Abstract
Most site-specific recombinases can be grouped into two structurally and mechanistically different classes. Whereas recombination by tyrosine recombinases proceeds with little movements by the proteins, serine recombinases exchange DNA strands by a mechanism requiring large quaternary rearrangements. Here we use site-directed crosslinking to investigate the conformational changes that accompany the formation of the synaptic complex and the exchange of DNA strands by the Hin serine recombinase. Efficient crosslinking between residues corresponding to the ‘D-helix’ region provides the first experimental evidence for interactions between synapsed subunits within this region and distinguishes between different tetrameric conformers that have been observed in crystal structures of related serine recombinases. Crosslinking profiles between cysteines introduced over the 35 residue E-helix region that constitutes most of the proposed rotating interface both support the long helical structure of the region and provide strong experimental support for a subunit rotation mechanism that mediates DNA exchange.
INTRODUCTION
Site-specific DNA recombination reactions are distributed widely in nature and have been recently adopted as a useful tool for genetic engineering (1). Unlike homologous recombination, site-specific DNA recombination reactions promote exchange of DNA strands at specific loci and usually in a highly precise manner (2). They control diverse processes such as transcription, replication, resolution of chromosome dimers, rearrangement of DNA segments into functional genes, transposition of DNA, and viral integration into and excision from host chromosomes. These reactions are catalyzed by a recombinase that is dedicated to a specific DNA rearrangement, but often additional host DNA binding proteins are required to assist in assembly of synaptic complexes. Most site-specific recombinases can be classified into two mechanistically distinct families known as serine or tyrosine recombinases, but some are related to the DD-E transposase-like enzymes (3–6). The structural organization of the synaptic complexes containing the DNA elements and proteins that are assembled by members of the different families are unrelated, as are the enzymological steps involved in cleaving and re-ligating the DNA strands into the recombinant configuration.
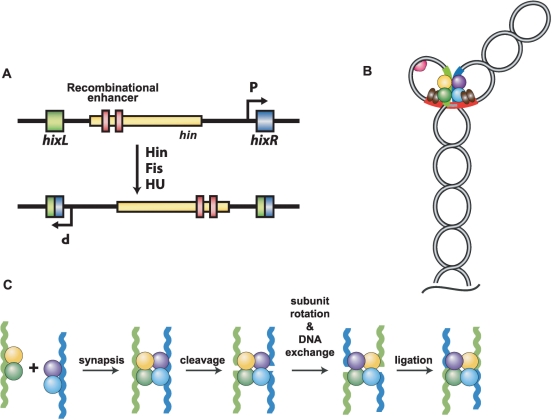
Hin is a member of the serine recombinase family (3), which is named because a conserved serine is the active site residue that catalyzes the DNA cleavage/joining reaction. Hin promotes inversion of a ∼1 kb DNA segment between two hix recombination sites within the chromosome of Salmonella sp (7). The flipping of the DNA segment reorients an internal promoter, which results in alternate expression of antigenically distinct flagellin protein genes (Figure 1A). Hin is a member of a subgroup of serine recombinases called DNA invertases that promote inversion reactions in different biological contexts. Members of another subgroup of serine recombinases, DNA resolvases, promote site-specific deletions of DNA (8). DNA invertases and resolvases are similar in size and share a similar ∼100 residue N-terminal catalytic domain linked to a ∼34 residue oligomerization helix (‘helix E’). The C-terminal DNA binding domains are more divergent at the sequence level, but crystal structures have shown that the 40–50 amino acid residue domains from γδ resolvase and Hin share a similar helix-turn-helix fold, which are positioned on opposite sides of the DNA duplex from the catalytic domain (9). Crystal structures of the catalytic domain of γδ resolvase have been determined in isolation, as a full-length dimer bound to a single recombination site and as tetrameric synaptic complexes (10–14). A goal of the present work is to establish the relationship between the resolvase tetrameric structures and the structure of Hin within active recombination complexes. A third class of serine recombinases promote phage integration/excision and DNA transposition reactions (3). The catalytic cores of these recombinases (15) are present within a much larger polypeptide chain.
Figure 1.
The Hin site-specific DNA inversion reaction from Salmonella sp. (A) Inversion of the chromosomal segment between the two hix recombination sites switches the orientation of an internal promoter. In one orientation, this promoter directs transcription of a flagellin gene and a negative regulator of an alternative flagellin gene located elsewhere in the chromosome. The recombinational enhancer with the two Fis binding sites (red) is depicted in its normal location within the N-terminal coding region of the hin gene. (B) Recombination intermediate (invertasome) formed during the native DNA inversion reaction. A tripartite complex containing a tetramer of Hin bound to the two hix sites, and two dimers of Fis (brown ellipses) bound to the enhancer assembles at the base of a supercoiled branch. Formation of this complex requires DNA supercoiling and is aided by HU (red half-sphere), which stabilizes short loops. (C) Recombination on linear DNA substrates by Fis-independent Hin mutants. Hin dimers bound to DNA fragments containing hix recombination sites assemble into a tetrameric synaptic complex. Each subunit of Hin cleaves a DNA strand at the center of hix to form a serine-phosphodiester bond with the 5′ end of the broken DNA. Data in this article and elsewhere (10,20) indicate that one set of synapsed subunits can rotate relative to the other to position the DNA strands in the recombinant configuration. Ligation of the DNA ends completes the reaction. Reactions performed in the presence of ethylene glycol and without Mg2+ generate stable cleaved synaptic complexes that support subunit rotation but not ligation.
A distinguishing feature of reactions catalyzed by DNA invertases like Hin is the requirement for the auxiliary protein Fis and a cis-acting DNA segment called a recombinational enhancer (7). The Fis-bound enhancer segment contains two binding sites for the Fis dimer and assembles together with Hin dimers bound to the two hix recombination sites to form a catalytically competent invertasome structure (Figure 1B) (16). The assembly of this tripartite structure requires DNA supercoiling, and in the case of Hin, is stimulated by the HU DNA bending protein. A class of ‘gain-of-function’ Hin mutants, represented by Hin–H107Y, is able to catalyze recombination without any accessory factors (17). Unlike the native enzyme, Hin–H107Y can assemble oligonucleotide substrates containing hix sites into stable synaptic complexes that promote recombination in vitro (Figure 1C).
Multiple lines of evidence from Hin and resolvase systems have demonstrated that the active synaptic complex contains the recombining DNA segments on the outside of the protein core (10,18–20). Within this complex the four recombinase subunits cleave both strands of DNA within the center of each hix site to form a phosphoserine linkage with the 5′ end of the cleaved DNA. The repositioning of the DNA strands into the recombinant configuration is proposed to occur by a translocation of a pair of synapsed subunits within the recombinase tetramer (10,20–23). The crystal structures of the γδ resolvase tetramer in a synaptic complex with DNA provide a plausible structural mechanism by which a ‘subunit rotation’ process could occur (10,11). The synapsed pairs of subunits are separated by a largely flat and aliphatic interface, which could form a compatible surface for rotation. Nevertheless, such a process would have to display remarkable fidelity in order to prevent dissociation of synapsed subunits, which would lead to double strand DNA breaks.
We have used site-directed protein crosslinking strategies to probe the structure and dynamic nature of Hin synaptic complexes and to provide further experimental support for the subunit rotation mechanism. The experiments are guided by molecular models of Hin tetramers based on the recent crystal structures of γδ resolvase synaptic complexes. These structures, together with a recent structure from a more distantly related serine integrase TP901, provide evidence for at least two structurally distinct tetrameric conformers. We begin by evaluating the proximity of residues within the D helices as these are predicted to be proximal to each other only in one of the tetrameric forms. A 30–50 Å movement would be required to associate these residues during remodeling from the initial interaction of dimers to the fully remodeled tetramer. We also probe crosslinking efficiencies at cysteines introduced throughout the predicted E-helix region that forms most of the tetramer interface and the surface that engages in subunit rotation to promote DNA strand exchange. The cumulative data support a model for the recombinationally active Hin tetramer that closely fits the X-ray structures of one of the tetrameric conformers of resolvase and provide strong experimental support for exchange of DNA strands by a subunit rotation mechanism.
