Abstract
LysR-type transcriptional regulators (LTTRs) form the largest family of bacterial regulators acting as both auto-repressors and activators of target promoters, controlling operons involved in a wide variety of cellular processes. The LTTR, CrgA, from the human pathogen Neisseria meningitidis, is upregulated during bacterial–host cell contact. Here, we report the crystal structures of both regulatory domain and full-length CrgA, the first of a novel subclass of LTTRs that form octameric rings. Non-denaturing mass spectrometry analysis and analytical ultracentrifugation established that the octameric form of CrgA is the predominant species in solution in both the presence and absence of an oligonucleotide encompassing the CrgA-binding sequence. Furthermore, analysis of the isolated CrgA–DNA complex by mass spectrometry showed stabilization of a double octamer species upon DNA binding. Based on the observed structure and the mass spectrometry findings, a model is proposed in which a hexadecameric array of two CrgA oligomers binds to its DNA target site.
INTRODUCTION
The largest family of one-component DNA-binding transcription factors found in bacteria are the LysR-type transcriptional regulators (LTTRs). LTTRs regulate the expression of a wide variety of genes, including operons involved in amino acid metabolism, oxidative stress, degradation of aromatic compounds and bacterial virulence (1). They comprise an N-terminal DNA-binding domain (DBD) containing a winged helix turn-helix-motif (wHTH) joined by a long linker helix (LH), involved in oligomerization, to a C-terminal regulatory domain (1,2). The regulatory domains of LTTRs share a similar fold to periplasmic substrate-binding proteins (3) and constitute the sensor part of the molecule, binding effector molecules (1,4) or responding directly to redox-active compounds through the thiol groups of cysteine residues (5). LTTRs are dual function regulators acting as both auto-repressors and activators of target promoters, frequently of genes co-located with the LTTRs in the chromosome (1). Typically, an LTTR binds to a 50–60-bp stretch of DNA that comprises the recognition-binding site (RBS), containing an LTTR consensus-binding motif (T-N11-A), and an activation-binding site (ABS), which is usually in close proximity to the –35 promoter element. Transcriptional activation of the target gene is commonly dependent on the interaction of the LTTR with an effector (1). This is often a substrate or metabolite of the regulated gene, which functions as a classical allosteric regulator, though LTTRs appear to remain bound to their cognate DNA targets under both inducing and non-inducing conditions. Reports of differential DNase protection of the ABS (6–8) and promoter DNA relaxation in the presence of an effector suggest that the LTTR undergoes a conformational change upon effector binding (7,8), which leads to either a quantitative and/or qualitative change in DNA binding. The molecular details of these events remain unclear since structural characterization of full-length LTTRs has proved challenging, largely due to their poor solubility (9,10). In fact, the structure of only one full-length LTTR has been reported to date, that of CbnR from Ralstonia eutropha NH9, which regulates an operon involved in the degradation of chlorocatechols (2). The crystal structure shows that CbnR assembles, as a dimer of dimers, into a homo-tetramer, which corresponds to the oligomeric state that has been reported for a number of other LTTRs (8,9,11,12). In contrast to full-length LTTRs, several regulatory domain structures of LTTRs have been described, including CysB (3), BenM, CatM (4) and Cbl (13). Examination of these structures has confirmed earlier mutagenesis data (11,14,15) that the site of effector interaction is located at the interface between the two subdomains of the C-terminal domain, termed RD-I and RD-II (4,5). Interaction with effectors has been shown to be associated with a conformational change in the C-terminal domain though how this is transmitted to the DBD is not known (4,5).
Thus, despite the central role of LTTRs in the regulation of numerous metabolic processes in bacteria, their mechanism of action remains poorly understood at a structural level. To address the relationship between LTTR structure and function, we have focused on contact-regulated gene A (CrgA), an LTTR from Neisseria meningitidis, which has been implicated in host–pathogen interactions. CrgA was initially identified as being induced upon contact of N. meningitidis with human epithelial cells (16) and appears to be part of a group of genes that are co-ordinately upregulated during initial adhesion (17). CrgA is an autorepressor and activates the expression of the divergently orientated gene, modulator of drug activity gene (mdaB), a putative quinone NADPH oxidoreductase, suggesting that it may be involved in the response of Neisseria to oxidative stress (18,19). Here, we report the crystal structure of CrgA representing the second full-length structure of an LTTR to be described. Unexpectedly, the structure of CrgA showed the assembly of the protein into an octameric ring-like structure, in marked contrast to the tetrameric form of CbnR, which has become accepted as the usual oligomeric state of LTTRs. Cluster analysis of LTTR sequences showed that CrgA was associated with a distinct subset of LTTRs and may therefore define a novel subclass of regulators that function as octamers. Non-denaturing mass spectrometry (MS) and sedimentation velocity analytical ultracentrifugation (AUC) were used to analyze the stoichiometry and relative stability of apo CrgA and holo CrgA in complex with its DNA target. On the basis of the stoichiometry of CrgA:DNA complexes detected and the octameric crystal structure, we propose a model of how CrgA interacts with its DNA target sequence and discuss the wider significance of these findings for transcriptional regulation by the LTTR family.
MATERIALS AND METHODS
Crystallography and structure solution
The crystallization of CrgA and data collection methods have been reported previously (20). The structure of the selenomethionine derivative of the regulatory domain (residues 89–300) of CrgA was determined by multiple-wavelength anomalous dispersion (MAD) methods. Crystals of RD–CrgA were grown from protein at 3.9 mg ml–1 in 20 mM Tris pH 7.5, 200 mM NaCl, 1mM TCEP, using an additive screen (Hampton Research) optimization procedure in 200 mM ammonium acetate, 25% (w/v) polyethylene glycol 3350, 0.1 M bis–Tris pH 5.5 (21). The SHELX program suite was used to solve the selenium substructure (22) from data collected from Crystal I. SOLVE/RESOLVE (23) were used for the refinement of selenium positions and phase extension to a resolution of 2.3 Å using data collected from a second crystal. The model was built manually using COOT (24) and refined with CNS (25) using simulated annealing and positional refinement with main chain NCS restraints followed by individual isotropic B factor refinement. The final stages of refinement were carried out with REFMAC (26) from the CCP4 Suite (27) using TLS followed by restrained refinement. The structure validation programs PROCHECK (28) and RAMPAGE (29) were used to assess the refined structures. The final refined model has an R factor of 20.1% (Rfree 25.8%) with all residues in the allowed regions and 99.2% of residues in the favored region of the Ramachandran plot.
The FL–CrgA protein was concentrated to 8.2 mg–1 in 20 mM citrate, 200 mM NaCl supplemented with 200 mM NDSB-256 (Hampton). Crystals of form A were grown in 10% (v/v) 2-methyl-2-4-pentanediol in 0.1 M sodium acetate pH 5.0. Crystals of form B were grown in 0.4 M ammonium sulfate, 0.8 M lithium sulfate and 0.08 M tri-sodium citrate pH 5.6. The full-length structure of CrgA (Crystal form A) was determined to a resolution of 3.2 Å by molecular replacement using MOLREP (30). A native Patterson and MOLREP indicated the presence of a pseudo-translation. Using the refined regulatory domain of CrgA as the model an initial solution containing six molecules was found. Residues 1–87 from the CbnR structure (PDB code 1IZ1) was used in a further molecular replacement experiment to locate four DBD pairs of CrgA. The two remaining regulatory domains were placed manually using the position of the DBD pairs as a guide. Each CrgA monomer was split into three NCS restraint groups, DBD, LH and regulatory domain. Density modification and averaging was carried out with GAP (Jonathan M. Grimes and David I Stuart, unpublished program). The model was refined using rigid body refinement and then subjected to iterative rounds of manual rebuilding using COOT and TLS and restrained refinement in REFMAC. The final model has an R factor of 21.8% (Rfree 28.6%) with >95% of the residues in the favored region and 0.3% in the disallowed regions of the Ramachandran plot (29).
The structure was determined for crystal form B using MOLREP with a dimeric unit of Crystal form A as the model. After rigid body refinement with REFMAC the R factor was 29.3% (Rfree 40.0%) for data between 3.8 and 30 Å. No further refinement was carried out due to the lower resolution.
The data collection and refinement statistics are presented in Tables 1 and 2. Representative electron density for the refined models of CrgA, are shown in Supplementary Figure S5.
Table 1.
