Abstract
Even though nearly every human gene has at least one alternative splice form, very little is so far known about the structure and function of resulting protein products. It is becoming increasingly clear that a significant fraction of all isoforms are products of noisy selection of splice sites and thus contribute little to actual functional diversity, and may potentially be deleterious. In this study, we examine the impact of alternative splicing on protein sequence and structure in three datasets: alternative splicing events conserved across multiple species, alternative splicing events in genes that are strongly linked to disease and all observed alternative splicing events. We find that the vast majority of all alternative isoforms result in unstable protein conformations. In contrast to that, the small subset of isoforms conserved across species tends to maintain protein structural integrity to a greater extent. Alternative splicing in disease-associated genes produces unstable structures just as frequently as all other genes, indicating that selection to reduce the effects of alternative splicing on this set is not especially pronounced. Overall, the properties of alternative spliced proteins are consistent with the outcome of noisy selection of splice sites by splicing machinery.
INTRODUCTION
Bioinformatic analysis of expressed sequence tag (EST) sequences as well as microarray experiments shows that at least 78% of all human genes undergo alternative splicing, producing an average of four isoforms for every gene (1–4). Three broad hypotheses have been proposed to explain the prevalence of alternative splicing in higher Eukaryotes. The first hypothesis is that alternative splicing generates functional diversity by producing alternative protein products (5–7). The second is that alternative splicing acts as a regulation mechanism to control the level of useful gene products, by means of changing the fraction of functional and non-functional transcripts (8–10). The third hypothesis is that alternative transcripts are non functional, partly as a result of stochastic noise in the splicing machinery (2,6).
There are many well-established examples of alternative splicing with diverse functional consequences at the protein level, supporting the first hypothesis. Almost all types of molecular functions have been reported, including antagonists to the action of a major isoform (11), modified ligand-binding specificity (12), altered subcellular location (13) and reduced protein halflife (14).
There are also examples in support of the second hypothesis, alternative splicing that is functional at the message level, and acting as a mechanism for switching off or downregulating the expression of a protein (9). Some well-established examples are Drosophila sex-lethal (Sxl) (15), mdm2 (16), ABCC4 (17), MID1 (18), hUPF2 (19). Regulation via nonsense-mediated decay (NMD) has also been suggested, although a recent microarray based survey by Pan et al. indicates that only a small fraction of premature stop codon transcripts are substantially regulated in this way (20). Nevertheless, it remains the case that the vast majority of alternative transcripts have no known function at any level, leaving the possibility that the third hypothesis, most splicing is in some sense non-functional noise, is the predominant explanation.
The extent to which the non-functional noise is a contributing factor to the protein diversity remains largely unexplored. Although there have been numerous bioinformatics studies that have looked at the impact of alternative splicing on proteins, none have been focused explicitly on testing noise hypothesis (21–27). Furthermore, due to differences in methodology it is hard to reconcile conclusions reached by various studies. While there is evidence, which shows that disruption of protein domains is less frequent than expected by chance (24), and that there is a greater tendency to conserve reading frames, there is also clear evidence that very few alternative events are conserved across species and that a large fraction of resulting proteins are unstable (26) or need to undergo large conformational changes to assume stable folds (28).
We have shown in a previous work that a number of non-trivial properties of the distributions of isoform abundance and diversity are consistent with most alternative splicing being the consequence of noise in the splicing process (29). In this study, we test this hypothesis on the protein level. Using random exon deletions as a control, we examine the effect of observed deletions on structural properties such as exposed hydrophobic area, loss of 3D contacts and length of the gap created in the polypeptide chain. Splicing in monogenic disease genes is also investigated, since these are likely to be under the strongest selection pressure to maintain function.
We find that, on average, alternative splicing events have a markedly deleterious effect on protein domain structure, similar to that found for random exon deletions, and so are unlikely to encode for alternative protein function. Disease-associated genes do not show special sensitivity to alternative splicing, indicating that there is no strong selection to remove deleterious changes introduced by alternative splicing. Overall, our conclusion is that the majority of alternative proteins are structurally unstable and if expressed will be without function, consistent with the noisy splicing hypothesis.
MATERIALS AND METHODS
Data sources
The human genome sequence (30) was downloaded from NCBI (NCBI Human Genome Build 35). The transcript data were obtained from Refseq (31) (Release 17; May 2006; 29475 sequences), Unigene (31) (May 2006; 6586000 sequences) and H-InvDB (41) (Release 3.0;449186 sequences). The location of genes on chromosomes was taken from Refseq database annotation. Information about homologous genes in other species was obtained from the NCBI Homologene Database (31) (Release 48; May 2006). For each gene, all sequences were aligned to a human genomic contig using the sim4 algorithm (32) and then checked for alignment errors (see list of rules below).
Alignment quality control
The following five rules are used to identify and reject sequences containing alignment and sequencing errors.
All implied splice sites must conform to the spliceosome pattern—‘GT/AG’.
All exons must have >90% identity with the corresponding genomic sequence.
Alignment to genomic sequence must not contain any missing segments.
The sequence around exon junctions (six nucleotides into each exon) must have 100% identity with the corresponding contig.
The cDNA must not contain any introns of size <30 nucleotides.
Two additional filters were applied to minor isoforms: (a) Minor isoforms must share at least one exon with the corresponding major isoform (overlap of >1 nucleotide). (b) Minor isoforms must not contain an intron retention event relative to the major isoform.
Selection of major isoforms
For each gene, we identified one of the cDNAs as the major isoform. We first created a list of introns and all sequences that are associated with those introns. For each intron, we calculate the number of EST sequences and the number of unique EST libraries that contain the resulting exon–exon bridge. We then use these data with the rules below to choose a major isoform.
mRNA-only Rule: only full-length mRNAs are selected.
Pairwise Library Rule: if there is more than one full-length isoform, each candidate isoform is compared with all others candidates. In each comparison, for each exon–exon bridge a score of 1 is assigned to the isoform with the greater number of EST libraries with at least one occurrence of that bridge. Isoforms are then sorted by total score, larger scores ranking higher.
