Abstract
DT40 is a B-cell lymphoma-derived avian cell line widely used to study cell autonomous gene function because of the high rates with which DNA constructs are homologously recombined into its genome. Here, we demonstrate that the power of the DT40 system can be extended yet further through the use of RNA interference as an alternative to gene targeting. We have generated and characterized stable DT40 transfectants in which both topo 2 genes have been in situ tagged using gene targeting, and from which the mRNA of both topoisomerase 2 isoforms can be conditionally depleted through the tetracycline-induced expression of short hairpin RNAs. The cell cycle phenotype of topo 2-depleted DT40 cells has been compared with that previously reported for other vertebrate cells depleted either of topo 2α through gene targeting, or depleted of both isoforms simultaneously by transient RNAi. In addition, the DT40 knockdown system has been used to explore whether excess catenation arising through topo 2 depletion is sufficient to trigger the G2 catenation (or decatenation) checkpoint, proposed to exist in differentiated vertebrate cells.
INTRODUCTION
DT40 is an avian B-cell line that, because of its hyper-recombination phenotype, has gained widespread popularity as a model system for studying cell autonomous gene function (1,2). In this work, we demonstrate that DT40's powerful genetic toolbox can be extended even further through the use of RNA interference (RNAi) (3) to stably and conditionally KD selected target gene products.
DT40 cell lines have been established that allow for the depletion of topoisomerase 2 (topo 2). In vertebrates there are two isoforms of this enzyme (α and β), encoded by separate genes (4,5). While topo 2α and 2β are enzymatically very similar, the C-terminal domains of the enzymes differ extensively in amino acid sequence and post-translational modification. The two forms have distinct patterns of expression: topo 2α, which is cell-cycle regulated, has levels that are highest in G2/M, and is essential for the survival of proliferating cells. In contrast, levels of topo 2β are independent of proliferative status and it is dispensable at the cellular level (5–14). Topo 2β is not normally able to compensate for loss of 2α, although it has been shown that cultured human cells can be rescued from the lethal effects of 2α depletion by 2β if levels of the beta isoform are very high (15). Thus, although topo 2α is likely to be the major form of topo 2 responsible for decatenation and chromosome segregation in proliferating cells, the contribution of the two isoforms has not yet been fully established (16,17). While in human HT1080 cells, a conditional null mutant for topo 2α has been established (18), the inefficiency of gene targeting in most vertebrate cells has restricted the availability of systems allowing for the depletion of both isoforms simultaneously. Even in DT40, the establishment of a cell line conditionally null for both genes would require five rounds of gene targeting (because of the presence of the 2β gene on chromosome 2, which is trisomic in these cells). Here, we show that it is possible to establish vertebrate cells from which both isoforms can be conditionally depleted through stable and inducible short hairpin RNA (shRNA) expression (19). Although for topo 2, as for many chicken proteins, antibody availability is limited, the ease of in situ tagging in DT40 has allowed this possible limitation to be circumvented.
In order to validate the use of stable, short hairpin-activated, gene silencing for gene function studies in DT40, the cell- cycle phenotype of DT40 cells depleted either of topo 2α, or of both isoforms simultaneously, has been compared with that previously reported for other vertebrate cells [depleted either of topo 2α (6,18,20) or topo 2β (14,20) through gene targeting, or depleted of both isoforms by transient RNAi (21,22)]. In addition, the DT40 KD system has been used to explore whether excess catenation arising through topo 2 depletion is sufficient to trigger the G2 catenation checkpoint, proposed to exist in differentiated vertebrate cells (23–26).
MATERIALS AND METHODS
Antibodies
The antibodies and antibody kits used were: anti-tetracycline (Tet)-Repressor rabbit polyclonal antibody TET01 (Mo Bi Tec); anti-GFP (Abcam); anti-Flag M2 monoclonal antibody (Sigma); anti-BrdU MoBu-1 (Abcam); anti-phospho-Histone H3 (Ser10) (Abcam); Annexin V-FITC Apoptosis detection reagent (Abcam); anti-δH2AX(Ser139), mouse monoclonal JBW301 (Upstate); mouse monoclonal (DM1A) α-tubulin (Abcam) and mouse monoclonal (AC-15) β-actin (Sigma) (western loading controls). Secondary antibodies used for indirect immunofluorescence (IF) were: swine anti-rabbit FITC (Dako) and goat anti-mouse Texas Red (Molecular Probes).
Cell culture and treatments
DT40 cells were routinely grown in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum, 1% chicken serum, 10–5 M β-mercaptoethanol, penicillin, streptomycin (all from Invitrogen-Gibco), at 37°C. To induce expression from 2×TetO2-containing promoters, cells were grown in medium containing 2.5 µg ml–1 doxycycine (dox) (Sigma), with the medium being renewed every second day. For stable transfection of DNA, cells were electroporated using standard conditions: 1 × 107 cells in 800 µl phosphate-buffered saline [PBS(A)] using a BioRad Pulser with 30 µg linearized plasmid DNA at 25 μF and 550 V (Sale 2007). After 24 h, the cells were plated into selective medium in flat-bottomed 96-well dishes. Biochemical selections were as follows: puromycin (Sigma), 0.5 µg ml–1; blasticidin S (MP Biomedicals), 30 µg ml–1; zeocin (Invitrogen-Gibco), 1 mg ml–1; G418SO4 (Invitrogen-Gibco), 2 mg ml–1.
