Abstract
Terminal deoxynucleotidyltransferase (Tdt) and DNA polymerase μ (pol μ) are two eukaryotic highly similar proteins involved in DNA processing and repair. Despite their high sequence identity, they differ widely in their activity: pol μ has a templated polymerase activity, whereas Tdt has a non-templated one. Loop1, first described when the Tdt structure was solved, has been invoked as the major structural determinant of this difference. Here we describe attempts to transform Tdt into pol μ with the minimal number of mutations in and around Loop1. First we describe the effect of mutations on six different positions chosen to destabilize Tdt Loop1 structure, either by alanine substitution or by deletion; they result at most in a reduction of Tdt activity, but adding Co++ restores most of this Tdt activity. However, a deletion of the entire Loop1 as in pol λ does confer a limited template-dependent polymerase behavior to Tdt while a chimera bearing an extended pol μ Loop1 reproduces pol μ behavior. Finally, 16 additional substitutions are reported, targeted at the two so-called ‘sequence determinant’ regions located just after Loop1 or underneath. Among them, the single-point mutant F401A displays a sequence-specific replicative polymerase phenotype that is stable upon Co++ addition. These results are discussed in light of the available crystal structures.
INTRODUCTION
Pol μ is a recently described mammalian polymerase belonging to the polX family (1,2). Since its discovery (3,4) its exact function(s) and physiological substrate(s) have been the subject of intense research. Quite early on, it has been implicated in the non-homologous end joining (NHEJ) process that is responsible for repairing double strand breaks in the DNA (5,6). Indeed, it was demonstrated that pol μ physically interacts with Ku 70/80, XRCC4 and Ligase IV (7), as shown earlier for Tdt (8). In addition, pol μ has been shown (successively) to be able to incorporate ribonucleotides as well as deoxynucleotides (9,10), to function as a translesion polymerase (i.e. to bypass defects in the DNA such as Pt-DNA adduits or AAF-adduits), and to be error-prone, especially prone to slippage (frameshift) errors (11–14).
Strikingly, it was shown that pol μ accommodates gaps/breaks in the template strand, namely that it can still direct a template-based DNA synthesis provided that there is ‘micro-homology’ upstream the templating base, with at least one Watson-Crick base pair (15). This establishes a ‘gradient of template dependence’ in the different members of the polX family, going from pol β and pol λ to pol μ and TdT. Pol β is thought to be mainly involved in the nucleotide-excision or base-excision repair processes with 1 or 2 nt gaps in the primer strand. On the sequence level pol β and pol λ form a subgroup different from pol μ and Tdt, but they also have some translesion polymerase activity (16). The exact role of each of these polX members is difficult to establish as there appears to be some functional redundancy between them (5,6,15,17). Other members of the polX family identified in yeasts also have unusual slippage properties and the ability to bypass defects in DNA (18–21).
Until recently, the closest homologous structure available for pol μ was terminal deoxynucleotidyltransferase (Tdt), whose structure has been solved in our laboratory in 2002 (22). The physiological role of Tdt is to add nucleotides in the variable regions of immunoglobulins during V(D)J recombination. Tdt is well known to be unable to perform template-based DNA synthesis. This was readily explained once the Tdt structure was revealed (22), because the so-called Loop1 sterically interferes with the path of a putative template strand. In principle pol μ 3D structure should be amenable to homology modeling techniques, as it has about 42% sequence identity with Tdt. Pol μ does possess a Loop1 of length similar to the one of Tdt, but its sequence differs widely from that of Tdt and its length (16–18 aa) prevents a reliable prediction of its structure by homology modeling techniques.
More recently the crystal structure of mouse pol μ itself was solved as a complex with a template–primer duplex (23): it turns out that Loop1 is invisible in the electron density map from residues 370 to 384, meaning that it is essentially disordered. To appreciate the inherent flexibility of this loop we generated dozens of possible Loop1 conformations using the Knapp algorithm (24) as implemented in our website PDB_Hydro (25) starting from pol μ structure. They differ widely, mainly because they are completely exposed to the solvent region and there is no real way to distinguish between them (they have a very similar energy). Loop1 may of course become ordered once the true substrate is bound, for instance a gapped template–primer duplex. Clearly, this is a situation where modeling studies must be supplemented by as much experimental data as possible.
Both pol λ (26–29) and pol β (30–32) have been the subject of intense structural characterization in the recent years but both of them lack Loop1 altogether (Figure 1). Therefore, the challenge remains to try to understand the structural basis of the functional role of Loop1 in pol μ in the absence of a known ordered structure of Loop1 deposited in the PDB.
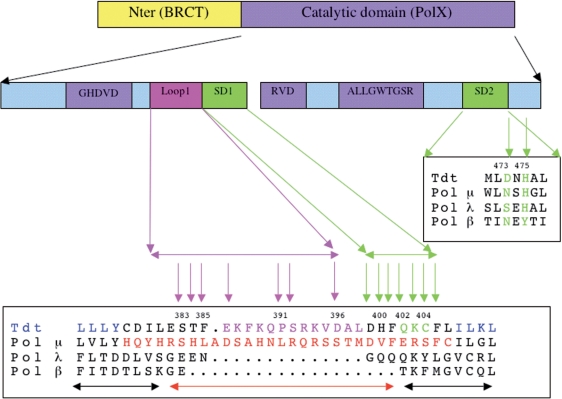
Figure 1.
General organization of Tdt sequence and related polX members. Top: domain organization with the N-terminal BRCT-like domain and the C-terminal polX domain. Bottom: sequence motifs and determinants in Tdt core domain are indicated with the following color code: Motifs C and A (HDVD and DVR) are in magenta as well as the ALLGWTGS sequence, all known to be involved in the dNTP-binding site (22). Loop1 is in red, while SD1 and SD2 regions are in green. The SD1 and SD2 regions are defined as those strictly conserved regions in all Tdt sequences and all pol μ sequences, with length at least two contiguous amino acids, and that differ from one family to the other. The sequences of mouse Tdt, mouse pol mu, and human pol λ and human pol β are shown aligned for selected regions, namely the Loop1 + SD1 and SD2 regions. The chimera construct (Tdt and pol μ Loop1) can be read by following sequentially the blue–red–blue colors. The peptide suppressed in the Δ13 construct is indicated in magenta in the Tdt sequence. The Loop1 mutations are shown with magenta vertical arrows while the SD1 (and beyond) and SD2 regions mutations are shown with green arrows. The pol μ chimera construct of Juarez et al. (2006) in (33) is shown with a red horizontal arrow, with the two neighboring β strand limits indicated with black horizontal arrows.
Here we address this problem by using molecular biology techniques with the aim of switching activities (i.e. making pol μ from Tdt) through the minimum number of mutations. This is in effect the reverse of the experiments reported by Blanco and colleagues (33) on pol μ, which were able to confer Tdt activity to a pol μ by grafting just its Loop1. In the following we will describe activity assays of 28 mutants of Tdt and we will often refer to ‘Tdt activity’ and ‘pol μ activity’; by this we simply mean an activity identical (in the same conditions) to wild-type Tdt or pol μ, respectively (provided as controls in the same figure).
The initial strategy was to destabilize the structure of Loop1, in order to render it more prone to be displaced by the template strand. Mutations were designed to target those residues that were previously described to contribute to the stability of Tdt Loop1 structure (22). First, substitutions by alanine were tried, then cumulative deletions. Both strictly conserved and less highly conserved residues were targeted. Then a larger deletion inspired by pol λ and a chimera with pol μ Loop1 were tried. Finally, the study was further extended to two adjacent regions that are called sequence-determinant regions (SD1 and SD2) located downstream of Loop1 (SD1) and underneath it (SD2): they involve stretches of residues that are strictly conserved in all known Tdt sequences and all known pol μ sequences but differ one from the other [see Figures 1 and 2 of (22)].
Figure 2.