Lowest Number of Libraries Rule: if application of rule (ii) results in two or more isoforms with the same score, isoforms are further ranked on the basis of the least supported exon–exon bridge in each. For each exon–exon bridge in an isoform, the number of EST libraries containing at least one sequence with that bridge is determined. The smallest number of libraries supporting any of the bridges is noted, and isoforms are ranked by these values.
Lowest Number of EST Observations Rule: if application of rule (iii) results in two or more isoforms with the same rank, the procedure of rule (iii) is repeated, now ranking based on the minimal number of ESTs supporting a bridge in each isoform, rather than the minimal number of libraries
Longest Isoform Rule: if application of rule (iv) still results in two or more isoforms with the same rank, the longest isoform is chosen.
Length bias primarily arises from rule (v), simply choosing the longest isoform. But this rule is only applied in cases where the first four do not result in a clear choice, a relatively rare event. If this rule is omitted, arbitrarily selecting from the same ranked isoforms after application of rule (iv), the same isoform is selected in 88% of cases.
Application of rule (ii) may also be somewhat biased since in comparing two isoforms, a score is given for every pair of introns, and longer isoforms tend to have more introns. If both rule (v) and rule (ii) are dropped, the same isoform is selected for 75% of genes. Note that the drop from 88% to 75% greatly exaggerates the length bias of rule (ii), since in most cases selection is based on the library count criterion, independent of length.
Reconstruction of exon structure for EST sequences
The full exon structure of EST-derived isoforms must be predicted before these isoforms can be translated. We make the assumption that missing exon structure is identical to the exon structure observed in the mRNA sequence of the major isoform. On this basis, the major isoforms are used as templates to expand each EST to a full-length transcript. For 3′ EST sequences, we copy exon structure starting at the 5′ end of the major isoform until we find an overlap with an exon in the EST sequence. Since EST sequences typically end at an arbitrary location within an exon, the overlapping exon is discarded and replaced with the corresponding exon from the major isoform. An equivalent process is used to extend 5′ EST sequences. For internal EST sequences, the exon structure is copied from both ends of the major isoform.
Location of translation initiation site
Although typically the first ‘AUG' in an mRNA sequence is used to initiate translation, this is not always correct (33), and the exact location of protein translation initiation is generally not known. To make sure that our protein translations are plausible, we find the longest translation with the translation initiation site supported by other species. This is done by first finding all possible translations of an isoform. The translations are sorted according to distance to the 5′ end of the transcript, and the first 20 amino acids of each implied translation are searched against a database of all N-terminal 20 amino-acid sequences compiled from 40 Eukaryotic Refseq genomes (540000 sequences). The translation initiation sites are sorted based on the number of hits to other species, and the one with the most hits is selected.
Approximately 55% of all isoforms had translations that could be confirmed in more than two species, the remaining 45% were found only in human. As a check of this procedure, we compared our translations of Refseq sequences with Refseq annotated translations and found 95% agreement between the two sets. In some cases a single mRNA transcript can produce a variety of protein sequences through leaky translation (34). In our analysis, we assume that each isoform produces a single protein sequence.
Protein splicing fragments
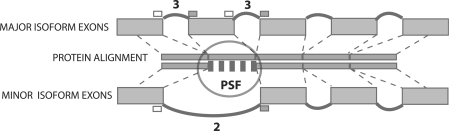
Protein translations of isoforms were aligned using the genomic coordinates of the underlying exons. This is done by first generating an mRNA alignment between major and minor isoforms pairs to genomic sequence, and then using the mRNA alignment to generate protein alignment. Protein splicing fragments (PSFs) are defined as regions within the protein alignment that differ. The alternative splicing event(s) that are responsible for producing differences in protein sequence are identified by looking for alternative introns in the region underlying each PSF (Figure 1).
Figure 1.
Identification of PSFs. Protein sequences corresponding to the major and a minor isoform of a gene are aligned, and regions in the alignment that differ identified. Deletions are defined as missing fragments in minor isoforms. Replacement is defined as a substitution of identical size. Truncations and elongations are substitutions that change the length of a fragment. Numbers above each intron bridge are conservation scores, the number of species in which this or a homologous bridge is found. Here, the alternative intron has a score of 2, indicating it was detected in human and one other species (the maximum possible conservation score is 11).
Conservation of splice junctions
We search a 40-nucleotide sequence around each splice junction (20 into each exon) against all EST and mRNA sequences of all homologous genes from other species. Gene homology information was obtained from the NBCI Homologene Database (31). Transcripts of homologous genes were obtained from the Unigene database. All 40 nucleotides must align with a minimum E-score of 0.01 and no more than two gaps to the corresponding fragment in the homologous transcript. We define the conservation score for each junction as the number of other species that had at least one significant hit to that junction. For example, if a junction was detected in mouse, rat and human, it would receive a conservation score of 3. This procedure was repeated for all exon–exon junctions from all isoforms in all genes.
Conservation score for PSFs
Using the mapping between exon structure and protein translation, we find the subset of introns in the major and minor isoforms underlying each PSF. By comparing genomic coordinates of minor and major introns within each PSF's intron subset, we identify all alternative intron pairs responsible for production of the PSF. The conservation score for a PSF is taken to be the maximum species conservation score of all the minor isoform introns. For example, if an exon insertion event resulted in an Insertion PSF with two alternative splice junctions and the first junction is supported by two species while the second one is supported by three, the conservation score is 3.
Mapping of PSFs to structure
A PSI-BLAST (35) position specific matrix (PSSM) is complied for the protein sequence of each major isoform, by searching against the Uniprot (36) database for three rounds. The PSSMs were then used to search the RCSB (37) protein sequence database for potential homologous templates using PSI-BLAST with an E-score cutoff of 10e-5. The location of each exon in the 3D structure is obtained by mapping the protein segment corresponding to the exon onto the alignment. The PSF coverage score was calculated as the fraction of all residues in the PSF that are covered by a structural template. Only PSFs that are 95% covered by a structural template were used in this study.