Diffraction data and refinement statistics for the regulatory domain of CrgA
| SeMet RD-CrgA | I |
II | ||
|---|---|---|---|---|
| Data collection | Peak | Remote | Inflection | Peak |
| X-ray source | BM14 | BM14 | ||
| Detector | MAR225 CCD | MAR225 CCD | ||
| Space group | P21 | P21 | ||
| Unit-cell parameters | ||||
| a (Å) | 51.59 | 51.64 | ||
| b (Å) | 65.49 | 65.58 | ||
| c (Å) | 63.31 | 63.50 | ||
| β (°) | 105.27 | 105.02 | ||
| Wavelength (Å) | 0.9780 | 0.9065 | 0.9785 | 0.9795 |
| Resolution range (Å) | 50.0–2.76 | 50.0–2.29 | ||
| Last resolution shell (Å) | 2.86 –2.76 | 2.37–2.29 | ||
| Unique reflections | 10 553 (1058) | 8620 (235) | 8096 (167) | 15 951 (636) |
| Completeness (%) | 99.9 (99.5) | 80.8 (22.1)a | 75.9 (15.7)b | 85.8 (34.8)c |
| Redundancy | 7.4 (6.6) | 4.0 (1.5) | 3.5 (1.3) | 4.1 (1.8) |
| Average I/σ (I) | 20.1 (4.2) | 14.1 (3.3) | 13.2 (3.8) | 18.0 (2.1) |
| Rmerged | 0.108 (0.422) | 0.097 (0.286) | 0.097 (0.214) | 0.073 (0.299) |
| d”/σe | 2.48 (1.27) | 1.38 (0.87) | 1.38 (0.91) | |
| Refinement | ||||
| Resolution (Å) | 30–2.30 | |||
| No of reflections | 14 781 | |||
| Rwork/Rfree | 20.1/25.8 | |||
| Number of atoms | ||||
| Protein | 3139 | |||
| Water | 203 | |||
| Other | 2 | |||
| Rmsd bonds (Å) | 0.006 | |||
| RMSD angles (°) | 1.0 | |||
| B Wilson (Å2) | 34 | |||
| Mean B factor | 30.7 | |||
| Protein | 30.5 | |||
| Water | 33.2 | |||
| Other | 39.6 | |||
| Ramachandran plot | ||||
| Most favored (%) | 99.5 | |||
| Allowed (%) | 0.5 | |||
Values in parentheses are for data in the highest resolution shell.
a,b,cData were processed into the corners of the detector and are essentially complete to 3.5 Å, 3.8 Å, 2.7 Å, respectively.
dRmerge = ∑hkl ∑i | Ii(hkl) –〈I(hkl) |/∑hkl ∑i Ii(hkl).
eAnomalous signal/noise ratio as calculated by SHELXC, with data from resolution 50–8.0 Å and 5–4.2 Å respectively.
Table 2.
Diffraction data and refinement statistics for full-length CrgA
| Native FL-CrgA | Crystal form A | Crystal form B |
|---|---|---|
| X-ray source | ID14–4 | |
| Space group | P212121 | P21 |
| Detector | Q315r ADSC CCD | |
| Unit-cell parameters | ||
| a (Å) | 104.64 | 123.17 |
| b (Å) | 119.09 | 204.99 |
| c (Å) | 250.20 | 124.80 |
| β (°) | 93.10 | |
| Wavelength (Å) | 0.9765 | 0.9765 |
| Resolution range (Å) | 30.0–3.20 | 30.0– 3.80 |
| Last resolution shell (Å) | 3.31–3.20 | 3.94–3.80 |
| Unique reflections | 43 934 (4398) | 60 401 (5853) |
| Completeness (%) | 83.8a (85.3) | 99.6 (97.2) |
| Redundancy | 3.7 (2.7) | 3.7 (3.3) |
| Average I/σ(I) | 17.8 (1.66) | 10.4 (4.10) |
| Rmergeb | 0.061 (0.512) | 0.138 (0.365) |
| Refinement | ||
| Resolution (Å) | 29.9–3.2 | |
| No of reflections | 40 596 | |
| Rwork/Rfree | 21.8/28.6 | |
| Number of atoms | 18304 | |
| Rmsd bonds (Å) | 0.006 | |
| RMSD angles (°) | 0.9 | |
| B Wilson (Å2) | 96.5 | |
| Mean B factor | 83.5 | |
| Ramachandran plot | ||
| Most favoured (%) | 95.6 | |
| Allowed (%) | 4.2 | |
| Disallowed (%) | 0.3 |
Values in parentheses are for data in the highest resolution shell.
aRmerge = ∑hkl ∑i | Ii(hkl)–〈I(hkl) |/∑hkl ∑i Ii(hkl).
bResolution shell 3.6–3.79 Å is 27.2% complete due to ice rings.
The atomic coordinates of the regulatory domain and full-length CrgA structures have been deposited in the Protein Data Bank under the accession codes 3HHF and 3HHG, respectively.
Bioinformatics and structure analysis
Structural neighbors of CrgA were identified using Dali (31). To find suitable structures for modeling the interaction of CrgA with DNA, all the structures in the Protein Data Bank solved by X-ray crystallography containing both protein and DNA were selected for a pair-wise protein structural comparison using secondary-structure matching (SSM) (32). The interface properties were analyzed using protein interfaces, surfaces and assemblies service PISA at European Bioinformatics Institute (33). The residue conservation of CrgA was calculated using Consurf (34). The effector pocket was analyzed with the Computed atlas of surface topography of proteins CASTp (35). The structural figures were produced using Pymol (36).
Preparation of CrgA, CrgA–DNA complex and EMSA
CrgA protein was prepared as previously described in 20 mM Tris pH7.5, 200 mM NaCl (20). DNA of the 63-bp CrgA sequence footprint (ACTTTATAATTTAAAAGTG CAAAAATAAGAAAACACTTTTGCGTCAAATGAAATAATCAGATG), the 30-bp CrgA site 1 (ACTTTATAATTTAAAAGTGCAAAAATAAGA) the 30-bp CrgA site 2 (CACT TTTGCGTCAAATGAAATAATCAGATG) (18) and the 32-bp control DNA (AAATTTTCAAAATCAAACGAGCTCATTACAAC) were prepared from oligonucleotides, either PAGE purified for MS or OligoGold standard for EMSA, purchased from Eurogentec. A fluorescence-based EMSA Kit (Invitrogen) was used according to the manufacturer's instructions (37) to analyze the binding of CrgA to DNA. EMSA reactions of 10 µl were made up in 150 mM KCl, 0.1 mM dithiothreitol, 0.1 mM EDTA, 10 mM Tris, pH 7.4. The amount of DNA in each reaction was kept constant at 2.5 pmol for experiments with CrgA or 1.5 pmols for experiments with the R55Q CrgA protein. The molarity of CrgA, calculated for an octameric species was varied up to a 4× molar excess over the amount of DNA. After the addition of 2 µl of 6× EMSA-loading buffer, the samples were loaded on to a non-denaturing polyacrylamide 4–20% TBE gel and electrophoresed in 0.5× TBE running buffer at ∼120 V for 1.5 h. The gel was stained for DNA with SYBR Green EMSA gel stain (Invitrogen).
The CrgA:DNA complex was formed by elution of CrgA in the presence of excess DNA of the 63-bp CrgA sequence footprint, from a 16/60 Hiload superdex 200 column (GE healthcare).
Mass spectrometry
Nano-electrospray MS and IM-MS of CrgA, DNA and CrgA:DNA complexes were acquired using either a Qstar XL (MDS Sciex, Applied Biosystems, Concord, ON, Canada) or a Synapt HDMS (Waters, Manchester, UK) under conditions optimized for the transmission of non-covalent protein complex interactions using previously described protocols (38,39). Apo and holo protein was buffer exchanged using 5 kDa molecular weight cut-off Vivaspin columns (Vivascience) into 200 mM ammonium acetate, pH 6.8, in order to mimic the ionic strength of the original buffer. DNA was exchanged into 200 mM ammonium acetate using micro bio-spin 6 columns (BioRad). Typical instrument parameters, in positive ion mode, on the Qstar XL for apo and holo CrgA were: ion spray voltage 1.2–1.65 kV, de-clustering potential 200–250 V, focusing potential 200 V, de-clustering potential 2 15V, quadrupole voltage (Q0) 100–200 V, collision gas (CAD) 12, ion release delay 6 and ion release width 5. Experiments were acquired at instrument base pressure of 6.5 mbar.
Typical instrument parameters, in positive ion mode, on the Qstar XL for apo DNA were: ion spray voltage 1.2 kV; de-clustering potential 100 V; focusing potential 150 V; de-clustering potential 2 15 V; quadrupole voltage (Q0) 50 V; collision gas (CAD) 3; ion release delay 6 and ion release width 5.
Typical instrument parameters for IM-MS, using a Synapt HDMS, in positive mode, were: capillary voltage 1.7 kV; sample cone 40 V; extractor cone 0 V; trap collision energy 10 V; transfer collision energy 10 V; bias voltage 20V; backing pressure 4.3 mbar; trap pressure 5.3 × 10−2 mbar; T-wave drift cell pressure 5.1 × 10−1 mbar and ToF pressure 1.8 × 10–6 mbar. For activation experiments voltages were increased gradually to sample cone 200 V (in 20 V steps), extraction cone 10 V (in 2 V steps), trap collision energy 200 V (in 20 V steps) and bias voltage 180 V (in 20 V steps).
Data were acquired and processed using Analyst QS (Applied Biosystems, Concord, ON, Canada) and Masslynx software 4.1 with driftscope (Waters, Milford MA, USA). All spectra are shown with minimal smoothing. Spectra were calibrated externally using a 100 mg ml–1 solution of caesium iodide solution.