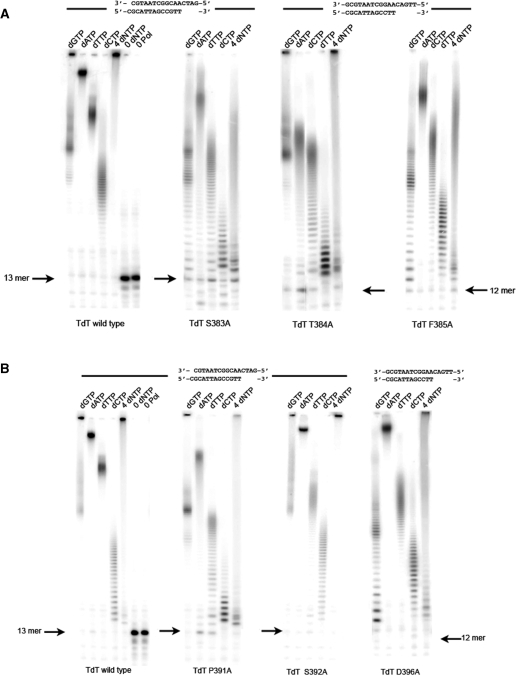
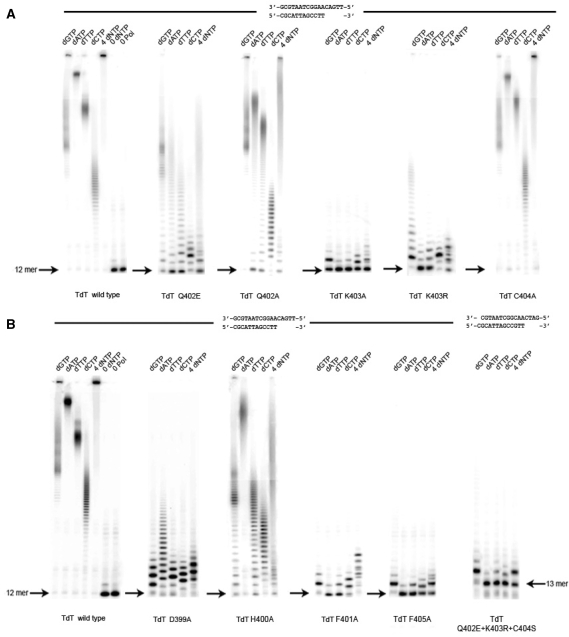
Substitution Tdt Mutants in Loop1. (A) Catalytic activity without CoCl2 but in the presence of 4 mM MgCl2 (from left to right) of wild-type Tdt, S383A, T384A and F385A mutants is shown for each dNTP and a mixture of all dNTPs, using the oligonucleotide duplex indicated on top as a primer. (B) The activity without CoCl2 but in the presence of 4 mM MgCl2 (from left to right) of Tdt wild-type, P391L, S392A and D396A mutants is shown for each dNTP and a mixture of all dNTPs, all using the oligonucleotide duplex indicated on top as a primer.
A still unresolved issue in our case comes from the role of Co++ (or Mn++), which is known to accelerate greatly the template-independent nucleotidyltransferase activity of TdT, especially for pyrimidine incorporation (34,35). Although this effect has been known for many years, its structural basis is still unknown and will be only briefly discussed here. Nevertheless, we will report systematically activity assays of all mutants described here both with and without added Co++. Even though neither Mn++ nor Co++ are present at the mM concentration in physiological conditions, it has been shown for Tdt that both divalent ions mimick well mixtures of Mg++ and Zn++ in concentrations close to that encountered in physiological conditions, as they can play the role of both Mg++ and Zn++ (36); they are also easier to use and dose than Zn++, which can stick to surface residues and/or cysteines when present in the mM range concentration, resulting in non-specific effects. We have shown in control experiments that adding 1 mM Co++ or Mn++ have qualitatively comparable effects both on Tdt and pol μ, but Co++ is quantitatively more effective than Mn++: in particular, 1 mM of Co++ is enough to switch on a significant part of the intrinsic template-independent nucleotidyltransferase activity of wild-type pol μ (Supplementary Figure S1).
MATERIALS AND METHODS
Cloning of TdT mutants
Single point mutations were obtained by a PCR-based method using the Site-Directed Mutagenesis kit from Finnzymes. This mutagenic protocol uses the Phusion High-Fidelity DNA polymerase and comprises three steps: (i) PCR amplification of target plasmid with two 5′-phosphorylated primers purified with reverse phase high performance liquid chromatography (RP-HPLC) and purchased from Sigma-Proligo; (ii) purification (Wizard SV Gel and PCR Clean-Up System, Promega) and circularization by ligation with Quick T4 DNA Ligase (New England Biolabs) overnight at 16°C of mutated PCR products; (iii) transformation into Escherichia coli DH5α by electroporation. All constructions were confirmed by sequencing (GATC Biotech).
Construction of the chimera and the long deletion
The starting material was mouse Tdt as cloned in a Novagen pET28 vector (37). In the first step, 29 aa of mouse Tdt Loop1 were deleted, corresponding to amino-acid residues (378–406) CDILESTFEKFKQPSRKVDALDHFQKCFL. This construction was obtained by PCR using the Site-Directed Mutagenesis kit from Finnzymes with 5′-phosphorylated primers (30 nt) that border the deleted area on both sides. In the second step, after DNA sequencing to confirm the deletion, 30 amino-acid residues (HQYHRSHLADSAHNLRQRSSTMDVFERSFC) of pol μ Mouse Loop1 were added. The insertion was achieved in two PCR steps. The 5′-phosphorylated oligonucleotides were:
Chimera1_Fwd: 5′TTGGCAGACTCAGCCCACAACCTGATTCTGAAGCTGGACCACGGG
Chimera1_Rev: 5′ATGGCTGCGGTGGTACTGGTGGTACAAAAGCAACCCCTGCTGC
Chimera2_Fwd: 5′TGTCTTTGAGAGGAGTTTCTGCTTCTGAAGCTGGACCACGGGAGA
Chimera2_Rev: 5′CATGGTGGAGCTCCGCTGCCGCAGGTTGTGGGCTGAGTCTGCCAA
After each step each construct was confirmed by DNA sequencing.
The Δ13 construct, which transforms CDILESTFEKFKQPSRKVDALDHFQKCFL into CDILEST-ADHFQKCFL, was also built using the following oligonucleotides:
Delta-13_Fwd: 5′ CGACATCTTAGAGTCAACCGCCGACCATTTCCAGAAATGC
Delta-13_Rev: 5′ GCATTTCTGGAAATGGTCGAGGGTTGACTCTAAGATGTCG
Purification of Tdt mutants
After getting the mouse Tdt chimera clone, BL21-Gold DE(3) pLysS Escherichia coli cells were transformed. The bacterial culture (2 l) was grown at 37°C until OD600 0.6–0.7. After addition of 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG), cultures were incubated at 16°C overnight (16 h) and harvested by centrifugation at 4000 rpm during 15 min (37).
The pellet was suspended in 100 ml containing 50 mM Tris–HCl pH 8, 500 mM NaCl, 5 mM Imidazole, supplemented with tablets protease inhibitor cocktail (Complete, EDTA-Free, Roche), lysed by sonication on ice and centrifuged at 15 000 rpm during 30 min. The supernatant was filtered through a 0.45 µm filter, loaded at a rate of 1 ml/min on a 5 ml Nickel-affinity resin (HisTrapHP, GE Healthcare). Fractions containing the chimera were pooled, dialyzed in Buffer A [50 mM NaOAc, pH 4.9, 50 mM Mg(OAc)2, 50 mM (NH4)2SO4, 200 mM NaCl, 10% Glycerol and 5 mM DTT] and then loaded on a ion exchanger HiLoad 16/10 SP Sepharose column (GE Healthcare) equilibrated in Buffer A (38). Proteins were eluted with 2 M NaCl gradient, pooled and dialyzed in Buffer B [50 mM MES, pH 6.5, 50 mM Mg(OAc)2, 200 mM KCl, 5 mM DTT, 10% Glycerol]. Dialyzed fractions were concentrated with Amicon Ultra10K (Millipore) and stored at −20°C after addition of 50% glycerol. Purified proteins were analyzed by SDS–polyacrylamide gel electrophoresis, using a 12% acrylamide gel and molecular mass standards from BioRad (Low Range). Gels were stained with Coomassie Blue R-250. All mutants of Tdt were purified in the same way.