Calculation of structural properties
We deleted the atomic coordinates of protein fragments corresponding to deletion PSFs from the structural templates and calculated various statistics. CCP4 (38) was used to calculate the exposed surface area of all atoms in the original templates and for all atoms in the modified templates formed by deletion of PSFs. The newly exposed area is calculated as the sum over all atoms that were previously buried but now exposed (buried defined as zero surface area). The newly exposed hydrophobic area is the sum of all contributions from carbon atoms to newly exposed area. The number of lost contacts is the number of contacts in an original template that no longer exist in the modified template (contact defined as distance between atoms of less than 6Å). Deletion end-to-end distance was calculated as the distance between Cα atoms of residues at each end of a deletion.
RESULTS
Difference in protein sequences of isoforms
We first examine the effect of alternative splicing at the protein sequence level. We compiled an initial set of 85136 isoforms from 19743 genes using sequence data obtained from the Refseq (39), Unigene (40) and Hinv (41) databases. All isoforms in this set were subjected to quality control checks to ensure that each splice junction is valid. An additional 18995 isoforms were removed because of uncertainty in the alignment at the 5′ or 3′ ends. Transcripts from genes with no observed alternative isoforms were also eliminated, leaving 10972 genes with two or more isoforms each. There were a total of 55217 isoforms, of which 40% (20998) were derived from full-length mRNAs, and the remaining 60% (34219) were derived from EST sequences. Partial isoforms derived from ESTs were completed by copying missing exon structure from the corresponding major isoform (see ‘Methods’ section for details of all these procedures).
All validated isoforms were translated to provide the corresponding protein sequences, allowing for possible errors in N-terminal position. For each gene, one isoform was selected as the major isoform, using the procedure described in ‘Methods’ section. Note that while the choice of major isoform is occasionally ambiguous, for the vast majority of cases, it is straightforward. To obtain an overall measure of protein sequence length differences between minor and major isoforms, we subtracted the length of the major isoform from that of each minor isoform of the same gene and plotted the histogram of length differences (Figure 2). Approximately 20% of all minor isoforms had the same protein sequence length as the major isoform, mostly due to alternative splicing outside the coding regions (not shown in the plot). Most minor isoforms are significantly shorter than major isoforms (∼70%) and only 9% of all minor isoforms are longer than major isoforms. Care was taken to minimize any bias towards selecting longer isoforms as the major ones, so that this effect is not an artifact. The signal is dominated by protein length differences of more than 100 amino acids (∼43%).
Figure 2.
Distribution of differences in amino-acid sequence length. Distribution of differences in amino-acid sequence length between proteins coded for by major and minor isoforms. Most minor isoforms are substantially shorter than the major isoforms.
We repeated this analysis on the subset of isoforms derived only from full-length mRNAs. We found that both datasets have approximately the same distribution of length differences. The full-length mRNA dataset has an increased fraction of longer minor isoforms (∼17% rather than ∼9%) and a correspondingly smaller fraction of same length isoforms, but the fraction of shorter major isoforms remains the same in both sets. Thus the shortness of the minor isoforms is not an artifact of incorrect reconstruction from ESTs. Total 10160 isoforms are predicted to be subject to the NMD degradation pathway (prediction based on the ‘50 nucleotide rule') (8). Removal of these results in a reduction in the fraction of isoforms more than 100 amino acids shorter than the major one (43–36%). The same effect is observed in the full-length mRNA-only subset.
In order to analyze the differences between isoforms further, we aligned the implied protein translation of each minor isoform to that of the corresponding major isoform. The alignment is preformed in two steps. First, we align the major and minor isoforms' exons to a common genome reference frame. Second, we use the resulting mRNA alignment to create aligned protein translations. Figure 1 illustrates the procedure.
We term each continuous stretch of difference within an alignment between two isoforms a PSF. On the one hand, in cases where alternative splicing lies outside the coding region, the protein alignment will be identical and thus no PSF will be produced. On the other hand, if there are multiple alternative splicing events within a coding region, multiple PSFs will be generated from a single major/minor comparison. For example, in the tropomyosin 2 (TPM2) gene (Figure 3), there are two alternative splicing differences between the major and minor isoforms, generating two PSFs.
Figure 3.
Example of PSFs in TPM2. The exon alignment of two isoforms shows two PSFs. In this case, both are the result of exon swaps (an exon in one isoform is replaced by a different one in the other isoform), and in each instance, the replacement exon is the same length as the major isoform one. The PSF in the middle of the sequence is thus classified as an internal replacement, and the one at the 3′ end is a C-terminal replacement.
The PSFs were classified into three broad categories: deletion, insertion and substitution. Substitutions were further classified into three classes: perfect replacement—a fragment is replaced with another fragment of the same size; truncation—a fragment is replaced with a smaller fragment; and elongation—a fragment is replaced with a larger fragment. We also classified PSFs into four broad classes based on location: N-terminal, internal, C-terminal and not classified. The last category is reserved for differences that extend over the entire length of the alignment. As an example, consider the alignment between two isoforms of TPM2 gene in Figure 3, where thereare two replacements, one creating an internal 26 residue long substitution, the other also a 26 residue replacement, at the C terminus.
The results of the PSF classification are summarized in Table 1. As already implied by the whole protein length comparisons, the majority of fragments are deletions and truncations (87.6% of all fragments). Many of the C-terminal truncations are produced as a result of premature stop codons due to frame shifts. Many of these are predicted to be degraded by NMD by the ‘50 nucleotide rule' (8,42), although this prediction might not be correct: as noted earlier, recent data from Pan et al., suggest that a significant fraction of transcripts with premature stop codons will not be affected by NMD (20). The most common types of deletions are N-terminal deletions. These are largely generated through alternative promoters, and strictly speaking, should be considered separately from other alternative splicing events, since different machinery is involved. As far as is known, there is no quality control mechanism similar to NMD for deletions on the N-terminal ends of proteins, thus we would expect that these proteins are actually produced. Replacement of a protein fragment with another protein fragment of exactly the same size is the least common type of fragment (0.4%). Although rare, these events are highly expressed (supported by large numbers of EST observations) and we will show in the next section that these events exhibit the strongest conservation signal across multiple species.
Table 1.