For transient Cre recombinase expression [allowing for the excision, and re-use, of floxed selectable marker cassettes, circular (non-linearized) Cre expression plasmid (pBS185, Invitrogen, (27)] was electroporated as above. Cells were subcloned by plating into non-selective medium at limiting dilutions (0.1–10 cells/well) in 96-well plates; the drug-sensitivity of colonies was tested to confirm excision.
Small interfering (si) RNA (10–20 µg) (MWG) was transiently transfected into 5 × 106 DT40 cells using an Amaxa Nucleofector (Solution T, program B-23) according to the manufacturer's recommended protocol.
Cells were irradiated with 4 Gy using a Torrex 150 X-ray cabinet. Cells were treated with the following as appropriate: 1 μM ICRF-193 (EuroMedex), 0.5 μg ml–1 nocodazole (Sigma).
In situ targeting (IST): constructs and transfection
A targeting construct allowing for the introduction of the Flag epitope at the C-terminus of one topo 2α allele was generated as follows:
the 5′ and 3′ homology blocks were PCR amplified from genomic DT40 DNA using Takara LA Taq polymerase and the sequence of cloned fragments confirmed.
The 1.3-kb 5′ homology block was designed to end at the stop codon of the open reading frame (ORF), with a single base change converting the TGA into TGT. The 5′ homology block was cloned into pBSII SK using KpnI and SalI to generate pBS′5T2A.
Sixty-nucleotide oligos encompassing two copies of the Flag epitope with SalI and BamHI overhangs, were designed to produce an in-frame topo 2α:Flag fusion protein.
The 1.4-kb 3′ homology block, extending from the stop codon of topo 2α, was PCR amplified with BamHI and NotI overhangs.
A three-way ligation was undertaken with pBS5′T2A (linearized using SalI and NotI), 2 × Flag (SalI–BamHI) and the 3′ homology block (BamHI–NotI).
Finally a floxed β-actin promoter-driven puromycin resistance (puroR) gene was cloned as a BamHI fragment into the unique BamHI site lying between the Flag cassette and the 3′ homology block.
A targeting construct allowing for the introduction of enhanced-green fluorescent protein (eGFP) at the C-terminus of one topo 2β allele was generated using a similar strategy:
The 2.9-kb 5′ homology block was designed to end at the stop codon of the ORF, with a single base change converting the TAA into TAC. The 5′ homology block was cloned into pBSII SK using KpnI and SalI to generate pBS′5T2B.
The ORF of eGFP was PCR amplified with SalI and BamHI overhangs, designed, upon SalI ligation, to produce an in-frame topo 2β:eGFP fusion protein.
The 1.8-kb 3′ homology block, extending from the stop codon of topo 2β, was PCR amplified with BamHI and NotI overhangs.
A three-way ligation was undertaken with pBS5′T2B (linearized using SalI and NotI), eGFP (SalI–BamHI) and the 3′ homology block (BamHI–NotI).
Finally, a floxed β-actin promoter-driven puroR gene was cloned as a BamHI fragment into the unique BamHI site lying between the eGFP cassette and the 3′ homology block.
For electroporation (BioRad Pulser), constructs were NotI-linearized. Genomic DNA from puromycin-resistant clones was screened by PCR using:
- a forward primer lying beyond the 5′ homology block:
- topo 2α 5′-CTGCTTGTCGCTTCTCTTTGTGATG-3′
- topo 2β 5′-CGTAAGCATCAGTTCTTCACTCTC-3′
- a reverse primer, based either on eGFP:
- 5′-GTCGTCCTTGAAGAAGATGGTGCG-3′ or, on the puroR sequence:
- 5′-GAGATGGGGAGAGTGAAGCAGAAC-3′
Selection of target sequences for RNAi
To identify effective targets, potential sequences were tested using transient transfection of 10–20 μg siRNAs by nucleofection (Amaxa). KD effectiveness was estimated after 2–3 days by monitoring (i) the effect on cell proliferation (for topo 2α targets) and (ii) by indirect IF to detect, as appropriate, the eGFP- or Flag-tagged isoform in fixed cells. The selected target sequences were then cloned as 60-nt oligos into pSUPERIOR (target sequence in bold):
topo 2α
5′ GATCCCC GGAGTTGGTCCTGTTCTCA TTCAAGAGA TGAGAACAGGACCAACTCC TTTTTA 3′
which targets nts 2106–2124 of the ORF.
topo 2β
5′ GATCCCC GAAGGAGCCTGGTACTAGA TTCAAGAGA TCTAGTACCAGGCTCCTTC TTTTTA 3′
which targets nts 3939–3957 of the ORF.
Although some alternatively spliced variants of both isoforms have been identified, these affect the N-terminal region of the enzyme (28); the selected target sequences are predicted to silence all known transcripts.
RNAi expression constructs and transfection
An RNAi expression construct, in which the insert is insulated in order to minimize position effects after integration, was generated as follows:
Oligonucleotides for shRNA generation were annealed and cloned, using their BglII and HindIII overhangs, into pSUPERIOR (Oligoengine).
A fragment containing two copies of the 250-bp ‘core’ chicken hypersensitive site-4 (HS4) insulator was released from pNI-CD (29) (kindly provided by Dr G. Felsenfeld) using KpnI and cloned into the KpnI site of pBluescript (BS)II SK (Stratagene). A second ∼575-bp HS4 core insulator fragment was isolated from pNI-CD using EcoRI and BamHI and sticky-end ligated into the same vector, generating pBSTCF.
