the graph have been removed (from 50 to 150) to fit all the data into the figure. B: normalized microarray signal intensities for each gene indicated directly above in A.
For each technique, we normalized the larger signal (e.g., spinal cord) by the smaller signal (e.g., LMN) to obtain a fold change for a given gene across the two samples. If the spinal cord signal was larger, the fold change was deflected in the positive direction along the y-axis. If the LMN signal was larger, the fold change was deflected in the negative direction along the y-axis. Figure 7 demonstrates that 5 of the 10 genes examined in this manner yielded similar fold changes with both methods such that the fold changes obtained for a given gene were within a factor of 2 of one another (apolipoprotein D, Peripherin1, VAchT, preproenkephalin1, and Adcyap1). On the other hand, the fold changes associated with the remaining five genes (MBP, D2R, D3R, D4R, and Mobp) varied significantly with the protocol.
The fold changes for MBP showed the largest discrepancy between the two methods: a 166-fold change was detected between spinal cord and LMN samples using qRT-PCR vs. a 2.5-fold change using Affymetrix gene chips. This difference is easily explained by the probe set included on the Affymetrix chip. As discussed earlier, the gene for MBP is alternately spliced to produce many proteins, some expressed only in oligodendrocytes (classic MBP) and others expressed in both MNs and oligodendrocytes (golli) (14, 29, 43). Unlike the previously described primer-probe set used in qRT-PCR, the Affymetrix MBP probe sets contain oligonucleotides that are common to both golli and classic MBP transcripts. Therefore, the MBP signal obtained with Affymetrix gene chips cannot be used as an indicator of oligodendrocyte contamination in captured neuronal populations. These data highlight the need for an appreciation of the probe sequence on the microarray when interpreting discrepancies.
The reasons for the inconsistencies associated with D2R, D3R, D4R, and Mobp are less obvious. This type of result could be obtained if the measure of gene expression on the chip was not linear over the target range we examined (7). For example, a given probe set on a chip could saturate. In our experiments, the signals on a chip range from 7 to 22,000. Figure 7B indicates that the signal intensities for all five genes were well below the maximal intensity; thus it is unlikely that probe set saturation can explain this discrepancy. Alternatively, if the concentration of a transcript is below the linear range of detection, nonspecific background noise could significantly contribute to the total measure, so that at very low expression levels (i.e., low-abundance transcripts), a small signal-to-noise ratio could prevent accurate measurement of signal intensity. Figure 7B illustrates that the D2R, D3R, D4R, and Mobp signals in isolated LMN are 91, 55, 138, and 53, respectively, whereas in whole spinal cord, the signal intensities are 111, 64, 177, and 224, respectively. Thus the low expression levels in one or both samples could be below the linear range of detection for these probe sets, such that they represent mostly background noise.
The fold changes observed in Fig. 7 are consistent with those that would be predicted from previous literature. Some of the genes that are enriched in spinal cord have been demonstrated previously to be glial specific (MBP and Mobp), or more highly expressed in dorsal horn, as opposed to the ventral horn where MNs are located (preproenkephalin) (1, 54). Similarly, genes that appear to be enriched in MNs were as anticipated. 1) MNs are cholinergic, and VAchT transports acetylcholine into synaptic vesicles. 2) Peripherin is an intermediate filament protein that has been used as a motoneuron marker (12, 47).
As a further test of the robustness of the expression profiles, we examined which genes were the most highly enriched or depleted in whole spinal cord relative to captured LMNs, to determine whether the changes were consistent with what is known in the literature. The 10 genes with the highest fold changes are shown in Table 4. Five of the ten genes are enriched in LMNs. The β-isoform of protein phosphatase 1 catalytic subunit (Ppp1cb) has previously been shown to be highly expressed in rat cervical LMNs using immunocytochemistry (55). Similarly, phosphoprotein F1–20 is known to be a neuronal-specific, synapse-associated protein (53, 64). The type I sodium channel -polypeptide (SCN1A) is a neuronal- specific sodium channel subunit (11). Protocadherin-α4 is a neuronal-specific protein localized at synaptic junctions (25). Syndecan-binding protein (syntenin) has previously been shown to be expressed in fetal kidney, liver, lung, and brain (65) and is a PDZ protein that can associate with glutamate receptors (19). However, to our knowledge, there are no reports on its expression and distribution in spinal cord.
Table 4.