MATERIALS AND METHODS
Molecular models of Hin
Hin closed tetramer conformation models were generated based on the γδ resolvase tetramer X-ray structures 2GM4 (3.5 Å) and 1ZR4 (3.4 Å) (10,11). Phyre (24) was used to create the initial subunit structure for the 2GM4 model, and Modeller (25) was used to generate 100 models corresponding to 1ZR4. Most of the models generated by Modeller fit the catalytic domain of resolvase well, but the trajectories of the E helices beyond residue 118 varied considerably. The selected model had the conformation of its α-helix E most closely matching those of 1ZR4 and the lowest RMSD to 1ZR4. The Hin subunit models were docked onto DNA by aligning with the respective 2GM4 (chain B) and 1ZR4 (chain A) template structures using LSQMAN. The 2.4 Å X-ray structure of the Hin DNA binding domain (1IJW) (26) was then substituted in place of the C-terminal residues 143–190 and the peptide segment between 142 and 149 was manually adjusted using COOT (27) to optimize the fit. Model tetramers were generated by superimposing (LSQMAN) the DNA bound subunits onto the respective 2GM4 and 1ZR4 tetramers. The structures were subjected to iterative cycles of energy minimization by simulated annealing using CNS (28), analysis using PROCHECK (29) and visual inspection, and adjustment of rotamer conformations using COOT. In some cases, rotamers were selected that most closely matched those in resolvase. The final tetramer models of 2GM4 and 1ZR4 have no residues in the generously allowed or disallowed regions of the Ramachandran plot. Alignment of all peptide backbone atoms of the 2GM4 Hin model and resolvase tetramer give an RMSD of 1.9 Å and an RMSD of 1.0–1.1 Å over the respective peptide chains from residues 1–134. Alignment of backbone atoms of the 1ZR4 Hin model and resolvase tetramer give an overall RMSD of 3.1 Å and an RMSD of 0.8–1.1 Å over respective peptide chains from residues 1–134.
A symmetric Hin ‘open’ tetramer conformation model was generated based on chains A and B of the 2.1 Å asymmetric resolvase tetramer structure 2GM5 (11) as follows. A model of a Hin subunit based on chain A of 2GM5 was created with Modeller and aligned to chains A and B of 2GM5. A symmetric ‘open’ tetramer of Hin was created by duplicating the Hin model, and aligning to chains C and D (closed subunits within 2GM5) along the helix E from residues 100–120. A minor manual adjustment of the chains C and D unit alleviated clashes in the loop region between residues 91–96. The 2GM5 X-ray structure only extends to residue 123. In order to generate an open tetramer model of the complete protein, the 2GM4-based Hin model was aligned to the 2GM5-based Hin model over the E-helix. The N-terminal portion from the 2GM5 model was merged with the C-terminal portion of the 2GM4 model at residue 120. The final model was refined and energy minimized in a similar manner as described for the closed conformation models.
Mutagenesis and purification of Hin mutants
Codon changes specifying cysteines in helix D of hin gene cloned in pET11a were introduced by a modification of the QuikChange method (Stratagene), and those within the E-helix region were introduced by a two-step PCR method (30). The Hin–H107Y gene that served as the template was described in Sanders and Johnson (17). Most Hin mutant proteins were purified from inclusion bodies using either a slow dialysis re-folding procedure described previously (17) or by a rapid dilution method. In the latter method, 5 mg of Hin from purified inclusion bodies were solubilized in 1 ml of 7 M guanidine–HCl, 20 mM HEPES (pH 7.5), 0.1 mM EDTA, 50 mM CHAPS, 10 mM DTT and 20% glycerol and added at a rate of 2 ml/h into 167 ml of buffer (30 μg/ml final Hin concentration) containing 0.5 M urea, 25 mM HEPES (pH 7.5), 0.5 M NaCl, 10 mM DTT, 20% glycerol, 7.5 mM CHAPS and 0.05 mM MgCl2. This was then diluted with buffer containing 0.5 M urea, 25 mM HEPES (pH 7.5), 5 mM DTT, 20% glycerol and 1 mM EDTA such that the final concentration of NaCl was 0.375 M. The renatured Hin was collected by binding to 1 ml heparin–Sepharose (GE Life Sciences). The column was thoroughly washed with HB (20 mM HEPES (pH 7.5), 5 mM DTT, 4 mM CHAPS, 0.1 mM EDTA, and 20% glycerol) containing 0.5 M NaCl, followed by 10 ml of HB + 0.6 M NaCl, and the Hin was eluted with HB + 1.2 M NaCl and stored at −20°C in HB + 1 M NaCl and 50% glycerol.
In vitro recombination and site-directed Hin crosslinking assays
Hin-catalyzed DNA cleavage and inversion reactions were performed as described (31) using our standard supercoiled plasmid substrate pMS551 (32). DNA-cleaved synaptic complexes were assembled on hixL-containing oligonucleotides or supercoiled plasmids under Mg2+-free ethylene glycol conditions (17,33). Typically, 25–50 ng Hin–H107Y cysteine mutants were incubated at 37°C with 3′-32P-labeled 36 bp duplex oligonucleotides (0.2 pmol) or plasmid DNA (0.1 pmol) in a 25 μl reaction containing 20 mM HEPES (pH 7.5), 80 mM NaCl, 4 mM CHAPS, 2 mM EDTA, 30% ethylene glycol, 10 μg poly-glutamate and 15 ng Fis protein for plasmids. Crosslinking on oligonucleotide substrates was performed as detailed in (20). Typically, synaptic complexes were exposed to 0.4 mM crosslinker (see below), and after 1 min, the crosslinking reaction was quenched with 50 mM DTT and the reaction was applied to a 10% native polyacrylamide gel containing 10% glycerol in 45 mM TBE buffer. Cleaved synaptic complexes were eluted from the gel slices and then applied to 10% or 12.5% SDS–PAGE gels. To evaluate ligation, crosslinked or control DNA-cleaved synaptic complexes were excised from the native polyacrylamide gel and soaked in buffer with MgCl2 (17). SDS was added to 1% after 30 min and the products were eluted from the gel and subjected to SDS–PAGE. Crosslinking reactions on plasmids (pRJ2372) were performed similarly except the reactions were quenched with 50 mM DTT plus 0.05% diethylpyrocarbonate and ethanol precipitated. pRJ2372 contains EcoR1 sites 50 bp from the center of each hix site so digestion with EcoR1 releases the hix sites (see Figure 6B). The EcoR1 sites were labeled with α-32P-dATP using Klenow polymerase and the products were subjected to SDS–PAGE. For ligation assays, the crosslinked or control complexes were quenched with DTT only and were then diluted with 20 mM HEPES (pH 7.5), 80 mM NaCl and 10 mM MgCl2 such that the ethylene glycol concentration was reduced to 5%. After 15 s the reaction was stopped with 0.05% diethylpyrocarbonate and treated as above. Cysteine-specific crosslinkers (Pierce) with varying spacer lengths between sulfhydryl reactive groups were: bis-maleimidoethane (BMOE, 8 Å), bis-maleimidobutane (BMB, 11 Å), and bis-maleimidohexane (BMH, 16 Å). Oxidation to evaluate disulfide bond formation utilized 0.4 mM diamide (Sigma).
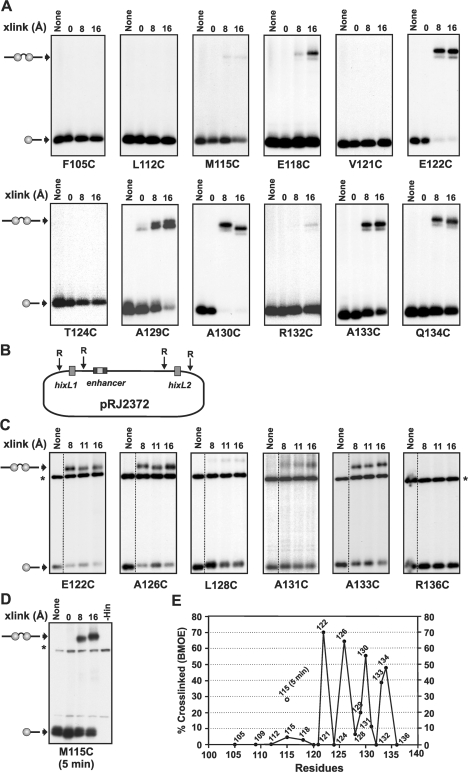
Figure 6.