The ∼300-bp H1 Tet-repressible promoter:shRNA cassette was removed from pSUPERIOR using SalI and EcoRI and inserted into the polylinker of pBSTCF, such that the shRNA expression cassette is flanked on both sides by tandemly oriented insulator elements.
To confer blasticidin resistance, a floxed β-actin promoter-driven blasticidin resistance marker (30) was cloned into the BamHI site of pBSTCF. Alternatively, for Zeocin-resistance, the HS4 insulated shRNA expression cassette was removed from pBSTCF using Pvu2 and blunt-ligated into the EcoRV site of pRSVZeo [kindly provided by Dr M. Nakanishi (31)].
For electroporation, shRNA expression constructs were NotI-linearized.
RNA extraction, reverse transcription and real-time PCR
Total RNA was prepared from cells, after 0 and 5 days doxycycline (dox) exposure, using Trizol, and 5 μg reverse transcribed using oligo dT (Superscript III First Strand Synthesis System, Invitrogen). Real-time PCR reactions were done for topo 2 (α or β) (target genes) and GAPDH (reference gene) using SYBR Green Supermix on an IQ5 system (BioRad). Primer sequences used were:
Topo 2α 5′-ACTGTGGAGACTGCGTGTC-3′ and 5′-CTCCGAGACATCAGTGCTAC-3′
Topo 2β 5′-AGCCTCCAACTGTGGTATC-3 and 5′-GAATGTTTGCCACCTGC-3′
GAPDH 5′-ACCCCAGCAACATCAAATGG-3′ and
5′-CACCAGTGGACTCCACAACATACT-3′
Following validation experiments, the Comparative CT (ΔΔCT) method (32) was used to compare topo 2 gene expression in dox-treated relative to untreated samples for each cell line.
Tet-inducible KD-resistant (KDR) topo 2α expression: construct and transfection
Topo 2α cDNA was synthesized from DT40 cells using the Superscript III CellsDirect cDNA synthesis system (Invitrogen). cDNA across the ORF was generated by PCR amplification of several smaller sub-fragments using Accuzyme DNA polymerase (Bioline) and subcloning in pSC-A (StrataClone PCR cloning kit, Stratagene). The fidelity of each subclone was checked by sequencing, before ligation into pBSII SK, to generate the complete ORF. The fidelity of all junctions was again confirmed by sequencing.
Site-directed mutagenesis, using a QuikChange multisite-directed mutagenesis kit (Stratagene), was used to generate a KDR form of topo 2α through the introduction of two silent base changes within the shtopo 2α target sequence (a T to C transition at nt position 2110 of the ORF and a C to G transversion at nt position 2115) using the following oligonucleotide (with mutations in bold):
5′ CATCAACAAGGAGCTGGTGCTGTTCTCAAACTC 3′.
After confirmation by sequencing, the mutated (KDR) topo 2α ORF was cloned 3′ of eGFP in peGFP-C1 (BD Biosciences Clontech), as a SalI fragment, so as to encode an inframe eGFP:topo 2α fusion protein. The chimeric ORF was removed using NheI and ApaI and placed under a Tet-inducible CMV promoter in pcDNA4/TO (Invitrogen). Stable transfectants were isolated by electroporation of FspI-linearized pcDNA4/TO eGFP:KDRtopo2α followed by zeocin selection.
Flow cytometry and cell-cycle analyses
Mitotic index (MI) determination was undertaken by flow cytometry for detection of phospho Ser10 histone H3 (33). To monitor progression, through G2 into M, cells were trapped in M phase by the addition of 0.5 μg ml−1 nocodazole for 8 h. DNA synthesis was monitored through uptake of bromodeoxyuridine (BrdU) and its detection by flow cytometery using anti-BrdU (34). Cell death was quantitated by flow cytometry using Annexin V-FITC Apoptosis detection reagent and propidium iodide (PI) counterstain, as recommended by the manufacturer (Abcam).
Indirect IF and microscopy
Cells were either cytospun (ThermoShandon) onto, or allowed to loosely attach to, poly-l-lysine-coated slides. Indirect IF with anti-GFP antibody (1:500) was used to monitor topo 2β levels in Beaft6.2-derived lines (levels being too faint to visualize directly), while endogenous topo 2α levels were monitored using anti-Flag M2 antibody (1:500). Cells were fixed in methanol, with antibody dilutions and washes undertaken in KCM (120 mM KCL, 20 mM NaCl, 10 mM Tris–HCl pH 8.0, 0.5 mM EDTA, 0.01% Triton X-100). For the detection of δH2AX cells were fixed in 4% paraformaldehyde (35).
Secondary antibodies (1:100) were swine anti-rabbit FITC (Dako), rabbit anti-mouse FITC (Dako) or goat anti-mouse Texas Red (Molecular Probes). DNA was counterstained with DAPI (0.5 μg ml–1) and mounted in Vectorshield (Vector Labs).
Images were captured using the Cytovision system (Applied Imaging) with a Zeiss Axioskop 2 microscope.