Genes with highest fold changes between isolated LMN and whole spinal cord
| Affy Probe Set | Fold Change | GenBank | Gene |
|---|---|---|---|
| 100088_at | −19.8* | M27073 | protein phosphatase 1, catalytic subunit, beta isoform |
| 100455_at | −17.9* | M83985 | phosphoprotein (F1-20); synaptosomal-associated protein 91 |
| 94201_at | −16.2* | L42339 | sodium channel, voltage-gated, type I, alpha polypeptide; sodium channel 3 |
| 160610_at | −16* | D86916 | protocadherin alpha 4; CNR1 |
| 93017_at | −13.9* | AF077527 | syndecan binding protein; syntenin |
| 92353_at | 15.9† | Y18723 | protease, serine, 18 (Prss 18); secreted serine protease (BSSP). |
| 95471_at | 11.8† | U22399 | Cdk-inhibitor p57KIP2 (KIP2); cyclin-dependent kinase inhibitor 1C (P57) |
| 103548_at | 8.8† | D14423 | preprotachykinin B (PPT-B) |
| 102405_at | 7.2† | M31811 | myelin-associated glycoprotein |
| 93750_at | 6.8† | J04953 | gelsolin |
Negative fold changes indicate enrichment in the lateral MN (LMN) vs. whole spinal cord sample (LMN/spinal cord).
Positive fold changes indicate enrichment in the whole spinal cord vs. the LMN sample (spinal cord/LMN). As stated in MATERIALS AND METHODS, whole spinal cord data were obtained from targets made from 10 ng of input RNA.
The remaining five genes in Table 4 exhibit decreased expression levels in LMNs relative to whole spinal cord. Preprotachykinin B (PPT-B) is mainly expressed in the dorsal horn of the spinal cord (40, 61), which is consistent with the fact that LMNs reside in the ventral horn. The proteins for three of the remaining four genes have previously been shown to be enriched in oligodendrocytes, including myelin-associated glycoprotein (MAG) (48), gelsolin (32, 34, 57), and protease serine 18 (Prss18) (37, 63). The fifth downregulated gene, Cdk-inhibitor p57KIP2, is highly expressed in skeletal muscle, brain, heart, lungs, and eye (38), but to our knowledge, its distribution in spinal cord is unknown. Thus the genes showing the greatest enrichment or depletion in LMNs are consistent with the literature.
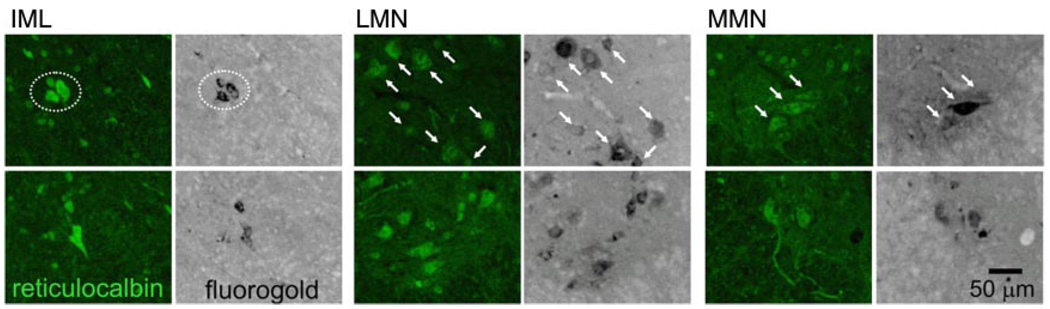
Finally we examined differences between MN populations at the protein level using ICC, as described in materials and methods. According to Supplemental Table S1, reticulocalbin is 2.4-fold higher in IML vs. LMN, 1.7-fold higher in IML vs. MMN, and 1.4-fold higher in MMN vs. LMN. Figure 8 shows representative sections of Fluoro-Gold-labeled MNs stained with an antibody against reticulocalbin. Consistent with the expression profiling data, all MNs appear to express reticulocalbin. Densitometric measurements from this experiment show that reticulocalbin is 1.8-fold greater in IML vs. LMN (P < 0.001), 1.5-fold greater in IML vs. MMN (P < 0.001), and 1.2-fold higher in MMN vs. LMN (P < .05). Thus the levels of protein in the different cell types are consistent with differences in transcript levels reported in the expression profiles.
Fig. 8.
Reticulocalbin protein levels vary across cell types as predicted by expression profiles. Cryostat sections of Fluoro-Gold-injected mice were stained with anti-reticulocalbin as described. Cell types are indicated at top. For each cell type, the top and bottom panels represent 2 independent sections; the left panels show reticulocalbin staining and the right panels Fluoro-Gold staining for the same section. For each cell type, MNs at top are demarcated by dotted circles (IML) or arrows (MMN and LMN). MNs are not indicated in the bottom sections.