Classification of PSFs
| Not classified | Cterm | Internal | Nterm | Fraction (%) | |
|---|---|---|---|---|---|
| Truncation | 3000 | 9425 | 1252 | 2336 | 47.2 |
| Deletion | 0 | 278 | 4603 | 8359 | 39 |
| Elongation | 369 | 1070 | 507 | 537 | 7.3 |
| Insertion | 0 | 25 | 1597 | 444 | 6.1 |
| Replacement | 1 | 39 | 68 | 35 | 0.4 |
| Fraction (%) | 9.9 | 31.9 | 23.6 | 34.5 | 100 |
Subsequences affected by splicing are classified by the effect on length (truncation—a shorter fragment than in the major isoform, deletion—removal of a fragment, elongation—a longer fragment than in the major isoform, insertion—insertion of a fragment and replacement—replacement of a fragment with another of the same length) and by location in the open reading frame (not classified, C terminal, internal and N terminal). The majority of PSFs belong to the deletion and truncation categories.
Conserved alternative splicing subsets
To obtain a splicing subset enriched for function, we compiled a list of alternative splicing events conserved across multiple species. Although it is reasonable to assume that cross-species conservation is correlated to functionality, there are two caveats to be borne in mind. First, the conserved subset is biased towards genes expressed in high abundance in other species, since they are more likely to be detected in EST experiments. Second, high abundance genes are also likely to produce more alternative isoforms as a result of noise (29), so some of the isoforms are likely to be non-functional. Nevertheless, as noted earlier, several evolutionary trends have been shown to be correlated to conservation of alternative splicing events.
To find conserved alternative splicing events, we searched sequences of all exon–exon junctions for significant hits in transcripts of homologous genes in other species (obtained from Homologene, see ‘Methods’ section), and defined the conservation score for each junction as the number of species that had at least one significant hit to that junction. We then defined the conservation score for each PSF as the maximum conservation score from all of alternative splice junctions that underlie that PSF. The distribution of PSF conservation scores from all isoforms except those predicted to be subject to NMD is shown in Table 2.
Table 2.
Cross-species conservation of PSFs
| Human only | Two species | Three species | More than four species | |
|---|---|---|---|---|
| Replacement | 0.4 | 0.6 | 6.2 | 9.8 |
| Deletion | 50.6 | 38.7 | 27.6 | 29.4 |
| Truncation | 34.2 | 29.6 | 29.2 | 21.6 |
| Elongation | 7.7 | 15.7 | 26.9 | 33.3 |
| Insertion | 7 | 15.4 | 10 | 5.9 |
| Total (%) | 100 | 100 | 100 | 100 |
| N (PSFs) | 23 287 | 1761 | 438 | 51 |
Splicing effects most likely to be functional (replacement, and elongation and insertion) are markedly enhanced in the conserved sets.
Alternative splicing where a protein fragment is replaced by another of the same length shows strongest conservation. These represent 9.8% of all PSFs conserved across four or more species—a 25-fold increase from the 0.4% value in the human-only subset. The fraction of insertions and elongations also increases with increasing conservation, while deletions and truncations decrease. The most obvious explanation for these observations is that deletions and truncations have a greater tendency to disrupt protein structure and are thus less likely to be conserved. As we observed in the distribution of length changes (Figure 2), deletions and truncations tend to remove a large numbers of residues, typically more than a 100. Of course, perfect replacements are least likely to affect protein structure, since they preserve protein length. Insertions and elongations tend to change the length by fewer than 25 residues, and are thus also less likely to disrupt structure. This effect is investigated further in the next section.
The relationship between change in length and conservation across species is shown in Figure 4A. Figure 4B shows the relationship between length change and minor isoform abundance. The assumption here is that more abundant isoforms are more likely to be functional, providing another means of examining the relationship between structure properties and function. Abundance is defined as the number of EST observations for alternative splice junctions underlying the PSF. In cases where there are multiple alternative splice junctions underlying a PSF, we use the junction with the highest EST count as the measure of abundance. This is an approximation since it does not take into account EST sampling biases, but it is expected to be correlated to the actual number of copies of the isoform in the sample. Figure 4 clearly shows that change in length is highly correlated to both abundance and conservation across species. Small changes are both conserved and abundant, suggesting that they are more likely to be functional.
Figure 4.
Cross-species conservation and abundance versus PSFs length change. PSFs were divided into 120 groups, with at least 200 PSFs within each group, and mean cross-species conservation scores and abundance were calculated. Large changes in length are typically only found in unconserved splice forms and at low abundance.
Properties of disease gene subsets
We have also analyzed the frequency of alternative splicing events in a subset of genes strongly associated with human diseases. Disease-associated genes were obtained from three databases: OMIM (43) (2241 genes), HGMD (44) (834 genes) and Genetests (45) (1000 genes). We also compiled a CORE set of 530 genes found in all three databases. Although the exact mechanism leading to disease is different in each case, in general, the cause can be attributed to reduced total activity of a protein product as a result of a mutation of some kind.
The extent of change to protein sequence as a result of alternative splicing is far greater than the change due to a typical single amino acid mutation, and as a consequence the likelihood of disruption of function is also greater. But unlike a deleterious amino acid mutation, which will affect all transcripts of a gene, alternative splicing only affects a fraction of all transcripts. As long as the fraction of alternative transcripts does not exceed some level where the normal function of a gene is seriously affected there will be no pressure to reduce deleterious changes. If these assumptions are correct, disease genes should undergo alternative splicing with the same frequency as all other genes.
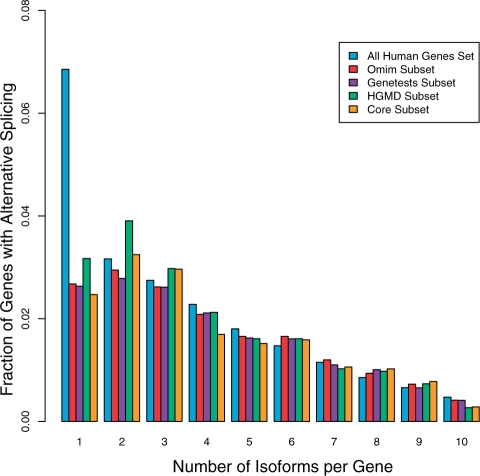
Figure 5 shows the distribution of the number of isoforms per gene for all human genes in our database (19743 genes, 85136 isoforms) and for the various subsets of disease-associated ones. Except for the first set of bars (single isoform, i.e. no alternative spicing), all the sets show nearly identical distributions. Abundance of alternative isoforms (as measured by fraction of all transcripts per gene that are alternative) is nearly identical (8% all vs ∼7% disease). We also looked at the distribution of overall length change between major and minor forms of proteins in disease-associated genes, which also shows no significant differences between gene sets. We did not find significant differences in predicted NMD fraction, or the types and locations of PSFs. Based on these observations, we conclude that pressure to reduce the frequency, or severity of impact of alternative splicing events is the same for disease genes as non-disease genes.