Purification of polymerase mu
A truncated version of mouse pol μ lacking the N-terminal domain and identical to the one described in (23) was cloned in pET16b with a TEV cleavage site (ENLYFQGS) between the His-Tag and the beginning of the sequence (MPAYACQR). Bacterial cell culture of transformed BL21-Gold DE(3) pLysS was similar to that described for Tdt, except that cell culture was continued after induction by IPTG at 20°C for pol μ. Cell lysis and all purification steps were carried out at 4°C, and involved first a HisTrap column, cleavage by the TEV protease, and repurification on the HisTrap column to get rid of uncleaved pol μ and tagged TEV protease. Finally the resulting cleaved protein (checked by mass spectrometry) was purified on a Superdex 75 gel filtration column and dialyzed into 25 mM Tris–HCl pH 8, 200 mM NaCl, 5% glycerol. It was then concentrated up to 10 mg/ml and stored at 4°C.
Polymerase activity tests
Primer strand was 5′-labeled with γ-32P-ATP (GE-Healthcare, 3000 Ci/mM) and T4 polynucleotide kinase (New England Biolabs) during 1 h at 37°C; the labeling reaction was stopped by inactivating the kinase at 75°C for 10 min. The polymerization reaction typically involved 0.05 µM duplex, 2 µM polymerase and 250 µM dNTPs (Qiagen). It was stopped after 30 min at 37°C by the addition of 10 mM EDTA and 95% formamide and the products of the reaction were analyzed by gel electrophoresis on a 20% acrylamide gel, 8 M urea. The 0.4 mm wide gel was run for 4–5 h at 40 V/cm and scanned by a PhosphorImager Storm 860 (Molecular Dynamics, Amersham).
For activity tests, the protein is typically diluted to the final desired concentration using a 10× reaction buffer: for pol μ, this 10× buffer contains Tris–HCl 500 mM pH 7.1, TCEP 10 mM and MgCl2 10 mM; for Tdt, it contains Tris–HCl 250 mM pH 6.6, Na-cacodylate 2M, MgCl2 40 mM and BSA 2.5 mg/ml. For activity tests in the presence of Co++ or Mn++, everything was similar except for the addition of CoCl2 (or MnCl2) at the final concentration of 1 mM.
In order to quantify the sequence specificity of nucleotide incorporation we integrated the intensity in the spot corresponding to the non-elongated labeled primer strand for the expected (Iright) and wrongly incorporated (Iwrong) nucleotides. We then calculated the quantity Q = 1 – Iright/<Iwrong>, where <Iwrong> is the mean value of Iwrong for the three misincorporated nucleotides.
Oligonucleotides
Oligonucleotides were purchased from Eurogentec (Belgium), dissolved in Tris 50 mM pH 8, 1 mM EDTA, 30 mM NaCl and annealed by heating during 5 min up to 90°C; slow cooling to room temperature was allowed to take place overnight. All concentrations were estimated by UV absorbance using an absorption coefficient ε at 260 nm as given by Eurogentec.
Mass spectrometry controls
The molecular weight of the purified fragment after digestion by TEV protease was determined by mass spectrometry (SELDI–TOF) after capture on various chemical surfaces (ProteinChip Arrays) using the Ciphergen's technology (BioRad).
RESULTS
Substitution mutations aimed at destabilizing Loop1 conformation as seen in Tdt crystal structure
A number of single point mutants of Loop1 of Tdt have been generated in order to convert Tdt into a replicative polymerase on the basis of its three-dimensional structure (22). The general organization of sequence motifs along Tdt sequence is shown in Figure 1, along with a blow-up of the sequence of related polX in and around Loop1. The first mutations were designed to target those residues (T384 and S392) that were previously described to contribute to the stability of Tdt Loop1 structure through a specific network of hydrogen-bonds (22). Single point mutations into an alanine were performed (T384A, S392A), purified and tested for activity. The results show that both mutants still display a Tdt-like activity but no pol μ activity (i.e. template-dependent) on a normal template–primer duplex (Figure 2). The double mutant TS→AA was also constructed with similar results (data not shown).
Looking at the multialignment of all known Tdt sequences and all known pol μ sequences, the position P391 stands out as a potential sequence determinant of the sub-families [see Figure 2 in (22)]. Therefore, the mutant P391L, also likely to destabilize Loop1 conformation in Tdt, was expressed, purified and tested. However, this mutation did not change the phenotype (Figure 2B).
Next, the mutation S383A was also done because this position is the only one to be strictly conserved among all known Tdt sequences in Loop1: this time, the phenotype shows a much reduced Tdt-like activity for all dNTPs and also when all four dNTPs are added in the reaction mixture (Figure 2), but still no real template-directed synthesis.
For the sake of completeness, two additional mutants were constructed and tested, namely F385A and D396A. The first one involves a position that is not strictly conserved but is always of an hydrophobic amino-acid type and the second is always a carboxylic acid. On the structural level (22), F385 is in interaction with F401, whose mutation has a very strong effect on Tdt activity (see below). D396A mutants give a phenotype very similar to the wild-type one, while F385A resembles the S383A phenotype, with a much reduced Tdt-like activity (Figure 2).
In all cases, the addition of 1 mM Co++ restores full TdT activity to the mutant (see Table 1, where the effect of all the mutants described here are summarized, with or without Co++ added).
Table 1.
Summary of Tdt mutants measured activity
| Mutation (region) | Observed phenotype wild-type (wt), reduced (−), much reduced (−−) or very much reduced (−−−) Tdt activity | Effect of 1 mM Co++ in Re-establishing full (+) or partial (+/−) template independent Tdt-like activity or none (−) | Remarks (The default enzyme concentration is 2 μM) |
|---|---|---|---|
| S383A (Loop1) | − | + | More pronounced effect at 0.5 μM |
| T384A (Loop1) | wt/− | + | |
| F385A (Loop1) | − | + | More pronounced effect at 0.5 μM |
| P391L (Loop1) | wt/− | + | Sequence as in pol μ |
| S392A (Loop1) | wt | + | Sequence as in pol μ |
| D396A (Loop1) | wt/− | + | |
| Δ1384 (Loop1) | − | + | |
| Δ1392 (Loop1) | − | + | |
| Δ2384/392 (Loop1) | −− | + | |
| Δ3384/391/392 (Loop1) | −−−/Pol | + | More pronounced effect at 0.5 μM |
| Deletion/Loop1 | Pol | +/− | Deletion as in pol λ |
| Chimera/Loop1 | Pol | +/− | |
| D399A (extended SD1) | −−−/Pol | + | More pronounced effect at 0.5 μM |
| H400A (extended SD1) | − | + | |
| F401A (extended SD1) | Pol | − | Template-dep sequence specific pol. |
| Q402E (SD1) | −− | + | |
| Q402A (SD1) | wt | + | |
| QKC=>ERS (SD1) | Pol (stuck) | +/− | Sequence as in pol μ |
| K403A (SD1) | Pol (stuck) | +/− | |
| K403R (SD1) | |||
| C404A (SD1) | wt | + | |
| F405A (extended SD1) | Pol (stuck) | +/− | |
| Pol μ (control) | Pol | +/− | Control (pol μ) |
| D473A (SD2) | Pol | +/− | Less pronounced effect at 0.5 μM |
| N474A (SD2) | wt/− | + | |
| H475A (SD2) | −− | +/− | |
| 473DNH475=>ANA (SD2) | −−−/Pol | +/− | Template-dep. if 4 nt added |
| 473DN474=>NS (SD2) | − | + | Sequence as in pol μ |
| ERS+NS (SD1+SD2) | Pol (stuck) | +/− | Sequence as in pol μ |
In summary, destabilizing the structure of Loop1 as seen in Tdt crystal structure is not enough to switch its activity from a template-independent one to a template-directed one.
Deletion point mutations in Loop1 and full Loop1 deletion as in pol λ
Assuming that the length of Tdt was important for its stability and function, we constructed different deletion mutants at the positions described above (Δ384 and Δ392), as well as the corresponding double deletion. Again, we see no switch of activity (Figure 3A), but the double mutant Δ2384/392 is clearly less active than the single deletion mutants. To confirm this trend we also expressed the triple deletion mutant Δ3384/391/392, where an additional deletion was made at position P391. The enzyme indeed becomes even less active, up to the point where it actually only incorporates a few nucleotides (instead of several hundreds) when individual dNTPs are added, with a clear preference for 1-2 G or 2-3 C. This is also true when adding the 4 nt dNTPs, where synthesis stops after 4 incorporations. However, it switches back to fully active again upon addition of Co++ (Table 1 and Figure 3B).
Figure 3.