Analytical ultracentrifugation
Sedimentation velocity experiments were performed in a Beckman Optima XL-I analytical ultracentrifuge, as previously described (40). The experiment was performed at 40 000 rpm in an An60Ti rotor at 20°C. The partial specific volume used was 0.73 ml g–1. The buffer density was 1 g ml–1 and the viscosity 0.01002 poise. Protein samples were prepared in 20 mM Tris pH 7.5, 200 mM NaCl. The sedimentation profiles of samples of CrgA (1.9 µM) alone and in the presence of an equimolar amount of the 63 bp DNA probe were observed over time using interference optics, since the signal in the absorbance trace was dominated by that from the DNA. Analysis by the c(s,f/f0) method used the software SEDFIT (41,42). The c(s) is a method for model-independent analysis of velocity boundary profiles whereby an apparent distribution of sedimentation coefficients, apparent as a Gaussian distribution, is shown by each sedimenting species or set of species (a set of species if interconverting rapidly on the timescale of the experiment). The Gaussian distribution derives from the effects of diffusion on the apparent sedimentation coefficient, as affected by the changing concentration of the sedimenting species in the boundary. c(s,f/f0) Distributions are two dimensional plots of the relationship between sedimentation coefficient and frictional ratio (f/f0), which can be inferred from the degree of broadening in the boundary. The proportions of species observed with different S values were calculated by integrating the areas under the peaks of two dimensional plots.
The function c(s,f/f0) can then be used to derive functions such as c(s,M), dependence of sedimentation coefficient and weight, and c(s,Rs), dependence of sedimentation coefficient and Stokes radius. The first 30 interference scans were analyzed for both samples, allowing for frictional ratios up to 2.5. A resolution of 50 was used for the calculations. Sedimentation coefficients were computed for atomic models using the program HYDROPRO (43).
RESULTS
Overall structure of CrgA
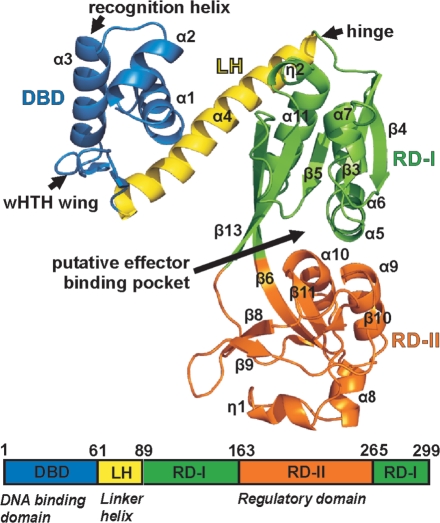
We first solved the structure of the regulatory domain of the protein (residues 89–300) using selenomethionine-substituted protein by multiple wavelength anomalous dispersion to 2.3 Å resolution. This model was then used to determine the structures of the full-length protein in both orthorhombic and monoclinic crystal forms by molecular replacement to resolutions of 3.2 Å and 3.8 Å, respectively. As expected, each CrgA subunit of the full-length protein comprises an N-terminal DBD, connected via a long LH to a C-terminal regulatory domain (Figure 1). The DBD contains a wHTH motif (α1, α2, α3, β1 and β2) that is comparable to that of other wHTH superfamily members superimposing with a root mean-square deviation (RMSD) of 2.3–3.0 Å (for 53–58 common Cα residues) (Supplementary Figure S1). As expected the position of the α2–α3 (residues 21–41) is extremely well conserved while the position and length of the wing vary within the family. The regulatory domain contains two subdomains RD-I (residues 89–162 and 265–299) and RD-II (residues 163–264). RD-I consists of a core of four parallel and one anti-parallel β strands (β3, β4, β5, β6 and β13) sandwiched between α helices (α5–α6, α7 and α11) and a 310 helix (η2). The β6 and β13 strands cross over between the two subdomains. RD-II contains three α helices (α8, α9 and α10), a 310 helix (η1) and a series of β strands (β7–β12) (Figure 1). The putative effector pocket, located at the interface of RD-I and RD-II, is large with a solvent-accessible surface area of 603 Å2 and a volume of 1072 Å3 (35). The residues lining the pocket are predominantly non-polar (66%) including four phenylalanine residues (residues F164, F193, F242 and F268) suggesting that the potential effector is largely hydrophobic in nature. The only clue as to the identity of the effector, which we presume modulates the activity of CrgA, is the observation that treatment of Neisseria with α-methylene–γ-butyrolactone (MBL) has been shown to induce CrgA-dependent expression of mdaB (18). However in E. coli MBL is not a direct substrate of the MdaB enzyme and in a thermal shift (ThermoFluor) assay, that can be used to assess ligand binding (44), we observed no evidence of a direct interaction between MBL and CrgA suggesting that MBL is acting indirectly (data not shown).
Figure 1.
CrgA fold and domain architecture.
Novel octameric assembly of CrgA
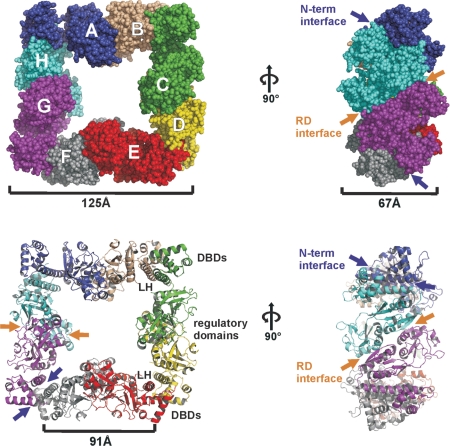
Although the fold of each CrgA subunit is similar to that observed for CbnR, the overall assembly of the complex is strikingly different. CbnR is tetrameric whereas CrgA assembles into square-like hollow octameric rings with dimensions of 125 × 125 × 67 Å. The regulatory domains are sandwiched between the DBDs that are located in pairs (DBD pairs) at the four corners of the disc. Viewed from above, the hollow of the octamer has a cross-like shape with a distance of ∼95 Å between opposing LHs and ∼50 Å between opposing regulatory domains (Figure 2). On first inspection the octamer, which occupies one asymmetric unit, appears to have 4-fold symmetry. However, there are significant differences in the orientation of the LHs and DBDs within the eight subunits, which break this apparent symmetry. Due to the nature of the assembly, each CrgA subunit contacts just two of the other subunits. Each subunit forms an interface with one adjacent subunit, principally through their equivalent N-terminal domains, (residues 1–89, N-term interface) and to a second subunit, through their regulatory domains (RD interface) (Figure 2). The sizes of these two interfaces are similar, each burying an average surface area of ∼1300 Å2. The RD interface contains two sets of equivalent interactions, between residues of the RD-I and RD-II on the adjacent chain while the N-terminal interface is primarily formed by dimerization of the LHs through intermolecular anti-parallel coiled coil interactions (Figures 4A and 6D).
Figure 2.
Octameric assembly of CrgA found in the full-length structure.
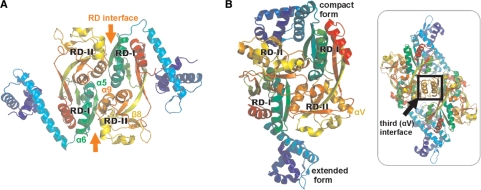
Figure 4.
Comparison of CrgA and CbnR. (A) Dimeric unit of CrgA. (B) Dimeric unit of CbnR (PDB code 1IZ1) and CbnR tetramer (Insert).
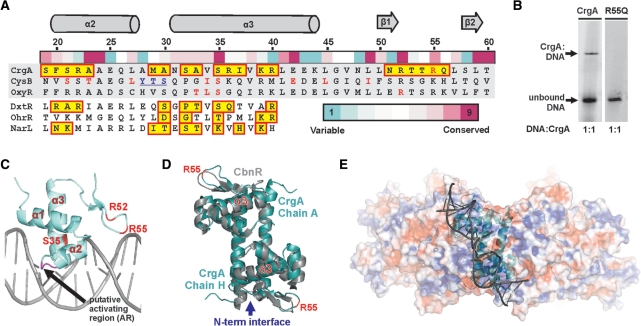
Figure 6.