Western blot
Cells were harvested, washed in cold PBS and lysed on ice in TG lysis buffer (20 mM Tris (pH 7.5), 137 mM NaCl, 1 mM EGTA, 1% (v/v) Triton X-100, 10% (v/v) glycerol, 1.5 mM MgCl2, 1 mM Na3VO4, 1 mM PMSF, 20 μM leupeptin, 10 μg/ml aprotinin and 50 mM NaF). Lysates were cleared by centrifugation at 13 000 g and transferred to fresh tubes on ice. Total protein was determined by Bradford assay (Bio-Rad). Equal amounts of cell lysate were fractionated on SDS–PAGE gels (Hoefer Mighty Small system) and transferred to methanol soaked PVDF membranes at 300 mA for 1 h in a mini-transblot apparatus (BioRad). Membranes were incubated in Tris-buffered saline with 0.1% (v/v) Tween-20 (TBST) plus 5% (w/v) powdered milk (TBST + milk) for at least 1 h before probing with anti-sera. Primary antibody incubations were performed in TBST + milk at 4°C overnight followed by three washes of TBST. Anti-GFP and anti-Flag antibodies were used at 1:2000. Secondary antibody incubation was performed in TBST + milk for 1 h at room temperature followed by five washes TBST. Antibody-antigen complexes were detected using ECL according to the manufacturer's instructions (Amersham).
Statistical analyses
A two-tailed t-test (unpaired) was used to assess the statistical significance of the observed differences in cell-cycle data in Figures 6 and 8.
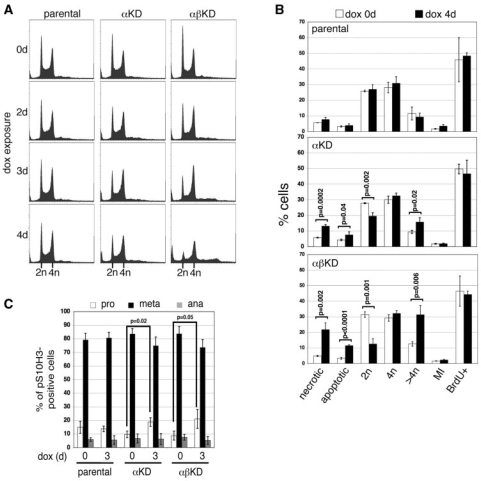
Figure 6.
(A) Flow cytometric analysis of nuclear DNA content of the parental, αKD and αβKD cell lines over a 4-day exposure to 2.5 μg ml–1 dox: x-axis, propidium iodide fluorescence; y-axis, events (total events 30 000). (B) Summary graph of flow cytometric cell cycle data collected from the parental, αKD and αβKD cell lines at 0 (white bars) and 4 days (black) dox. Data represents the mean (±SD) based on ≥3 independent experiments. (C) Breakdown of M phase cells (into prophase, prometa/metaphase and anaphase) at 0 and 3 days dox based on microscopic examination. Data points are the mean (±SD) of three experiments and are based on >300 pS10H3-positive cells.
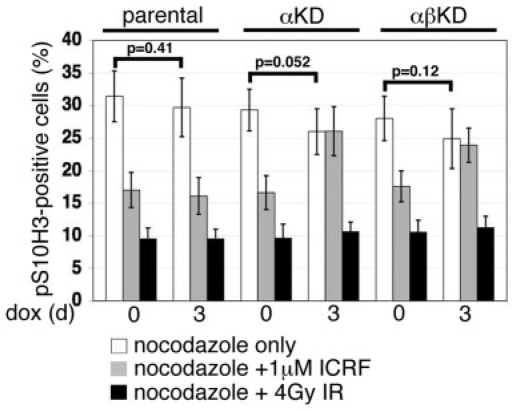
Figure 8.
Summary graph of the accumulation of cells staining positive for the mitotic marker pS10H3 (quantified by flow cytometry) after 8 h in the presence of nocodazole, in cultures treated with dox for 0 or 3 days (based on nine independent experiments) (white bars). The impact of 1 μM ICRF-193 (present throughout the 8-h period with nocodazole) on progression into M phase is shown alongside (three independent experiments) (grey bars). To address whether topo 2 depletion has any effect on the ability of DT40 cells to arrest in G2 in response to DSBs, cultures were exposed to 4 Gy of X-irradiation immediately prior to nocodazole addition (three independent experiments) (black bars). Values correspond to the means (±SD).
RESULTS
In situ tagging of both topo 2 isoforms in DT40
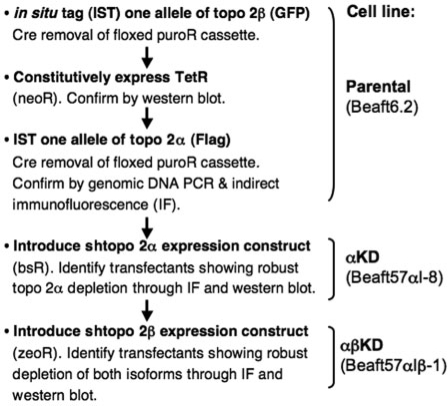
To simplify the detection of the two isoforms of chicken topo 2, a DT40 cell line was established in which one allele of each had been in situ tagged at its C-terminus. In addition, this cell line (designated Beaft6.2 and referred to here after as the parental cell line) expresses the Tetracycline repressor protein (TetR), allowing for tet/dox-inducible shRNA expression. The generation of this cell line was achieved by the following steps (Figure 1):
one allele of the topo 2β isoform was in situ tagged with GFP (Figure 2A);
transient Cre recombinase expression was then used to remove a floxed puroR selection cassette;
a TetR expression construct (pcDNA3/TR, Invitrogen) was introduced and expression confirmed by western blot of stable transfectants (data not shown);
one allele of topo 2α was in situ tagged with the Flag epitope (Figure 2B). This involved a second targeting construct, with the floxed puroR cassette being re-cycled (30).
Figure 1.
Stepwise-strategy for the generation of stable conditional topo 2α and 2β knockdown DT40 cell lines.