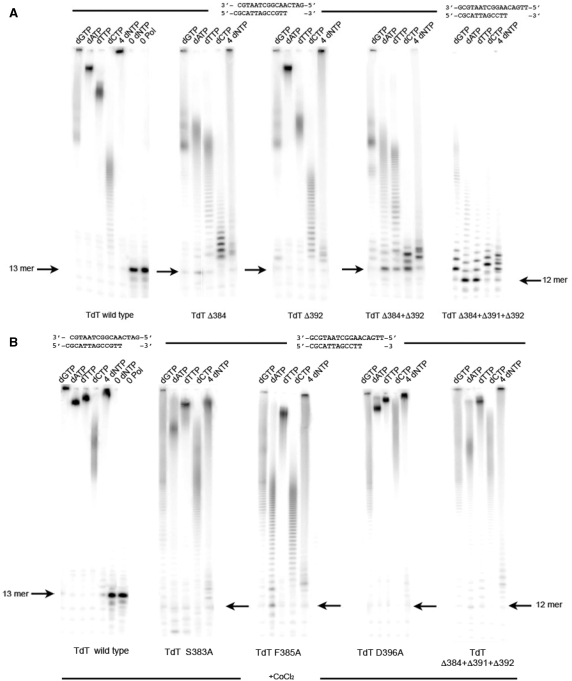
Deletion Tdt Mutants in Loop1. (A) The activity without CoCl2 but in the presence of 4 mM MgCl2 of (from left to right) wild-type Tdt, Δ384, Δ392, the double mutant Δ2383/392 and triple mutant Δ3383/391/392 is shown for each dNTP and a mixture of all dNTPs. (B) The effect of Co++ addition (1 mM) to the reaction buffer on the activity of selected mutants described in Figure 2 and Figure 3A.
In the next step, an attempt was made to mimic pol λ by deleting most of Loop1: 13 residues were deleted, as guided by the multialignement of the corresponding sequences (Figure 1). The corresponding enzyme now displays templated polymerase behavior: it adds one (expected) G, but no or very little of A or T (Figure 4A). It also makes errors in that a C (or two) is also efficiently incorporated. When all four dNTPs are provided for, it incorporates up to only two or three bases, instead of the five expected ones. Upon addition of Co++, one observes partial restoration of template-independent nucleotidyltransferase activity that is comparable to the corresponding basal activity of pol μ in the presence of Co++ (Figure 4B).
Figure 4.
Mimicking pol μ and pol λ with Tdt. (A) The activity without CoCl2 but in the presence of 4 mM MgCl2 of wild-type Tdt and Tdt Chimera containing the pol μ Loop1, as well as the Δ13 mutant (as in pol λ) is shown for each dNTP and a mixture of all dNTPs. (B) The effect of Co++ addition (1 mM) to the reaction buffer is shown for the same mutants.
In conclusion, gradual deletions in Loop1 showed diminished Tdt activity, while mimicking pol λ with a very short Loop1 resulted in an error-prone polymerase activity.
Because the enzyme concentration used in the assay was rather high (2 µM) and because we were concerned that our results might depend on it, all activity tests were done again at a lower enzyme concentration (500 nM). All described above for Loop1 mutants’ results are essentially conserved except for S383 and F385, where the reduction of Tdt activity is stronger (Supplementary Figure S2). This is also true for Δ3384/391/392.
Conferring pol μ activity to the Tdt framework through pol μ Loop1 grafting (chimera)
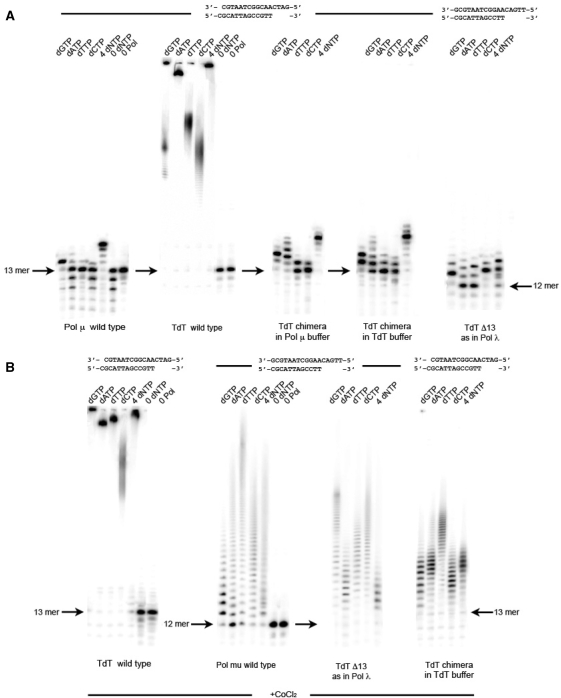
In a recent paper Juarez and collegues (33) described how to transfer non-templated nucleotidyltransferase activity to pol μ simply by exchanging their Loop1 in a pol μ framework. Here we report the reverse experiment, namely the transfer of pol μ template-dependent activity onto Tdt by merely switching their Loop1. A new construct was tried that involved 29 residues from pol μ in the Loop1 region (Figure 1) that were incorporated in two steps, starting from a Tdt mutant lacking 29 residues around Loop1. Incidentally, this deletion mutant showed little or no expression under conditions used for all Tdt other mutant constructs and could not be tested for activity. In Figure 4A it can be seen that the chimera is devoid of any Tdt activity. Rather, its activity is comparable to pol μ templated polymerase activity, both in Tdt or pol μ reaction buffers. We can analyze in more details the specificity (template-instructed) of elongation: when only one dNTP is furnished the expected G is incorporated, but a second G incorporation is also seen; also, depending on the buffer, one, two or even three A can be added; overall, the mutant makes more errors than wild-type pol mu, but its behavior in presence of all four dNTPs is exactly what is expected, namely it stops correctly at the end of the template.
If one adds Co++, one sees a reactivation of the template-independent nucleotidyltransferase activity (Figure 4B). However, there is only partial restoration of Tdt activity in this case, which is comparable with the activity of wild-type pol μ in the presence of Co++. The same behavior is seen at a reduced concentration of 500 nM (Supplementary Figure S2).
Mutating the first ‘sequence determinant’ SD1 region (and beyond)
The multialignement of all known pol μ sequences and all known Tdt sequences reveals two blocks of sequences completely conserved in each sub-family but not identical to each other [Figure 2 of (22)], which we call here the two ‘sequence determinant’ regions SD1 and SD2 (Figure 1). The SD1 contains the QKC tripeptide, just outside of Loop1. As it is part of our grafted sequence that confers pol μ behavior to Tdt, we decided to mutate it separately (Figure 5A). The triple mutant QKC→ERS does incorporate the correct nucleotide (G) when in presence of dGTP, and does not incorporate efficiently any of the three other dNTPs. When in presence of all four dNTPs it adds only one nucleotide, but then stops completely. Therefore we characterize this mutant not as a pol μ, but rather as a ‘stuck’ polymerase; to explain this effect, we speculate that one of the products of the reaction (elongated primer or PPi) may be an inhibitor or cannot be released.
Figure 5.
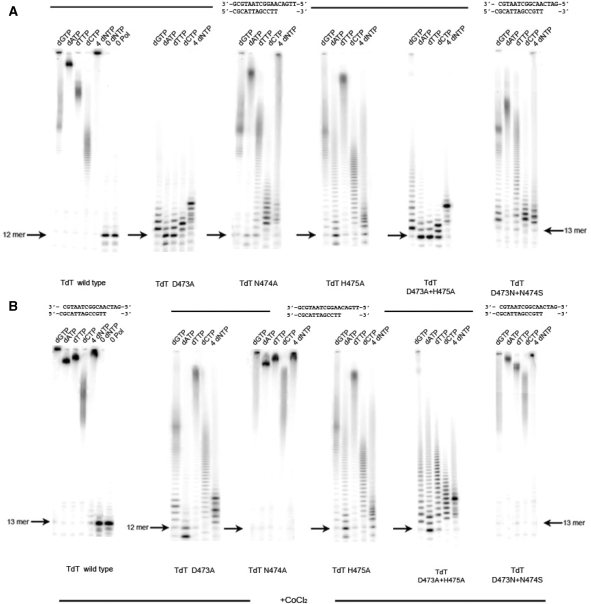
SD1 region mutants (QKC). (A) The activity without CoCl2 but in the presence of 4 mM MgCl2 of wild-type Tdt, Q402E, Q402A, K403R, K403A and C404A mutants is shown for each dNTP and a mixture of all dNTPs. (B) The activity without CoCl2 but in the presence of 4 mM MgCl2 of Tdt wild-type, along with the triple mutant in SD1 switching QKC into ERS (as in pol μ), D399A, H400A, F401A and F405A mutants is shown for each dNTP and a mixture of all dNTPs. The effect of Co++ addition (1 mM) to the reaction buffer is shown in Supplementary Figure S2.