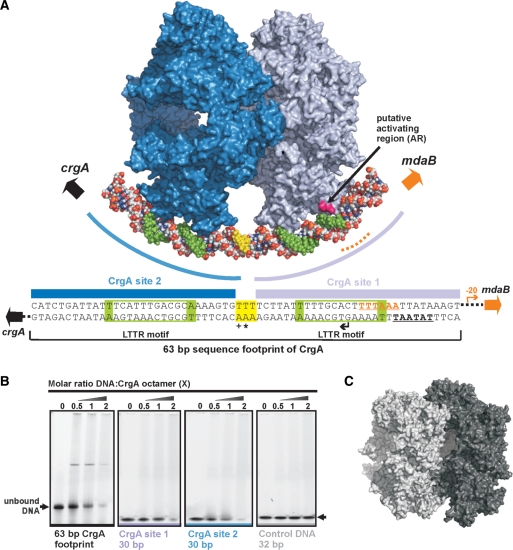
DNA binding and recognition in CrgA. (A) Sequence alignment of the DBD of CrgA with two further LTTR family members [E. coli, CysB and OxyR) and the three HTH proteins (DtxR from Corynebacterium diphtheriae (56), OhrR from Bacillus subtilis (55) and NarL from E. coli (54)] used for modeling. Residues boxed in yellow, contact the DNA in the co-structures of DtxR (PDB code 1F5T), OhrR (PDB code 1Z9C) and NarL (PDB code 1ZG5) or are predicted to contact DNA in CrgA. Residues important for DNA binding identified by mutational analysis in CysB, OxyR and CrgA are shown in red (11,57). Residues underlined form the activating region identified in CysB (64). The residue conservation scores of CrgA obtained with Consurf, based on 70 LTTRs from the Swissprot database, are shown above the alignment. Only residues of the structurally equivalent α2 and α3 are shown for DtxR, OhrR and NarL. (B) EMSA on CrgA wild type (2.5 pmol) and CrgA R55Q mutant (1.5 pmol) mixed in a 1:1 molar ratio with the reported 63-bp CrgA DNA footprint (18,48). (C) One DBD of CrgA with modeled DNA showing selected residues discussed in this report. (D) Structural comparison of a DBD and LH pair from CrgA and CbnR (Cα RMSD 1.1 Å2 for 86 equivalent residues). R55 of CrgA is labeled for orientation purposes in panel E. (E) Electrostatic surface potential of the CrgA octamer showing one DNA-binding pair with modeled DNA (DxtR, PDB code 1F5T).
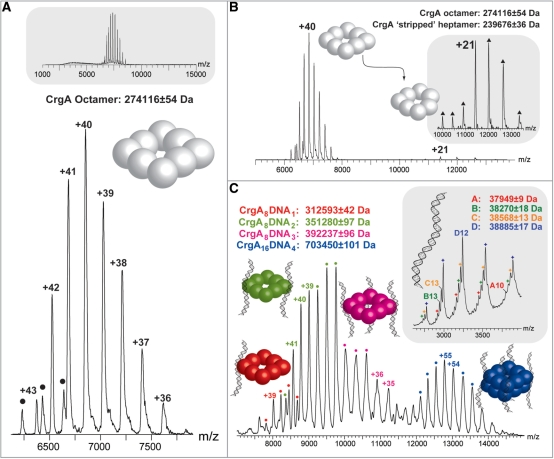
The question arises as to whether the octameric assembly of CrgA observed in the crystal structure is relevant to the protein in solution and hence the biologically active form of the protein. Therefore, the protein was analyzed by non-denaturing nano-electrospray mass spectrometry (MS) to determine the precise oligomeric state of CrgA in solution,. The results showed an intense and well-resolved charge state series, from +36 to +43, in the 6500–8500 m/z range, which corresponded to an octameric assembly for CrgA (Figure 3A). The insert in Figure 3A confirms the presence of one dominant oligomeric state over a large m/z range. Additionally, a small amount of hexameric CrgA was detected by MS, (hexamer peaks are identified by black circles). The measured mass of the octamer was 273 542 ± 25 Da, which is slightly greater than the predicted mass of 272 919 Da. This discrepancy in mass is explained by the inability to achieve complete species desolvation of the complex with the ‘soft’ ionization methods required to maintain subunit interactions. This results in the retention of water and buffer molecules within the protein complex ions that are detected (45). The octameric stoichiometry was confirmed by activating the complex using collision-induced dissociation (CID) (46). This results in a proportion of the octamer dissociating into a heptamer as a monomer is unfolded and ejected from the intact assembly carrying with it a disproportionate amount of the overall charge of the complex (Figure 3B insert). Due to the greater activation of the complex during CID experiments, enhanced peak resolution is achieved, enabling a more accurate mass measurement and confirming the octameric assembly of CrgA. As described below, the results of AUC experiments also confirmed the octameric state of the CrgA protein in solution (Figure 5A).
Figure 3.
Analysis of CrgA and CrgA–DNA complexes by non-denaturing mass spectrometry. The predominant charge state series in the nano-electrospray mass spectrum shown in (A) corresponds to the +36 to +43 charge states of CrgA (3 μM) with an octameric stoichiometry. A small amount of hexameric CrgA was also observed (indicated by black circles) and the insert at the top of the panel confirms there are no other stoichiometries over a wide m/z range. (B) The CrgA octamer under activating MS conditions, resulting in the formation of a ‘stripped’ heptameric species (insert). (C) The mixed population of CrgA–DNA complexes observed by MS, these include CrgA8DNA1, CrgA8DNA2, CrgA8DNA3 and CrgA16DNA4. A spectrum of DNA (80 μM) alone is shown as an insert to (C). In this insert four charge state series were identified with charges states from +10 to +14. The full length DNA corresponds to series D, while the smaller masses (series A–C) likely correspond to various nucleic acid truncations of the DNA.
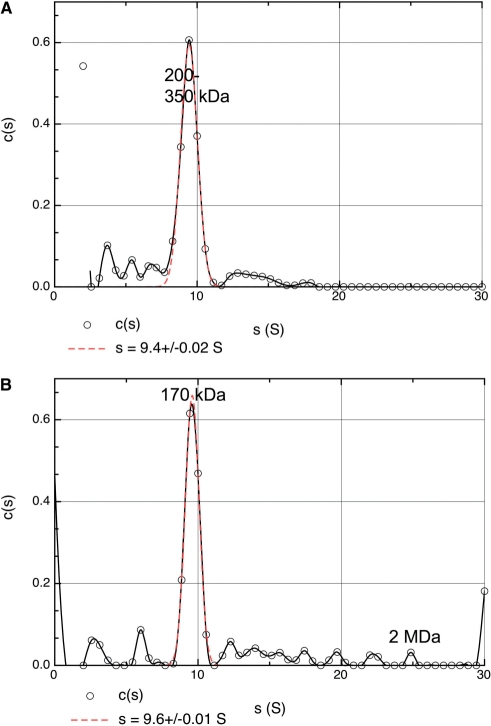
Figure 5.
Plots of sedimentation coefficient population distribution (c(s)) against s. Each clearly defined peak represents a single species, the main peak in both cases being fit with a Gaussian to allow for precise calculation of the s value (red dashed line). (A) CrgA. (B) CrgA with DNA.
Comparison of the assembly of CrgA and CbnR
The most notable difference between CrgA and CbnR is the orientation of their DBD and LH. In all eight subunits of CrgA the LH folds back toward the regulatory domain whereas in CbnR in the compact form (chains A and P), the LH folds back toward the regulatory domain. While in the extended form (chains B and Q), the LH extends away from the regulatory domain with a rotation of ∼80° (Figure 4). The hinge region of LTTRs, first identified in CbnR, largely controls the orientation of the DBD and LH. Cross-comparison (Supplementary Figure S2) of the eight chains of CrgA indicates that there is a significant amount of rotational freedom of the LH around the hinge but that there is no large pivot movement of the regulatory domain away from the DBD–LH as observed between the compact and extended forms of CbnR (2). In CrgA, the hinge (H88, E89, I90 P91, Q92 and G93) contains a PXG motif whereas in CbnR, there are two glycine residues (GXXG) in this region. The more constrained geometry of a proline compared to a glycine residue in the hinge of CrgA may cause it to have less structural freedom than CbnR, which can adopt two markedly different conformations. The fact that the PXG motif is conserved within close homologs, though not within the LTTR family as a whole, suggests that it may be functionally important.
Significantly, in CbnR there is a third type of interface formed between the two αV helices (residues S201 to H212) of RD-II in chains A and P (both in the compact form), which stabilizes the central core of the tetramer (2). In CrgA, this helix is replaced by a β strand (β8) preventing a comparable interface forming in this protein (Figure 4B). CrgA is atypical since 14/15 closest structural neighbors identified using a Dali search contain a helix at this position.
To probe the relationship between CrgA and other members of the LTTR family we generated a cluster map of 4843 LTTR sequences with the program CLANS which uses pair-wise BLAST similarity scores (47). The 2D cluster map contains four major groups with a large main cluster of 2928 sequences containing the tetrameric LTTR CbnR in addition to other characterized LTTRs including MetR and OxyR. CrgA forms part of a smaller cluster of 921 (19%) protein sequences, which includes the close E. coli homolog YafC (59% sequence identity) as well as proteins annotated as AaeR and PtxR (Supplementary Figure S3). This partitioning of LTTR sequences indicates that CrgA is not unique and we suggest that the octameric assembly of CrgA may be shared with other LTTRs within this cluster.
Although the overall oligomeric states of CrgA and CbnR are different, there are parallels in the way that the CrgA octamer and CbnR tetramer are held together. The DBD–LH module of these two LTTRs is particularly well conserved (Cα RMSD of 1.1 Å for 86 equivalent residues) and probably within the wider family (Figure 6D). Dimerization of the coiled coil controls the overall spacing and position of the two wHTH motif within an LTTR DNA-binding pair and will thus impart some of the general characteristics of an LTTR DNA-binding site, for example the length of the palindromic bases recognized. The RD interface does not show the same level of structural conservation between the two structures. Although 25/35 of the equivalent residues involved in formation of the CrgA RD interface are also found at the CbnR RD interface, in CbnR the two RD-IIs also interface with each other through residues 248–253 whereas in CrgA they are clearly separate.