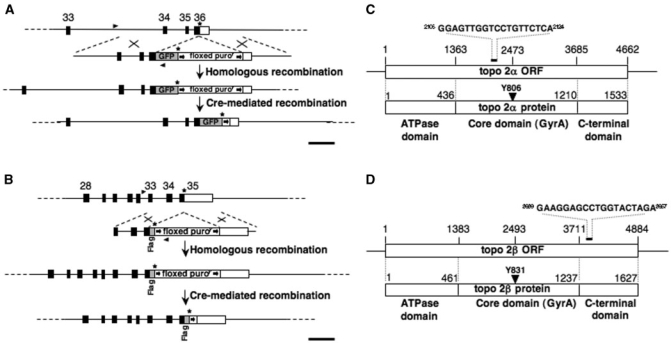
Figure 2.
(A) Map showing the 3′ region of the Gallus gallus topo 2β genomic locus and the GFP in situ tagging (IST) construct. A homologous recombination event will result in exon 36, which contains the stop codon, being disrupted, such that the portion encoding the final part of the 2β open reading frame is fused in-frame to sequence encoding eGFP, followed by a stop codon, a floxed puromycin-resistance gene cassette and sequence from the 3′ homology block. (B) Map showing the 3′ region of the chicken topo 2α genomic locus and the Flag IST construct. In the event of gene targeting, exon 35, which contains the stop codon, will be disrupted such that the protein-encoding portion of the exon is fused in-frame to a Flag epitope. This is immediately followed by a stop codon, the floxed puroR marker and sequence from the 3′ homology block. In each case, homologously recombined clones were initially identified by PCR of genomic DNA amplified using forward primers (arrowheads) lying 5′ to the homology blocks and with reverse primers based on the GFP or puroR genes. In targeted-tagged clones, the floxed puroR gene was removed by transient expression of Cre recombinase. Black boxes represent the position of exons. Exon numbering is based on the Gallus gallus reference sequence assembly 2.1. Scale bar, 1 kb. (C) Schematic showing the Gallus gallus topo 2α mRNA and its open reading frame (ORF), the various domains of the protein and the position of the RNAi target sequence (5). (D) Schematic showing the Gallus gallus topo 2β mRNA and ORF, the various domains of the protein and the position of the RNAi target sequence (5).
Generation of stable DT40 cell lines that can be conditionally depleted of topo 2
The in situ tagged/tetR-expressing DT40 (parental) line was stably retrofitted for the tet/dox-inducible production of topo 2α shRNA (Figures 1 and 2C). In this system, expression from the tet/dox-inducible H1 promoter is repressed by TetR binding to the Tet operator sequence (2xTetO2) in the absence of tet/dox (36).
The H1/2xTetO2 promoter:shtopo 2α cassette was cloned between HS4 insulator sequences in order to allow position site-independent expression (37–39). The vector, which also contains a β-actin promoter-driven bsR marker gene, was linearized, transfected into Beaft6.2 by electroporation and blasticidin-resistant clones expanded. Ten independent transfectants were screened at the RNA and protein level, by real-time PCR and indirect IF, respectively (Figure 3 and data not shown). Data from the two assays correlated well, validating the use of the in situ tag for routinely monitoring topo 2 depletion.
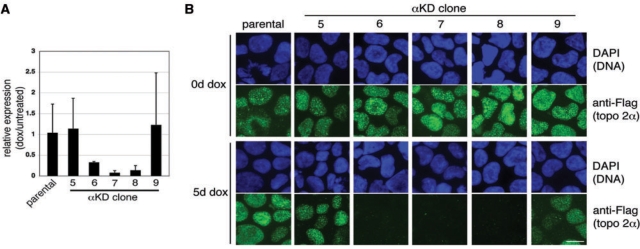
Figure 3.
(A) Real-time PCR was used to estimate the level of topo 2α mRNA knockdown in several transfectants, five of which are shown here. Each bar represents the expression level in cells exposed to dox (5 days) relative to that in untreated cells from the same cell line, calculated using the Comparative CT (ΔΔCT) method (mean ± SD, based on ≥2 independent experiments). (B) In parallel, the same transfectants were examined by indirect IF using anti-Flag (FITC) to detect protein from the in situ tagged topo 2α locus. DNA was counterstained with DAPI (blue). Scale bar, 10 μm.
One line displaying robust topo 2α depletion (KD clone 8, referred to subsequently as αKD) was re-transfected, using a shtopo 2β expression construct (Figure 2D) conferring zeocin-resistance. After an initial screen of several transfectants, one line showing robust topo 2β depletion was characterized further, with the KD of both isoforms being confirmed by indirect IF of fixed cells (Figure 4A) and western blotting (Figure 4B) (cell line designated αβKD).
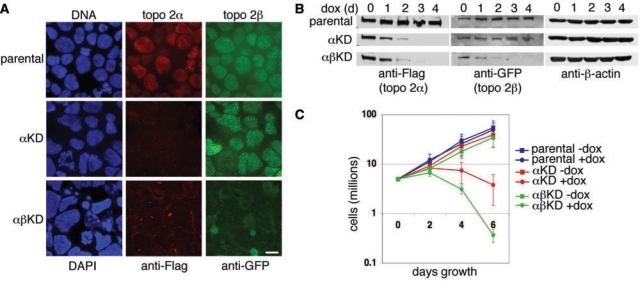
Figure 4.