Point mutations involving K403A, K403R, C404A mutants as well as Q402A and Q402E were then examined separately (Figure 5A). The C404A was shown to still display a Tdt wild-type phenotype. Q402E mutant is a Tdt with very much reduced activity but no template-dependent polymerase behavior, while Q402A is similar to wild type. The K403A point mutation however has a more drastic effect and displays a ‘stuck’ polymerase phenotype, with partial incorporation of one G (the expected nucleotide) or one C; then it stops. On the structural side, the sidechain of K403 points directly into the active site, contrary to Q402 and C404, which point away from it, inside the β sheet. K403R phenotype is even more drastic with clear preference for adding (several) G and C and very few of A and T. Adding Co++ reveals template-independent elongation activity for each separate dNTP; however, when adding all four dNTPs simultaneously in solution, both template-independent and template-dependent behaviors are clearly present (Supplementary Figure S3). Template-independent nucleotidyltransferase activity is fully restored upon Co++ addition for Q402E, Q402A, but only partly for K403A and QKC = >ERS mutants (Supplementary Figure S3).
On the basis of these results it was decided to mutate a few other residues beyond this QKC determinant sequence, both on its left and right borders, especially as these borders contain strictly conserved residues in the alignment of Tdt sequences (Figure 5B).
First, the effect of the two bordering mutations F401A and F405A has been assessed: the second mutant F405A has a phenotype very much alike the QKC =>ERS triple mutant, with templated polymerase behavior that stops after one nucleotide addition, but a non-templated nucleotide(s) addition activity is restored upon Co++ addition. Surprisingly, however, the first mutant F401A also has a truly templated polymerase phenotype; remarkably, this time it is resistant (stable with respect) to Co++ addition. F401A is our most striking result, with maximum change of phenotype and minimum mutation. Additional data probing the effect of changing the template sequence have been collected and show that this mutant is indeed a template-dependent polymerase (see below).
Second, we further investigated the effect of two more mutations beyond the QKC tripeptide, involving strictly conserved residues, namely D399A and H400A (Figure 5B). In the presence of separate dNTPs D399A has a much reduced Tdt-like activity (but less so with dATP) that is almost fully restored upon Co++ addition, while H400A mutant behaves as Tdt with a mildly reduced activity, also fully restored upon Co++ addition. In the presence of all four dNTPs, nucleotide addition seems to stop after four to five steps with D399A, as expected for a replicative polymerase given the length of the template overhang, whereas a purely Tdt-like behavior is observed with H400A in the presence of all four dNTPs (Supplementary Figure S3). The behavior of the D399A mutant in the presence of dATP prevents from qualifying it as a polymerase.
The second ‘sequence determinant’ region SD2
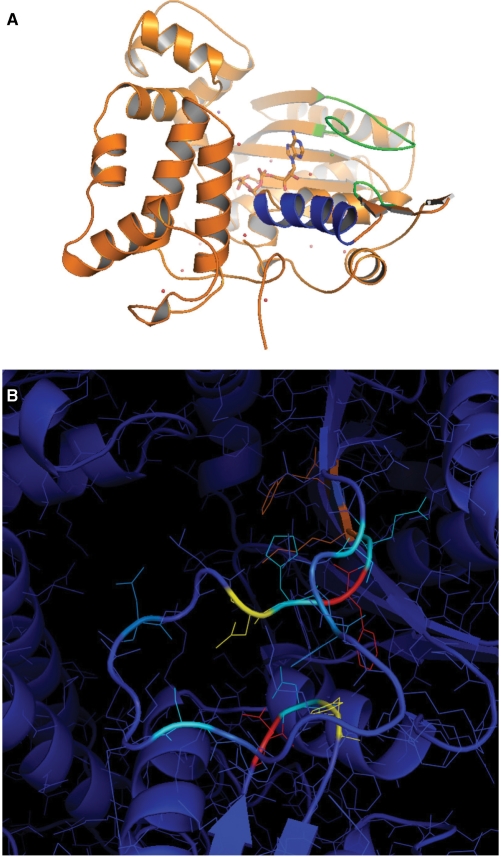
The effect of systematic substitutions of SD2 residues was also studied, both with the DN => NS double mutant (as in pol μ) as well as with the three separate D473A, N474A and H475A single mutants (Figure 6A). When looking at the structure of Tdt, it is apparent that the SD1 and SD2 regions are close in space (Figure 8A).
Figure 6.
SD2 region mutant (DNH). (A) The activity without CoCl2 but in the presence of 4 mM MgCl2 of Tdt wild-type, D473A, N474A and H475A, along with the double mutant switching DNH into NSH (as in pol μ) or into ANA, is shown for each dNTP and a mixture of all dNTPs. (B) The effect of Co++ addition (1 mM) to the reaction buffer is shown in the lower panel.
Figure 8.
Impact of mutations in a 3D framework. (A) Structural context of Loop1 and SD2 (green) as well as helix α14 (blue), just after the ALLGWTGS region, with respect to dNTP-binding site. SD1 is at the C-terminus of Loop1 and the beginning of the next β strand. (B) 3D representation of the effect of mutations. Each mutation described in Table 1 is colored according to its effect (blue = minimum; red = maximum).
Among all these mutants, D473A is the only one with the clearest templated polymerase phenotype, in that it correctly stops nucleotide addition after 5 nt have been incorporated, as dictated by the length of the template. It also shows preference of G (expected) or C addition (not expected) versus A or T in the presence of a single dNTP and can add at least two of them, very much alike the F401A mutant. However, template-independent nucleotidyltransferase activity is partly (and only partly) restored upon Co++ addition (Figure 6B), contrary to the F401A mutant where this activity is not seen.
The N474A mutant is comparable to a wild-type Tdt. H475A shows reduced Tdt activity when in presence of each dNTP separately, with an effect that is unusually pronounced for dATP addition, and template-length limited synthesis when in presence of all four dNTPs (presumably also due to inefficient dATP addition). This is also true of the DN→NS double mutant, where in this case it appears to be caused by the very limited incorporation of dCTP.
The effect of Co++ addition is strong for the DN/NS double mutant, restoring an almost wild-type Tdt activity (Figure 6B) but is very limited for D473A and H475A, especially for the lanes where only dATP is added, or where all four dNTPs are added.
The double mutant where both 473–475 positions where mutated to alanines was also analyzed: it shows basal (reduced) template-independent nucleotidyltransferase activity, both in the absence and in the presence of Co++ ions. When all four dNTPs are present in the reaction mixture, however, it shows clear template-dependent polymerase behavior.
The robustness of this data to the enzyme concentration was assessed for all SD1 and SD2 regions and the results of an activity assay when all four dNTPS are present, at an enzyme concentration of 500 nM, are shown in Supplementary Figure S3B. Except for a slightly different behavior of the D399A mutant, all the previous conclusions are maintained, especially concerning the F401A mutant.
Mutating both the SD1 and SD2 regions (as in pol μ)
In the absence of Co++ ions, the ERS + DS mutant (see Table 1) displays a template-dependent reduced activity, with some specificity: only G and C are incorporated, when G is expected. However, the results in the presence of all four dNTPs in solution show that the enzyme is not very efficient. Adding Co++ ions reveals basal template-independent polymerase activity for each separate lane concerning each dNTP, while adding all four dNTPs together shows clear template-dependent polymerase behavior. One possible explanation for this behavior is that the blunt-end product resulting from templated and sequence-directed elongation of the primer is not a substrate for this mutant (see also the 473ANA475 mutant).
