Abstract
Polypyrimidine tract-binding protein (PTB) is a splicing regulator that also plays a positive role in pre-mRNA 3′ end processing when bound upstream of the polyadenylation signal (pA signal). Here, we address the mechanism of PTB stimulatory function in mRNA 3′ end formation. We identify PTB as the protein factor whose binding to the human β-globin (HBB) 3′ UTR is abrogated by a 3′ end processing-inactivating mutation. We show that PTB promotes both in vitro 3′ end cleavage and polyadenylation and recruits directly the splicing factor hnRNP H to G-rich sequences associated with several pA signals. Increased binding of hnRNP H results in stimulation of polyadenylation through a direct interaction with poly(A) polymerase. Therefore, our results provide evidence of a concerted regulation of pA signal recognition by splicing factors bound to auxiliary polyadenylation sequence elements.
INTRODUCTION
The 3′ ends of most RNA polymerase II transcripts are generated by a nuclear co-transcriptional process that involves a site-specific endonucleolytic cleavage event followed by the addition of 15–200 adenylate residues (1–4).
In mammals, cleavage and polyadenylation requires the co-transcriptional assembly of a large complex consisting of at least six multimeric factors onto a bipartite core signal (pA signal), composed of the highly conserved AAUAAA hexamer situated 10–30 nucleotides upstream of the cleavage site and a more degenerate U- or GU-rich downstream sequence element (DSE). The pre-mRNA forms a stable complex through a cooperative interaction between AAUAAA hexamer-bound cleavage and polyadenylation specificity factor (CPSF) and DSE-bound cleavage stimulatory factor (CstF). Additional factors are also required such as cleavage factor I (CF I), cleavage factor II (CF II), poly(A) polymerase (PAP) and nuclear poly(A)-binding protein (PABPN1). These factors are both necessary and sufficient to reconstitute the 3′ end processing reaction in vitro (5,6). However, optimal in vivo processing not only depends upon the concerted recognition of the hexamer and DSE by the core 3′ end machinery but also utilizes additional auxiliary cis-acting sequence elements and trans-acting factors that have been reported to modulate processing efficiency.
Upstream sequence elements (USE) are U-rich regions identified in association with several cellular pA sites (7–10). A number of factors have been identified as USE-binding proteins, including splicing factors of the hnRNP and SR protein families and polyadenylation factors. Among the splicing factors, polypyrimidine tract-binding protein (PTB), a major hnRNP protein that plays multiple roles in mRNA metabolism, has been frequently found to associate with USEs. In particular, PTB has been shown to interact with the USE of complement C2 (C2), prothrombin (F2) and cyclooxigenase-2 (COX-2) pre-mRNAs where it enhances 3′ end processing efficiency (7,10,11). Contrastingly, PTB binding to the DSEs of the human α-globin and β-globin (HBB) pre-mRNA pA signals decreases the efficiency of 3′ end cleavage (12). A similar inhibitory function of DSE-bound PTB has been observed for the C2 pA signal, thus revealing that PTB may play a role in both activation and repression of polyadenylation (12). While the competition between PTB and CstF 64 for binding to the DSE can account for the general inhibitory function of PTB (12), the molecular mechanism of 3′ end processing stimulation by PTB bound to upstream enhancer elements remains unclear.
In contrast to USEs, auxiliary sequence elements located downstream of the pA site (AUX-DSEs) are less frequently characterized. The best described AUX-DSE is the G-rich sequence element (GRS) found downstream of the SV40 late (SVL) pA signal (13–16). The SVL GRS which serves as a binding site for the splicing and polyadenylation factor hnRNP H (13, 15), has been proposed to stimulate 3′ end processing by stabilizing the binding of CstF to the DSE (15,17–19). Similarly, GRSs which exhibit a stimulatory function on 3′ end cleavage through the binding of hnRNP H have been found associated with other pA signals (13,15,20,21).
The importance of auxiliary regulatory sequence elements for 3′ end processing is highlighted by the increasing number of human diseases in which this process is deregulated by mutations in cis-regulatory sequences (4). The HBB 3′ end processing reaction is highly susceptible to β-thalassemia causing mutations within both the core polyadenylation elements and the flanking regulatory sequences (4). We previously reported that β-thalassemia mutations within the last splice acceptor site of HBB pre-mRNA reduce the binding of the splicing factor U2AF65 and interfere with the efficiency of 3′ end cleavage (22).
In this report, we have investigated the mechanism by which the naturally occurring β-thalassemia term+6 (C to G) mutation, located 6 bases after the stop codon of the HBB 3′ UTR, reduces 3′ end cleavage efficiency (23). We show that the mutation resides in an evolutionary conserved pyrimidine tract and that it impairs the binding of PTB to this element. In agreement with the enhancing function of PTB in 3′ end processing, PTB tethered upstream of the HBB pA signal stimulates in vitro cleavage/polyadenylation reactions and enhances the RNA binding activity of hnRNP H. Increased binding of hnRNP H results in stimulation of polyadenylation through a direct interaction with PAP. Importantly, PTB enhances hnRNP H recruitment to other PTB-regulated pA signals, suggesting that the interaction between the two splicing factors plays a general role in pA signal recognition.
MATERIALS AND METHODS
RNA substrates
DNA templates for in vitro transcription were obtained by two rounds of PCR. The forward primers containing the r17 sequence upstream of the indicated gene-specific sequence and reverse primers used in the first PCR were the following: HBB pA signal, forward: 5′-AATTTCTATTAAAGGTTCCTT-3′ and reverse: 5′-GTTTGAACTAGCTCTTCATTTCTTTATG-3′; C2 pA signal, forward: 5′-ATGGAATTTCCCAGTTAT-3′ and reverse: 5′-GCTCTTGGAGTCATTCTGGC-3′; F2 pA signal, forward: 5′-CTAAAACTATGGTTCCCAAT-3′ and reverse: 5′-TCCCACCTCAGCCTCCCGAG-3′.
In the second round of PCR, the three PCR products were amplified using a forward primer containing the T7 promoter sequence upstream of the r17 sequence and reverse primers as in the first round of PCR. Capped, uniformly 32P-labeled RNAs used for cleavage, polyadenylation and UV cross-linking assays were obtained by in vitro transcription of these PCR products. The L3 pA signal was obtained by in vitro transcription of the R17-L3 linearized plasmid as previously described (22).
Cleavage and polyadenylation reactions
In vitro cleavage reactions were performed by incubating 32P-labeled RNA substrates with nuclear extracts (NE) for 90 min at 30°C in the presence of purified recombinant GST–R17 or GST–R17–PTB fusion proteins as previously described (24). The in vitro polyadenylation assays using NE were performed for 15 min at 30°C as for the cleavage reaction except that it was done without cordycepin and in the presence of 0.7 mM ATP and Mn2+. hnRNP H/F depleted NE were performed by three consecutive rounds of incubation of NE with 1 μg of streptavidine/agarose-bound SVL GRS for 1 h at 4°C. Alternatively, sequestration of hnRNP H/F was performed by addition of 1 μg of SVL GRS (or a control RNA) in the cleavage assay. Reconstituted polyadenylation reactions were performed for 15 min at 37°C as described in ref. (24). Analysis and quantification of cleavage/polyadenylation reactions after RNA extraction and resolution on a denaturing 6% polyacrylamide gel was done by PhosphorImager (Molecular Dynamics) analysis. Cleavage activity was calculated by dividing the amount of upstream cleavage product by the sum of cleavage plus precursor products. Polyadenylation efficiency was calculated by dividing the pre-cleaved product by the polyadenylated product.