(A) Indirect IF of topo 2α and topo 2β in the parental, αKD and αβKD cell lines after 6 days dox exposure. Cells were cytospun onto slides. Topo 2α was detected using anti-Flag antibody (Texas Red) and topo 2β using anti-GFP (FITC). DNA was counterstained using DAPI (blue). Scale bar, 10 μm. (B) Western blots showing robust knockdown, over 4 days dox exposure, of topo 2α in αKD cells and of both isoforms in αβKD cells. Topo 2α was detected using anti-Flag antibody and 2β using anti-GFP. β-actin was used as a loading control. (C) Growth curves of the parental (blue), αKD (red) and αβKD (green) cell lines grown in the presence (closed circles) and absence (closed squares) of 2.5 μg ml–1 dox for 6 days. Data points represent the mean (±SD) based on ≥3 independent experiments.
The effect of topo 2 depletion on cell proliferation
While depletion of topo 2α alone is lethal, with population growth slowing and then declining over 6 days of dox exposure, the simultaneous depletion of both isoforms results in an even more rapid decline in cell number (Figure 4C). Similar findings, of a more severe response following depletion of both isoforms than seen after depletion of 2α alone, have been reported in cultured human cells (21,22,40).
Rescue of the lethal topo 2α depletion phenotype by introduction of a KDRtopo 2α transgene
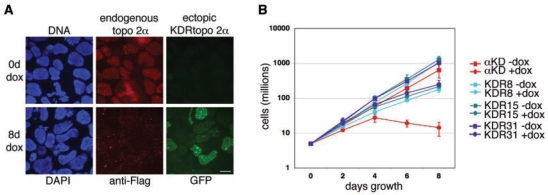
To demonstrate that dox-induced lethality is due to topo 2α depletion, the αKD cell line was engineered to express KDR topo 2α from a dox-inducible promoter. A CMV/2xTetO2 promoter-driven GFP:KDRtopo 2α transgene was introduced and stable zeocin-resistant transfectants characterized. Although the αKD cells already express a GFP fusion protein, from the in situ tagged endogenous topo 2β locus, the levels of this protein are barely detectable when visualized directly (i.e. without indirect IF using anti-GFP antibody) (Figure 5A). Indirect immunofluorescence to detect endogenous topo 2α, using anti-Flag antibody, revealed that upon dox addition, wild-type (WT) topo2α is depleted (Figure 5A). This is consistent with shtopo 2α being expressed from the H1/2xTetO2 promoter resulting in targeted degradation of WTtopo 2α mRNA. At the same time, the direct visualization of GFP in dox-treated cells, revealed levels much higher than in untreated cells, consistent with the induction of KDR topo 2α mRNA from the ectopic eGFP:KDRtopo 2α transgene (Figure 5A). Although GFP levels were variable from cell-to-cell across the clonal population (an observation made for several independently derived KDR clones), growth curves revealed that the induction of GFP:KDRtopo 2α, rescues αKD cells from dox-lethality, substantially restoring their growth rate (Figure 5B).
Figure 5.
(A) Detection of endogenous and ectopically expressed topo 2α in a knockdown-resistant (KDR) derivative of the αKD cell line. The KDR line was grown in the presence or absence of 2.5 μg ml–1 dox for 8 days, cells fixed and cytopsun onto slides. Endogenous topo 2α was detected indirectly using anti-Flag (Texas Red). The green signal of the ectopically expressed GFP:KDRtopo2α fusion protein was visualized directly (green). (Note: the background level of expression from the endogenous GFP-tagged topo 2β locus is extremely faint when visualized directly.) Scale bar, 10 μm. (B) Growth curves of the αKD cell line compared with three KDR clonal derivatives, grown in the presence or absence of 2.5 μg ml–1 dox for 8 days. Data points represent the mean (±SD) based on ≥3 independent experiments.
Aneuploidy, polyploidy and cell death in cells depleted of topo 2
Flow cytometry of the nuclear DNA content by PI staining revealed a decline in the 2N fraction following topo 2 depletion. While there was no marked effect on the 4N fraction, a significant increase in the accumulation of cells with a >4N content was detected over 4 days of dox exposure, especially in cells depleted of both isoforms (Figure 6A and B). A sub-2N peak was detectable by Day 4 in cells depleted of both topo 2α and 2β and Annexin V apoptosis assays confirmed a significant increase in cell death (apoptosis and necrosis) (Figure 6B).
The MI following topo 2 depletion
The MI of the viable cell population was determined by indirect IF using anti-phospho Ser10 histone H3 (pS10H3), and detection by microscopy and flow cytometry (Figure 6B and data not shown). Both approaches revealed a MI of ∼2% for all three cell lines. While the MI increased very slightly in cells exposed to dox over 4 days, this increase was also observed in the parental cells, suggesting that it is independent of topo 2 depletion.
The distribution of M phase (pS10H3-positive) cells in prophase, prometaphase/metaphase or anaphase was examined using microscopy. A slight increase in the fraction of M phase cells in prophase (P ≤ 0.05) and corresponding decrease in the numbers in both prometa/metaphase and (to a lesser extent) anaphase was observed in both topo 2 depletion lines at Day 4 of dox treatment, while no change was seen in the parental line (Figure 6C). This suggests that topo 2 depletion in DT40 cells, while not affecting the MI, may result in cells spending slightly longer in the early stages of M phase. Others have reported similar findings in HeLa cells (21).