DISCUSSION
The NIH has recently mandated the use of microarrays to produce a catalog of cell type transcriptomes. In this paper, we explore the feasibility of this notion and identify significant differences between three developmentally related neuronal cell types that ultimately innervate different targets: LMN, MMN, and IML. We found that 7% of the 11,552 genes examined were differentially expressed between columnar cell types, and most differentially expressed genes encoded proteins involved in signal transduction. Although 7% is small in an absolute sense, the number of differentially expressed genes may actually seem large when one considers that these neurons are developmentally related and functionally very similar. The fact that most differentially expressed genes are involved in signal transduction suggests that these structurally and functionally similar neurons differ mainly in their ability to receive, interpret, and respond to extracellular signals.
Given the caveat that all microarray analyses suffer from the fact that small signal-to-noise ratios may obscure significant changes in gene expression for low-abundance transcripts, this study indicates that our current expression profiling protocol for identified populations of neurons is sensitive and accurate. Genes known to be expressed in MNs are upregulated in the LMN population relative to whole spinal cord, and genes expressed in glia or dorsal horn neurons are correspondingly downregulated. This protocol should work well for other cell types as well; however, our data suggest that populations should only be compared if the amounts of input RNA are within a factor of five (approximately), and targets are produced with the same protocol. There may be several reasons why the representation of genes varies with the amount of input RNA. For example, in any polymerase-directed reaction, product length (and hence representation of a gene) varies with the concentrations of enzyme, primer, and template. Similarly, template interactions vary with the concentration of template. In addition, after the second-round RT, longer RNA templates are represented by several shorter cDNA fragments due to binding of multiple random hexamers along the length of a single template. This, followed by the use of oligo(dT) primer in the second-round SSS, selects for the amplification of only a subset of all fragments produced in the second-round RT (i.e., those with a poly A tract). This handicap would be more severe if it were not for the fact that the Affymetrix probe sets on the chips used here are obtained from the last 600 bases of a transcript (i.e., the portion of the transcript adjacent to the poly A tail).
Because there is a possibility of neighboring microglia and micro-blood vessel contamination, and there is heterogeneity in the characteristics and distributions of microglia (31) and micro-blood vessels (2), some differences in our neuronal populations are reflective of nonneuronal cells. Minor contamination of isolated neuronal populations by micro-blood vessel, microglia, and astrocytes limits the interpretation of the data. The problem is further confounded by the fact that proteins once thought to be unique to blood cells and/or microglia, like complement factors, have more recently been shown to be expressed by neurons (59). For certain proteins, in situ hybridization and/or immunocytochemistry may be required to determine the source of the differential expression.
Despite all the limitations associated with microarray analyses of identified cell types, the data provide an abundance of new and important information, including a catalog of expression levels for ~12,000 genes in 3 cell types (accession no. GSE2595; NCBI GEO). Not only does this catalog provide the first insight into the degree of molecular divergence between adult MN populations, but it is an important resource for scientists interested in the molecular underpinnings of motoneuron physiology. Such detailed molecular descriptions of functionally discrete populations of MNs can illuminate factors that influence MN function and may eventually improve our ability to target proteins for both future investigation and clinical therapies of MN disease.
Our study suggests that 7% of the genes are differentially expressed across the three cell types, and only 0.6% show a greater than twofold change. The signal intensity correlations among the three cell types was always >95%. On the other hand, in an earlier study, Erlander and colleagues (35) isolated large and small neurons from dorsal root ganglia (DRG) and compared their expression profiles using custom-made microarrays. According to their protocol, input RNA levels appeared to be within a factor of five for the two cell types. They found that out of 477 genes examined on the chip, ~8% showed a >1.5-fold difference and 4% showed a >2-fold difference between the two cell types. Furthermore, a comparison of large and small neurons yielded a much lower signal intensity correlation (R2 = 0.6789). Several factors could contribute to the differences in our results compared with this earlier study. For example, each study employed a different normalization procedure and microarray. Results are known to vary according to normalization procedures (Ref. 5; see also materials and methods) and microarrays (36). In addition to these technical considerations, large and small DRG neurons may simply be less closely related developmentally and functionally than the three classes of MNs. Indeed, it is not surprising that correlations vary with the cell types under study. In a later paper, Erlander’s group hybridized targets from single CA1 hippocampal neurons to arrays containing ~4,000 genes. A comparison of 11 arrays representing 11 individual CA1 neurons yielded an average R2 = 0.85, while one neuron yielded an average R2 = 0.7 (22).