Crosslinking of helix E cysteine mutants. (A) Crosslinking reactions were performed using 3′ 32P-labeled 36 bp hixL fragments. Crosslinked synaptic complexes were purified from native polyacrylamide gels, extracted and then subjected to SDS–PAGE. The A129C panel is from referece (20). (B) Diagram of pRJ2372. (C) Crosslinking reactions performed using supercoiled pRJ2372 in the presence of Fis. After 1 min crosslinking reactions, the plasmid was digested with EcoR1, which cleaves 50 bp on either side of each hix site, labeled with 32P-ATP, and subjected to SDS–PAGE. The labeled bands denoted with an asterisk and present in all lanes are from a DNA fragment released by EcoR1 cleavage. All crosslinking reactions in A and B were performed for 1 min using diamide (0 Å), BMOE (8 Å spacer), BMB (11 Å) or BMH (16 Å spacer). The Hin–M115C reaction in (D) was crosslinked for 5 min. Each cysteine was introduced into a Hin–H107Y background. (E) Plot of crosslinking efficiencies of cysteines as a function of location along helix E. Percentage crosslinked is the amount of crosslinked di-protomer/non-crosslinked + crosslinked Hin–DNA covalent complexes. Data is from 1 min BMOE crosslinking reactions on supercoiled DNA. The results of 5 min BMOE crosslinking reactions with M115C (D) is also denoted.
RESULTS
Molecular models of Hin complexes
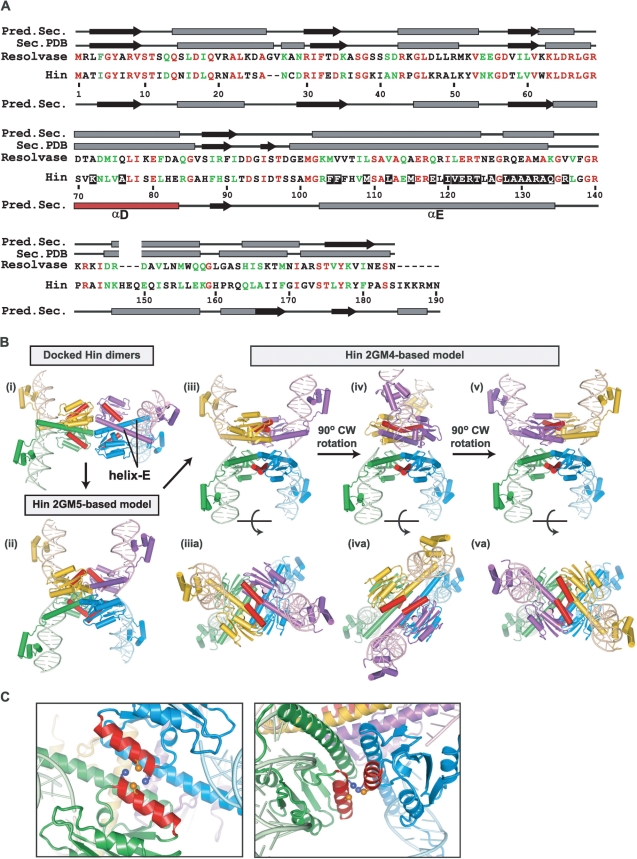
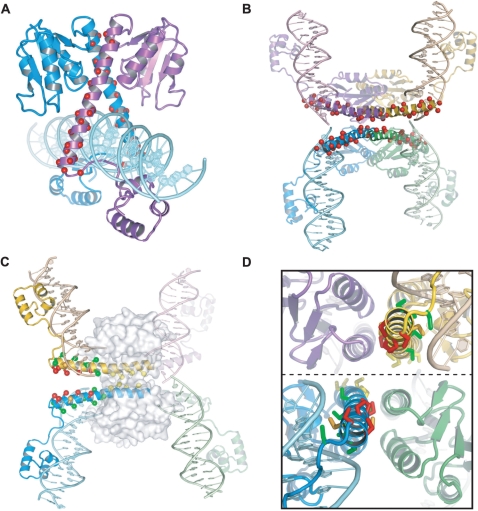
In the absence of atomic structures of the catalytic domain of the Hin DNA invertase, we have generated models of different quaternary structures of Hin based on crystal structures of γδ resolvase. Hin and γδ resolvase share 40/60% amino acid residue identity/similarity, over their N-terminal ∼100 amino acid catalytic domains and 41/69% identity/similarity over their ‘helix-E’ oligomerization regions (Hin residues 100–134) (Figure 2A). Multiple structure folding programs (34–38) applied to the Hin catalytic and oligomerization domains predict a secondary structure profile that generally matches the γδ resolvase structures, except for the region between Ala26 and Ile31 where a 2-residue gap exists. The model for the Hin dimer [Figure 2B, panel (i)] based on the γδ resolvase dimer catalytic domain [PDB code 1GDT; (12)] and C-terminal Hin DNA binding domain crystal structures [residues 139–190, PDB code 1IJW; (26)] has been described previously (9,31). The DNA in the Hin model is less bent than in the resolvase structure because of the small amount of bending induced by Hin dimer binding as estimated by gel electrophoresis. As discussed by Yang and Steitz (12), the dimer is in a catalytically inactive conformation because the active site Ser 10 residues are far from and sterically blocked from directly approaching the DNA cleavage sites by the C-terminal ends of the dimerization E helices.
Figure 2.
Molecular models of Hin synaptic complexes. (A) Amino-acid sequence alignment of Hin and γδ resolvase. Predicted and actual (Sec. PDB derived from the 1ZR4 γδ resolvase tetramer) secondary structure elements (β strands, black arrows; α-helices, grey rectangles) are denoted above and below the resolvase and Hin sequences, respectively. Residues substituted with cysteine that are analyzed in this work are highlighted with a black background. (B) (i) Two Hin dimers (yellow + green and blue + purple) bound to hix sites positioned in a manner that may reflect initial synaptic interactions. (ii) Model of a symmetric ‘open’ Hin tetramer based on one synapsed subunit pair of the 2GM5 resolvase tetramer. The approximate path of DNA is shown based on the cleaved DNA segments in 2GM4. However, the DNA is not expected to be cleaved, and the conformer shown could not bind uncleaved DNA without changes within the C-terminal end of helix E [see (11)]. (iii) Model of a ‘closed’ Hin tetramer bound to cleaved DNA based on the 2GM4 resolvase tetramer. (iv) The Hin 2GM4 model after a 90° clockwise rotation of a synapsed pair (yellow + purple) of subunits to generate the helix E-aligned conformer. (v) An additional 90° clockwise subunit rotation that positions the DNA for ligation in the recombinant configuration. Panels (iiia–va) of the 2GM4-based models show each structure rotated 90° about the x-axis. In each case, the D helices are highlighted in red. See the Supplementary Data movie for an animated depiction of the rotations represented in panels (iii–v). (C) Two views highlighting the D helices (red) from one synapsed pair of subunits in the 2GM4 Hin tetramer model showing cysteines substituted at residues 72 (blue) and 76 (orange). Sγ atoms are rendered as spheres.
Tetrameric Hin models bound to a cleaved synaptic complex were derived from the crystal structures of resolvase (PDB code 1ZR4) and of a resolvase-Hin chimera (PDB code 2GM4) in which Hin residues 96–105 were substituted in place of resolvase (10,11). These structures, which contain the active site serines linked to the 5′ ends of the cleaved DNAs, will be referred to as the closed synaptic tetramer conformation [Figure 2B, panel (iii)]. This conformation contains the aliphatic and flat interface that is primarily constituted by the E helices and is proposed to support the rotation of synapsed subunits together with their linked DNA strands to generate the recombinant DNA [Figure 2B, panels (iv) and (v)].