MS reveals a double octamer CrgA–DNA complex
Previously, DNase I protection assays of the intergenic region between crgA and the divergently transcribed gene, mdaB, identified two 30-bp CrgA DNA-binding sites (crgA sites 1 and 2) separated by 3 bp, each containing the characteristic LTTR-binding motif, T-N11-A (48). We have confirmed the specific interaction between purified CrgA and this 63-bp CrgA footprint in a gel-shift experiment. As observed by Deghmane et al. (16), there is not a complete shift of the probe by CrgA indicating that binding under the conditions of electrophoresis is relatively weak (Figure 6B). In order to determine the precise stoichiometry of the interaction between CrgA and the target DNA, MS was carried out on the isolated CrgA–DNA complex. A mixed population of CrgA–DNA complexes was detected. It was found that a single CrgA octamer could bind one, two or three lengths of DNA (Figure 3C). Furthermore, in the presence of DNA a CrgA 16mer was detected bound to a maximum of four DNA strands (Figure 3C). This double octamer species was not observed in the apo form suggesting that the presence of the DNA is required to stabilize an interaction between the two CrgA octameric rings. The peak intensities give an indication of the relative populations of the different protein–DNA species in solution; however, particularly for complexes with a similar composition, absolute solution populations maybe affected by subtle differences in the ionization properties as a result of each CrgA–DNA complexes’ unique stoichiometry (49,50). It is interesting that in the 16mer arrangement, all four DNA-binding sites for each octamer are occupied suggesting some co-operativity of DNA binding. In fact, as indicated by modeling, once one DNA binds orthogonally to two CrgA octamer rings the DNA-binding sites at the remaining three corners of each octamer are aligned such that it is more favorable for three additional strands to bind (see below). A nano-electrospray mass spectrum of the DNA strand alone is shown as an insert to Figure 3C. The predicted mass of the DNA is 38 788 Da and is in close agreement to the measured mass of 38 885 ± 1 Da (series D) calculated using the +14 to +10 charge states. The three additional charge states series (A–C) are likely to be the result of nucleic acid truncations.
In order to investigate further the effect of DNA binding on the CrgA protein ion mobility mass spectrometry (IM-MS) was used to compare the apo CrgA octamer with a holo CrgA octamer binding two DNA stands. IMMS separates ions not only by their m/z mass and charge but also by their drift time mobility through a nitrogen filled T-wave drift cell which is influenced by the species shape and size (51). Additionally, sufficient gas phase activation of a ring-like structure in a mass spectrometer typically results in structural collapse of the quaternary topology. This is then followed by an expansion of the protein complex. Minimal activation however allows the transmission of a ring-like protein complex with little compromise to the presence of the cavity (52). In the case of the apo CrgA octamer, a reduction and then increase drift time was observed for the lowest charge state ion (+28) of the protein complex (Supplementary Figure S4). This is consistent with a small collapse, assigned to the loss of the cavity, followed by much greater expansion as the complex is activated and unfolded, The holo CrgA octamer with two DNA strands exhibits a similar collapse for its lowest charge state (+30) but to a smaller degree than the +28 charge state of the apo CrgA octamer. The difference between the initial drift time and minimum drift time for the most collapsed structure, normalized for the charge of the ion, is smaller for the holo (22.80 ms) than for the apo (28.84 ms) form of the CrgA octamer. Moreover, the energy required to retain the most compact state for the DNA-bound CrgA octamer is greater for the holo form compared to the apo CrgA. Both the higher activation required to collapse the structure and lower level of collapse observed for the holo CrgA suggest that the DNA contributes to stabilization of the CrgA octamer. Moreover, these results suggest that the ring-like conformation of CrgA is maintained upon DNA binding.
Comparison of CrgA and CrgA-DNA complexes by analytical centrifugation
To complement the analysis by MS, sedimentation velocity experiments were carried out on CrgA either alone or in the presence of the same DNA probe used in MS. In the absence of DNA (Figure 5A), the sedimentation profile was dominated by a species of 9.4 S, accounting for 90% of the observed molecules, followed by a small tail at higher s values. This tail did not consist of well-defined peaks and indicated a range of interactions, which were either non-specific or very transitory. In the presence of DNA, a major peak of 9.6 S was observed together with a series of species, which were well resolved and extended to 25 S. The proportion of observed molecules in the 9.6 S peak was 74%. Calculation of apparent weight c(s,M) distributions for both samples (Supplementary data Figure S6) showed that the 9.4 S CrgA species observed in the absence of DNA corresponded to a diffused peak between 200 and 350 kDa whereas the major 9.6 S species in the sample with DNA gave a well-defined peak at 170 kDa (Figure 5). Peaks at higher values were also observed including a clear non-trivial peak at 2 MDa (Supplementary data figure S6) corresponding to the 25 S species, referred to above. The calculated sedimentation coefficients for an LTTR tetramer and octamer are 6.65 S and 11.03 S based on the atomic structures of CnBr [2] and CrgA, respectively. Therefore, we conclude that the major species observed for CrgA subjected to AUC in both the absence and presence of DNA is most likely to be an octamer consistent with the results of MS, described above. The slower sedimentation of CrgA, ∼9.5 S with or without DNA, is probably due to additional hydrodynamic drag resulting from the hollow nature of the octamer that is not allowed for in the atomic model-based calculation of the s value. Further drag is presumably introduced by the bound DNA since the added DNA does not increase the overall sedimentation coefficient though it does significantly improve the definition of the peak in the c(s,M) distribution.
Most interestingly, we observed that the addition of DNA resulted in a proportion (∼25%) of CrgA molecules forming higher molecular weight/sedimentation coefficient species. The fact that these oligomers were only observed in the presence of DNA strongly suggests that they were induced by DNA binding (Figure 5). They include species that presumably correspond to the hexadecamer observed by MS and also higher order assemblies up to 25 S. The calculated sedimentation coefficient for the CrgA hexadecamer is 17.75 S though the experimental S value may well be smaller as a result of extra hydrodynamic drag referred to above for the octamer. Therefore it is not possible to unambiguously designate one of the observed peaks as the hexadecamer. The failure of MS to detect any oligomers larger than the hexadecamer may be due to a combination of their low abundance and/or inefficient ionization compared to the octamer and hexadecamer. Differences in the size range of oligomers detected by MS compared to AUC have been reported in a study of amyloid fibril formation (53).
Interaction of CrgA with its DNA target site
The CrgA residues likely to contact the DNA were identified using modeling with the DNA co-structures of DtxR, OhrR and NarL (Figure 6A) (54–56). M29, A30, S32, A33, S35, R36, I37, K39 and R40 in the turn preceding or within the recognition helix (α3) are predicted to contact the major groove and residues of the wing are predicted to contact the minor groove of the target DNA. The conserved arginine residues present in the wing appear to have an important role in DNA binding in LTTRs. An R50 (equivalent to CrgA R52) mutant in OxyR and additionally an R55Q mutation in CrgA abolishe all DNA binding on electrophoretic mobility shift assay (EMSA) (Figure 6B and C) (57). In addition residues N4-E7 may also make contact with the DNA, though the nature of these interactions are difficult to predict as the orientation of the α1 helix is not well conserved within the wHTH family.
The DBD pairs of CbnR and CrgA are highly similar suggesting that individual DBD pairs of LTTRs bind to DNA in a similar way (Figure 6D). The overall length of DNA bound by a single LTTR DBD pair would be in the region of 70–80 Å (20–24 bp) with a distance of ∼33 Å (equivalent to 1 helical turn of DNA) between the two recognition helices (Cα of S32) and ∼63 Å between the two wings (Cα of R55). Almost all LTTRs bind to the T-N11-A motif and the more highly conserved residues of the wHTH are likely to contact these T/A bases (57). It follows that the 11-bp spacing of these bases indicates that the molecular determinant for LTTR consensus selectivity is likely to lie within the α-helical core of the DBD pair and not in the outer wing regions. A possible candidate for conferring this selectivity is the serine/threonine at residue 35, one of the most highly conserved residues in the recognition helix of the LTTR family (Figure 6A) and required for DNA binding in OxyR, CysB and GcvA (57–59). Furthermore, modeling suggests that in a DBD pair of CrgA, the two S35 residues would be capable of making base contacts to the major groove of DNA on opposite strands with ∼11 bp in between (Figure 6C). Perhaps, further indication of a role for this Ser/Thr in LTTR motif recognition is MauR, an LTTR that unusually contains an alanine rather than a Ser/Thr at this position binds to DNA lacking the classical LTTR motif (60).
The calculated electrostatic potential of the outer surface of the octamer showed localized regions of both positive and negative charge. Taken together, with the orientation of the wHTH motif within CrgA suggests that, unless there is a significant structural rearrangement upon DNA binding, the DNA would bind across the octameric disc at an angle of ∼60° (axis between adjacent corners) and not directly around the periphery of the disc (Figure 6E). The distance between DBD pairs (measured between S32) at adjacent corners is 91 Å. Therefore ∼26 bp or more of bridging DNA would be required to link these adjacent DBD pairs together, if they were to engage DNA sequence elements in the same stretch of DNA. However, the spacing of the two LTTR consensus sites within the 63-bp CrgA sequence footprint is just 15 bp, equivalent to ∼50 Å of DNA and would not be long enough to bridge two DBD pairs of a single CrgA octamer (18,48). We conclude from these observations that the two binding sites, which make up the 63-bp sequence footprint of CrgA cannot be bound by the same octamer.