Because somatic MNs (LMN and MMN) innervate muscle and visceral MNs (IML) control the viscera, we expected the greatest divergence to be between somatic and visceral MNs. We found that the largest divergence was between LMN and IML (6% of genes differentially expressed). However, MMNs were equally similar to LMN and IML (1.3 and 1.8% divergence, respectively). This could indicate that the MMN columnar cell type is much more heterogeneous than the other columnar cell types. In any event, these data highlight the fact that target innervation alone does not determine the genetic program of a cell.
While significant differences in transcript levels are relatively few, they appear to reside in genes that underpin all fundamental cell systems and processes. These include genes involved in signaling, transcription, somatodendritic mRNA transport, protein trafficking and the endomembrane system, nuclear transport, the cytoskeleton, the extracellular matrix, receptors and other proteins involved in cell-cell and cell-matrix interactions, mitochondria, ribosomes, proteosomes, and proteins involved in the transport and metabolism of lipids, proteins, carbohydrates, nucleotides, and small cations and anions. These global changes may reflect a coordinate regulation of all systems by the cell, and/or that all systems are interrelated. The genes encoding the signal transduction machinery seem to be the most finely tuned, with many small but significant changes occurring in all cell types.
By mining the data set for specific transcription factors, we discovered that the transcription factors that establish MN identity during embryonic development are also expressed in adult MNs. Not surprisingly, there were many differences between adult and embryonic expression patterns. The data suggest that new combinatorial codes operate in adult neurons to maintain differences between motor columns, although additional experiments are required to conclusively identify transcription factors that participate in the code.
In summary, microarray expression profiling of identified MN subpopulations provides broad insight into their molecular properties. Comparing motoneurons from the three different motor columns reveals that 813/11,552 (7%) genes studied are differentially expressed. Visceral motoneurons (IML) have the most uniquely differentially expressed genes, and the greatest differences in expression are found between IML and lateral motor columns. The data indicate that expression differences occur in genes associated with all fundamental cellular processes. However, most of the differentially expressed genes were involved in cell signaling, suggesting that, while the cells are functionally and structurally similar, they differentially receive and interpret extracellular signals. Finally, by mining the data for developmentally important transcription factors, we were able to show that, while many of the same transcription factors that operate throughout development to establish MN identity are also expressed in adult MNs, they most likely form different combinatorial codes to maintain differences between columnar cell types.
Supplementary Material
ACKNOWLEDGMENTS
We thank Ole Kiehn and Jesper Ryge for efforts, advice, and support during the initial stages of this project. In addition, we thank Merry Clark, Muhgda Vasireddi, Shruti Subramaniam, Stefan Clemens, Hong Zhu, Chuck Derby, Tim Cope, Richard Nichols, Stan Nakanishi, Susmita Datta, and Kim Woller for useful discussions on various aspects of the work.
GRANTS
This work was supported by National Institutes of Health Grants NS-40440 (S. Hochman) and NS-38770 (D. J. Baro) and National Science Foundation-Science and Technology Center Grant IBN-9876754.
Footnotes
The Supplemental Material for this article (Supplemental Tables S1 and S2) is available online at http://physiolgenomics.physiology.org/cgi/content/full/00109.2005/DC1.