Figure 5.
Fraction of genes with alternative splicing in disease-associated genes. All genes (19743 genes, 85136 isoforms), and three subsets of disease-related genes derived from OMIM (2241 genes), Genetests (1000 genes) and HGMD (834 genes). The Core set are genes present in all three databases (530 genes). All sets show a similar distribution of number of isoforms per gene, with the exception of the first set of bars, which represents the fraction of genes without alternative splicing (7% vs ∼2–3% in disease subsets).
Stability of protein structures produced by alternative splicing
For proteins with known or modelable 3D structure we can ask what fraction of alternative splicing events is likely to result in a stably folded protein product. For this purpose, possible templates for the major isoform of each gene were identified in the PDB, the exons were mapped on to the 3D structural coordinates, and regions corresponding to PSFs arising from internal deletions were removed (see ‘Methods’ section). The analysis was performed on the resulting set of modified protein structures. The impact of a deletion is measured in terms of the distance between the resulting chain ends, newly exposed hydrophobic area, total newly exposed area and number of residue–residue contacts lost. An example of mapping of alternative splicing to structural fragments in growth hormone 1 gene (GH1) is illustrated in Figure 6.
Figure 6.
Alternative splicing in GH1. The exon structure of the major isoform and six minor isoforms are shown as yellow bars. The location of deletions and truncations in the protein structure relative to the major isoform are highlighted in red. Isoforms A, B, C and D are classified as Internal Deletions. Isoforms E and F produce a frame shift and are classified as C-terminal Truncations. Isoform B was derived from EST sequence, and part of the exon structure (colored brown) was copied from the major isoform. The isoforms are sorted by severity of impact on structure.
There are no tools for accurate prediction of protein stability. However, deletion of a randomly chosen exon is very unlikely to result in a stable protein structure. We make use of this feature to generate a reference set of unstable structures, and compare properties of proteins produced by observed alternative splicing deletions with these. About 60% of all possible internal exon deletions result in a frame shift, and almost all of these are predicted to be degraded by NMD, thus they were not considered in our calculations.
The remaining 40% of in-frame deletions form a pool from which the reference set were selected. For each observed exon deletion included in the analysis, a random exon deletion with the same number of residues was found, generating a reference set with the same length distribution as the observed data. Just as with real deletions, random exon deletions were mapped to 3D structure coordinates, and regions of chain corresponding to the exon was removed. The final data sets consist of 1439 random deletions and 1439 real deletions (1085 splicing events observed only in human, 263 in two species, 76 in three species and 15 in four or more species).
The distributions of various structural features are shown in Figure 7. The random and full human-only sets have very similar distributions for all structural properties. We conclude from this that the large majority of alternatively spliced deletions, on the one hand, result in the production of unstable protein folds. On the other hand, it is immediately obvious that deletions that are conserved across multiple species tend to remove fewer residues, have a smaller end-to-end distance, lose fewer contacts and expose less total and hydrophobic surface area. That is, conserved deletions also tend to be conservative in term of structural impact, supporting the view that these sets are enriched for function compared with unconserved events.
Figure 7.
Comparison between random exon deletions and deletions observed in minor isoforms. Genes are divided into five sets: Random Exon Deletions, Human Only Deletions, Conserved across two, three and four or more species. (A) Number of residues deleted (amino acids). (B) Number of contacts lost. (C) Newly exposed surface area (Å2). (D) Newly exposed hydrophobic surface area (Å2). (E) Distance between Cα atoms at the ends of a deletion (Å). (F) Number of EST sequences that support the alternative splicing. For all the structural properties, the random and all human distributions are very similar, whereas minor isoforms found in multiple species exhibit more conservative structural changes.
DISCUSSION
Alternative splicing can generate a large number of isoforms starting from a single premessage mRNA. A well-known example of production of molecular diversity by alternative splicing is the Drosophila Dscam gene, which can potentially generate as many as 38000 isoforms (46). In human, most genes are alternatively spliced (47). Most (>70%) isoforms change protein coding regions, and therefore potentially produce novel protein products (25). On this basis, it is frequently argued that alternative splicing provides a mechanism for complex organisms such as human to generate a large number of novel molecular components from a relatively small number of genes (6).
The basic assumption in this view is that products of alternative splicing are functional. However, little is known about the protein sequences and resulting protein structure of alternative isoforms. In an effort to decipher the functionality of isoforms by other means, numerous bioinformatics studies have analyzed various properties. It has been found that a small fraction of alternative splicing shows clear signs of functionality, particularly those exhibiting tissue specificity (48), expression in high abundance (49) and cross-species conservation (50). Conserved isoforms especially tend to preserve protein coding frames and are less likely to be subject to NMD (24,51,52). Compared to the large body of literature on the effects of alternative splicing at the message sequence level, relatively little is known about its affect on protein structure and function.
There have been a few studies regarding the impact of alternative splicing on tertiary structure of proteins. Detailed analysis of structural characteristics of alternative isoforms from ENCODE project by Tress et al. (26) is largely in agreement with our conclusions—the vast majority of isoforms show little indication of being functional and stable. Homma et al. have analyzed the location of alternative splice sites and relation to SCOP domain boundaries. They also found that variants encoding unstable protein products tend to be species specific and are expressed at a significantly lower levels compared to stable variants (53). Wang et al. examined 3D models of alternative isoforms via threading and molecular dynamics, and concluded that at least in some cases isoforms are capable of producing proteins with stable conformations (28).