An asymmetric tetramer structure of the catalytic domain of γδ resolvase has also been reported (PDB code 2GM5) in which one synapsed pair of subunits is in an alternative conformation from that of the DNA bound tetramers in 1ZR4/2GM4 (11). We have modeled a symmetric Hin tetramer to reflect this alternative ‘open’ conformation [Figure 2B, panel (ii)]. The E helices are in a similar conformation in the open and closed tetramers, but the catalytic domains (residues 1–99) are in different relative positions. The positions of the DNA binding domains and E helices are incompatible with binding to uncleaved DNA, and the active site serines at residue 10 in the catalytic domain are far from DNA. The open tetramer conformation bears resemblance to a recent structure of the catalytic domain of a serine recombinase that catalyzes integration of phage TP901 (15). The TP901 integrase exhibits 37% and 28% sequence identity to Hin and γδ resolvase (not accounting for indels), respectively, over Hin residues 1–120 but contains an unrelated long C-terminal segment after the common helix-E region. As depicted in Figure 2B and discussed in Kamtekar et al. (11) and Yuan et al. (15), an attractive model is that the open tetramer represents an intermediate conformer along the pathway from initial synapsis of dimers to the closed conformation that is competent for DNA cleavage and subunit rotation.
Crosslinking between residues of helix D
Helix D of Hin is predicted to undergo a dramatic reorientation during the transition from the initial synapsis of dimers to the closed tetramer (Figure 2B and C). Residues within helix D of different subunit are far from each other in the dimer and open tetramer forms, but the N-terminal ends of helix D from synapsed dimers are predicted to be close and potentially interacting in the closed tetramer. To determine whether the closed conformation model reflects the structure of recombining Hin complexes we probed crosslinking between residues in the N-terminal end of helix D within cleaved synaptic complexes.
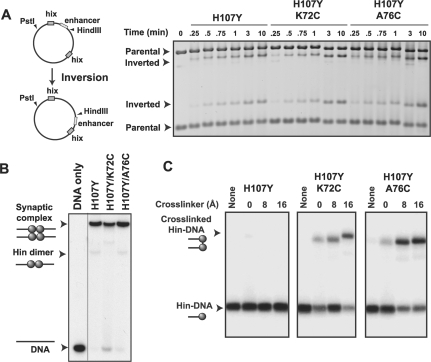
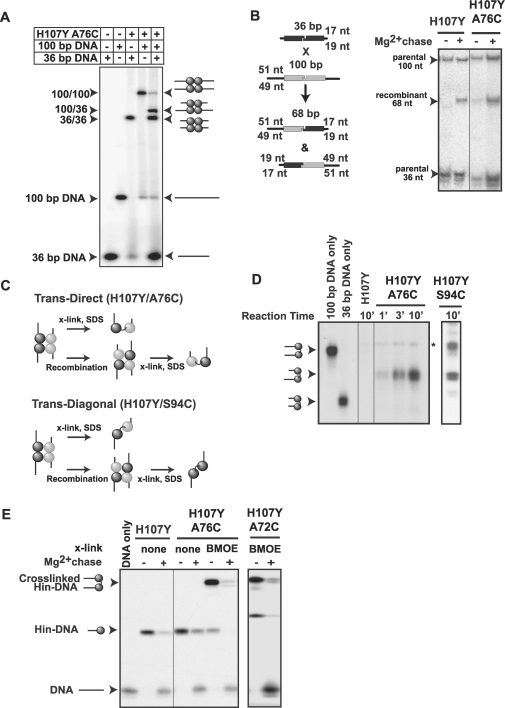
Cysteines were independently substituted for Lys72 and Ala76 in an otherwise wild-type and in a Hin–H107Y background. Hin–H107Y is a strong hyperactive mutant that can promote DNA exchange within stable, DNA-cleaved, synaptic complexes without the requirement for Fis, the enhancer, or DNA supercoiling (17). The cysteine mutants were purified and evaluated for their ability to promote DNA inversion in vitro. Hin–A76C exhibited Fis-activated inversion activity that was slightly elevated over wild-type Hin, whereas inversion by Hin–K72C was reduced to <10% wild-type rates (data not shown). Qualitative in vivo assays measuring transcription of a lacZ reporter gene as a function of Hin-catalyzed inversion (31) also showed that Hin–K72C was severely defective in promoting inversion and that Hin–A76C exhibited a slightly faster inversion rate. As shown in Figure 3A, combining K72C with the H107Y hyperactive mutation rescues the inversion defect; Hin–H107Y/A76C also exhibits similar Fis-activated inversion rates as Hin–H107Y.
Figure 3.
Activities and crosslinking of helix D mutants. (A) Inversion of DNA between the hix sites on pMS551 by Hin reorients the HindIII site to produce different sized fragments on an agarose gel when co-digested with Pst I (32). Time courses were performed with the indicated Hin mutant together with Fis and HU on supercoiled pMS551, and aliquots were taken at the times indicated. Hin–H107Y, H107Y/K72C and H107Y/A76C exhibit similar inversion kinetics. (B) Synaptic complex assembly on oligonucleotide substrates. Hin was incubated with 36 bp 3′ 32P-labeled DNA for 20 min and electrophoresed on a native polyacrylamide gel containing 10% glycerol to enhance synaptic complex formation. Each of the mutants forms large amounts of synaptic complexes. (C) Crosslinking of mutants with cysteines in helix D. Reactions were performed as in panel B and then subjected to crosslinking for 1 min. Synaptic complexes were excised from a native gel, eluted and crosslinked products were separated by SDS–PAGE. Crosslinkers were diamide (0 Å), BMOE (8 Å spacer) and BMH (16 Å spacer). The locations of non-crosslinked and crosslinked Hin–DNA(32P) covalent complexes are shown on the autoradiograph.
Encouraged that both cysteine substitutions in helix D were able to promote Fis-activated DNA inversion in the H107Y background, we evaluated their affects on formation of synaptic complexes. Hin–H107Y/K72C and Hin–H107Y/A76C were incubated for 20 min with 3′ 32P-labeled 36 bp hixL oligonucleotides under Mg2+-free ethylene glycol conditions, which promote formation of DNA cleaved synaptic complexes, and electrophoresed in a native polyacrylamide gel (Figure 3B). Both mutants accumulate tetrameric synaptic complexes at levels comparable to the Hin–H107Y control.
Synaptic complex reactions with Hin–H107Y, H107Y/K72C and H107Y/A76C were subjected to crosslinking reactions with diamide to induce direct disulfide bonds or bis-maleimide crosslinkers containing different length spacers for 1 min prior to gel electrophoresis. The cleaved synaptic complexes were eluted from the native gel and applied to SDS–polyacrylamide to separate crosslinked from non-crosslinked products (Figure 3C). As expected, Hin–H107Y forms only one product in the presence or absence of crosslinker, which corresponds to a 32P-labeled 17 nt cleaved DNA covalently bound to a Hin monomer. Crosslinking reactions with Hin–H107Y/K72C and H107Y/A76C generate an additional slower mobility species that reflects two Hin-(32P) 17 nt complexes crosslinked together. Cys72 gave up to 30% crosslinked protomers using diamide (direct), 70% with BMOE (8 Å spacer), and 75% with BMH (16 Å spacer). Cys76 gave up to 20% directly crosslinked protomers and 75% crosslinked protomers with BMOE or BMH (Figure 3C and data not shown).
Trans-direct orientation of subunits crosslinked between their D-helices
In order to map which subunits are proximal to each other over their D helices, cleaved synaptic complexes were assembled from different length DNA fragments. This approach enables Hin subunits bound to the separate hix sites to be differentially tagged by means of the different lengths of covalently associated DNA strands. In the experiment in Figure 4A, Hin–H107Y/A76C was incubated with 3′ 32P-labeled 36 or 100 bp hixL DNA fragments or a mixture of both fragments under Mg2+-free ethylene glycol conditions and electrophoresed in a native gel. The 100/36 band corresponds to a hetero-synaptic complex containing both fragments. The hetero-synaptic complex band was excised and incubated for 10 min with buffer containing Mg2+ to promote ligation followed by extraction and electrophoresis on a denaturing acrylamide gel to separate parental and recombinant DNA species. Figure 4B shows that Hin–H107Y/K76C forms 68 bp recombinant products at levels similar to H107Y, indicating that introduction of a cysteine at position 76 or 72 (not shown) does not interfere with DNA exchange activity in the oligonucleotide substrate reaction.