REFERENCES
- 1.Abraham KE, McGinty JF, Brewer KL. Spinal and supraspinal changes in opioid mRNA expression are related to the onset of pain behaviors following excitotoxic spinal cord injury. Pain. 2001;90:181–190. doi: 10.1016/s0304-3959(00)00402-4. [DOI] [PubMed] [Google Scholar]
- 2.Bajaj MS, Kuppuswamy MN, Manepalli AN, Bajaj SP. Transcriptional expression of tissue factor pathway inhibitor, thrombomodulin and von Willebrand factor in normal human tissues. Thromb Haemost. 1999;82:1047–1052. [PubMed] [Google Scholar]
- 3.Baro DJ, Levini RM, Kim MT, Willms AR, Lanning CC, Rodriguez HE, Harris-Warrick RM. Quantitative single-cell-reverse transcription-PCR demonstrates that A-current magnitude varies as a linear function of shal gene expression in identified stomatogastric neurons. J Neurosci. 1997;17:6597–6610. doi: 10.1523/JNEUROSCI.17-17-06597.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baugh LR, Hill AA, Brown EL, Hunter CP. Quantitative analysis of mRNA amplification by in vitro transcription. Nucleic Acids Res. 2001;29:E29. doi: 10.1093/nar/29.5.e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 6.Bonner RF, Emmert-Buck M, Cole K, Pohida T, Chuaqui R, Goldstein S, Liotta LA. Laser capture microdissection: molecular analysis of tissue. Science. 1997;278:1481–1483. doi: 10.1126/science.278.5342.1481. [DOI] [PubMed] [Google Scholar]
- 7.Chudin E, Walker R, Kosaka A, Wu SX, Rabert D, Chang TK, Kreder DE. Assessment of the relationship between signal intensities and transcript concentration for Affymetrix GeneChip arrays. Genome Biol. 2002;3 doi: 10.1186/gb-2001-3-1-research0005. RESEARCH0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Churchill GA. Fundamentals of experimental design for cDNA microarrays. Nat Genet. 2002;32 Suppl:490–495. doi: 10.1038/ng1031. [DOI] [PubMed] [Google Scholar]
- 9.Emmert-Buck MR, Bonner RF, Smith PD, Chuaqui RF, Zhuang Z, Goldstein SR, Weiss RA, Liotta LA. Laser capture microdissection. Science. 1996;274:998–1001. doi: 10.1126/science.274.5289.998. [DOI] [PubMed] [Google Scholar]
- 10.Eng LF, Ghirnikar RS, Lee YL. Glial fibrillary acidic protein: GFAP-thirty-one years (1969–2000) Neurochem Res. 2000;25:1439–1451. doi: 10.1023/a:1007677003387. [DOI] [PubMed] [Google Scholar]
- 11.Escayg A, MacDonald BT, Meisler MH, Baulac S, Huberfeld G, An-Gourfinkel I, Brice A, LeGuern E, Moulard B, Chaigne D, Buresi C, Malafosse A. Mutations of SCN1A, encoding a neuronal sodium channel, in two families with GEFS+2. Nat Genet. 2000;24:343–345. doi: 10.1038/74159. [DOI] [PubMed] [Google Scholar]
- 12.Escurat M, Djabali K, Gumpel M, Gros F, Portier MM. Differential expression of two neuronal intermediate-filament proteins, peripherin and the low-molecular-mass neurofilament protein (NF-L), during the development of the rat. J Neurosci. 1990;10:764–784. doi: 10.1523/JNEUROSCI.10-03-00764.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ginsberg SD, Che S. RNA amplification in brain tissues. Neurochem Res. 2002;27:981–992. doi: 10.1023/a:1020944502581. [DOI] [PubMed] [Google Scholar]
- 14.Givogri MI, Bongarzone ER, Campagnoni AT. New insights on the biology of myelin basic protein gene: the neural-immune connection. J Neurosci Res. 2000;59:153–159. [PubMed] [Google Scholar]
- 15.Goldsmith ZG, Dhanasekaran N. The microrevolution: applications and impacts of microarray technology on molecular biology and medicine. Int J Mol Med. 2004;13:483–495. [PubMed] [Google Scholar]
- 16.Golowasch J, Abbott LF, Marder E. Activity-dependent regulation of potassium currents in an identified neuron of the stomatogastric ganglion of the crab Cancer borealis. J Neurosci. 1999;19:RC33. doi: 10.1523/JNEUROSCI.19-20-j0004.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gustincich S, Contini M, Gariboldi M, Puopolo M, Kadota K, Bono H, LeMieux J, Walsh P, Carninci P, Hayashizaki Y, Okazaki Y, Raviola E. Gene discovery in genetically labeled single dopaminergic neurons of the retina. Proc Natl Acad Sci USA. 2004;101:5069–5074. doi: 10.1073/pnas.0400913101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guthrie S. Neuronal development: putting motor neurons in their place. Curr Biol. 2004;14:R166–R168. [PubMed] [Google Scholar]