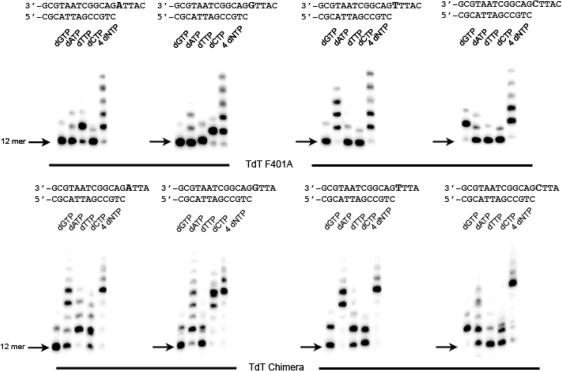
Sequence specificity of nucleotide incorporation for the chimera and F401A constructs
The sequence-dependent incorporation of nucleotide was investigated systematically for the best constructs obtained. In these experiments, dNTP incorporation is studied for all 4 nt, and the templating base is varied, while keeping everything else constant. This is shown in Figure 7 and can be further quantified by the Q ratio defined in ‘Materials and Methods’ section. For the chimera constant we find Q = 0.97, 0.96, 0.99, 0.97 for a templating base of C, T, A, or G, respectively. For the F401A mutant, we find Q = 0.74, 0.92, 0.96, 0.96 for the same templating bases. In both cases the sequence specificity is very good.
Figure 7.
Sequence specificity of F401A mutant and chimera construct.The incorporation of each dNTP is shown for four different oligonucleotides differing only by their templating base. Here the enzyme concentration was 500 nM.
DISCUSSION
The current understanding of the unique Tdt properties is that its lack of template-directed polymerase activity results from the sterical exclusion of the template strand by Loop1 (22). As the closely related pol μ has both a Loop1 and a template-directed polymerase activity, it may be inferred that Loop1 structures differ in detail in pol μ and TdT, perhaps due to their different sequences. The simplest hypothesis consistent with all data is that pol μ's Loop1 can easily be displaced by the incoming template strand, thereby allowing template-based replication, whereas Tdt's Loop1 structure cannot.
Two constructs described here indeed confirm that Loop1 is important in controlling the template-independent nucleotidyltransferase or templated replicating properties of Tdt and pol μ, respectively: a 13-residues long deletion mimicking pol λ and a 29-residue-long pol μ chimera construct [the reverse equivalent of the one described in (33) for pol μ] indeed showed templated polymerase behavior.
Furthermore, the fact that Loop1 is disordered in the presence of a regular primer–template duplex as seen in the pol μ recent structure (23) suggests that destructuring Loop1 might be sufficient to transform Tdt into pol μ. This hypothesis was tested here, using either single-point substitution(s) or deletion(s) at six different positions in Loop1, which were chosen based on the crystallographic structure of Tdt. None of the single-point substitution or deletion tried directly in Loop1 was sufficient to switch a Tdt into a pol μ. However, there were clear indications that increasing the number of deletions (up to three) could severely reduce the template-independent nucleotidyltransferase activity of Tdt (see also Supplementary Figure S2).
On the structural level, it is interesting to look at the structure of Loop1 region in pol μ (PDB code 2IHM), and to locate where exactly it becomes disordered in the electron density. The borders are (i) H369 of RSH, the equivalent of the T384 in Tdt and (ii) F387 of DVF (strictly conserved in Tdt at positions 399–401). Therefore, the disordered part of pol μ corresponds to that part of Loop1 that is absent in pol λ (Figure 1) and also the deleted part of Loop1 that we tested here in the mutant Δ13. In this context, it is worth mentioning the hypothesis of Juarez et al. (33), made before the structures of pol μ and pol λ were solved, who suggest that Loop1 is stabilized in a ‘closed’ conformation in Tdt through mainly hydrophobic interactions involving F401 (SD1), F385 (Loop1) and perhaps H475 (SD2). In other words, according to these authors, the observed well-defined structure of Loop1 might be more due to its anchoring to the rest of the structure through hydrophobic interactions, rather than due to a couple of hydrogen bonds involving T384 and S392 (22). This is indeed consistent with the work reported here, as discussed below.
Experiments have been carried out to assess whether or not mutations concerning the strictly conserved residues on the border of Loop1, such as F401 (SD1), or underneath it, such as H475 (SD2), are able to confer a pol μ activity to a TdT framework, when using regular DNA duplexes substrates. Here, the entire SD1 Region 399–405 has been systematically mutated.
Based on the Tdt structure and the description of its dNTP-binding site it was expected that F405A, K403A and D399A should have a stronger effect compared to H400 and C402, as they clearly line up the active site (Figure 8) on the same side of a β strand, while H400 and C402 do not. This is indeed what is observed.
More strikingly, we find that the mutation F401A in SD1 region has a spectacular effect while F385A effect in Loop1 is more modest but still one of the most significant effects among the Loop1 mutants, along with S383A, in accordance with the hypothesis described in (33). The effect of mutating F401 is puzzling as it is strictly conserved in all known Tdt and pol μ sequences and yet its mutation is sufficient to confer a pol μ phenotype to Tdt. A possible explanation is that the hydrophobic-binding pocket of the phenylalanine sidechain in Tdt structure governs the overall re-entry of Loop1 into the core beta sheet and is therefore essential for Loop1 conformation. What was not expected at all, however, is that F401A phenotype would be resistant to Co++ addition, because this residue is not in direct contact with the catalytic pocket (dNTP-binding site) nor with the primer-binding site. Clearly F401 is far away from the two catalytic divalent ions Mg++, which are thought to be exchangeable, both by Mn++ and Co++ (41), so it is hard to imagine how mutating it could have an influence on the modulation of the catalytic activity by Co++.
In any case, it is clear from both pol μ and pol λ structures (PDB codes 2IHM and 2PQF) that F401 (or its equivalent) is in a different conformation in these two structures, compared to the one seen in Tdt: its side chain actually occupies the site of the H475 side chain, that is in a different rotameric state compared to what is observed in Tdt structure. F385 is also in a different orientation in pol μ and Tdt, very close to H475. This may be interpreted as a sign of a structural rearrangement involving at least H475, F401, S383 and F385 during the catalytic cycle or during the ternary complex assembly.
Looking at the structures of both pol μ and pol λ as a complex with a DNA normal duplex, it is apparent that the so-called SD1 and SD2 regions interact together more clearly and closely in these two structures than in Tdt (PDB code 1JMS). More precisely, in pol μ H475 interacts directly with E402 (Tdt numbering). So the systematic mutations of residues 473–475 in the SD2 region then seemed a natural extension of our site-directed mutagenesis campaign in SD1 region. Furthermore, the role of SD2 (NEY) for pol β, as described in Figure 6 of (32), has been clearly established as being involved with the swinging of R258 in the RVD motif towards the catalytic D192, in the DVD motif, thereby providing an indirect link between the active site and the SD2 region. However, R258 is not conserved in Tdt, pol mu, nor pol λ. Here we observe that D473A indeed has template-directed replication behavior, with H475A also showing a polymerase-like activity but only when all nucleotides are added. This is again compatible with the suggestion of Juarez et al. (33). However, the differential effect of Co++ addition on the restoration of template-independent nucleotidyltransferase activity in these two mutants is difficult to explain as they are both located far away from the catalytic divalent ions-binding sites.
At this stage, we want to stress that the work described here involves mutating a region quite different from the ones most studied in related polX family members, which usually involve either the two catalytic DVD or RVD motifs or the ALLGWTGS region (colored in magenta in Figure 1): for instance H342 in pol μ and Tdt (23) and Y505 and F506 in pol λ (39). Mutations in the same regions in Tdt also result in phenotypes that differ strongly from the wild-type but will be reported elsewhere [see also (36) and (40) for Tdt mutations in the DVD or RVD regions]. We point out that the important role of SD1 and SD2 regions uncovered by the present site-directed mutagenesis study is new but compatible with was described earlier with pol β structures (32) and pol λ simulations (42).
Concerning the effect of Co++ on the activity, we obtained five types of different phenotypes that can be described as follows: (i) wild-type Tdt activity (ii) decreased Tdt activity, reactivated by Co++, (iii) ‘stuck’ polymerase, reactivated to Tdt-like nucleotidyltransferase (template-independent) by Co++, (iv) templated polymerase phenotype, switched back to Tdt-like by Co++ and (v) non-switchable templated polymerase phenotype (only one mutant, F401). So the situation is more complicated than the one described in Juarez et al. 2006 (33) for pol μ At this stage, the effect of Co++ is still not well understood. It is known that Co++ can play the roles of both Mg++ and Zn++; it is also known that Zn++ binding alters mainly the primer strand-binding constant and is not an intrinsic part of the protein (35), so it is rather difficult to explain its differential effect on mutations affecting residues located so far away from the known primer strand-binding site (see for instance the mutants S383A, F385A, F401A, F405A). In this respect, it is worth mentioning that the preferred divalent ion for pol λ is Mn++ and that this effect is still actively studied both experimentally (43) and computationally (44). Clearly, pol λ has a template-independent nucleotidyltransferase activity in the presence of Mn++ divalent ions (45).