Model of the CrgA–DNA complex
As mentioned above, the published DNA footprinting data for CrgA show that the protein interacts with two adjacent LTTR-binding motifs in the CrgA/MdaB promoter region (48). We have shown that CrgA assembles as an octamer and have argued from an examination of the crystal structure that geometric constraints preclude the simultaneous binding of both sites by the same CrgA octamer. The MS of CrgA–DNA complexes showed the binding of single octamers to DNA and sedimentation velocity experiments confirmed that single octamers are the predominant species observed in the presence of DNA. We conclude that in this case, only one of the two LTTR-binding motifs in the DNA are involved in CrgA binding. Consistent with this, EMSA assays showed that in addition to binding to the 63-bp CrgA DNA footprint, CrgA binds to the individual CrgA site 1 and site 2 sequences (Figure 7B). The mobilities of these 30-bp DNA fragments were clearly shifted by CrgA. No shift was observed for a randomized control oligonucleotide of similar GC base content, indicating that the interaction with the CrgA sites was sequence specific (Figure 7B). In contrast to the EMSA with the 63-bp sequence, no defined band corresponding to a CrgA–DNA complex for these shorter DNA fragments was observed. This possibly indicates that the presence and occupancy of both binding sites is required for the formation of a stable complex in the EMSA. In these experiments, we did observe some material that did not enter the gel, which may correspond to high-molecular-weight complexes. However, given the propensity of the protein to aggregate it could also include non-specifically aggregated material.
Figure 7.
Model of a hexadecameric array of two CrgA oligomers binding to the DNA target site. (A) Two CrgA octamers bound to the 63-bp CrgA DNA footprint (48). The first octamer (blue) binds to CrgA site 1 and the second octamer (gray) to site 2. The LTTR motifs (boxed in green in the sequence) are bound between the two recognition helices of a DBD pair. One strand of the DNA is coloured to represent the sequence elements depicted in the schematic. The LTTR motifs are indicated by green colouring of the DNA. The three base pairs between CrgA site 1 and 2 are shown in yellow. The bases overexposed to DMS methylation (+) or hypersensitive to DNase I (*) are indicated. The predicted transcription and translation start points are represented with bent and broad arrows, respectively. The predicted –10 promoter element for crgA is underlined. The location of the –35 promoter (underlined) element of mdaB is indicated with a dashed orange line. (B) EMSA of purified CrgA binding to its DNA target site. The amount of DNA was kept constant at 2.5 pmol per reaction and the molarity of CrgA was varied as indicated. (C) Asymmetric unit of Crystal form B of CrgA.
The MS of CrgA–DNA complexes showed in addition to the binding of single octamers to DNA, the unexpected stabilization of a double octameric species by DNA binding. Sedimentation velocity experiments also indicated that higher order oligomers of CrgA are induced by the addition of DNA. Intriguingly, in crystal form B of CrgA, two octamers were observed stacked together providing a view of how CrgA could be self-associated (Figure 7C). The DBD pairs of the two octameric rings found in the asymmetric unit were orientated in such a way that both octamers could bind to the same strand of DNA (Figure 7C). These observations lead us to propose a model whereby CrgA interacts with both LTTR-binding sites in the CrgA/MdaB promoter region and which therefore reconciles the octameric assembly state of CrgA and its interaction with DNA revealed by footprinting (48). In this model (Figure 7A) we envisage two CrgA octamers stacked against each other, in a comparable manner to that observed in the second crystal form, with DNA binding through a single DBD pair of the two octamers. The LTTR motifs are aligned between the two recognition helices of each pair with S35 contacting the conserved T/A bases. The model satisfies the 15-bp spacing between the two LTTR motifs and means that the three bases between CrgA site 1 and 2, which are hypersensitive to DNase, do not contact the protein. Our model does not rule out the possibility that individual CrgA octamers interact with the CrgA/MdaB promoter region in vivo as indicated by MS and AUC in vitro. However, as discussed below, the functional consequences in terms of promoter activation/repression under these circumstances would be different to the binding of a double since only single LTTR-binding sites would be engaged at a time.
DISCUSSION
A large variety of both global and specific transcription factors are deployed in bacteria to control gene expression in response to different environmental signals. One of the largest families of such proteins is the LTTRs, which function to both induce and repress transcription. Despite considerable progress in understanding the genetics and biochemistry of LTTRs there remains much to be learnt about the molecular details of how these proteins regulate transcription. In particular, the lack of a strong consensus DNA-recognition sequence has hindered efforts to unambiguously characterize the relationship between LTTRs and their DNA targets. Combining X-ray crystallography, MS and analytical ultracentrifugation, we have elucidated the structure of the LTTR, CrgA from N. meningitidis and have determined the stoichiometry of binding to DNA. Unlike other LTTRs, which form tetramers, we show that CrgA assembles into an octameric array, which can bind up to four 63-bp DNA oligonucleotides. On the basis of these observations, we have developed a model for the interaction of CrgA with its cognate promoter region, which involves the binding of two stacked CrgA octamers to adjacent recognition sites in the target DNA. This contrasts with the current model for tetrameric LTTRs in which the DBDs of a single LTTR located unilaterally on the tetramer are thought to engage adjacent binding sites (2). However, only in the case of CysB from Salmonella typhimurium has direct experimental evidence been obtained to support this stoichiometry (61).
In the non-induced state, the transcription of the LTTR is repressed and the divergent regulated gene is not actively transcribed. In response to some environmental cue, a conformational change in the LTTR, propagated through interactions in the C-terminal domain, results in a change in protein–DNA contact. This appears to be associated with relaxation of the angle of DNA bending within the promoter region and involves, in some cases, translocation of the protein from one binding site to another as observed by a change in length and position of the DNA footprints e.g. OxyR, (62) OccR (63) and AtzR (8). It is presumed that this in turn favors RNA polymerase engagement presumably by relieving some steric hindrance and may involve protein–protein interactions between the LTTR and RNA polymerase. In the regulation of cysP by cysB in E. coli, it has been shown that direct interaction between CysB with the C-terminal domain of RNAPα (αCTD) is required for the activation of transcription both in vitro and in vivo (64). Based on the above considerations and our model for the CrgA–DNA complex, we propose the following mechanism for how CrgA may switch on mdaB expression, while also repressing its own transcription. During initial adhesion between Neisseria and epithelial cells, expression of crgA is upregulated as part of a regulon, which responds to contact with host cells (17). Increased cellular levels of CrgA leads to the occupancy of both the LTTR-binding sites in the crgA–mdaB promoter region. This, in turn, would repress crgA transcription as RNA polymerase would be sterically blocked from accessing the crgA promoter, resulting in self-limiting autoregulated expression of CrgA. Results from DNA protection assays with dimethylsulfate (DMS) and DNase suggest that the promoter DNA undergoes a conformational change upon CrgA binding (48,65). From our model of a CrgA–DNA complex, occupancy of both CrgA-binding sites by the double octamer would cause the DNA to bind inhibiting transcription of mdaB. It is known that expression of mdaB is induced following cell adhesion (66) and also by treatment of cells with MBL, which has been shown to be dependent upon CrgA expression (18). For this to happen CrgA must respond to a further effector signal generated post-adhesion or following exposure to quinones, which induces a conformational change in the protein. This leads to a change in the disposition of the octamers with respect to each other and relaxation of the bound DNA, which in turn leads to the recruitment of RNA polymerase by CrgA and initiation of transcription of the mdaB gene. The proposed sequence of events is necessarily speculative but provokes follow-up experiments including exploring the role of DNA bending in the interaction of CrgA with its target promoters, the possible direct interaction with RNA polymerase and visualization of the assembly of the CrgA-dependent transcription complex. In conclusion, we have shown a new assembly state for the LTTR family of transcription factors, which indicates a role for multimerization in the interaction with DNA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
UK Medical Research Council and the Biotechnology Biological Research Council. MRC Research Studentship (to S.S.); EPSRC/RSC Analytical Chemistry Trust Fund studentship (to L.A.L.). Funding for open access charge: Medical Research Council UK.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank the beam line staff at ESRF BM14 and ID14.4 for assistance with data collection and R. Copley and S. Graham for help with bioinformatics.