- 19.Hirbec H, Perestenko O, Nishimune A, Meyer G, Nakanishi S, Henley JM, Dev KK. The PDZ proteins PICK1, GRIP, and syntenin bind multiple glutamate receptor subtypes. Analysis of PDZ binding motifs. J Biol Chem. 2002;277:15221–15224. doi: 10.1074/jbc.C200112200. [DOI] [PubMed] [Google Scholar]
- 20.Hobert O, Westphal M. Functions of LIM-homeobox genes. Trends Genet. 2000;16:75–83. doi: 10.1016/s0168-9525(99)01883-1. [DOI] [PubMed] [Google Scholar]
- 21.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 22.Kamme F, Salunga R, Yu J, Tran DT, Zhu J, Luo L, Bittner A, Guo HQ, Miller N, Wan J, Erlander M. Single-cell microarray analysis in hippocampus CA1: demonstration and validation of cellular heterogeneity. J Neurosci. 2003;23:3607–3615. doi: 10.1523/JNEUROSCI.23-09-03607.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kania A, Jessell TM. Topographic motor projections in the limb imposed by LIM homeodomain protein regulation of ephrin-A:EphA interactions. Neuron. 2003;38:581–596. doi: 10.1016/s0896-6273(03)00292-7. [DOI] [PubMed] [Google Scholar]
- 24.Kiehn O, Kullander K. Central pattern generators deciphered by molecular genetics. Neuron. 2004;41:317–321. doi: 10.1016/s0896-6273(04)00042-x. [DOI] [PubMed] [Google Scholar]
- 25.Kohmura N, Senzaki K, Hamada S, Kai N, Yasuda R, Watanabe M, Ishii H, Yasuda M, Mishina M, Yagi T. Diversity revealed by a novel family of cadherins expressed in neurons at a synaptic complex. Neuron. 1998;20:1137–1151. doi: 10.1016/s0896-6273(00)80495-x. [DOI] [PubMed] [Google Scholar]
- 26.Krassioukov AV, Bygrave MA, Puckett WR, Bunge RP, Rogers KA. Human sympathetic preganglionic neurons and motoneurons retrogradely labelled with DiI. J Auton Nerv Syst. 1998;70:123–128. doi: 10.1016/s0165-1838(98)00042-3. [DOI] [PubMed] [Google Scholar]
- 27.Labosky PA, Kaestner KH. The winged helix transcription factor Hfh2 is expressed in neural crest and spinal cord during mouse development. Mech Dev. 1998;76:185–190. doi: 10.1016/s0925-4773(98)00105-1. [DOI] [PubMed] [Google Scholar]
- 28.Ladle DR, Frank E. The role of the ETS gene PEA3 in the development of motor and sensory neurons. Physiol Behav. 2002;77:571–576. doi: 10.1016/s0031-9384(02)00907-1. [DOI] [PubMed] [Google Scholar]
- 29.Landry CF, Ellison JA, Pribyl TM, Campagnoni C, Kampf K, Campagnoni AT. Myelin basic protein gene expression in neurons: developmental and regional changes in protein targeting within neuronal nuclei, cell bodies, and processes. J Neurosci. 1996;16:2452–2462. doi: 10.1523/JNEUROSCI.16-08-02452.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lanuza GM, Gosgnach S, Pierani A, Jessell TM, Goulding M. Genetic identification of spinal interneurons that coordinate left-right locomotor activity necessary for walking movements. Neuron. 2004;42:375–386. doi: 10.1016/s0896-6273(04)00249-1. [DOI] [PubMed] [Google Scholar]
- 31.Lawson LJ, Perry VH, Dri P, Gordon S. Heterogeneity in the distribution and morphology of microglia in the normal adult mouse brain. Neuroscience. 1990;39:151–170. doi: 10.1016/0306-4522(90)90229-w. [DOI] [PubMed] [Google Scholar]
- 32.Legrand C, Ferraz C, Clavel MC, Rabie A. Immunocytochemical localisation of gelsolin in oligodendroglia of the developing rabbit central nervous system. Brain Res. 1986;395:231–235. doi: 10.1016/s0006-8993(86)80201-3. [DOI] [PubMed] [Google Scholar]
- 33.Lein ES, Zhao X, Gage FH. Defining a molecular atlas of the hippocampus using DNA microarrays and high-throughput in situ hybridization. J Neurosci. 2004;24:3879–3889. doi: 10.1523/JNEUROSCI.4710-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lena JY, Legrand C, Faivre-Sarrailh C, Sarlieve LL, Ferraz C, Rabie A. High gelsolin content of developing oligodendrocytes. Int J Dev Neurosci. 1994;12:375–386. doi: 10.1016/0736-5748(94)90021-3. [DOI] [PubMed] [Google Scholar]