In any case, our control experiments on the effect of Co++ onto pol μ nucleotidyltransferase activity are striking: on one hand they confirm the results of Juarez et al. (33) for the effect of Mn++ addition but on the other hand they indicate an even stronger effect for Co++ addition in stimulating the non-templated nucleotidyltransferase activity of pol μ that is highly reminiscent of the effect of Co++ onto Tdt activity (Supplementary Figure S1). Altogether, our results indicate that Tdt and pol μ may be much more functionally related than previously thought.
CONCLUSION
To summarize, the mutants generated in this work were all inspired and guided by the Tdt three-dimensional structure (22), and concerned the region called Loop1 (and its close vicinity). Both a chimera construct using pol μ Loop1 and a deletion of Loop1 inspired by pol λ were successfully transformed into a template-directed polymerase phenotype. We then tried to dissect this effect by using site-directed mutagenesis. Surprisingly, single-point mutants in Loop1 designed to destabilize Loop1 were unsuccessful. Extending the mutations to zones called SD1 and SD2 (strictly conserved but maximally different between pol μ and Tdt) produced some spectacular phenotypes (F401, F405, D473). However, as these positions are quite far away from the active site itself, it is rather difficult at the present time to explain these effects by the structure of the free protein itself (PDB code 1JMS) or any of its published binary complexes (PDB codes 1KDH, 1KEJ). To resolve this situation, we may invoke at least two explanations:
The conformation of Loop1 in Tdt structure may not completely reflect the situation in solution (and may be partly determined by the crystal packing).
There might be a substantial structural rearrangement upon binding all substrates: this would not be uncommon in the polX family, where poly(A) polymerase and terminal uridyltransferase recent structures showed indeed an unusual role for the primer strand in shaping the final structure of the active site (46,47).
Further experiments will be needed in order to distinguish between these two hypotheses.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
ARC [grant number 3155]. Funding for open access charge: ARC 3155.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank PT5 (I. Pasteur) and especially P. Beguin for his help in building several point mutations described here and J. d’Alayer for mass spectrometry controls.
REFERENCES
- 1.Holm L, Sander C. DNA polymerase β belongs to an ancient nucleotidyltransferase superfamily. Trends Biochem. Sci. 1995;20:345–347. doi: 10.1016/s0968-0004(00)89071-4. [DOI] [PubMed] [Google Scholar]
- 2.Aravind L, Koonin EV. DNA polymerase β-like nucleotidyltransferase: identification of three new families, classification and evolutionary history. Nucleic Acids Res. 1999;27:1609–1618. doi: 10.1093/nar/27.7.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aoufouchi S, Flatter E, Dahan A, Faili A, Bertocci B, Storck S, Delbos F, Cocea L, Gupta N, Weill JC, Reynaud CA. Two novel human and mouse DNA polymerases of the polX family. Nucleic Acids Res. 2000;28:3684–3693. doi: 10.1093/nar/28.18.3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Domínguez O, Ruiz JF, Laín de Lera T, García-Díaz M, González MA, Kirchhoff T, Martínez AC, Bernad A, Blanco L. DNA polymerase μ, homologous to Tdt, could act as a DNA mutator in eukaryotic cells. EMBO J. 2000;19:1731–1742. doi: 10.1093/emboj/19.7.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Delbos F, Aoufachi S, Faili A, Weill JC, Reynaud CA. Pol mu, pol λ and Tdt. Immunity. 2006;25:31–41. [Google Scholar]
- 6.Paull TT. Saving the ends for last: the role of pol μ in DNA end joining. Mol. Cell. 2005;5:294–296. doi: 10.1016/j.molcel.2005.07.008. [DOI] [PubMed] [Google Scholar]
- 7.Mahajan KN, Nick McElhinny SA, Mitchell BS, Ramsden DA. Association of DNA polymerase μ (pol μ) with Ku and ligase IV: role for pol μ in end-joining double-strand break repair. Mol. Cell Biol. 2000;22:5194–5202. doi: 10.1128/MCB.22.14.5194-5202.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mahajan KN, Gangi-Peterson L, Sorscher DH, Wang J, Gathy KN, Mahajan NP, Reeves WH, Mitchell BS. Association of terminal deoxynucleotidyl transferase with Ku. Proc. Natl Acad. Sci. USA. 1999;96:13926–13931. doi: 10.1073/pnas.96.24.13926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nick McElhinny SA, Ramsden DA. Polymerase μ is a DNA-directed DNA/RNA polymerase. Mol. Cell Biol. 2003;23:2309–2315. doi: 10.1128/MCB.23.7.2309-2315.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ruiz JF, Juárez R, García-Díaz M, Terrados G, Picher AJ, González-Barrera S, Fernández de Henestrosa AR, Blanco L. Lack of sugar discrimination by human pol μ requires a single glycine residue. Nucleic Acids Res. 2003;31:4441–4449. doi: 10.1093/nar/gkg637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Havener JM, Nick McElhinny SA, Bassett E, Gauger M, Ramsden DA, Chaney SG. Translesion synthesis past platinum DNA adducts by human DNA polymerase μ. Biochemistry. 2003;42:1777–1788. doi: 10.1021/bi0270079. [DOI] [PubMed] [Google Scholar]
- 12.Covo S, Blanco L, Livneh Z. Lesion bypass by human DNA polymerase μ reveals a template-dependent, sequence-independent nucleotidyl transferase activity. J. Biol. Chem. 2004;279:859–865. doi: 10.1074/jbc.M310447200. [DOI] [PubMed] [Google Scholar]
- 13.Duvauchelle JB, Blanco L, Fuchs RP, Cordonnier AM. Human DNA polymerase μ (Pol μ) exhibits an unusual replication slippage ability at AAF lesion. Nucleic Acids Res. 2002;30:2061–2067. doi: 10.1093/nar/30.9.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Y, Wu X, Yuan F, Xie Z, Wang Z. Highly frequent frameshift DNA synthesis by human polymerase μ. Mol. Cell Biol. 2001;21:7995–8006. doi: 10.1128/MCB.21.23.7995-8006.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nick McElhinny SA, Havener JM, Garcia-Diaz M, Juárez R, Bebenek K, Kee BL, Blanco L, Kunkel TA, Ramsden DA. A gradient of template dependence defines distinct biological roles for family X polymerases in Non-Homologous End Joining. Mol. Cell. 2005;19:357–366. doi: 10.1016/j.molcel.2005.06.012. [DOI] [PubMed] [Google Scholar]
- 16.Blanca G, Villani G, Shevelev I, Ramadan K, Spadari S, Hübscher U, Maga G. Human DNA polymerases λ and β show different efficiencies of translesion DNA synthesis past abasic sites and alternative mechanisms for frameshift generation. Biochemistry. 2004;43:11605–11615. doi: 10.1021/bi049050x. [DOI] [PubMed] [Google Scholar]
- 17.Capp JP, Boudsocq F, Besnard AG, Lopez BS, Cazaux C, Hoffmann JS, Canitrot Y. Involvement of DNA polymerase μ in the repair of a specific subset of DNA double-strand breaks in mammalian cells. Nucleic Acids Res. 2007;35:3551–3560. doi: 10.1093/nar/gkm243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pardo B, Ma E, Marcand S. Mismatch tolerance by DNA polymerase Pol4 in the course of NHEJ in S. cerevisiae. Genetics. 2006;172:2689–2694. doi: 10.1534/genetics.105.053512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Daley JM, Vander Laan RL, Suresh A, Wilson TE. DNA joint dependence of polX family polymerase action in NHEJ. J. Biol. Chem. 2005;280:29030–29037. doi: 10.1074/jbc.M505277200. [DOI] [PubMed] [Google Scholar]