Figure 4.
Recombination, crosslinking and ligation of helix D cysteine Hin mutants. (A) Assembly of hetero-synaptic complexes. Hin–H107Y/A76C was incubated with 36 or 100 bp 3′ 32P-labeled hixL substrates singly or together and electrophoresed on a native polyacrylamide gel containing 10% glycerol. Synaptic complexes containing one Hin dimer bound to the 36 bp and one Hin dimer bound to the 100 bp migrate between the 100 bp synaptic complex and 36 bp synaptic complexes. (B) DNA fragment recombination assay. Recombination between 36 and 100 bp substrates results in 68 bp products. Cleaved hetero-synaptic complexes were assembled and gel-isolated from a reaction as in A and incubated with Mg2+-containing buffer for 10 min to permit ligation. The gel-extracted DNA fragments were then electrophoresed in a denaturing polyacrylamide gel. Both Hin–H107Y and H107Y/A76C form the 68 bp recombinant product. (C) Schematic representation of crosslinking between subunits in a trans-direct or trans-diagonal configuration within the Hin tetramer. If crosslinking is trans-direct, as expected for Cys76, the crosslinked diprotomer will contain a subunit in a covalent complex with a cleaved 36 bp DNA linked to a subunit in a covalent complex with a cleaved 100 bp DNA regardless of whether subunit exchange occurs. If crosslinking is trans-diagonal, as previously shown for Cys94, a mixture of crosslinked diprotomers will be present because of subunit rotation (20). (D) Hin–H107Y/A76C crosslinks subunits in a trans-direct orientation. Cleaved hetero-synaptic complexes were assembled with 3′ 32P-labeled 100 bp and unlabeled 36 bp DNA substrates. The Hin reaction was subjected to 30 s BMOE crosslinking reactions followed by a DTT quench at 1, 3 and 10 min after addition of Hin. Reactions were incubated for a total of 20 min to accumulate synaptic complexes, which were isolated from a native gel and then separated on a 10% SDS gel. Hin–H107Y/A76C accumulates only crosslinked hetero-diprotomers linked to cleaved 36 and 100 bp DNA fragments. A control reaction electrophoresed on the same gel but employing Hin–H107Y/S94C generates crosslinked hetero-diprotomers with cleaved 36 and 100 bp and crosslinked homo-diprotomers with both subunits linked to cleaved 100 bp DNA fragments. The homo-diprotomer product with two cleaved unlabeled 36 bp DNA fragments is not visible on the autoradiograph. The light band denoted with an asterisk that is also present in the H107Y control lane is not crosslinking dependent. (E) DNA ligation by crosslinked complexes. Hin reactions with 36 bp hixL fragments were subjected to 1 min crosslinking with BMOE and purified on a native polyacrylamide gel. The synaptic complexes were excised and incubated with buffer containing Mg2+ to induce ligation followed by electrophoresis on a denaturing acrylamide gel. Ligation results in a loss of the crosslinked and uncrosslinked Hin–DNA covalent complexes and an increase in the regenerated 36 bp DNA band.
The model of the synaptic complex and DNA exchange by subunit rotation predicts that subunits bound to the same side of the two hix sites [in a 2D projection, see Figures 2B, panels (iii–v) and 4C] should be connected by their D helices, and that this subunit pair should form a rotating unit. Crosslinks between proximal cysteines within helix D will therefore be between subunits in a ‘trans-direct’ configuration, and no change in the 17–49 nt hetero-diprotomer crosslinked product will occur upon subunit rotation (Figure 4C, top panel). This contrasts with the crosslinking profile that was previously observed between subunits oriented in a trans-diagonal configuration (20). In this case crosslinking initially produces the 17–49 nt hetero-diprotomer, but homo-diprotomer products are also generated as subunit exchange occurs (Figure 4C, bottom panel).
Hin–H107Y/A76C was added to Mg2+-free ethylene glycol buffer containing unlabeled 36 bp hix DNA and 32P-labeled 100 bp hix DNA fragments and subjected to 30 s crosslinking reactions with BMOE (8 Å spacer) after 1, 3 and 10 min at 37°C. After quenching the crosslinking reactions with excess DTT, the incubation was continued for a total of 20 min to accumulate synaptic complexes. Hetero-synaptic complexes were extracted from a native gel as above and then subjected to SDS–PAGE (Figure 4D). At each time point, Hin–H107Y/A76C is exclusively crosslinked in a 17–49 nt hetero-diprotomer product. Because DNA exchange is occurring within these complexes during this time frame (Figure 4B), these results demonstrate that crosslinking at Cys76 within the Hin tetramer is in a trans-direct orientation. This kinetic profile contrasts with profiles observed with cysteines located in trans-diagonal orientation (e.g. Cys94, Figure 4D) where after 10 min both hetero- and homo-diprotomer crosslinked products are nearly equally represented (20).
We then asked whether cleaved synaptic complexes whose subunits are crosslinked in a trans-direct orientation remain competent for DNA ligation. Crosslinked (1 min with BMOE) Hin–H107Y/A76C synaptic complexes assembled on 36 bp hix DNA fragments were purified on native gels. The gel slices were incubated with buffer containing Mg2+ to promote ligation, and the extracted products were electrophoresed in an SDS gel (Figure 4E). Most of the crosslinked complexes had reversed the Hin–DNA covalent linkage and ligated the hix DNA with an efficiency that was similar to the Hin–H107Y control. Similar experiments with Hin–H107Y/K72C showed that complexes crosslinked at Cys72 were also capable of efficient ligation. We conclude that tetramers crosslinked between their interacting D helices remain catalytically active.
Crosslinking throughout the helix-E region
Helix E is a predicted long α-helical region between residues 100 and 134 that connects the catalytic domain with the DNA binding domains (Figure 2A and B). The helix-E region constitutes a major part of the interface between subunits in the dimer and between all four subunits in the tetramer. Moreover, much of the surface between rotating pairs of subunits during the translocation of subunits is over residues of the E helices. Earlier experiments demonstrated that cysteines substituted at residues 129 and 134 at the C-terminal end of helix E were able to efficiently crosslink with bis-maleimide crosslinkers containing spacers of ≥8 Å when assembled in cleaved synaptic complexes (20). In the model of the Hin tetramer, residues 129 and 134 from any of the subunits are too far apart to generate the observed crosslinks. Our favored model to explain these crosslinks is that one synapsed pair of subunits rotates relative to the other to align the C-terminal ends of the E helices of subunits appropriately for crosslinking. However, alternative or additional models that could contribute to the robust crosslinking can also be considered. One model is that the C-terminal half of the helix-E region may adopt a random coil conformation that could enable crosslinking with partners without extensive movements of the rest of the protein. In support of this idea, a number of previous structural studies have found that the C-terminal half of helix E is weakly structured. Examples include NMR and several crystal structures of γδ resolvase and a crystal structure of TP901 serine integrase, all in the absence of DNA, where the polypeptide chains beyond the equivalent of Hin residues 115–123 were disordered (11,13–15,39). A recent structure of a DNA complex of the Sin resolvase dimer also exhibits very different conformations for the polypeptide chains after the equivalent of Hin residue 120 (40).