UV crosslinking/IP
Purified GST-tagged R17, R17–PTB or PTB proteins were incubated with 32P-labeled transcripts corresponding to the HBB, C2, F2 or L3 pA signals under cleavage conditions for 30 min at room temperature. The reaction mixtures were then irradiated on ice with UV light (254 nm) in a Stratalinker (Stratagene) at 0.4 J/cm2 at 10 cm distance. Then, 50 units of RNAse ONE (Promega) was added and the reaction mixtures were incubated for 30 min at 37°C. SDS gel loading buffer was added and the samples were boiled for 2 min before fractionation on a 10% SDS–PAGE. For IP analysis, protein G sepharose was incubated with antibodies prior to addition of UV-crosslinked complexes in NETN buffer (20 mM Tris at pH 8.0, 100 mM NaCl, 0.5% NP-40, 0.5 mM EDTA). After 1 h at 4°C, bound proteins were washed six times with NETN buffer and eluted by adding SDS loading buffer to the beads. Antibodies used were hnRNP H/F (mAb 1G11, Abcam), PTB (mAb Bb7, Abcam), U2AF65 (mAb MC3;a gift of M. Carmo-Fonseca), CFIm (polyclonal; a gift of W. Keller), CstF 64 (mAb 3A1; a gift of C. McDonald).
Bioinformatical sequence motif search
In order to select genes with the PYR2 consensus motif [C/T]TTTC[C/T]TGCT in their 3′ ends, we developed three custom bioinformatics programs that extract genomic sequences, search these sequences for regular expressions and then partition the sequences based on their motif content/topology. The extraction used the UCSC human genome sequence (hg18) in conjunction with the transcript coordinates provided by the refFlat.txt SQL dump and the polyA prediction track file polyApredict.txt. Coordinates of transcripts and polyA sites were used to extract genomic sequences encompassing all 3′ UTR exons and downstream genomic sequences. A set of regular expressions were generated to match the sequence motifs of interest and to search the above-mentioned 3′ end sequence database. Finally, a program was developed to partition all sequences with matching motifs based on the order and proximity of the motifs in order to select the final set of genes that contained the desired motif topology.
Recombinant proteins
GST fusion proteins were cloned in the pGEX-2T E. coli expression vector. His-tagged hnRNP H and hnRNP F encoding plasmids were kindly provided by Douglas Black. All fusion proteins were expressed in E. coli at 20°C overnight and purified to homogeneity by glutathione agarose chromatography. The His-tagged bovine PAP (residues 1–694), hnRNP H and hnRNP F were purified to homogeneity by Ni2+–NTA chromatography as described in ref. (25). Recombinant purified PABP was kindly provided by Maria Carmo-Fonseca.
GST pull-down assays
GST pull-down assays were performed by incubating 1 μg of purified GST–R17 or GST–R17–PTB bound to 20 μl of glutathione agarose beads with 200 μg of HeLa NEs (Figure 3a) in NETN buffer (20 mM Tris at pH 8.0, 100 mM NaCl, 0.5% NP-40, 0.5 mM EDTA) for 60 min at 4°C. Beads were then washed five times, treated with 10 μg/ml RNAse A at room temperature for 30 min and washed again. Protein elution was performed by adding SDS loading buffer to the beads. Eluted proteins were resolved by 10% SDS–polyacrylamide gel electrophoresis (PAGE) and analyzed by western blot. GST pull-down assays shown in Figures 3b and 6g and h were performed as described above except that it was done with 1 μg of His-tagged hnRNP H, hnRNP F or PAP, and bound proteins were visualized by Coomassie blue staining.
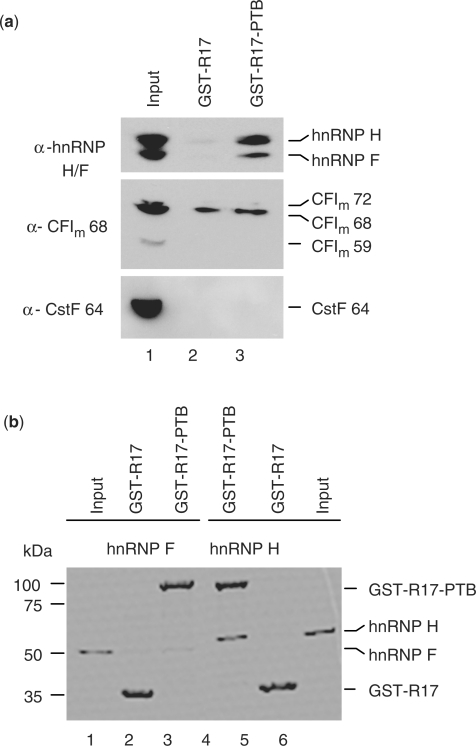
Figure 3.
PTB interacts directly with hnRNP H, but not with hnRNP F. (a) In vitro GST pull-down assay using GST-tagged R17–PTB or R17 proteins and NEs, visualized by SDS–PAGE and followed by western blot analysis using antibodies against hnRNP H/F, CFIm 68 or CstF 64. A 1/20th equivalent of the input NE is shown in lane 1. The experiments were carried out in the presence of RNase A, suggesting that the interaction was not mediated through RNA binding. (b) In vitro GST pull-down assay using GST-tagged R17–PTB or R17 proteins and recombinant His-tagged hnRNP F or hnRNP H, and this was analyzed by Coomassie staining of SDS–PAGE. The input lanes account for 10% of hnRNP H and hnRNP F used in the assay, respectively.
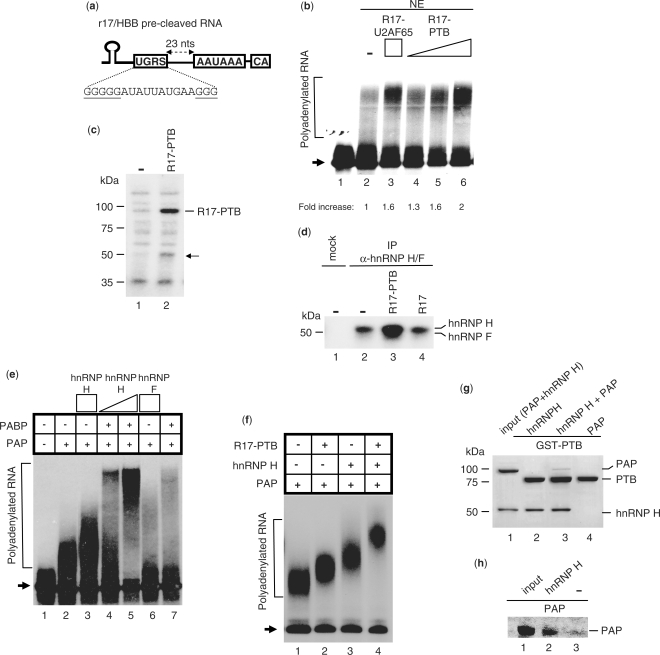
Figure 6.