REFERENCES
- 1.Schell MA. Molecular biology of the LysR family of transcriptional regulators. Annu. Rev. Microbiol. 1993;47:597–626. doi: 10.1146/annurev.mi.47.100193.003121. [DOI] [PubMed] [Google Scholar]
- 2.Muraoka S, Okumura R, Ogawa N, Nonaka T, Miyashita K, Senda T. Crystal structure of a full-length LysR-type transcriptional regulator, CbnR: unusual combination of two subunit forms and molecular bases for causing and changing DNA bend. J. Mol. Biol. 2003;328:555–566. doi: 10.1016/s0022-2836(03)00312-7. [DOI] [PubMed] [Google Scholar]
- 3.Tyrrell R, Verschueren KH, Dodson EJ, Murshudov GN, Addy C, Wilkinson AJ. The structure of the cofactor-binding fragment of the LysR family member, CysB: a familiar fold with a surprising subunit arrangement. Structure. 1997;5:1017–1032. doi: 10.1016/s0969-2126(97)00254-2. [DOI] [PubMed] [Google Scholar]
- 4.Ezezika OC, Haddad S, Clark TJ, Neidle EL, Momany C. Distinct effector-binding sites enable synergistic transcriptional activation by BenM, a LysR-type regulator. J. Mol. Biol. 2007;367:616–629. doi: 10.1016/j.jmb.2006.09.090. [DOI] [PubMed] [Google Scholar]
- 5.Choi H, Kim S, Mukhopadhyay P, Cho S, Woo J, Storz G, Ryu S. Structural basis of the redox switch in the OxyR transcription factor. Cell. 2001;105:103–113. doi: 10.1016/s0092-8674(01)00300-2. [DOI] [PubMed] [Google Scholar]
- 6.Picossi S, Belitsky BR, Sonenshein AL. Molecular mechanism of the regulation of Bacillus subtilis gltAB expression by GltC. J. Mol. Biol. 2007;365:1298–1313. doi: 10.1016/j.jmb.2006.10.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ogawa N, McFall SM, Klem TJ, Miyashita K, Chakrabarty AM. Transcriptional activation of the chlorocatechol degradative genes of Ralstonia eutropha NH9. J. Bacteriol. 1999;181:6697–6705. doi: 10.1128/jb.181.21.6697-6705.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Porrua O, Garcia-Jaramillo M, Santero E, Govantes F. The LysR-type regulator AtzR binding site: DNA sequences involved in activation, repression and cyanuric acid-dependent repositioning. Mol. Microbiol. 2007;66:410–427. doi: 10.1111/j.1365-2958.2007.05927.x. [DOI] [PubMed] [Google Scholar]
- 9.Smirnova IA, Dian C, Leonard GA, McSweeney S, Birse D, Brzezinski P. Development of a bacterial biosensor for nitrotoluenes: the crystal structure of the transcriptional regulator DntR. J. Mol. Biol. 2004;340:405–418. doi: 10.1016/j.jmb.2004.04.071. [DOI] [PubMed] [Google Scholar]
- 10.Verschueren KH, Addy C, Dodson EJ, Wilkinson AJ. Crystallization of full-length CysB of Klebsiella aerogenes, a LysR-type transcriptional regulator. Acta. Crystallogr. D. 2001;57:260–262. doi: 10.1107/s0907444900016851. [DOI] [PubMed] [Google Scholar]
- 11.Lochowska A, Iwanicka-Nowicka R, Plochocka D, Hryniewicz MM. Functional dissection of the LysR-type CysB transcriptional regulator. Regions important for DNA binding, inducer response, oligomerization, and positive control. J. Biol. Chem. 2001;276:2098–2107. doi: 10.1074/jbc.M007192200. [DOI] [PubMed] [Google Scholar]
- 12.Rosario CJ, Bender RA. Importance of tetramer formation by the nitrogen assimilation control protein for strong repression of glutamate dehydrogenase formation in Klebsiella pneumoniae. J. Bacteriol. 2005;187:8291–8299. doi: 10.1128/JB.187.24.8291-8299.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stec E, Witkowska-Zimny M, Hryniewicz MM, Neumann P, Wilkinson AJ, Brzozowski AM, Verma CS, Zaim J, Wysocki S, Bujacz GD. Structural basis of the sulphate starvation response in E. coli: crystal structure and mutational analysis of the cofactor-binding domain of the Cbl transcriptional regulator. J. Mol. Biol. 2006;364:309–322. doi: 10.1016/j.jmb.2006.06.033. [DOI] [PubMed] [Google Scholar]
- 14.Cebolla A, Sousa C, de Lorenzo V. Effector specificity mutants of the transcriptional activator NahR of naphthalene degrading Pseudomonas define protein sites involved in binding of aromatic inducers. J. Biol. Chem. 1997;272:3986–3992. doi: 10.1074/jbc.272.7.3986. [DOI] [PubMed] [Google Scholar]
- 15.Jorgensen C, Dandanell G. Isolation and characterization of mutations in the Escherichia coli regulatory protein XapR. J. Bacteriol. 1999;181:4397–4403. doi: 10.1128/jb.181.14.4397-4403.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deghmane AE, Petit S, Topilko A, Pereira Y, Giorgini D, Larribe M, Taha MK. Intimate adhesion of Neisseria meningitidis to human epithelial cells is under the control of the crgA gene, a novel LysR-type transcriptional regulator. EMBO J. 2000;19:1068–1078. doi: 10.1093/emboj/19.5.1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morelle S, Carbonnelle E, Nassif X. The REP2 repeats of the genome of Neisseria meningitidis are associated with genes coordinately regulated during bacterial cell interaction. J. Bacteriol. 2003;185:2618–2627. doi: 10.1128/JB.185.8.2618-2627.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ieva R, Alaimo C, Delany I, Spohn G, Rappuoli R, Scarlato V. CrgA is an inducible LysR-type regulator of Neisseria meningitidis, acting both as a repressor and as an activator of gene transcription. J. Bacteriol. 2005;187:3421–3430. doi: 10.1128/JB.187.10.3421-3430.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hong Y, Wang G, Maier RJ. The NADPH quinone reductase MdaB confers oxidative stress resistance to Helicobacter hepaticus. Microb. Pathog. 2008;44:169–174. doi: 10.1016/j.micpath.2006.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sainsbury S, Ren J, Saunders NJ, Stuart DI, Owens RJ. Crystallization and preliminary X-ray analysis of CrgA, a LysR-type transcriptional regulator from pathogenic Neisseria meningitidis MC58. Acta. Crystallogr. Sect. F. 2008;64:797–801. doi: 10.1107/S1744309108024068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Walter TS, Diprose JM, Mayo CJ, Siebold C, Pickford MG, Carter L, Sutton GC, Berrow NS, Brown J, Berry IM, et al. A procedure for setting up high-throughput nanolitre crystallization experiments. Crystallization workflow for initial screening, automated storage, imaging and optimization. Acta. Crystallogr. D. Biol. Crystallogr. 2005;61:651–657. doi: 10.1107/S0907444905007808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schneider TR, Sheldrick GM. Substructure solution with SHELXD. Acta. Crystallogr. D. 2002;58:1772–1779. doi: 10.1107/s0907444902011678. [DOI] [PubMed] [Google Scholar]
- 23.Terwilliger T. SOLVE and RESOLVE: automated structure solution, density modification and model building. J. Synchrotron. Radiat. 2004;11:49–52. doi: 10.1107/s0909049503023938. [DOI] [PubMed] [Google Scholar]
- 24.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta. Crystallogr. D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 25.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta. Crystallogr. A. 1991;47(Pt 2):110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 26.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta. Crystallogr. D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 27.Collaborative Computational Project,N. The CCP4 suite: programs for protein crystallography. Acta. Crystallogr. D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 28.Laskowski RA, Macarthur MW, Moss DS, Thornton JM. Procheck – a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993;26:283–291. [Google Scholar]
- 29.Lovell SC, Davis IW, Arendall WB, 3rd, de Bakker PI, Word JM, Prisant MG, Richardson JS, Richardson DC. Structure validation by Calpha geometry: phi, psi and Cbeta deviation. Proteins. 2003;50:437–450. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
- 30.Lebedev AA, Vagin AA, Murshudov GN. Model preparation in MOLREP and examples of model improvement using X-ray data. Acta. Crystallogr. D. 2008;64:33–39. doi: 10.1107/S0907444907049839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Holm L, Sander C. Mapping the protein universe. Science. 1996;273:595–603. doi: 10.1126/science.273.5275.595. [DOI] [PubMed] [Google Scholar]
- 32.Krissinel E, Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta. Crystallogr. D. 2004;60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- 33.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 34.Landau M, Mayrose I, Rosenberg Y, Glaser F, Martz E, Pupko T, Ben-Tal N. ConSurf 2005: the projection of evolutionary conservation scores of residues on protein structures. Nucleic Acids Res. 2005;33:W299–W302. doi: 10.1093/nar/gki370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dundas J, Ouyang Z, Tseng J, Binkowski A, Turpaz Y, Liang J. CASTp: computed atlas of surface topography of proteins with structural and topographical mapping of functionally annotated residues. Nucleic Acids Res. 2006;34:W116–W118. doi: 10.1093/nar/gkl282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.DeLano W. Alto, CA, USA: DeLano Scientific; 2002. The PyMol Molecular Graphics system. [Google Scholar]