Entry into S phase
To examine whether topo 2-depleted cells continue to cycle up until cell death, cells were exposed to BrdU for 30 min at 0 and 4 days dox exposure. Uptake of BrdU and the total DNA content of cells were monitored both by microscopy and flow cytometry using anti-BrdU and PI (Figure 6B). No differences in the fraction of cells synthesizing DNA were detected in any of the cell lines as a result of dox exposure.
Anaphase bridging and aberrant cytokinesis
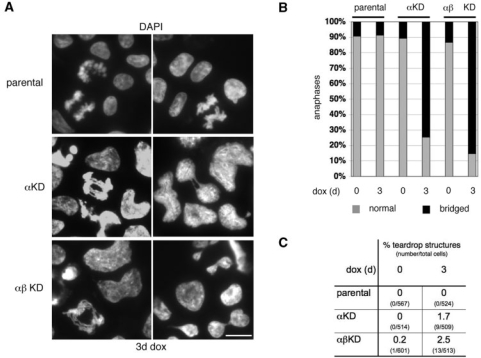
Microscopic examination revealed that the majority of anaphase cells showed extensive chromatin bridging after topo 2 (α alone, or α plus β) depletion over 3 days of dox exposure (Figure 7A and B). In addition, DAPI-staining revealed the occasional appearance of aberrant teardrop structures in cells depleted of topo 2α, suggestive of abnormal cytokinesis (Figure 7A and C).
Figure 7.
(A) Representative examples of anaphase chromatin bridging and teardrop structures suggestive of aberrant cytokineses in αKD and αβKD cells depleted of topo 2 (3 days dox). DNA has been stained with DAPI. Scale bar, 10 μm. (B) Summary graph of the frequency of anaphase bridging after 3 days dox. Data points are based on three experiments, with a total of >150 anaphases examined. (C) Detection of occasional ‘teardrop’ structures in topo 2-depleted (3 days dox) DT40 cells.
The effect of topo 2 depletion and chemical inhibition on progression from G2 into M phase
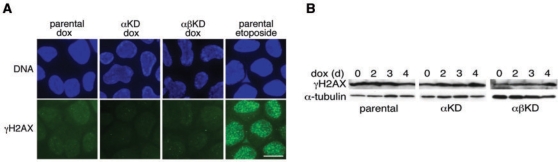
The progression of cells from G2 into M phase was monitored at 0 and 3 days dox exposure. For the last 8 h, cells were incubated in medium containing nocodazole, fixed and flow cytometric detection of anti-pS10H3 used to monitor their accumulation in M phase. A small, but not significant (P > 0.05), decrease in the accumulation of cells in M phase was detected in all three cell lines after dox exposure (Figure 8). This suggests that depletion of topo 2α (or of both isoforms together) does not activate a G2 checkpoint. This is consistent with the lack of any change in the MI of the asynchronously growing cell populations (Figure 6B) and with the lack of any detectable effect on levels of DNA damage (as determined by phosphorylated histone H2AX, γH2AX) (Figure 9A and B).
Figure 9.
(A) As a measure of levels of DSBs after 3 days dox exposure, parental, αKD and αβKD cells were examined by indirect IF for phosphorylated histone H2AX (γH2AX)-containing foci (FITC) in the cell nuclei. As a positive control, the parental cell line was exposed briefly to the topo 2 poison etoposide (10 μM, 30 min). DNA is counterstained with DAPI. Scale bar, 10 μm. (B) Western blot of γH2AX levels over a 4-day time course of dox exposure. α-tubulin is the loading control.
For comparison, we examined the effect of chemically inhibiting topo 2 on progression into M phase. In the parental cell line Beaft6.2, irrespective of dox exposure, the topo 2 inhibitor ICRF-193 induces a robust G2 delay, resulting in a ∼50% decrease in the M phase fraction (Figure 8). This effect is absent in cells depleted of topo 2, consistent with ICRF-193 acting through topo 2 alone (41). No difference was seen in the ICRF-193 response between cells depleted of topo 2α alone compared to those depleted of both isoforms together, suggesting that, in this rapidly proliferating chicken cell line, ICRF-193 is exerting its effect on G2 progression primarily through topo 2α (42).
To determine whether topo 2-depleted cells remain competent to mount a G2 checkpoint delay, at least in response to DNA damage, cells grown in the presence of dox for 0 or 3 days were subject to 4 Gy of X-irradiation (IR) and the accumulation of pS10H3-positive cells over the following 8 h monitored. All three cell lines show a robust G2 delay in response to IR-induced DNA damage, which appears unaffected by depletion of topo 2 (Figure 8).
DISCUSSION
Where functional analysis demands complete loss of gene expression gene targeting, to generate knockout alleles, is the method of choice. However, even in DT40 cells, gene targeting can be laborious. Moreover if, like topo 2α, the gene-of-interest (g-o-i) is essential a conditional gene knockout has to be generated. Typically, this necessitates the introduction of a repressible rescue construct that maintains cell viability after gene disruption. Subsequent repression (or deletion) of the rescue gene then reveals the full knockout phenotype. Even assuming that cell viability is maintained through ectopic expression of the rescuing transgene (which is not always the case), the generation of conditional knockouts is time consuming. Ideally, any strategy to study an essential gene in the DT40 cell line would meet the following criteria: (i) depletion of the gene product must be conditional; (ii) depletion must proceed through the endogenous gene, obviating the need for a rescue construct; and (iii) it should be possible without the time consuming and costly need to raise an antibody against the protein-of-interest.