PTB promotes polyadenylation by increasing the binding of hnRNP H to a GRS located upstream of the HBB pA signal. (a) Illustration of the r17/HBB pre-cleaved substrate containing a GRS element upstream of the pA signal (named UGRS). (b) In vitro polyadenylation assays using the 32P-labeled r17/HBB pre-cleaved substrate and NEs in the absence or presence of R17–U2AF65 (10 pmol) or increasing amounts of R17–PTB (2.5, 5 and 10 pmol). Normalized pA efficiency is shown. (c) UV crosslinking of NE proteins using the 32P-labeled r17/HBB pre-cleaved substrate with (5 pmol) or without R17–PTB. The arrow indicates the 50-kDa protein whose binding to the HBB pre-cleaved RNA is increased upon R17–PTB addition. (d) IP of NE proteins UV crosslinked to the 32P-labeled r17/HBB pre-cleaved substrate in the absence or presence of R17–PTB (5 pmol) or R17 (5 pmol) using the hnRNP H/F antibody. (e) Reconstituted polyadenylation assays using the 32P-labeled r17/HBB pre-cleaved substrate, PAP (0.1 pmol) and PABPN1 (1.2 pmol), in the presence of increasing amounts of hnRNP H (2 and 4 pmol) or hnRNP F (4 pmol). Lane 1: input RNA; arrow: non-polyadenylated RNA substrate. (f) Reconstituted polyadenylation assays using the 32P-labeled r17/HBB pre-cleaved substrate and PAP (0.1 pmol), in the presence of R17–PTB (4 pmol) and/or hnRNP H (4 pmol). Arrow: non-polyadenylated RNA substrate. (g) GST pull-down assay to test the interaction between GST-tagged PTB and PAP in the absence or presence of hnRNP H. The input lane accounts for 10% of hnRNP H and PAP used in the assay. (h) IP pull down assay with hnRNP H and PAP using the hnRNP H/F antibody. The input lane accounts for 10% of PAP used in the assay.
RESULTS
PTB binds to a pyrimidine tract upstream of the HBB pA signal
Previous data have shown that the term+6 mutation in the HBB 3′ UTR causes β-thalassemia by interfering with correct 3′ end formation (23). Analysis of the sequence surrounding the mutation revealed that it resides within a region containing a long series of pyrimidine residues, which represents a potential PYR tract (named PYR2, Figure 1a). This pyrimidine-rich sequence closely resembles the HBB IVSII PYR tract (named PYR1; Figure 1a) at the IVSII 3′ splicing site that binds U2AF65 and is involved in 3′ end processing (22,24).
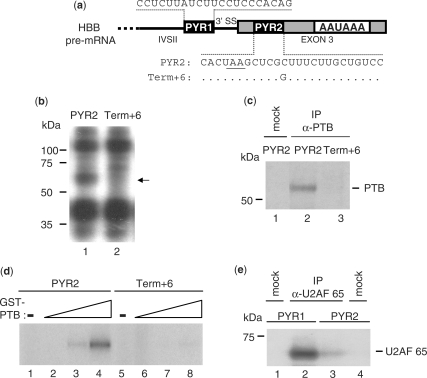
Figure 1.
PTB binding to a pyrimidine tract within the HBB 3′ UTR is impaired by the β-thalassemia term+6 mutation. (a) Schematic representation of the 3′ terminal region of the HBB pre-mRNA showing the sequence of two pyrimidines tracts, PYR1 and PYR2, located at the last intron (IVSII) 3′ splice site (3′SS) and in the terminal exon (exon 3), respectively. The sequence difference between the PYR2 and term+6, which contains the β-thalassemia C to G mutation at 6 nucleotides after the stop codon (underlined), is shown. (b) UV crosslinking using 5′ 32P-labeled RNA oligonucleotides corresponding to the PYR2 or term+6 RNAs in the presence of NEs and resolution by SDS–PAGE. The 55-kDa protein UV crosslinked to the PYR2 but not to the term+6 RNA is indicated by the arrow. (c) IP of the UV-crosslinked complexes described in (b) with the PTB antibody (Bb7) followed by SDS–PAGE analysis. (d) UV crosslinking of GST-tagged PTB (2.5, 1.25 or 0.6 pmol) to the PYR2 or term+6 RNAs. Lanes 1 and 5: UV crosslinking in the absence of PTB. (e) IP following UV crosslinking of NEs to 5′ 32P-labeled PYR1 or PYR2 with the U2AF65 antibody followed by SDS–PAGE analysis.
In order to determine the protein factor(s) that bind to PYR2 and that may be involved in the mechanism by which the term+6 mutation affects 3′ end processing, we performed UV crosslinking and immunoprecipitation (IP) experiments. UV crosslinking using a 32P-end labeled RNA oligonucleotide corresponding to PYR2 (PYR2 RNA) in the presence of HeLa cell NEs produced three major bands (Figure 1b). The term+6 mutation within PYR2 (term+6 RNA) abolished the binding of a protein of 55 kDa. This band was identified as PTB by IP of the UV-crosslinked complexes with the PTB antibody (Figure 1c). No IP bands were detectable using either the mutated RNA or a mock IP (Figure 1c), indicating that binding of PTB to PYR2 RNA was eliminated by the term+6 mutation.
Next, we performed UV crosslinking experiments using recombinant, purified PTB. Even the highest concentrations of PTB were only able to bind to the wild-type PYR2 RNA (Figure 1d). As U2AF65 has been found by us (22) and others (11) to regulate 3′ end processing by binding to upstream pyrimidine-rich regulatory elements, we next determined whether this splicing factor was able to bind to PYR2 RNA. UV crosslinking/IP experiments with the U2AF65 antibody gave a strong immunoprecipitated complex with PYR1 RNA, while only a faint band was detectable with PYR2 RNA (Figure 1e). This suggests that U2AF65 more closely associates with the PYR1 compared to the PYR2 RNA.
Taken together, these results demonstrate that the HBB term+6 mutation impairs the binding of PTB to the PYR2 element and that PTB, but not U2AF65, may be the factor involved in the mechanism of 3′ end processing deregulation induced by this genetic defect.
PTB bound upstream of the pA signal stimulates 3′-end cleavage
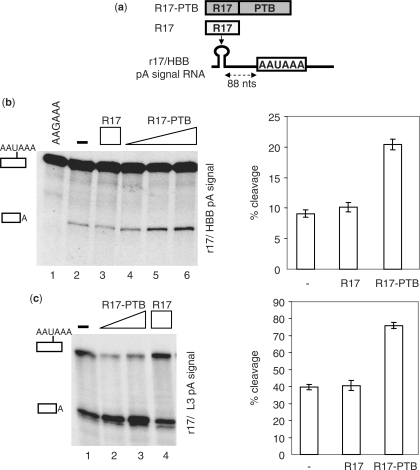
PTB has been shown to play an inhibitory function in the HBB 3′ end processing by competing with the cleavage factor CstF 64 for the U-rich DSE (12). We therefore investigated the function of PTB bound upstream of the HBB pA signal by tethering a R17/MS2-PTB fusion protein to a 32P-labeled HBB pre-mRNA containing a high-affinity R17 binding site (named r17) in place of PYR2 (Figure 2a) and using NEs. No cleavage product was observed with an RNA substrate containing a AAUAAA to AAGAAA mutation in the HBB pA signal (Figure 2b). Addition of R17–PTB, but not R17 alone, increased cleavage efficiency in a dose-dependent manner up to 2-fold (Figure 2b). Similar results were obtained using the r17/adenovirus L3 (L3) pA signal substrate (Figure 2c), suggesting a more general positive function of PTB in 3′ end cleavage when bound upstream of a pA signal.