In order to investigate the structure of the putative helix E segment in Hin at the time of crosslinking and further test the subunit rotation model, cysteines were substituted at 21 positions throughout the region [denoted in Figure 5A (dimer model) and B (closed tetramer model)]. We reasoned that a helical pattern of crosslinking efficiency would provide evidence that the segment (extending to 134) was folded into an α-helix at the time of crosslinking and would provide further support for formation of a rotational conformer in which the E helices were aligned. The cysteines were introduced into the Fis-independent Hin–H107Y background to enable crosslinking using labeled oligonucleotide substrates as described above. Most of these cysteine mutants retained good activity in assembling cleaved DNA complexes on supercoiled DNA in both the presence and absence of Fis (Table 1). Thirteen formed sufficient amounts of synaptic complexes on oligonucleotide substrates to evaluate crosslinking efficiencies. One minute crosslinking reactions were performed on these mutants, and the complexes were purified on native gels followed by SDS–PAGE to evaluate crosslinking, as described for the helix D cysteine mutants (Figure 6A, Table 1). One minute crosslinking reactions were also performed on Fis/enhancer-activated reactions employing supercoiled plasmids. After crosslinking on plasmid substrates, the DNA was digested with EcoR1, which cleaves 50 bp on either side of both recombination sites, end-filled with radiolabeled nucleotides, and the products electrophoresed in SDS gels. The increased activity by Fis allowed us to obtain cysteine crosslinking data on six additional residues where cysteine mutants failed to form synaptic complexes in oligonucleotide reactions (Figure 6B, Table 1). Where data on both substrates were obtained, relative crosslinking efficiencies with different length crosslinkers were similar.
Figure 5.
Cysteine substitutions in the helix-E region. (A) Hin dimer model with the locations of the cysteine substitutions denoted with red spheres. (B) Hin tetramer model (GM4-based) with the locations of the cysteine substitutions denoted with red spheres. (C) Hin tetramer model after a 90° clockwise rotation. Substituted residues have been converted to cysteine with Sγ atoms denoted as spheres. Red designates efficient crosslinking [residues 134, 133, 130, 126 and 122 listed from the C-terminal (left) end]; orange designates crosslinking (residues 131 and 129); green designates poor or no crosslinking (residues 136, 132, 128, 124, 121 and 118); yellow designates cysteines that exhibit poor or no crosslinking and are within the catalytic core and excluded from solvent (residues 115, 112, 109, 105) (Table 1 and Figure 6). The catalytic core is rendered as a transparent surface. (D) End-on view looking down the E helices from the C-terminal ends of the yellow and blue subunits after a 90° subunit rotation. To obtain this view, the image in C was rotated about the y-axis. The dotted line denotes the rotating interface. Coloring is the same as for C. Note that rotations of 80°–105° are required to optimally position individual cysteine pairs for crosslinking. See the Supplementary Data movie for an animated view of the rotations and crosslinking.
Table 1.
Activities and crosslinking efficiencies of helix E cysteine Hin mutants
| Mutanta | Cleavageb |
Crosslinking |
||
|---|---|---|---|---|
| +Fis | −Fis | oligoc | plasmidd | |
| F104C | + | − | no SC | nd |
| F105C | +++ | +++ | − | − |
| M109C | +++ | ± | no SC | − |
| L112C | ++ | ++ | − | − |
| M115C | +++ | +++ | + | + |
| E118C | +++ | +++ | + | + |
| I120C | ± | − | no SC | − |
| V121C | +++ | +++ | − | − |
| E122C | + | + | +++ | +++ |
| R123C | − | − | no SC | nd |
| T124C | +++ | ++ | − | − |
| A126C | + | ± | no SC | +++ |
| L128C | +++ | +++ | no SC | + |
| A129C | +++ | +++ | ++ | ++ |
| A130C | + | + | +++ | +++ |
| A131C | + | + | no SC | ++ |
| R132C | +++ | +++ | − | − |
| A133C | +++ | +++ | ++ | +++ |
| Q134C | +++ | +++ | ++ | +++ |
| R136C | +++ | ++ | no SC | − |
aAll cysteine substitutions were coupled with H107Y.
bRelative amounts of DNA cleavage products generated by the Hin mutant after a 10 min incubation with supercoiled pMS551 in the presence or absence of Fis. Hin–H107Y generates >75% of substrates with double strand cleavages at each hix site, which is designated (+++). (++) indicates cleavage products are reduced to 25%–75%; (+) cleavage products reduced to 5%–25%; (±) detectable activity but <5% of Hin–H107Y; (−) no activity.
cCrosslinking efficiencies from reactions performed using oligonucleotide substrates where synaptic complexes were subjected to 1 min crosslinking reactions and then purified by native PAGE. No SC indicates the mutant did not form a sufficient amount of synaptic complex to obtain data. Relative efficiencies are from reactions using BMOE (8 Å linker). (+++), (++), and (+) indicate >35%, 10%–35% and 3%–10% of the Hin–DNA covalent complexes are crosslinked, respectively. (−) indicates trace or no detectable crosslinking.
dRelative crosslinking efficiencies from Fis-activated Hin reactions performed on supercoiled pRJ2372. Crosslinking reactions were for 1 min and directly analyzed by SDS–PAGE. nd indicates no data available because of low reactivity by the Hin mutant. Relative efficiencies are as designated for the oligonucleotide reactions.
A subset of cysteines in the helix-E region between residues 122 and 134, which extends from the core of the Hin catalytic domain, exhibited robust crosslinking. The Sγ of these residues are modeled to range from 30 to 45 Å apart in the DNA-aligned conformer and thus could not crosslink. However, as shown in Figure 5C and D, an 80–100° clockwise (or counterclockwise, not shown) rotation of synapsed subunit pairs about the flat interface aligns these residues appropriately for crosslinking with the bis-maleimide reagents. The residues that support high efficiency crosslinking exhibit an α-helical phasing pattern (Figure 6E), and all have their side chains oriented towards each other across the flat interface after subunit rotation when the region is modeled as an α-helix extending to residue 134 (Figure 5C and D). Cysteines at residues 122, 126, 130, 133 and 134 (highlighted in red) are the most active, generating 35–70% crosslinked di-protomers after a 1 min crosslinking reaction with BMOE (8 Å). These residues are predicted to have their side chains after rotation oriented the most favorably for crosslinking of all the cysteines evaluated between positions 122 and 136.
The distances between the Sγ atoms located at the C-terminal end of helix E in the Hin models or resolvase structures exceeds the 8 Å linker length of BMOE, even when each is rotated to be optimally aligned for crosslinking. However, crosslinking can be rationalized by the fact that the C-terminal ends of helix E in resolvase adopt different curvatures in different crystal forms (11). We have also modeled the Hin tetramer such that the E helices emanate in a straighter trajectory from the catalytic core; helix E-aligned conformers of this model have each of the five efficiently crosslinking cysteines within 8 Å of each other. Thus, movements that shift the ends of the helix E closer to each other are probably reasonable, and indeed, a closer approach may even be stabilized in the helix E-aligned conformer because of interactions between residues. We conclude that the region of Hin between residues 122 and 134 adopts a helical conformation at the time of crosslinking and that a rotational conformer forms within cleaved synaptic complexes in which at least the C-terminal third of the E helices between subunit pairs are positioned across from each other.
Cysteines at residues 105, 109, 112 and 115 exhibited low or no detectable crosslinking, even though they can be modeled to be appropriately oriented to generate crosslinks in a helix E-aligned conformer (Figures 5C, D and 6, Table 1). These residues are located within the core of the tetramer (Figure 5C), suggesting that their poor crosslinking efficiency is because they are not accessible to the crosslinking reagent. Significant amounts of crosslinked protomers do accumulate on supercoiled DNA with Hin–H107Y/M115C, if the reactions are incubated for longer times with the crosslinking reagent (Figure 6D). Cys105 and Cys112, which are more buried within the core, appear refractory to crosslinking even in long reactions. Chemical modification of cysteines is known to depend on their solvent exposure in addition to spatial proximity, and this feature has been used to identify buried regions of proteins [for a recent review see (41)].