It is clear that the impact of alternative splicing on protein structure, stability and function remains poorly understood. Although there seems to be a general agreement that alternative splicing events conserved across species probably result in stable and functional protein products, there seems to be no consensus for species–specific isoforms. In other work we argue that a large fraction of isoforms are products of occasional splicing mistakes in selection of splice sites (29). That hypothesis is supported by observations that the increase in number of isoforms increases with expression level and number of introns in a gene; that most isoforms are expressed at low abundance levels and that few show clear tissue specificity (54,55). The main principle of the noise hypothesis is that large error rates can be tolerated as long as adequate levels of functional products are produced and toxic effects on the system are avoided. If these requirements are satisfied there will be no further selection pressure to reduce the frequency of alternative splicing, and a great diversity of isoforms can be generated. A prediction of this model is that most non-conserved isoforms will be non-functional, and therefore will tend to disrupt the implied proteins structure in a random manner.
At the sequence level, we find that many alternative isoforms are predicted to produce proteins that are significantly smaller than the corresponding major isoforms. Removing isoforms that are predicted to be subject to NMD does not change this outcome. These findings are in a qualitative agreement with other studies regarding the impact of alternative splicing on protein sequences (28). Wang et al. have analyzed alternative isoforms annotated in the SWISS-PROT database and found that deletions account for 57% of all annotated events, while insertions account for only 5% of all annotated splicing events.
Using conservation as a proxy for functionality, we find that small changes in sequence length are more likely to be conserved. Replacements that do not change the protein length show the strongest conservation signal. These observations make sense, since the smaller the change, the less likely it is to be disruptive to protein structure, increasing the likelihood of maintaining function or possibly generating new function. At the level of 3D structure, we compared the impact of in-frame deletions introduced by alternative splicing to that of randomly selected in-frame exon deletions. Random deletions are unlikely to result in a stable protein fold, and so provide a reference set for testing the viability of deletions observed in real isoforms. We find that isoforms observed only in human show the same distribution as the random ones, for all structural parameters. Deletions that are conserved across multiple species tend to be more structurally conservative—the distances between ends of deletions tend to be smaller, they expose less hydrophobic surface and lose fewer contacts. From this observation, we conclude that most species-specific isoforms are unlikely to result in stable conformations.
Our analysis of disease genes did not reveal any surprising results. If alternative splicing had a negative impact on the normal functions of these genes, we should have observed strong selection pleasure to reduce the frequency and severity of such events, since in this set protein function is tightly coupled to fitness. No such pressure was observed. The distributions of number of alternative isoforms, fractional abundance of alternative transcripts, number of NMD isoforms and protein length changes in disease genes were nearly identical to those for all genes. If any pressure exists to reduce frequency of alternative splicing, it is not particularly pronounced in this set of genes.
The evidence presented in this study strongly suggests that the majority of alternative isoforms do not code for functional protein products and have little impact on phenotype, yet they are common, with nearly every gene producing several alternatives. These trends are consistent with noise in the splicing process due to stochastic fluctuations of various splicing factors. Noise is an inherit part of any complex biological process, and selection forces will have optimized the fidelity of the splicing system so as to produce sufficient levels of the functional components and to reduce harmful effects from non-functional products. Evidently these requirements are satisfied at a significant level of noise. An accidental positive aspect of the high level of noise is that it provides an additional pool of variability in which novel functional forms can be discovered.
FUNDING
National Institutes of Health (P01 GM57890). Funding for open access charge: grant number P01 GM57890.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Steve Mount and Arlin Stoltzfus for helpful discussions.
REFERENCES
- 1.Mironov AA, Fickett JW, Gelfand MS. Frequent alternative splicing of human genes. Genome Res. 1999;9:1288–1293. doi: 10.1101/gr.9.12.1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kan Z, States D, Gish W. Selecting for functional alternative splices in ESTs. Genome Res. 2002;12:1837–1845. doi: 10.1101/gr.764102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Modrek B, Resch A, Grasso C, Lee C. Genome-wide detection of alternative splicing in expressed sequences of human genes. Nucleic Acids Res. 2001;29:2850–2859. doi: 10.1093/nar/29.13.2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Johnson JM, Castle J, Garrett-Engele P, Kan Z, Loerch PM, Armour CD, Santos R, Schadt EE, Stoughton R, Shoemaker DD. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science. 2003;302:2141–2144. doi: 10.1126/science.1090100. [DOI] [PubMed] [Google Scholar]
- 5.Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- 6.Graveley BR. Alternative splicing: increasing diversity in the proteomic world. Trends Genet. 2001;17:100–107. doi: 10.1016/s0168-9525(00)02176-4. [DOI] [PubMed] [Google Scholar]