Figure 2.
PTB stimulates in vitro 3′ end cleavage in NEs. (a) Illustration of the ‘tethering’ assay consisting in binding R17–PTB or R17 fusion proteins to the high-affinity R17 binding site (r17) inserted in place of the PYR2 element located 88 nucleotides upstream of the HBB pA signal. (b) In vitro cleavage reactions using the r17/HBB pA signal substrate containing the wild-type (AAUAAA) or the mutated (AAGAAA) hexamer. 32P-labeled transcript was incubated with NE in the presence of an increasing amount (3.5, 7 or 14 pmol) of R17–PTB or R17 (14 pmol). (c) Same as (b), with the 32P-labeled RNA containing the R17 binding site upstream of the L3 pA signal and R17–PTB (7 or 14 pmol) or R17 (14 pmol) proteins. Identities of the uncleaved and cleaved products are shown on the left. The percentage of cleavage in the absence or in the presence of 14 pmol of R17–PTB or R17 proteins is shown on the right of (c) and (d), respectively.
PTB interacts directly with hnRNP H
In order to dissect the molecular mechanism whereby PTB stimulates 3′ end processing, we wished to identify proteins with which PTB interacts. As PTB and hnRNP H form a cooperative assembly on the splicing downstream control sequence element of the c-src pre-mRNA (26), we tested the interaction between PTB and hnRNP H family regulators of 3′ end processing, including hnRNP H/H′ and hnRNP F. To this end, we performed a glutathione S-transferase (GST) pull-down experiment using GST–R17–PTB and NEs. Western blot analysis with an hnRNP H/F antibody detected both hnRNP proteins, suggesting an interaction between PTB and both hnRNP H and F (Figure 3a). No protein–protein interactions were found between PTB and two other 3′ cleavage/polyadenylation factors, CstF 64 and CFIm, which are often targeted by 3′ end processing regulators (Figure 3a). GST pull-down assays using only recombinant proteins revealed that hnRNP H was able to bind to GST–R17–PTB but not to the control GST–R17 fusion protein; no binding was observed for hnRNP F (Figure 3b).
We therefore conclude that PTB binds directly to hnRNP H. The interaction between PTB and hnRNP F shown in Figure 3a is probably indirect and mediated by the ability of hnRNP H to heterodimerize with hnRNP F (27).
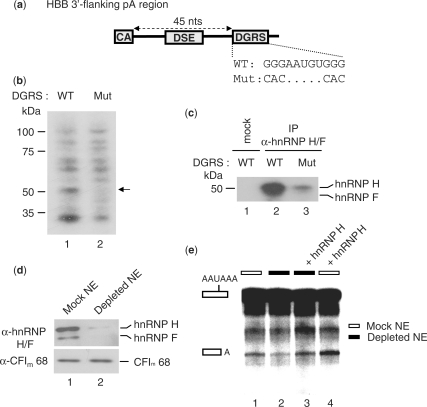
HnRNP H regulates 3′ end formation of HBB pre-mRNA by binding to a G-rich AUX-DSE
The results shown in Figure 3 raise the possibility that the interaction between PTB and hnRNP H may be involved in the stimulatory effect of PTB on 3′ end processing. If so, we would expect that hnRNP H is able to modulate HBB pre-mRNA 3′ end processing efficiency by binding to a GRS flanking the core pA signal. Analysis of the HBB region downstream of the DSE revealed the presence of a putative GRS located 45 nucleotides downstream of the cleavage site (named DGRS, Figure 4a). To ascertain whether hnRNP H binds to this sequence element, we performed UV crosslinking assays using 32P-labeled RNA substrates corresponding to the HBB 3′ flanking region containing either wild-type (WT) or mutant (Mut) DGRS in the presence of NEs. A comparison of the UV crosslinking patterns of WT and Mut substrate RNAs revealed a specific band at 55 kDa that disappeared with the G-to-C/A mutation (Figure 4b). IP of the crosslinked complexes with the hnRNP H/F antibody showed that the 55-kDa band corresponds to hnRNP H/F and that the G-to-C/A mutation mostly abolished hnRNP H/F binding to the DGRS (Figure 4c).
Figure 4.
HnRNP H stimulates 3′ end cleavage at the HBB pA signal by binding to a downstream GRS element. (a) Illustration of the HBB region downstream of the cleavage site (CA) and including a GRS located 45 nucleotides downstream of the cleavage site (DGRS). The sequence difference between the wild-type (WT) and mutated DGRS (Mut) is shown. (b) UV crosslinking of NE proteins to the uniformly 32P-labeled r17/HBB pA signal RNA substrate containing the WT or Mut DGRS sequence. The 50-kDa protein crosslinked to the WT but not to the Mut RNA substrate is indicated by the arrow. (c) IP of UV-crosslinked complexes described in panel (b) with the hnRNP H/F antibody followed by SDS–PAGE analysis. (d) NEs depleted of endogenous hnRNP H/F without (mock) or with (depleted) a biotinylated RNA oligonucleotide corresponding to the SVL GRS. NEs were analyzed by SDS–PAGE followed by western blot analysis. (e) In vitro cleavage assays using the 32P-labeled r17/HBB substrate in the presence of mock-depleted NEs (white boxes) or NEs depleted of hnRNP H/F (black boxes), supplemented or not with recombinant hnRNP H (1 pmol). The low cleavage efficiency is due to the depletion procedure. Identities of the uncleaved and cleaved products are shown on the left.
We next investigated the function of hnRNP H in HBB 3′ end cleavage by performing in vitro cleavage assays using NEs that had been either mock or depleted of endogenous hnRNP H/F and add-back experiments using recombinant hnRNP H or F. The endogenous hnRNP H/F was efficiently depleted by incubation of NEs with a biotinylated RNA containing the SVL GRS and streptavidin–agarose (Figure 4d). As shown in Figure 4e, depletion of hnRNP H/F reduced 3′ end cleavage compared to mock-depleted extracts. Adding back recombinant hnRNP H restored 3′ end cleavage in depleted extracts (Figure 4e), while raising the level of hnRNP H in mock extracts increased the formation of the 3′ end cleavage product (Figure 4e). Unlike hnRNP H, add-back experiments with hnRNP F did not restore 3′ end processing in depleted extracts nor did increasing the amount of hnRNP F reduce 3′ end cleavage in mock NEs (Supplementary Figure S1). Similar results were obtained by performing 3′ end cleavage assays in the presence of SVL GRS RNAs that sequester hnRNP H/F from the 3′ end processing complex (Supplementary Figure S2).