DISCUSSION
Serine recombinases can be divided into a subfamily of small recombinases, exemplified by DNA invertases and resolvases, and a subfamily of much larger recombinases, exemplified by serine integrases and transposases (3). The 100–120 residue catalytic domains that are usually connected to a long helical (‘helix E’) region are relatively conserved between the different serine recombinases, but the additional polypeptide segments can vary considerably. Three structurally distinct snapshots of tetrameric catalytic domains plus helix E regions are currently available for serine recombinases: (i) the DNA-cleaved structures of γδ resolvase, which are referred here as being in a ‘closed’ conformer (10,11), (ii) an asymmetric tetramer of the catalytic domain of γδ resolvase without DNA, in which one of pair of synapsed subunits is in a closed conformation like the DNA-bound form and one is in an ‘open’ conformation where the rotational positions of the catalytic domains are different (11), and (iii) a structure of the catalytic domain of TP901 serine integrase without DNA where both synaptic subunit pairs are in an open conformation (15). In all three tetrameric crystal structures, the long E helices, which comprise much of the subunit interface of the original dimers, are reconfigured into a four helix bundle. The N-terminal half of the E helices of each newly formed synaptic (rotational) pair of subunits are associated such that their C-terminal ends point away from each other. The antiparallel helix E unit from one synaptic pair of subunits is oriented almost perpendicularly to the other with a flat and hydrophobic interface separating the pairs. In the closed tetramer, the helix E interface could enable rotation of one subunit pair relative to the other, thereby repositioning the covalently linked DNA ends into the recombinant configuration. An additional feature of the closed conformation is that interactions between synapsed subunits occur over their D helices, but these are not predicted to change during the DNA exchange reaction.
In this article, we use site-directed protein crosslinking to evaluate interactions along both the D and E helices within recombinationally active synaptic complexes of the Hin DNA invertases. Our results provide strong evidence that the Hin subunits assemble into a recombination complex that closely resembles the closed resolvase tetramer crystal structures. We also provide strong experimental support for the rotation of subunit pairs about the helix E interface. A movie correlating the observed crosslinks with the proposed movements of subunits is provided in the Supplementary Data. Taken together, we conclude that Hin forms a tetrameric molecular swivel that exchanges DNA strands during the process of site-specific DNA inversion.
Synaptic interactions between residues within the D helices
Interactions between subunits involving residues of helix D was unexpected prior to the resolvase tetramer structures but turns out to be a distinguishing feature of the closed tetramer conformation. Hin residues 72 and 76 are predicted to be sufficiently close to crosslink in the closed conformer but are 15–25 Å apart in the open tetramer model and >35 Å apart in initially synapsing dimers. Joining of the D helices appears to be coupled to positioning the catalytic domains appropriately to initiate DNA cleavage and for generating a flat interface throughout the tetramer that could support subunit rotation. We find that Hin synaptic complexes that are competent for recombination exhibit robust crosslinking between cysteines at positions 72 or 76, and we show that the crosslinks are formed between subunits bound to different hix sites in a manner consistent with a linked rotating subunit pair. Moreover, Hin tetramers crosslinked between their D helices remain active for DNA ligation, indicating that the closed conformation is catalytically active. As discussed by Kamtekar et al. (11), the existing structures are not informative as to how ligation occurs because even though the DNA ends are linearly aligned, the attacking 3′OH is about 15 Å from the serine-phosphate ester. Thus, additional movements within the context of a tetramer with tethered D helices would have to occur.
Interactions involving residues in helix D may directly or indirectly regulate Hin catalytic activity. Substitutions in residue 72 can lead to poor inversion activity (cysteine, this report) or hyperactivity (glutamate, J.K.H., unpublished results). Lys72Glu enables Hin to form stable synaptic complexes with oligonucleotide substrates. Other hyperactivating mutations within helix D that lead to variable amounts of Fis-independent inversion include Val71Ala, Lys76Cys (this report), and Glu80Arg.
The helix E rotating interface
Most of the rotating interface is comprised of residues 100–115 from the four E helices, which form a completely aliphatic surface in the tetramer. We found that cysteines between residues 105 and 115 are refractory to crosslinking, even though they are predicted to be positioned appropriately. The lack of crosslinking (or slow crosslinking in the case of Cys115) is consistent with a hydrophobic interface that is impermeable to small molecules. Earlier studies of Hin and γδ resolvase showed that cysteines positioned at the C-terminal protruding ends of helix E (residue 134) could crosslink within synaptic complexes active for DNA exchange (10,20). Since these cysteines are located up to 50 Å apart in the crystal structures, the most likely explanation for these crosslinks is that synapsed subunit pairs rotated within the tetramer to position the cysteines appropriately for crosslinking. A clockwise rotational movement is predicted to result in crosslinks between subunits bound to different recombination sites (i.e. generate crosslinked heterodimers), which was confirmed for Hin (20). Because formation of crosslinks between the ends of helix E constitutes the strongest evidence for subunit rotation, we quantitatively analyzed crosslinking at 18 positions throughout the helix-E region in Hin synaptic complexes. Cysteines substituted within the exposed C-terminal third of helix E (residues 122–134, note that 136 is predicted to be in the linker connecting helix E to the DNA binding domain) exhibited robust crosslinking in an α-helically phased manner. Modeling reveals that clockwise rotations from 80° (for Cys122 and 129) to 100° (for Cys131 and 134) are required to achieve optimal positioning for crosslinking of the active cysteines. These results significantly add to the evidence supporting subunit rotation and rule out a possible model in which the C-terminal end of the helix-E region is disordered at the time of crosslinking.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [GM038509 to R.C.J. and predoctoral fellowship GM07104 to M.M.M]. Funding for open access charge: National Institutes of Health (GM038509).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Stuart Sievers and Michael Sawaya for advice on the modeling.
Footnotes
The authors wish it to be known that, in their opinion, the first three authors should be regarded as joint First Authors.
REFERENCES
- 1.Craig NL, Craigie R, Gellert M, Lambowitz AM. Mobile DNA II. Washington, DC: ASM Press; 2002. [Google Scholar]
- 2.Grindley ND, Whiteson KL, Rice PA. Mechanisms of site-specific recombination. Annu. Rev. Biochem. 2006;75:567–605. doi: 10.1146/annurev.biochem.73.011303.073908. [DOI] [PubMed] [Google Scholar]
- 3.Smith M, Thorpe H. Diversity in the serine recombinases. Mol. Microbiol. 2002;44:299–307. doi: 10.1046/j.1365-2958.2002.02891.x. [DOI] [PubMed] [Google Scholar]
- 4.Nunes-Duby SE, Kwon HJ, Tirumalai RS, Ellenberger T, Landy A. Similarities and differences among 105 members of the Int family of site-specific recombinases. Nucleic Acids Res. 1998;26:391–406. doi: 10.1093/nar/26.2.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Swalla BM, Gumport RI, Gardner JF. Conservation of structure and function among tyrosine recombinases: homology-based modeling of the lambda integrase core-binding domain. Nucleic Acids Res. 2003;31:805–818. doi: 10.1093/nar/gkg142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tobiason DM, Buchner JM, Thiel WH, Gernert KM, Karls AC. Conserved amino acid motifs from the novel Piv/MooV family of transposases and site-specific recombinases are required for catalysis of DNA inversion by Piv. Mol. Microbiol. 2001;39:641–651. doi: 10.1046/j.1365-2958.2001.02276.x. [DOI] [PubMed] [Google Scholar]