- 37.Jing D, Beechem JM, Patton WF. The utility of a two-color fluorescence electrophoretic mobility shift assay procedure for the analysis of DNA replication complexes. Electrophoresis. 2004;25:2439–2446. doi: 10.1002/elps.200405994. [DOI] [PubMed] [Google Scholar]
- 38.Hernandez H, Robinson CV. Determining the stoichiometry and interactions of macromolecular assemblies from mass spectrometry. Nat. Protoc. 2007;2:715–726. doi: 10.1038/nprot.2007.73. [DOI] [PubMed] [Google Scholar]
- 39.Ruotolo BT, Benesch JLP, Sandercock AM, Hyung SJ, Robinson CV. Ion mobility-mass spectrometry analysis of large protein complexes. Nat. Protoc. 2008;3:1139–1152. doi: 10.1038/nprot.2008.78. [DOI] [PubMed] [Google Scholar]
- 40.Wang L, Gilbert RJ, Atilano ML, Filipe SR, Gay NJ, Ligoxygakis P. Peptidoglycan recognition protein-SD provides versatility of receptor formation in Drosophila immunity. Proc. Natl Acad. Sci. USA. 2008;105:11881–11886. doi: 10.1073/pnas.0710092105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brown PH, Schuck P. Macromolecular size-and-shape distributions by sedimentation velocity analytical ultracentrifugation. Biophys. J. 2006;90:4651–4661. doi: 10.1529/biophysj.106.081372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schuck P, Rossmanith P. Determination of the sedimentation coefficient distribution by least-squares boundary modeling. Biopolymers. 2000;54:328–341. doi: 10.1002/1097-0282(20001015)54:5<328::AID-BIP40>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 43.Garcia De La Torre J, Huertas ML, Carrasco B. Calculation of hydrodynamic properties of globular proteins from their atomic-level structure. Biophys. J. 2000;78:719–730. doi: 10.1016/S0006-3495(00)76630-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nettleship JE, Brown J, Groves MR, Geerlof A. In: Structural Proteomics: High-throughput Methods. Kobe B, Guss M, Huber T, editors. Vol. 426. Australia: Humana Press; 2008. pp. 299–318. [Google Scholar]
- 45.McKay AR, Ruotolo BT, Ilag LL, Robinson CV. Mass measurements of increased accuracy resolve heterogeneous populations of intact ribosomes. J. Am. Chem. Soc. 2006;128:11433–11442. doi: 10.1021/ja061468q. [DOI] [PubMed] [Google Scholar]
- 46.Benesch JL, Robinson CV. Mass spectrometry of macromolecular assemblies: preservation and dissociation. Curr. Opin. Struct. Biol. 2006;16:245–251. doi: 10.1016/j.sbi.2006.03.009. [DOI] [PubMed] [Google Scholar]
- 47.Frickey T, Lupas A. CLANS: a Java application for visualizing protein families based on pairwise similarity. Bioinformatics. 2004;20:3702–3704. doi: 10.1093/bioinformatics/bth444. [DOI] [PubMed] [Google Scholar]
- 48.Deghmane AE, Giorgini D, Larribe M, Alonso JM, Taha MK. Down-regulation of pili and capsule of Neisseria meningitidis upon contact with epithelial cells is mediated by CrgA regulatory protein. Mol. Microbiol. 2002;43:1555–1564. doi: 10.1046/j.1365-2958.2002.02838.x. [DOI] [PubMed] [Google Scholar]
- 49.Chen X, Westphall MS, Smith LM. Mass spectrometric analysis of DNA mixtures: instrumental effects responsible for decreased sensitivity with increasing mass. Anal. Chem. 2003;75:5944–5952. doi: 10.1021/ac030127h. [DOI] [PubMed] [Google Scholar]
- 50.Twerenbold D, Gerber D, Gritti D, Gonin Y, Netuschill A, Rossel F, Schenker D, Vuilleumier JL. Single molecule detector for mass spectrometry with mass independent detection efficiency. Proteomics. 2001;1:66–69. doi: 10.1002/1615-9861(200101)1:1<66::AID-PROT66>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 51.Pringle SD, Giles K, Wildgoose JL, Williams JP, Slade SE, Thalassinos K, Bateman RH, Bowers MT, Scrivens JH. An investigation of the mobility separation of some peptide and protein ions using a new hybrid quadrupole/travelling wave IMS/oa-ToF instrument. Int. J. Mass Spectrom. 2007;261:1–12. [Google Scholar]
- 52.Ruotolo BT, Giles K, Campuzano I, Sandercock AM, Bateman RH, Robinson CV. Evidence for macromolecular protein rings in the absence of bulk water. Science. 2005;310:1658–1661. doi: 10.1126/science.1120177. [DOI] [PubMed] [Google Scholar]
- 53.Smith AM, Jahn TR, Ashcroft AE, Radford SE. Direct observation of oligomeric species formed in the early stages of amyloid fibril formation using electrospray ionisation mass spectrometry. J. Mol. Biol. 2006;364:9–19. doi: 10.1016/j.jmb.2006.08.081. [DOI] [PubMed] [Google Scholar]
- 54.Maris AE, Kaczor-Grzeskowiak M, Ma Z, Kopka ML, Gunsalus RP, Dickerson RE. Primary and secondary modes of DNA recognition by the NarL two-component response regulator. Biochemistry. 2005;44:14538–14552. doi: 10.1021/bi050734u. [DOI] [PubMed] [Google Scholar]
- 55.Hong M, Fuangthong M, Helmann JD, Brennan RG. Structure of an OhrR-ohrA operator complex reveals the DNA binding mechanism of the MarR family. Mol. Cell. 2005;20:131–141. doi: 10.1016/j.molcel.2005.09.013. [DOI] [PubMed] [Google Scholar]
- 56.Chen CS, White A, Love J, Murphy JR, Ringe D. Methyl groups of thymine bases are important for nucleic acid recognition by DtxR. Biochemistry. 2000;39:10397–10407. doi: 10.1021/bi0009284. [DOI] [PubMed] [Google Scholar]
- 57.Kullik I, Stevens J, Toledano MB, Storz G. Mutational analysis of the redox-sensitive transcriptional regulator OxyR: regions important for DNA binding and multimerization. J. Bacteriol. 1995;177:1285–1291. doi: 10.1128/jb.177.5.1285-1291.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Colyer TE, Kredich NM. Residue threonine-149 of the Salmonella typhimurium CysB transcription activator: mutations causing constitutive expression of positively regulated genes of the cysteine regulon. Mol. Microbiol. 1994;13:797–805. doi: 10.1111/j.1365-2958.1994.tb00472.x. [DOI] [PubMed] [Google Scholar]
- 59.Jourdan AD, Stauffer GV. Mutational analysis of the transcriptional regulator GcvA: amino acids important for activation, repression, and DNA binding. J. Bacteriol. 1998;180:4865–4871. doi: 10.1128/jb.180.18.4865-4871.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Delorme C, Huisman TT, Reijnders WN, Chan YL, Harms N, Stouthamer AH, van Spanning RJ. Expression of the mau gene cluster of Paracoccus denitrificans is controlled by MauR and a second transcription regulator. Microbiology. 1997;143(Pt 3):793–801. doi: 10.1099/00221287-143-3-793. [DOI] [PubMed] [Google Scholar]
- 61.Hryniewicz MM, Kredich NM. Stoichiometry of binding of CysB to the cysJIH, cysK, and cysP promoter regions of Salmonella typhimurium. J. Bacteriol. 1994;176:3673–3682. doi: 10.1128/jb.176.12.3673-3682.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Toledano MB, Kullik I, Trinh F, Baird PT, Schneider TD, Storz G. Redox-dependent shift of OxyR-DNA contacts along an extended DNA-binding site: a mechanism for differential promoter selection. Cell. 1994;78:897–909. doi: 10.1016/s0092-8674(94)90702-1. [DOI] [PubMed] [Google Scholar]
- 63.Akakura R, Winans SC. Constitutive mutations of the OccR regulatory protein affect DNA bending in response to metabolites released from plant tumors. J. Biol. Chem. 2002;277:5866–5874. doi: 10.1074/jbc.M110555200. [DOI] [PubMed] [Google Scholar]
- 64.Lochowska A, Iwanicka-Nowicka R, Zaim J, Witkowska-Zimny M, Bolewska K, Hryniewicz MM. Identification of activating region (AR) of Escherichia coli LysR-type transcription factor CysB and CysB contact site on RNA polymerase alpha subunit at the cysP promoter. Mol. Microbiol. 2004;53:791–806. doi: 10.1111/j.1365-2958.2004.04161.x. [DOI] [PubMed] [Google Scholar]
- 65.Deghmane AE, Giorgini D, Maigre L, Taha MK. Analysis in vitro and in vivo of the transcriptional regulator CrgA of Neisseria meningitidis upon contact with target cells. Mol. Microbiol. 2004;53:917–927. doi: 10.1111/j.1365-2958.2004.04167.x. [DOI] [PubMed] [Google Scholar]
- 66.Dietrich G, Kurz S, Hubner C, Aepinus C, Theiss S, Guckenberger M, Panzner U, Weber J, Frosch M. Transcriptome analysis of Neisseria meningitidis during infection. J. Bacteriol. 2003;185:155–164. doi: 10.1128/JB.185.1.155-164.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.