Collectively, our results show that the 3′ end processing of the HBB pA signal can be modulated by hnRNP H binding to the GRS downstream of the cleavage site.
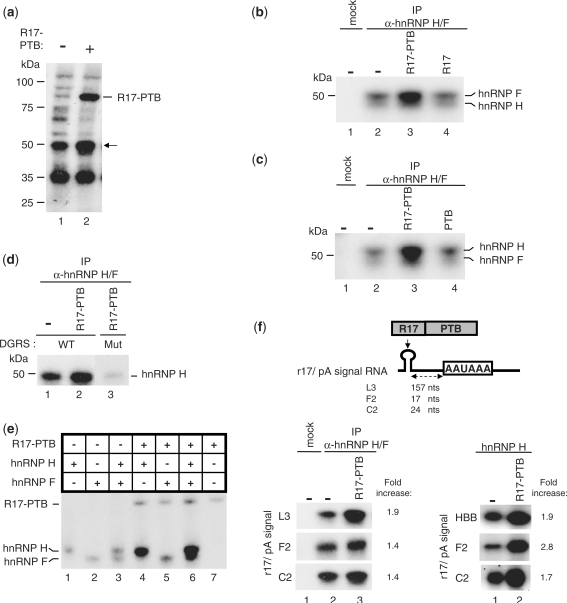
PTB bound upstream of the pA signal recruits hnRNP H to the pre-mRNA
To investigate whether PTB was able to recruit hnRNP H to HBB transcripts, we analyzed hnRNP H RNA binding activity upon addition of R17–PTB using UV crosslinking/IP experiments. The 32P-labeled r17/HBB pA signal RNA was incubated in NEs followed by the addition of the recombinant R17–PTB. The UV crosslinking pattern showed an increase in the binding of a 55-kDa species upon addition of R17–PTB (Figure 5a). This band was identified as hnRNP H/F by IP of the crosslinked proteins with the hnRNP H/F antibody (Figure 5b). Addition of R17–PTB (Figure 5b), but not of R17 (Figure 5b) or of PTB alone (Figure 5c), resulted in an enhanced hnRNP H crosslinking only in the presence of a WT DGRS (Figure 5d), suggesting that the physical interaction between PTB and hnRNP H creates a molecular link across the HBB pA signal.
Figure 5.
PTB facilitates hnRNP H binding to the HBB pA signal. (a) UV crosslinking using NEs with the 32P-labeled r17/HBB substrate in the absence or presence of R17–PTB. The identity of the 50-kDa protein whose binding to the RNA is increased upon R17–PTB addition (indicated by an arrow) was tested in (b) by IP of the UV-crosslinked complexes with the antibody against hnRNP H/F. SDS–PAGE analysis of the immunoprecipitated complexes UV crosslinked to the 32P-labeled r17/HBB substrate in the absence or presence of R17–PTB or R17. (c) UV crosslinking/IP as described in (b) except for lane 3 in which PTB replaced R17. The poor quality of the migration of the crosslinking reaction is due to the presence of polyvinylalcohol in the reaction. (d) UV crosslinking/IP as described in (b) but using a 32P-labeled r17/HBB substrate containing the DGRS WT or Mut (as described in Figure 4). (e) UV crosslinking of recombinant hnRNP H and/or hnRNP F to the 32P-labeled r17/HBB substrate in the absence or presence of the R17–PTB protein as indicated (upper panel). (f) (Left panel) UV crosslinking/IP of hnRNP H/F from NEs to 32P-labeled RNA substrates containing the r17 moiety upstream of the L3, F2 or C2 pA signals (illustrated in the upper part) in the absence or presence of R17–PTB. (Right panel) UV crosslinking of hnRNP H with or without R17–PTB and using the 32P-labeled r17/HBB, C2 or F2 pA signals.
In order to better characterize the effect of PTB on the RNA-binding activity of both hnRNP H and F, we performed UV crosslinking assays using only the r17/HBB pA signal substrate and recombinant proteins. Addition of R17–PTB strongly increased the binding of hnRNP H to the HBB substrate (Figure 5e). The effect of R17–PTB on hnRNP F binding to the RNA is less pronounced with hnRNP F alone but stronger in the presence of hnRNP H (Figure 5e). We therefore conclude that PTB bound upstream of the HBB pre-mRNA pA signal directly stimulates the binding of hnRNP H and indirectly helps recruit hnRNP F through heterodimerization with hnRNP H.
We next asked whether PTB was able to recruit hnRNP H on other pA signals that are modulated by PTB and possess a GRS downstream of the cleavage site. We performed UV crosslinking/IP assays using NEs and 32P-labeled RNA substrates corresponding to the C2 and F2 pA signals containing the r17 moiety in place of the natural PTB-binding site (Figure 5f). Since the L3 pA signal can be regulated by PTB (Figure 2c), we also tested the r17/L3 pA substrate. As shown in Figure 5f (left panel), R17–PTB bound to sequences upstream of several pA signals stimulated hnRNP H binding to the RNA. This stimulatory effect was stronger when performing UV crosslinking of the r17/C2 or the r17/F2 substrates using only recombinant hnRNP H and R17–PTB (Figure 5f, right panel). Overall, these results suggest a more general stimulatory effect of PTB on the hnRNP H-RNA interaction.
PTB simulates polyadenylation by recruiting hnRNP H on a GRS located upstream of the pA signal
We next analyzed whether PTB was able to influence polyA addition through its recruitment of hnRNP H to GRSs upstream of the pA signal. This hypothesis is based on previous studies that suggest an implication of PTB or hnRNP H in polyadenylation (7,10,11). In addition, we identified a putative hnRNP H binding site between PYR2 and the pA signal of the HBB pre-mRNA (named UGRS, Figure 6a).
To verify this hypothesis, we performed in vitro polydenylation reactions with NEs using a 32P-labeled r17/HBB pre-cleaved substrate (Figure 6a). We first tested whether PTB functions as a regulator of the polyadenylation reaction. Addition of recombinant PTB stimulated addition of adenosine residues in a dose-dependent fashion (Figure 6b) to an extent similar to that obtained in the presence of the 3′ end processing regulator U2AF65 (Figure 6b) (24).
To investigate the possible involvement of the PTB–hnRNP H interaction in the stimulatory effect of PTB on polyadenylation, we checked if PTB stimulated the interaction between hnRNP H and the pre-cleaved substrate using UV crosslinking/IP experiments with NEs. As expected, the UV crosslinking pattern of the pre-cleaved substrate was remarkably distinct in the 50-kDa region (Figure 6c), from that generated with the substrate including the HBB 3′ flanking region (Figure 5a). However, the addition of R17–PTB still resulted in an increased crosslinked protein at this molecular size (Figure 6c). IP of the crosslinked complexes with the hnRNP H/F antibody confirmed that hnRNP H is the protein factor whose binding is potentiated by addition of R17 linked to PTB (Figure 6d).