- 20.Bebenek K, Garcia-Diaz M, Patishall SR, Kunkel TA. Biochemical properties of Saccharomyces cerevisiae DNA polymerase IV. J. Biol. Chem. 2005;280:20051–20058. doi: 10.1074/jbc.M501981200. [DOI] [PubMed] [Google Scholar]
- 21.González-Barrera S, Sánchez A, Ruiz JF, Juárez R, Picher AJ, Terrados G, Andrade P, Blanco L. Characterisation of SpPol4, a unique pol X-family DNA polymerase in Schizosaccharomyces pombe. Nucleic Acids Res. 2005;33:4762–4774. doi: 10.1093/nar/gki780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Delarue M, Boulé JB, Lescar J, Expert-Bezançon N, Jourdan N, Sukumar N, Rougeon F, Papanicolaou C. Crystal structure of a template-independent DNA polymerase: murine terminal deoxynucleotidyltransferase. EMBO J. 2002;21:427–439. doi: 10.1093/emboj/21.3.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moon AF, Garcia-Diaz M, Bebenek K, Davis BJ, Zhong X, Ramsden DA, Kunkel TA, Pedersen LC. Structural insight into the substrate specificity of DNA polymerase μ. Nat. Struct. Mol. Biol. 2007;14:45–53. doi: 10.1038/nsmb1180. [DOI] [PubMed] [Google Scholar]
- 24.Knapp EW. Long time dynamics of a polymer with rigid body monomer units relating to a protein model: Comparison with the Rouse model. J. Comput. Chem. 1992;13:793–798. [Google Scholar]
- 25.Azuara C, Lindahl E, Koehl P, Orland H, Delarue M. PDB_Hydro: incorporating dipolar solvents with variable density in the Poisson-Boltzmann treatment of macromolecule electrostatics. Nucleic Acids Res. 2006;34:W38–W42. doi: 10.1093/nar/gkl072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garcia-Diaz M, Bebenek K, Krahn JM, Blanco L, Kunkel TA, Pedersen LC. A structural solution for the DNA polymerase λ-dependent repair of DNA gaps with minimal homology. Mol. Cell. 2004;13:561–572. doi: 10.1016/s1097-2765(04)00061-9. [DOI] [PubMed] [Google Scholar]
- 27.Garcia-Diaz M, Bebenek K, Krahn JM, Kunkel TA, Pedersen LC. A closed conformation for the Pol λ catalytic cycle. Nat. Struct. Mol. Biology. 2005;12:97–98. doi: 10.1038/nsmb876. [DOI] [PubMed] [Google Scholar]
- 28.Garcia-Diaz M, Bebenek K, Krahn JM, Pedersen LC, Kunkel TA. Structural analysis of strand misalignment during DNA synthesis by a human DNA polymerase. Cell. 2006;124:331–342. doi: 10.1016/j.cell.2005.10.039. [DOI] [PubMed] [Google Scholar]
- 29.Garcia-Diaz M, Bebenek K, Krahn JM, Pedersen LC, Kunkel TA. Role of the catalytic metal during polymerization by DNA polymerase λ. DNA Repair. 2007;6:1333–1340. doi: 10.1016/j.dnarep.2007.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sawaya MR, Pelletier H, Kumar A, Wilson SH, Kraut J. Crystal structure of rat DNA polymerase β: evidence for a common polymerase mechanism. Science. 1994;264:1930–1935. doi: 10.1126/science.7516581. [DOI] [PubMed] [Google Scholar]
- 31.Pelletier H, Sawaya MR, Kumar A, Wilson SH, Kraut J. Structures of ternary complexes of rat DNA polymerase β, a DNA template-primer, and ddCTP. Science. 1994;264:1891–903. [PubMed] [Google Scholar]
- 32.Sawaya MR, Prasad R, Wilson SH, Kraut J, Pelletier H. Crystal structures of human DNA polymerase β complexed with gapped and nicked DNA: evidence for an induced fit mechanism. Biochemistry. 1997;36:11205–11215. doi: 10.1021/bi9703812. [DOI] [PubMed] [Google Scholar]
- 33.Juárez R, Ruiz JF, Nick McElhinny SA, Ramsden D, Blanco L. A specific loop in human DNA polymerase μ allows switching between creative and DNA-instructed synthesis. Nucleic Acids Res. 2006;34:4572–4582. doi: 10.1093/nar/gkl457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kato K-I, Moura-Gonzalves J, Houts GE, Bollum FJ. Deoxynucleotide-polymerizing enzymes of calf thymus gland. II. Properties of the terminal deoxynucleotidyltransferase. J. Biol. Chem. 1969;242:2780–2789. [PubMed] [Google Scholar]
- 35.Chang LMS, Bollum FJ. Multiple roles of divalent cation in the terminal deoxynucleotidyltransferase reaction. J. Biol. Chem. 1990;265:17436–17440. [PubMed] [Google Scholar]
- 36.Yang B, Gathy KN, Coleman MS. Mutational analysis of residues in the nucleotide binding domain of human terminal deoxynucleotidyl transferase. J. Biol. Chem. 1994;269:11859–11868. [PubMed] [Google Scholar]
- 37.Boulé JB, Johnson E, Rougeon F, Papanicolaou C. High-level expression of murine terminal deoxynucleotidyl transferase in Escherichia coli grown at low temperature and overexpressing argU tRNA. Mol. Biotechnol. 1998;10:199–208. doi: 10.1007/BF02740839. [DOI] [PubMed] [Google Scholar]
- 38.Sukumar N, Boulé JB, Expert-Bezançon N, Jourdan N, Lescar J, Rougeon F, Papanicolaou C, Delarue M. Crystallization of the catalytic domain of murine terminal deoxynucleotidyl transferase. Acta Crystallogr D. 2000;56:1662–1664. doi: 10.1107/s090744490001297x. [DOI] [PubMed] [Google Scholar]
- 39.Shevelev I, Blanca G, Villani G, Ramadan K, Spadari S, Hübscher U, Maga G. Mutagenesis of human DNA polymerase λ essential roles of Tyr505 and Phe506 for both DNA polymerase and terminal transferase activities. Nucleic Acids Res. 2003;31:6916–6925. doi: 10.1093/nar/gkg896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Boulé JB, Rougeon F, Papanicolaou C. Terminal deoxynucleotidyl transferase indiscriminately incorporates ribonucleotides and deoxyribonucleotides. J. Biol. Chem. 2001;276:31388–31393. doi: 10.1074/jbc.M105272200. [DOI] [PubMed] [Google Scholar]
- 41.Pelletier H, Sawaya MR, Wolfle W, Wilson SH, Kraut J. A structural basis for metal ion mutagenicity and nucleotide selectivity in human DNA polymerase β. Biochemistry. 1996;35:12762–12777. doi: 10.1021/bi9529566. [DOI] [PubMed] [Google Scholar]
- 42.Foley MC, Arora K, Schlick T. Sequential side-chain residue motions transform the binary into the ternary state of DNA polymerase λ. Biophys. J. 2006;91:3182–3195. doi: 10.1529/biophysj.106.092080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Blanca G, Shevelev I, Ramadan K, Villani G, Spadari S, Hübscher U, Maga G. Human DNA polymerase λ diverged in evolution from DNA polymerase β toward specific Mn(++) dependence: a kinetic and thermodynamic study. Biochemistry. 2003;42:7467–7476. doi: 10.1021/bi034198m. [DOI] [PubMed] [Google Scholar]
- 44.Cisneros GA, Perera L, García-Díaz M, Bebenek K, Kunkel TA, Pedersen LG. Catalytic mechanism of human DNA polymerase λ with Mg2+ and Mn2+ from ab initio quantum mechanical/molecular mechanical studies. DNA Repair. 2008;7:1824–1834. doi: 10.1016/j.dnarep.2008.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ramadan K, Maga G, Shevelev IV, Villani G, Blanco L, Hübscher U. Human DNA polymerase λ possesses terminal deoxyribonucleotidyl transferase activity and can elongate RNA primers: implications for novel functions. J. Mol. Biol. 2003;328:63–72. doi: 10.1016/s0022-2836(03)00265-1. [DOI] [PubMed] [Google Scholar]
- 46.Stagno J, Aphasizheva I, Aphasizhev R, Luecke H. Dual role of the RNA substrate in selectivity and catalysis of terminal uridylyltransferase. Proc. Natl Acad. Sci. USA. 2007;104:14634–14639. doi: 10.1073/pnas.0704259104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Balbo PB, Bohm A. Mechanism of poly(A) polymerase: structure of the enzyme-MgATP-RNA ternary complex and kinetic analysis. Structure. 2007;15:1117–1131. doi: 10.1016/j.str.2007.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.