- 35.Luo L, Salunga RC, Guo H, Bittner A, Joy KC, Galindo JE, Xiao H, Rogers KE, Wan JS, Jackson MR, Erlander MG. Gene expression profiles of laser-captured adjacent neuronal subtypes. Nat Med. 1999;5:117–122. doi: 10.1038/4806. [DOI] [PubMed] [Google Scholar]
- 36.Marshall E. Getting the noise out of gene arrays. Science. 2004;306:630–631. doi: 10.1126/science.306.5696.630. [DOI] [PubMed] [Google Scholar]
- 37.Matsui H, Kimura A, Yamashiki N, Moriyama A, Kaya M, Yoshida I, Takagi N, Takahashi T. Molecular and biochemical characterization of a serine proteinase predominantly expressed in the medulla oblongata and cerebellar white matter of mouse brain. J Biol Chem. 2000;275:11050–11057. doi: 10.1074/jbc.275.15.11050. [DOI] [PubMed] [Google Scholar]
- 38.Matsuoka S, Edwards MC, Bai C, Parker S, Zhang P, Baldini A, Harper JW, Elledge SJ. p57KIP2, a structurally distinct member of the p21CIP1 Cdk inhibitor family, is a candidate tumor suppressor gene. Genes Dev. 1995;9:650–662. doi: 10.1101/gad.9.6.650. [DOI] [PubMed] [Google Scholar]
- 39.Merchenthaler I. Neurons with access to the general circulation in the central nervous system of the rat: a retrograde tracing study with fluorogold. Neuroscience. 1991;44:655–662. doi: 10.1016/0306-4522(91)90085-3. [DOI] [PubMed] [Google Scholar]
- 40.Moussaoui SM, Le Prado N, Bonici B, Faucher DC, Cuine F, Laduron PM, Garret C. Distribution of neurokinin B in rat spinal cord and peripheral tissues: comparison with neurokinin A and substance P and effects of neonatal capsaicin treatment. Neuroscience. 1992;48:969–978. doi: 10.1016/0306-4522(92)90285-a. [DOI] [PubMed] [Google Scholar]
- 41.Polo-Parada L, Bose CM, Landmesser LT. Alterations in transmission, vesicle dynamics, and transmitter release machinery at NCAM-deficient neuromuscular junctions. Neuron. 2001;32:815–828. doi: 10.1016/s0896-6273(01)00521-9. [DOI] [PubMed] [Google Scholar]
- 42.Polo-Parada L, Bose CM, Plattner F, Landmesser LT. Distinct roles of different neural cell adhesion molecule (NCAM) isoforms in synaptic maturation revealed by analysis of NCAM 180 kDa isoform-deficient mice. J Neurosci. 2004;24:1852–1864. doi: 10.1523/JNEUROSCI.4406-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pribyl TM, Campagnoni CW, Kampf K, Ellison JA, Landry CF, Kashima T, McMahon J, Campagnoni AT. Expression of the myelin basic protein gene locus in neurons and oligodendrocytes in the human fetal central nervous system. J Comp Neurol. 1996;374:342–353. doi: 10.1002/(SICI)1096-9861(19961021)374:3<342::AID-CNE3>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 44.Prinz AA, Billimoria C, Marder E. An alternative to hand-tuning conductance-based models: construction and analysis of databases of model neurons. J Neurophysiol. 2003;90:3998–4015. doi: 10.1152/jn.00641.2003. [DOI] [PubMed] [Google Scholar]
- 45.Prinz AA, Bucher D, Marder E. Similar network activity from disparate circuit parameters. Nat Neurosci. 2004;7:1345–1352. doi: 10.1038/nn1352. [DOI] [PubMed] [Google Scholar]
- 46.Rafuse VF, Polo-Parada L, Landmesser LT. Structural and functional alterations of neuromuscular junctions in NCAM-deficient mice. J Neurosci. 2000;20:6529–6539. doi: 10.1523/JNEUROSCI.20-17-06529.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Richards LJ, Murphy M, Dutton R, Kilpatrick TJ, Puche AC, Key B, Tan SS, Talman PS, Bartlett PF. Lineage specification of neuronal precursors in the mouse spinal cord. Proc Natl Acad Sci USA. 1995;92:10079–10083. doi: 10.1073/pnas.92.22.10079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schachner M, Bartsch U. Multiple functions of the myelin-associated glycoprotein MAG (siglec-4a) in formation and maintenance of myelin. Glia. 2000;29:154–165. doi: 10.1002/(sici)1098-1136(20000115)29:2<154::aid-glia9>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 49.Shah V, Drill E, Lance-Jones C. Ectopic expression of Hoxd10 in thoracic spinal segments induces motoneurons with a lumbosacral molecular profile and axon projections to the limb. Dev Dyn. 2004;231:43–56. doi: 10.1002/dvdy.20103. [DOI] [PubMed] [Google Scholar]