If the increased binding of hnRNP H to the RNA is responsible for the stimulation of pA addition by PTB, then it should be possible to reproduce the stimulatory effect by increasing the amount of recombinant hnRNP H in a polyadenylation assay. To test this possibility, we performed reconstituted polyadenylation assays using only recombinant PAP, PABPN1, hnRNP H or F and R17–PTB. As shown in Figure 6e, the polyadenylation activity of PAP alone or in the presence of PABPN1 is increased by the addition of hnRNP H. Conversely, when the polyadenylation reaction occurred in the presence of hnRNP F, the polyadenylation activity of PAP was significantly reduced (Figure 6e). While additional experiments are needed to further investigate the inhibitory activity of hnRNP F on polyadenylation, our results confirm that hnRNP H plays a role in stimulating pA addition. We next tested whether this stimulatory effect can be further potentiated in the presence of R17–PTB. R17–PTB was able to modify PAP activity only slightly (Figure 6f) suggesting that the stimulation of pA addition by R17–PTB in NEs does not rely on a direct effect of PTB on PAP. Interestingly, when both R17–PTB and hnRNP H were included in the reconstituted polyadenylation assay, the stimulatory activity of hnRNP H on poly(A) tail elongation was further increased compared to hnRNP H alone (Figure 6f). A possible explanation for these results is that PTB recruits PAP indirectly by means of its direct interaction with hnRNP H. This hypothesis implies that hnRNP H and PAP interact directly. We tested this possibility by performing GST pull-down assays using recombinant GST–PTB, hnRNP H and PAP and found that PTB was able to interact with hnRNP H but not with PAP alone (Figure 6g). Conversely, when both PAP and hnRNP H were present in the assay, PAP was pulled down by PTB (Figure 6g), suggesting that the binding of PTB to PAP is mediated by hnRNP H. To confirm this possibility using the same proteins as in the functional and pull-down assays, we immunoprecipitated hnRNP H and checked whether PAP was found in the IP fraction. As shown in Figure 6h, PAP was pulled down only in the presence of hnRNP H.
Taken together, these results show that PTB bound upstream of the HBB pA signal facilitates the association of hnRNP H with a GRS that in turn recruits PAP thereby stimulating pA addition.
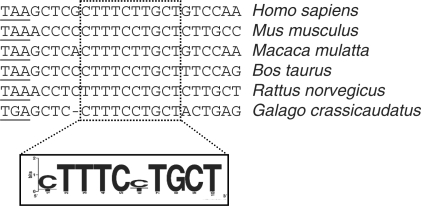
The HBB PYR2 element is conserved and can be found in other pA signals
Our results highlight the importance of the PYR2 element for efficient HBB pre-mRNA 3′ end processing. The term+6 β-thalassemia causing mutation within this element reduces both the binding of PTB and 3′ end processing efficiency. A sequence alignment of the 3′ ends of vertebrate HBB genes revealed that the nucleotide affected by the β-thalassemia mutation and the sequence surrounding this base are highly conserved among higher vertebrates (Figure 7). The sequence surrounding the mutation is the pyrimidine-rich region (9 pyrimidines out of 10 nucleotides of the most strongly conserved element) with only two pyrimidine transitions. The upstream, conserved element is also positionally preserved. To gain insight into the possible involvement of this sequence in the control of 3′ end formation of other mRNAs, we analyzed if other human mRNAs contain the sequence [C/T]TTTC[C/T]TGCT upstream of the pA signal. We identified 60 genes that contain this element in their pA signal region. Interestingly, 56 out of the 60 identified genes contained at least one GGG downstream/upstream of the pA signal (Supplementary Table S1).
Figure 7.
Eukaryotic conservation of the PTB-bound pyrimidine-rich element within the HBB 3′ UTR as demonstrated by multiple sequence alignment.
DISCUSSION
Bioinformatic analyses of the sequence surrounding the pA signal identified, in addition to the core elements, auxiliary over-represented cis-acting sequences, including U-rich upstream motifs and GRSs positioned both upstream and downstream of the pA signal (28). Both cis-elements have an enhancing function in 3′ end formation through the binding of trans-acting proteins, including two splicing regulators, PTB and hnRNP H. PTB interacts with the U-rich USE and stimulates 3′-end cleavage by an unknown mechanism (7,10,11), whereas hnRNP H binds to the GRS located at the AUX-DSE and promotes 3′ end formation by increasing the binding of CstF64 to the core pA signal (15,17–19). Our in vitro functional studies show that PTB and hnRNP H positively regulate both steps of HBB pre-mRNA 3′ end processing by binding to an upstream U-rich element and two GRS sequences positioned on either side of the pA signal, respectively. More importantly, we demonstrate that PTB bound to the upstream element facilitates the interaction between hnRNP H and the two GRSs proximal to the HBB pA signal and that this physical interaction plays an enhancing function in both steps of 3′ end formation. Thus, we provide evidence of a concerted regulation of pA signal recognition by splicing factors bound to auxiliary polyadenylation sequence elements.
On the basis of three main findings, we propose that this is a general mechanism of 3′ end processing regulation. First, we have found that the HBB pyrimidine U-rich element is associated with 60 pA signals and that 92% of these pA signals also possess a GRS upstream and/or downstream of the core element (Supplementary Table S1). This result suggests that PTB bound to the pyrimidine U-rich element may function to modulate the 3′ end processing efficiency of other pA signals and that hnRNP H may be involved in this stimulatory effect. Second, the sequence analysis of several PTB-regulated pA signals (including the C2, F2, proximal COX-2 and CT/CGRP exon 4 pA signals) has revealed that all of these pA signals contain a GRS in the proximity of the core pA element. Moreover, in the case of the C2 and F2 pA signals, we show that PTB stimulates hnRNP H binding to RNA (Figure 5f). Finally, we show that the strong L3 pA signal can also be positively regulated by PTB bound upstream of the AAUAAA hexamer and that this stimulatory effect is associated with increased binding of hnRNP H, presumably to a G-rich motif present in the functional AUX-DSE (17; our observation).
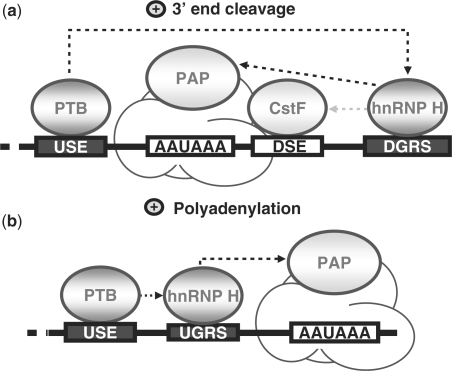
One clue to the mechanism by which PTB stimulates 3′ end processing via recruitment of hnRNP H comes from the observation that all of the pA signals considered in this study are associated with weak hnRNP H binding sites. Indeed, hnRNP H was identified as the protein factor that strongly binds to the SVL GRS. This element has the propensity to form G-quadruplex structures (29), and RNA secondary structures have been shown to play a role in polyadenylation (30,31). In addition, mutational analysis of the SVL GRS revealed that mutations disrupting the G-quadruplex structure not only affect the function of the GRS in the 3′ end processing of the SVL late pA signal but also reduce the GRS–hnRNP H interaction (19). Both GRS elements of the HBB pA signal do not form G-quadruplex structures according to bioinformatic prediction (QGRS mapper, (32) and our experimental observations (A.D., unpublished data), indicating that they may represent weak binding sites for hnRNP H. However, previous studies have shown that hnRNP H is able to stimulate 3′ end processing even when bound to weak binding sites and suggest that an accessory factor is needed to strengthen hnRNP H association and potentiate its stimulatory function (13). In agreement with this hypothesis and with the general view that auxiliary cis-acting motifs aid in the use of suboptimal polyadenylation sequence elements, we propose that PTB, bound to upstream auxiliary elements, exerts its stimulatory function by associating hnRNP H with suboptimal GRS elements and facilitating the assembly of the polyadenylation machinery (Figure 8).