- 7.Maniatis T, Tasic B. Alternative pre-mRNA splicing and proteome expansion in metazoans. Nature. 2002;418:236–243. doi: 10.1038/418236a. [DOI] [PubMed] [Google Scholar]
- 8.Lewis B, Green R, Brenner S. Evidence for the widespread coupling of alternative splicing and nonsense-mediated mRNA decay in humans. Proc. Natl Acad. Sci. USA. 2003;100:189–192. doi: 10.1073/pnas.0136770100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hillman R, Green R, Brenner S. An unappreciated role for RNA surveillance. Genome Biol. 2004;5:R8. doi: 10.1186/gb-2004-5-2-r8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rehwinkel J, Letunic I, Raes J, Bork P, Izaurralde E. Nonsense-mediated mRNA decay factors act in concert to regulate common mRNA targets. RNA. 2005;11:1530–1544. doi: 10.1261/rna.2160905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arinobu Y, Atamas SP, Otsuka T, Niiro H, Yamaoka K, Mitsuyasu H, Niho Y, Hamasaki N, White B, Izuhara K. Antagonistic effects of an alternative splice variant of human IL-4, IL-4delta2, on IL-4 activities in human monocytes and B cells. Cell Immunol. 1999;191:161–167. doi: 10.1006/cimm.1998.1431. [DOI] [PubMed] [Google Scholar]
- 12.Miki T, Bottaro DP, Fleming TP, Smith CL, Burgess WH, Chan AM, Aaronson SA. Determination of ligand-binding specificity by alternative splicing: two distinct growth factor receptors encoded by a single gene. Proc. Natl Acad. Sci. USA. 1992;89:246–250. doi: 10.1073/pnas.89.1.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matthews GD, Gould RM, Vardimon L. A single glutamine synthetase gene produces tissue-specific subcellular localization by alternative splicing. FEBS Lett. 2005;579:5527–5534. doi: 10.1016/j.febslet.2005.08.082. [DOI] [PubMed] [Google Scholar]
- 14.Scott RP, Eketjall S, Aineskog H, Ibanez CF. Distinct turnover of alternatively spliced isoforms of the RET kinase receptor mediated by differential recruitment of the Cbl ubiquitin ligase. J. Biol. Chem. 2005;280:13442–13449. doi: 10.1074/jbc.M500507200. [DOI] [PubMed] [Google Scholar]
- 15.Penalva LO, Sanchez L. RNA binding protein sex-lethal (Sxl) and control of Drosophila sex determination and dosage compensation. Microbiol. Mol. Biol. Rev. 2003;67:343–359. doi: 10.1128/MMBR.67.3.343-359.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Veldhoen N, Metcalfe S, Milner J. A novel exon within the mdm2 gene modulates translation initiation in vitro and disrupts the p53-binding domain of mdm2 protein. Oncogene. 1999;18:7026–7033. doi: 10.1038/sj.onc.1203182. [DOI] [PubMed] [Google Scholar]
- 17.Lamba JK, Adachi M, Sun D, Tammur J, Schuetz EG, Allikmets R, Schuetz JD. Nonsense mediated decay downregulates conserved alternatively spliced ABCC4 transcripts bearing nonsense codons. Hum. Mol. Genet. 2003;12:99–109. doi: 10.1093/hmg/ddg011. [DOI] [PubMed] [Google Scholar]
- 18.Winter J, Lehmann T, Krauss S, Trockenbacher A, Kijas Z, Foerster J, Suckow V, Yaspo M-L, Kulozik A, Kalscheuer V, et al. Regulation of the MID1 protein function is fine-tuned by a complex pattern of alternative splicing. Hum. Genet. 2004;114:541–552. doi: 10.1007/s00439-004-1114-x. [DOI] [PubMed] [Google Scholar]
- 19.Wittmann J, Hol EM, Jäck H-M. hUPF2 silencing identifies physiologic substrates of mammalian nonsense-mediated mRNA decay. Mol. Cell Biol. 2006;26:1272–1287. doi: 10.1128/MCB.26.4.1272-1287.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pan Q, Saltzman AL, Kim YK, Misquitta C, Shai O, Maquat LE, Frey BJ, Blencowe BJ. Quantitative microarray profiling provides evidence against widespread coupling of alternative splicing with nonsense-mediated mRNA decay to control gene expression. Genes. Dev. 2006;20:153–158. doi: 10.1101/gad.1382806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kriventseva EV, Koch I, Apweiler R, Vingron M, Bork P, Gelfand MS, Sunyaev S. Increase of functional diversity by alternative splicing. Trends Genet. 2003;19:124–128. doi: 10.1016/S0168-9525(03)00023-4. [DOI] [PubMed] [Google Scholar]
- 22.Liu S, Altman RB. Large scale study of protein domain distribution in the context of alternative splicing. Nucleic Acids Res. 2003;31:4828–4835. doi: 10.1093/nar/gkg668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Resch A, Xing Y, Modrek B, Gorlick M, Riley R, Lee C. Assessing the impact of alternative splicing on domain interactions in the human proteome. J. Proteome Res. 2004;3:76–83. doi: 10.1021/pr034064v. [DOI] [PubMed] [Google Scholar]
- 24.Yeo GW, Nostrand EV, Holste D, Poggio T, Burge CB. Identification and analysis of alternative splicing events conserved in human and mouse. Proc. Natl Acad. Sci. USA. 2005;102:2850–2855. doi: 10.1073/pnas.0409742102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takeda J-I, Suzuki Y, Nakao M, Barrero RA, Koyanagi KO, Jin L, Motono C, Hata H, Isogai T, Nagai K, et al. Large-scale identification and characterization of alternative splicing variants of human gene transcripts using 56,419 completely sequenced and manually annotated full-length cDNAs. Nucleic Acids Res. 2006;34:3917–3928. doi: 10.1093/nar/gkl507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tress ML, Martelli PL, Frankish A, Reeves GA, Wesselink JJ, Yeats C, Olason PL, Albrecht M, Hegyi H, Giorgetti A, et al. The implications of alternative splicing in the ENCODE protein complement. Proc. Natl Acad. Sci. USA. 2007;104:5495–5500. doi: 10.1073/pnas.0700800104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xing YL. Colloquium Paper: evidence of functional selection pressure for alternative splicing events that accelerate evolution of protein subsequences. Proc. Natl Acad. Sci. USA. 2005;102:13526. doi: 10.1073/pnas.0501213102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang P, Yan B, Guo J-T, Hicks C, Xu Y. Structural genomics analysis of alternative splicing and application to isoform structure modeling. Proc. Natl Acad. Sci. USA. 2005;102:18920–18925. doi: 10.1073/pnas.0506770102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Melamud E, Moult J. Stochastic noise in splicing machinery. Nucleic Acids Res. 2009;37:4873–4886. doi: 10.1093/nar/gkp471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 31.Wheeler DL, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, DiCuccio M, Edgar R, Federhen S, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2006;34:D173–D180. doi: 10.1093/nar/gkj158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Florea L, Hartzell G, Zhang Z, Rubin GM, Miller W. A computer program for aligning a cDNA sequence with a genomic DNA sequence. Genome Res. 1998;8:967–974. doi: 10.1101/gr.8.9.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kozak M. Regulation of translation in eukaryotic systems. Annu. Rev. Cell Biol. 1992;8:197–225. doi: 10.1146/annurev.cb.08.110192.001213. [DOI] [PubMed] [Google Scholar]