- 50.Sharma K, Leonard AE, Lettieri K, Pfaff SL. Genetic and epigenetic mechanisms contribute to motor neuron pathfinding. Nature. 2000;406:515–519. doi: 10.1038/35020078. [DOI] [PubMed] [Google Scholar]
- 51.Shirasaki R, Pfaff SL. Transcriptional codes and the control of neuronal identity. Annu Rev Neurosci. 2002;25:251–281. doi: 10.1146/annurev.neuro.25.112701.142916. [DOI] [PubMed] [Google Scholar]
- 52.Sockanathan S, Perlmann T, Jessell TM. Retinoid receptor signaling in postmitotic motor neurons regulates rostrocaudal positional identity and axonal projection pattern. Neuron. 2003;40:97–111. doi: 10.1016/s0896-6273(03)00532-4. [DOI] [PubMed] [Google Scholar]
- 53.Sousa R, Tannery NH, Zhou S, Lafer EM. Characterization of a novel synapse-specific protein. I. Developmental expression and cellular localization of the F1-20 protein and mRNA. J Neurosci. 1992;12:2130–2143. doi: 10.1523/JNEUROSCI.12-06-02130.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Spike RC, Todd AJ, Johnston HM. Coexistence of NADPH diaphorase with GABA, glycine, and acetylcholine in rat spinal cord. J Comp Neurol. 1993;335:320–333. doi: 10.1002/cne.903350303. [DOI] [PubMed] [Google Scholar]
- 55.Strack S, Kini S, Ebner FF, Wadzinski BE, Colbran RJ. Differential cellular and subcellular localization of protein phosphatase 1 isoforms in brain. J Comp Neurol. 1999;413:373–384. [PubMed] [Google Scholar]
- 56.Tamura S, Morikawa Y, Iwanishi H, Hisaoka T, Senba E. Expression pattern of the winged-helix/forkhead transcription factor Foxp1 in the developing central nervous system. Gene Expr Patterns. 2003;3:193–197. doi: 10.1016/s1567-133x(03)00003-6. [DOI] [PubMed] [Google Scholar]
- 57.Tanaka J, Sobue K. Localization and characterization of gelsolin in nervous tissues: gelsolin is specifically enriched in myelin-forming cells. J Neurosci. 1994;14:1038–1052. doi: 10.1523/JNEUROSCI.14-03-01038.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Thaler JP, Koo SJ, Kania A, Lettieri K, Andrews S, Cox C, Jessell TM, Pfaff SL. A postmitotic role for Isl-class LIM homeodomain proteins in the assignment of visceral spinal motor neuron identity. Neuron. 2004;41:337–350. doi: 10.1016/s0896-6273(04)00011-x. [DOI] [PubMed] [Google Scholar]
- 59.Thomas A, Gasque P, Vaudry D, Gonzalez B, Fontaine M. Expression of a complete and functional complement system by human neuronal cells in vitro. Int Immunol. 2000;12:1015–1023. doi: 10.1093/intimm/12.7.1015. [DOI] [PubMed] [Google Scholar]
- 60.Wang G, Scott SA. An early broad competence of motoneurons to express ER81 is later sculpted by the periphery. J Neurosci. 2004;24:9789–9798. doi: 10.1523/JNEUROSCI.3409-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Warden MK, Young WS., III Distribution of cells containing mRNAs encoding substance P and neurokinin B in the rat central nervous system. J Comp Neurol. 1988;272:90–113. doi: 10.1002/cne.902720107. [DOI] [PubMed] [Google Scholar]
- 62.William CM, Tanabe Y, Jessell TM. Regulation of motor neuron subtype identity by repressor activity of Mnx class homeodomain proteins. Development. 2003;130:1523–1536. doi: 10.1242/dev.00358. [DOI] [PubMed] [Google Scholar]
- 63.Yokoi T, Yamamoto N, Tada T, Fujita M, Moriyama A, Matsui H, Takahashi T, Togari H, Kato T, Asai K. Developmental changes and localization of mouse brain serine proteinase mRNA and protein in mouse brain. Neurosci Lett. 2002;323:133–136. doi: 10.1016/s0304-3940(02)00122-2. [DOI] [PubMed] [Google Scholar]
- 64.Zhou S, Tannery NH, Yang J, Puszkin S, Lafer EM. The synapse-specific phosphoprotein F1–20 is identical to the clathrin assembly protein AP-3. J Biol Chem. 1993;268:12655–12662. [PubMed] [Google Scholar]
- 65.Zimmermann P, Tomatis D, Rosas M, Grootjans J, Leenaerts I, Degeest G, Reekmans G, Coomans C, David G. Characterization of syntenin, a syndecan-binding PDZ protein, as a component of cell adhesion sites and microfilaments. Mol Biol Cell. 2001;12:339–350. doi: 10.1091/mbc.12.2.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.