- 7.Johnson RC. In: Mobile DNA II. Craig NL, Craigie R, Gellert M, Lambowitz AM, editors. Washington, DC: ASM Press; 2002. pp. 230–271. [Google Scholar]
- 8.Grindley NDF. In: Mobile DNA II. Craig NL, Craigie R, Gellert M, Lambowitz AM, editors. Washington, DC: ASM Press; 2002. pp. 272–302. [Google Scholar]
- 9.Feng J-A, Dickerson RE, Johnson RC. Proteins which promote DNA inversion and deletion. Cur. Opin. Struct. Biol. 1994;4:60–66. [Google Scholar]
- 10.Li W, Kamtekar S, Xiong Y, Sarkis GJ, Grindley ND, Steitz TA. Structure of a synaptic gamma delta resolvase tetramer covalently linked to two cleaved DNAs. Science. 2005;309:1210–1215. doi: 10.1126/science.1112064. [DOI] [PubMed] [Google Scholar]
- 11.Kamtekar S, Ho RS, Cocco MJ, Li W, Wenwieser SV, Boocock MR, Grindley ND, Steitz TA. Implications of structures of synaptic tetramers of gamma delta resolvase for the mechanism of recombination. Proc. Natl Acad. Sci. USA. 2006;103:10642–10647. doi: 10.1073/pnas.0604062103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang W, Steitz TA. Crystal structure of the site-specific recombinase gamma delta resolvase complexed with a 34 bp cleavage site. Cell. 1995;82:193–207. doi: 10.1016/0092-8674(95)90307-0. [DOI] [PubMed] [Google Scholar]
- 13.Sanderson MR, Freemont PS, Rice PA, Goldman A, Hatfull GF, Grindley ND, Steitz TA. The crystal structure of the catalytic domain of the site-specific recombination enzyme gamma delta resolvase at 2.7 Å resolution. Cell. 1990;63:1323–1329. doi: 10.1016/0092-8674(90)90427-g. [DOI] [PubMed] [Google Scholar]
- 14.Rice PA, Steitz TA. Refinement of gamma delta resolvase reveals a strikingly flexible molecule. Structure. 1994;2:371–384. doi: 10.1016/s0969-2126(00)00039-3. [DOI] [PubMed] [Google Scholar]
- 15.Yuan P, Gupta K, Van Duyne GD. Tetrameric structure of a serine integrase catalytic domain. Structure. 2008;16:1275–1286. doi: 10.1016/j.str.2008.04.018. [DOI] [PubMed] [Google Scholar]
- 16.Heichman KA, Johnson RC. The Hin invertasome: protein-mediated joining of distant recombination sites at the enhancer. Science. 1990;249:511–517. doi: 10.1126/science.2166334. [DOI] [PubMed] [Google Scholar]
- 17.Sanders ER, Johnson RC. Stepwise dissection of the Hin-catalyzed recombination reaction from synapsis to resolution. J. Mol. Biol. 2004;340:753–766. doi: 10.1016/j.jmb.2004.05.027. [DOI] [PubMed] [Google Scholar]
- 18.Leschziner AE, Grindley NDF. The architecture of the gamma delta resolvase crossover site synaptic complex revealed by using constrained DNA substrates. Mol. Cell. 2003;12:775–781. doi: 10.1016/s1097-2765(03)00351-4. [DOI] [PubMed] [Google Scholar]
- 19.Nollmann M, He J, Byron O, Stark WM. Solution structure of the Tn3 resolvase-crossover site synaptic complex. Mol. Cell. 2004;16:127–137. doi: 10.1016/j.molcel.2004.09.027. [DOI] [PubMed] [Google Scholar]
- 20.Dhar G, Sanders ER, Johnson RC. Architecture of the Hin synaptic complex during recombination: the recombinase subunits translocate with the DNA strands. Cell. 2004;119:33–45. doi: 10.1016/j.cell.2004.09.010. [DOI] [PubMed] [Google Scholar]
- 21.Stark WM, Sherratt DJ, Boocock MR. Site-specific recombination by Tn3 resolvase: topological changes in the forward and reverse reactions. Cell. 1989;58:779–790. doi: 10.1016/0092-8674(89)90111-6. [DOI] [PubMed] [Google Scholar]
- 22.Kanaar R, Klippel A, Shekhtman E, Dungan JM, Kahmann R, Cozzarelli NR. Processive recombination by the phage Mu Gin system: implications for the mechanisms of DNA strand exchange, DNA site alignment, and enhancer action. Cell. 1990;62:353–366. doi: 10.1016/0092-8674(90)90372-l. [DOI] [PubMed] [Google Scholar]
- 23.Wasserman SA, Dungan JM, Cozzarelli NR. Discovery of a predicted DNA knot substantiates a model for site-specific recombination. Science. 1985;229:171–174. doi: 10.1126/science.2990045. [DOI] [PubMed] [Google Scholar]
- 24.Bennett-Lovsey RM, Herbert AD, Sternberg MJ, Kelley LA. Exploring the extremes of sequence/structure space with ensemble fold recognition in the program Phyre. Proteins. 2008;70:611–625. doi: 10.1002/prot.21688. [DOI] [PubMed] [Google Scholar]
- 25.Eswar N, Webb B, Marti-Renom MA, Madhusudhan MS, Eramian D, Shen MY, Pieper U, Sali A. Comparative protein structure modeling using Modeller. Curr. Protoc. Bioinformatics. 2006 doi: 10.1002/0471250953.bi0506s15. Chapter 5, Unit 5.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chiu TK, Sohn C, Dickerson RE, Johnson RC. Testing water-mediated DNA recognition by the Hin recombinase. EMBO J. 2002;21:801–814. doi: 10.1093/emboj/21.4.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D. Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 28.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D. Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 29.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Cryst. 1993;26:283–291. [Google Scholar]
- 30.Landt O, Grunert HP, Hahn U. A general method for rapid site-directed mutagenesis using the polymerase chain reaction. Gene. 1990;26:125–128. doi: 10.1016/0378-1119(90)90351-q. [DOI] [PubMed] [Google Scholar]
- 31.Haykinson MJ, Johnson LM, Soong J, Johnson RC. The Hin dimer interface is critical for Fis-mediated activation of the catalytic steps of site-specific DNA inversion. Curr. Biol. 1996;6:163–177. doi: 10.1016/s0960-9822(02)00449-9. [DOI] [PubMed] [Google Scholar]
- 32.Johnson RC, Simon MI. Hin-mediated site-specific recombination requires two 26 bp recombination sites and a 60 bp recombinational enhancer. Cell. 1985;41:781–791. doi: 10.1016/s0092-8674(85)80059-3. [DOI] [PubMed] [Google Scholar]
- 33.Johnson RC, Bruist MF. Intermediates in Hin-mediated DNA inversion: a role for Fis and the recombinational enhancer in the strand exchange reaction. EMBO J. 1989;8:1581–1590. doi: 10.1002/j.1460-2075.1989.tb03542.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rost B, Sander C. Prediction of protein secondary structure at better than 70% accuracy. J. Mol. Biol. 1993;232:584–599. doi: 10.1006/jmbi.1993.1413. [DOI] [PubMed] [Google Scholar]
- 35.King RD, Sternberg MJ. Identification and application of the concepts important for accurate and reliable protein secondary structure prediction. Protein Sci. 1996;5:2298–2310. doi: 10.1002/pro.5560051116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Garnier J, Gibrat JF, Robson B. GOR method for predicting protein secondary structure from amino acid sequence. Methods Enzymol. 1996;266:540–553. doi: 10.1016/s0076-6879(96)66034-0. [DOI] [PubMed] [Google Scholar]
- 37.Frishman D, Argos P. Incorporation of non-local interactions in protein secondary structure prediction from the amino acid sequence. Protein Eng. 1996;9:133–142. doi: 10.1093/protein/9.2.133. [DOI] [PubMed] [Google Scholar]
- 38.Geourjon C, Deleage G. SOPMA: significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments. Comput. Appl. Biosci. 1995;11:681–684. doi: 10.1093/bioinformatics/11.6.681. [DOI] [PubMed] [Google Scholar]
- 39.Pan B, Deng Z, Liu D, Ghosh S, Mullen GP. Secondary and tertiary structural changes in gamma delta resolvase: comparison of the wild-type enzyme, the I110R mutant, and the C-terminal DNA binding domain in solution. Protein Sci. 1997;6:1237–1247. doi: 10.1002/pro.5560060612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mouw KW, Rowland SJ, Gajjar MM, Boocock MR, Stark WM, Rice PA. Architecture of a serine recombinase–DNA regulatory complex. Mol. Cell. 2008;30:145–155. doi: 10.1016/j.molcel.2008.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bass RB, Butler SL, Chervitz SA, Gloor SL, Falke JJ. Use of site-directed cysteine and disulfide chemistry to probe protein structure and dynamics: applications to soluble and transmembrane receptors of bacterial chemotaxis. Methods Enzymol. 2007;423:25–51. doi: 10.1016/S0076-6879(07)23002-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.