Figure 8.
Illustration of mechanistic models for the stimulatory functions of PTB in 3′ end processing. (a) PTB and 3′ end cleavage. We propose that PTB facilitates the binding of hnRNP H to the DGRS located at the AUX-DSE, which in turn, stimulates the assembly of the 3′ end cleavage machinery through the recruitment of CstF or PAP. (b) PTB and polyadenylation. We propose that PTB stabilizes the association of hnRNP H with a UGRS located upstream of the pA signal where it can increase the processivity of PAP. Black arrows indicate the functional interactions shown in this study while the grey arrow refers to the previously reported ability of hnRNP H to increase CstF binding to the DSE element (14,15,17–19).
The occurrence of a molecular and functional link between PTB and hnRNP H bound to their cis-regulatory elements may be critical for pA signals that inefficiently recruit the polyadenylation machinery due to U/GU-rich DSEs that are suboptimal for CstF binding, such as the DSEs from C2 and F2. For both pA signals, PTB bound to the USE stimulates both 3′ end processing efficiency (7,11) and hnRNP H binding (Figure 5). Increased association of hnRNP H stimulates the association of a stable complex between CstF64 and the DSE (14,15,17–19). Unlike the F2 and C2 pA signals, the HBB pA signal consists of a consensus CPSF binding site and a U-rich DSE and is therefore very efficient in terms of 3′ end processing. For this strong pA signal, the function of cis-auxiliary sequences is to further improve the cleavage/polyadenylation reaction, as shown for the pyrimidine tract near the last intron 3′ splice site (24) or for the heterologous F2 USE inserted upstream of the HBB pA signal (11). The β-thalassemia term+6 mutation reduces 3′ end efficiency by 22–30% (23), in agreement with the moderate ability of cis-regulatory sequences to modulate the efficiency of pA signals. As expected for a pA signal with a strong DSE, the PTB-induced increase in hnRNP H binding does not result in an enhanced binding of CstF 64 to the HBB cleavage substrate (data not shown). Consequently, the stimulatory function of hnRNP H might involve other components of the polyadenylation machinery. Our results show that hnRNP H stimulates pA addition in reconstituted polyadenylation assays (Figure 6e and f) and that it interacts with PAP (Figure 6g and h), implying that a PAP–hnRNP H interaction plays a role in the stimulatory function of PTB. Since PAP is often involved in both 3′ end cleavage and polyadenylation processes (33), a possible mechanism is that PTB stimulates the binding of hnRNP H to GRSs and that hnRNP H, in turn, enhances PAP activity in both steps of mRNA 3′ end formation (Figure 8). According to this model and considering that the recruitment of PAP to pA signals is a late event in the assembly of the pA machinery, modulation of PAP association through PTB–hnRNP H concerted regulation can only moderately modify HBB cleavage efficiency. Since CstF can also enhance polyadenylation when bound to upstream cis-elements (7), it is possible that hnRNP H stimulates CstF binding to the upstream region and promotes pA addition. Although it will be important to study interactions between hnRNP H and other components of the polyadenylation machinery, we propose that hnRNP H primarily influences 3′ end processing activity through PAP and CstF and that these interactions constitute the main determinant of PTB-mediated 3′ end processing regulation.
We have previously shown that β-thalassemia causing mutations within the 3′ splice site pyrimidine tract interfere with correct HBB pre-mRNA 3′ end processing by reducing the ability of U2AF65 to recruit CF Im to the pA signal (22, 24). In this study, we demonstrate that the term+6 mutation in the HBB 3 UTR, which reduces 3′ end processing, impairs the binding of PTB to a pyrimidine U-rich element. Our results show that a PTB-induced increase in binding of hnRNP H to GRS elements can account for the positive function of PTB in 3′ end processing. Based on our results, we propose that disease-causing mutations not only disrupt the interactions between an mRNA and a trans-acting factor, but also disable the formation of synergistic complexes that regulate and perform 3′ end processing reactions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
French National Institute for Health and Medical Research, the Institut Claudius Regaud, the Fondation de France; and by the Fondation pour la recherche médicale (Equipe FRM, soutenue par la Fondation Recherche Médicale) for work in the laboratory of S.V. Funding for open access charge: INSERM.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank D. Black for hnRNP H and F plasmids, M. Carmo-Fonseca for purified PABP and MC3 antibody, C. McDonald for CstF 64 antibody and W. Keller for CFIm 68 antibody.
REFERENCES
- 1.Proudfoot NJ, Furger A, Dye MJ. Integrating mRNA processing with transcription. Cell. 2002;108:501–512. doi: 10.1016/s0092-8674(02)00617-7. [DOI] [PubMed] [Google Scholar]
- 2.Rosonina E, Kaneko S, Manley JL. Terminating the transcript: breaking up is hard to do. Genes Dev. 2006;20:1050–1056. doi: 10.1101/gad.1431606. [DOI] [PubMed] [Google Scholar]
- 3.Mandel CR, Bai Y, Tong L. Protein factors in pre-mRNA 3′-end processing. Cell Mol. Life Sci. 2008;65:1099–1122. doi: 10.1007/s00018-007-7474-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Danckwardt S, Hentze MW, Kulozik AE. 3′ end mRNA processing: molecular mechanisms and implications for health and disease. EMBO J. 2008;27:482–498. doi: 10.1038/sj.emboj.7601932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Takagaki Y, Ryner LC, Manley JL. Four factors are required for 3′-end cleavage of pre-mRNAs. Genes Dev. 1989;3:1711–1724. doi: 10.1101/gad.3.11.1711. [DOI] [PubMed] [Google Scholar]
- 6.Wahle E. Purification and characterization of a mammalian polyadenylate polymerase involved in the 3′ end processing of messenger RNA precursors. J. Biol. Chem. 1991;266:3131–3139. [PubMed] [Google Scholar]
- 7.Moreira A, Takagaki Y, Brackenridge S, Wollerton M, Manley JL, Proudfoot NJ. The upstream sequence element of the C2 complement poly(A) signal activates mRNA 3′ end formation by two distinct mechanisms. Genes Dev. 1998;12:2522–2534. doi: 10.1101/gad.12.16.2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brackenridge S, Proudfoot NJ. Recruitment of a basal polyadenylation factor by the upstream sequence element of the human lamin B2 polyadenylation signal. Mol. Cell Biol. 2000;20:2660–2669. doi: 10.1128/mcb.20.8.2660-2669.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Natalizio BJ, Muniz LC, Arhin GK, Wilusz J, Lutz CS. Upstream elements present in the 3′-untranslated region of collagen genes influence the processing efficiency of overlapping polyadenylation signals. J. Biol. Chem. 2002;277:42733–42740. doi: 10.1074/jbc.M208070200. [DOI] [PubMed] [Google Scholar]