- 34.Wang X-Q, Rothnagel JA. 5′-untranslated regions with multiple upstream AUG codons can support low-level translation via leaky scanning and reinitiation. Nucleic Acids Res. 2004;32:1382–1391. doi: 10.1093/nar/gkh305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu CH, Apweiler R, Bairoch A, Natale DA, Barker WC, Boeckmann B, Ferro S, Gasteiger E, Huang H, Lopez R, et al. The Universal Protein Resource (UniProt): an expanding universe of protein information. Nucleic Acids Res. 2006;34:D187–D191. doi: 10.1093/nar/gkj161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Deshpande N, Addess KJ, Bluhm WF, Merino-Ott JC, Townsend-Merino W, Zhang Q, Knezevich C, Xie L, Chen L, Feng Z, et al. The RCSB Protein Data Bank: a redesigned query system and relational database based on the mmCIF schema. Nucleic Acids Res. 2005;33:D233–D237. doi: 10.1093/nar/gki057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Winn MD, Ashton AW, Briggs PJ, Ballard CC, Patel P. Ongoing developments in CCP4 for high-throughput structure determination. Acta Crystallogr. D Biol. Crystallogr. 2002;58:1929–1936. doi: 10.1107/s0907444902016116. [DOI] [PubMed] [Google Scholar]
- 39.Pruitt K, Tatusova T, Maglott D. NCBI Reference Sequence (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2004;33:501. doi: 10.1093/nar/gki025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wheeler DL, Church DM, Federhen S, Lash AE, Madden TL, Pontius JU, Schuler GD, Schriml LM, Sequeira E, Tatusova TA, et al. Database resources of the National Center for Biotechnology. Nucleic Acids Res. 2003;31:28–33. doi: 10.1093/nar/gkg033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Imanishi T, Itoh T, Suzuki Y, O'Donovan C, Fukuchi S, Koyanagi KO, Barrero RA, Tamura T, Yamaguchi-Kabata Y, Tanino M, et al. Integrative annotation of 21,037 human genes validated by full-length cDNA clones. PLoS Biol. 2004;2:e162. doi: 10.1371/journal.pbio.0020162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nagy E, Maquat L. A rule for termination-codon position within intron-containing genes: when nonsense affects RNA abundance. Trends Biochem. Sci. 1998;23:198–199. doi: 10.1016/s0968-0004(98)01208-0. [DOI] [PubMed] [Google Scholar]
- 43.Hamosh A, Scott AF, Amberger JS, Bocchini CA, McKusick VA. Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res. 2005;33:D514–D517. doi: 10.1093/nar/gki033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stenson PD, Ball EV, Mort M, Phillips AD, Shiel JA, Thomas NST, Abeysinghe S, Krawczak M, Cooper DN. Human Gene Mutation Database (HGMD): 2003 update. Hum. Mutat. 2003;21:577–581. doi: 10.1002/humu.10212. [DOI] [PubMed] [Google Scholar]
- 45.Pagon RA, Tarczy-Hornoch P, Baskin PK, Edwards JE, Covington ML, Espeseth M, Beahler C, Bird TD, Popovich B, Nesbitt C, et al. GeneTests-GeneClinics: genetic testing information for a growing audience. Hum. Mutat. 2002;19:501–509. doi: 10.1002/humu.10069. [DOI] [PubMed] [Google Scholar]
- 46.Chen BE, Kondo M, Garnier A, Watson FL, Püettmann-Holgado R, Lamar DR, Schmucker D. The molecular diversity of Dscam is functionally required for neuronal wiring specificity in Drosophila. Cell. 2006;125:607–620. doi: 10.1016/j.cell.2006.03.034. [DOI] [PubMed] [Google Scholar]
- 47.Xing Y, Lee C. Alternative splicing and RNA selection pressure-evolutionary consequences for eukaryotic genomes. Nat. Rev. Genet. 2006;7:499–509. doi: 10.1038/nrg1896. [DOI] [PubMed] [Google Scholar]
- 48.Xu Q, Modrek B, Lee C. Genome-wide detection of tissue-specific alternative splicing in the human transcriptome. Nucleic Acids Res. 2002;30:3754–3766. doi: 10.1093/nar/gkf492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pan Q, Bakowski MA, Morris Q, Zhang W, Frey BJ, Hughes TR, Blencowe BJ. Alternative splicing of conserved exons is frequently species-specific in human and mouse. Trends Genet. 2005;21:73–77. doi: 10.1016/j.tig.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 50.Nurtdinov RN, Artamonova I, Mironov AA, Gelfand MS. Low conservation of alternative splicing patterns in the human and mouse genomes. Hum. Mol. Genet. 2003;12:1313–1320. doi: 10.1093/hmg/ddg137. [DOI] [PubMed] [Google Scholar]
- 51.Sorek R, Shamir R, Ast G. How prevalent is functional alternative splicing in the human genome? Trends Genet. 2004;20:68–71. doi: 10.1016/j.tig.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 52.Alissa R, Xing Y, Alekseyenko A, Modrek B, Lee C. Evidence for a subpopulation of conserved alternative splicing events under selection pressure for protein reading frame preservation. Nucleic Acids Res. 2004;32:1261–1269. doi: 10.1093/nar/gkh284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Homma K, Kikuno RF, Nagase T, Ohara O, Nishikawa K. Alternative splice variants encoding unstable protein domains exist in the human brain. J. Mol. Biol. 2004;343:1207–1220. doi: 10.1016/j.jmb.2004.09.028. [DOI] [PubMed] [Google Scholar]
- 54.Modrek B, Lee C. Alternative splicing in the human, mouse and rat genomes is associated with an increased frequency of exon creation and/or loss. Nat. Genet. 2003;34:177–180. doi: 10.1038/ng1159. [DOI] [PubMed] [Google Scholar]
- 55.Pan Q, Shai O, Misquitta C, Zhang W, Saltzman AL, Mohammad N, Babak T, Siu H, Hughes TR, Morris QD, et al. Revealing global regulatory features of mammalian alternative splicing using a quantitative microarray platform. Mol. Cell. 2004;16:929–941. doi: 10.1016/j.molcel.2004.12.004. [DOI] [PubMed] [Google Scholar]