- 10.Hall-Pogar T, Liang S, Hague LK, Lutz CS. Specific trans-acting proteins interact with auxiliary RNA polyadenylation elements in the COX-2 3′-UTR. RNA. 2007;13:1103–1115. doi: 10.1261/rna.577707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Danckwardt S, Kaufmann I, Gentzel M, Foerstner KU, Gantzert AS, Gehring NH, Neu-Yilik G, Bork P, Keller W, Wilm M, et al. Splicing factors stimulate polyadenylation via USEs at non-canonical 3′ end formation signals. EMBO J. 2007;26:2658–2669. doi: 10.1038/sj.emboj.7601699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Castelo-Branco P, Furger A, Wollerton M, Smith C, Moreira A, Proudfoot N. Polypyrimidine tract binding protein modulates efficiency of polyadenylation. Mol. Cell Biol. 2004;24:4174–4183. doi: 10.1128/MCB.24.10.4174-4183.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arhin GK, Boots M, Bagga PS, Milcarek C, Wilusz J. Downstream sequence elements with different affinities for the hnRNP H/H′ protein influence the processing efficiency of mammalian polyadenylation signals. Nucleic Acids Res. 2002;30:1842–1850. doi: 10.1093/nar/30.8.1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bagga PS, Ford LP, Chen F, Wilusz J. The G-rich auxiliary downstream element has distinct sequence and position requirements and mediates efficient 3′ end pre-mRNA processing through a trans-acting factor. Nucleic Acids Res. 1995;23:1625–1631. doi: 10.1093/nar/23.9.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bagga PS, Arhin GK, Wilusz J. DSEF-1 is a member of the hnRNP H family of RNA-binding proteins and stimulates pre-mRNA cleavage and polyadenylation in vitro. Nucleic Acids Res. 1998;26:5343–5350. doi: 10.1093/nar/26.23.5343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qian ZW, Wilusz J. An RNA-binding protein specifically interacts with a functionally important domain of the downstream element of the simian virus 40 late polyadenylation signal. Mol. Cell Biol. 1991;11:5312–5320. doi: 10.1128/mcb.11.10.5312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen F, Wilusz J. Auxiliary downstream elements are required for efficient polyadenylation of mammalian pre-mRNAs. Nucleic Acids Res. 1998;26:2891–2898. doi: 10.1093/nar/26.12.2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Veraldi KL, Arhin GK, Martincic K, Chung-Ganster LH, Wilusz J, Milcarek C. hnRNP F influences binding of a 64-kilodalton subunit of cleavage stimulation factor to mRNA precursors in mouse B cells. Mol. Cell Biol. 2001;21:1228–1238. doi: 10.1128/MCB.21.4.1228-1238.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alkan SA, Martincic K, Milcarek C. The hnRNPs F and H2 bind to similar sequences to influence gene expression. Biochem. J. 2006;393:361–371. doi: 10.1042/BJ20050538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dalziel M, Nunes NM, Furger A. Two G–rich regulatory elements located adjacent to and 440 nucleotides downstream of the core poly(A) site of the intronless melanocortin receptor 1 gene are critical for efficient 3′ end processing. Mol. Cell Biol. 2007;27:1568–1580. doi: 10.1128/MCB.01821-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oberg D, Fay J, Lambkin H, Schwartz S. A downstream polyadenylation element in human papillomavirus type 16 L2 encodes multiple GGG motifs and interacts with hnRNP H. J. Virol. 2005;79:9254–9269. doi: 10.1128/JVI.79.14.9254-9269.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Millevoi S, Geraghty F, Idowu B, Tam JL, Antoniou M, Vagner S. A novel function for the U2AF 65 splicing factor in promoting pre-mRNA 3′-end processing. EMBO Rep. 2002;3:869–874. doi: 10.1093/embo-reports/kvf173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sgourou A, Papachatzopoulou A, Psiouri L, Antoniou M, Zoumbos N, Gibbs R, Athanassiadou A. The beta-globin C–>G mutation at 6 bp 3′ to the termination codon causes beta-thalassaemia by decreasing the mRNA level. Br. J. Haematol. 2002;118:671–676. doi: 10.1046/j.1365-2141.2002.03627.x. [DOI] [PubMed] [Google Scholar]
- 24.Millevoi S, Loulergue C, Dettwiler S, Karaa SZ, Keller W, Antoniou M, Vagner S. An interaction between U2AF 65 and CF I(m) links the splicing and 3′ end processing machineries. EMBO J. 2006;25:4854–4864. doi: 10.1038/sj.emboj.7601331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gunderson SI, Beyer K, Martin G, Keller W, Boelens WC, Mattaj LW. The human U1A snRNP protein regulates polyadenylation via a direct interaction with poly(A) polymerase. Cell. 1994;76:531–541. doi: 10.1016/0092-8674(94)90116-3. [DOI] [PubMed] [Google Scholar]
- 26.Markovtsov V, Nikolic JM, Goldman JA, Turck CW, Chou MY, Black DL. Cooperative assembly of an hnRNP complex induced by a tissue-specific homolog of polypyrimidine tract binding protein. Mol. Cell Biol. 2000;20:7463–7479. doi: 10.1128/mcb.20.20.7463-7479.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chou MY, Rooke N, Turck CW, Black DL. hnRNP H is a component of a splicing enhancer complex that activates a c-src alternative exon in neuronal cells. Mol. Cell Biol. 1999;19:69–77. doi: 10.1128/mcb.19.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu J, Lutz CS, Wilusz J, Tian B. Bioinformatic identification of candidate cis-regulatory elements involved in human mRNA polyadenylation. RNA. 2005;11:1485–1493. doi: 10.1261/rna.2107305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zarudnaya MI, Kolomiets IM, Potyahaylo AL, Hovorun DM. Downstream elements of mammalian pre-mRNA polyadenylation signals: primary, secondary and higher-order structures. Nucleic Acids Res. 2003;31:1375–1386. doi: 10.1093/nar/gkg241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Graveley BR, Fleming ES, Gilmartin GM. RNA structure is a critical determinant of poly(A) site recognition by cleavage and polyadenylation specificity factor. Mol. Cell Biol. 1996;16:4942–4951. doi: 10.1128/mcb.16.9.4942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu C, Alwine JC. Secondary structure as a functional feature in the downstream region of mammalian polyadenylation signals. Mol. Cell Biol. 2004;24:2789–2796. doi: 10.1128/MCB.24.7.2789-2796.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kikin O, Zappala Z, D’Antonio L, Bagga PS. GRSDB2 and GRS_UTRdb: databases of quadruplex forming G-rich sequences in pre-mRNAs and mRNAs. Nucleic Acids Res. 2008;36:D141–148. doi: 10.1093/nar/gkm982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takagaki Y, Ryner LC, Manley JL. Separation and characterization of a poly(A) polymerase and a cleavage/specificity factor required for pre-mRNA polyadenylation. Cell. 1988;52:731–742. doi: 10.1016/0092-8674(88)90411-4. [DOI] [PubMed] [Google Scholar]