For many investigations, in a wide range of model systems, the KD of gene products, using the technically more straightforward approach of RNAi, has been highly informative. Here, for the first time, we demonstrate that it is possible to use RNAi by means of shRNA to stably and effectively deplete gene products from DT40 cells, thus extending the tools available for manipulating this widely used vertebrate cell line. We found that by flanking the shRNA expression cassette with two tandemly orientated copies of the 250-bp ‘core’ element of the chicken HS4 insulator (37–39) we were able to generate, with remarkable ease, stable DT40 cell lines capable of inducible depletion of topoIIα and topoIIβ to a level sufficiently low to recapitulate the known knockout phenotype.
In addition, by using RNAi in conjunction with the in situ tagging of endogenous genes through gene targeting, we have developed a strategy for studying gene function that meets all three of the criteria outlined above. In the future, we envisage that in situ tagging, with epitopes such as GFP, will allow any g-o-i to be depleted through the use of generic shRNA expression constructs. In situ tagging of all endogenous alleles would be readily achievable through the recycling of selectable markers and targeting constructs, while KD of the selected gene's product through its epitope tag, using well-characterized shRNAs, would remove the need to identify effective, gene-specific, target sequences. Furthermore, use of the tet-inducible system for shRNA induction offers the possibility of simultaneously inducing expression of a modified, or mutant, KDR transgene in the absence of endogenous gene product.
Our strategy has been validated using the type 2 topoisomerases, with the phenotype observed following topo 2 depletion being consistent with that described following both transient KD in human cells (21,22), and the knockout of topo 2α in mouse or human cells (6,18,20,40). Further evidence for the observed phenotype being due specifically to the targeted degradation of topo 2 mRNAs is provided by the demonstration that αKD cells can be rescued from dox-induced lethality by expression of a KDR-form of topo 2α. Failure to fully restore the starting growth rate under these conditions could be due to a variety of factors:
the ‘rescuing’ topo 2α is fused at its N-terminus to GFP, which may compromise functionality;
the level of topo 2α protein may be suboptimal (too much, as well as too little, topo 2α protein is known to be detrimental) (43);
expression of the KDRtopo 2α protein is uncoupled from the cell cycle (since the ectopic copy is driven by a CMV promoter); or
the shRNAs may exert other, off-target, effects.
Although, as predicted, depletion of topo 2α alone is lethal in DT40 cells, the severity of the KD phenotype was enhanced by depletion of both isoforms simultaneously. Similar observations have been reported following transient KD of both topo 2 isoforms in HeLa cells (21,22) and the transient KD of topo 2β from topo 2α-conditionally null mutant HT1080 cells (40), suggesting that topo 2β contributes to sister chromatid decatenation in these cell lines.
While topo 2-depleted DT40 cells show extensive anaphase chromatin bridging and aberrant cytokineses, the cells do not arrest at any particular point in the cell cycle. Instead they continue to cycle, incorporating BrdU and accumulating in M phase at rates comparable to normal, becoming increasingly polyploid, with eventual cell death. Similar observations have been reported for human HT1080 cells conditionally null mutant for topo 2α (18). Consistent with this lack of any significant G2 delay in vertebrate cells, Saccharomyces cerevisiae cells also progress through S, G2 and M with normal kinetics in the absence of top2 protein (44). Since topo 2 is responsible for the removal of catenanes arising during DNA replication, the failure of topo 2 depletion to trigger the so-called G2 ‘catenation checkpoint’, in either vertebrate cells or budding yeast, raises questions about the nature of this checkpoint and whether failed decatenation is, in itself, sufficient to activate it.
The idea of a G2 checkpoint that responds to the presence of excess catenation emerged largely from observations made on cells exposed to bisdioxopiperazines, such as ICRF-193. ICRF-193 traps both DNA duplexes inside the topo2 dimer, blocking enzyme turnover (45–47) and has been shown to delay progression through G2 in many differentiated cell types (23–26). Since this inhibitor was originally considered not to induce DSBs, the idea emerged of a G2 ‘catenation checkpoint’, distinct from the DNA damage pathways (24). However, recent observations have raised questions regarding this response. Firstly, there is uncertainty about what actually triggers the delay—excess catenation (and if so whether both full- and/or hemi-catenanes can act as a trigger), or something else? It has been shown that as well as generating excess catenanes, inhibition of topo 2 by ICRF-193 traps the DNA in an immobile clamp altering chromatin topology (48), while several reports suggest that ICRF-193 does induce low levels of DSBs (49–51). Secondly, vertebrate cells normally enter mitosis (and even anaphase) with catenanes and/or hemi-catenanes persisting between sister chromatids (52–54), suggesting that any G2/M checkpoint would need to be set to respond only to catenation levels above a permissible threshold. Since DT40 cells expressing topo 2 show a robust G2 delay in response to ICRF-193, possible explanations for the lack of a response following topo 2 depletion might be that:
the KD mutant cells are so compromised after several days exposure to dox they are unable to mount a delay.
the ICRF-193-induced delay is triggered by a topo 2-dependent, but catenation-independent, effect of the inhibitor.
excess catenation requires topo 2 protein itself to signal a G2 delay.
a response occurs, but is masked by the gradual and asynchronous depletion of topo 2 across the KD cell population.
While the observation that DT40 cells depleted of topo 2 can still slow down progression into M phase in response to X-irradiation argues against the first point, we are currently unable to distinguish between the latter possibilities. The recent development of systems allowing for rapid depletion of protein from vertebrate cells should allow the last point to be addressed (55). If no G2 delay is revealed in such a system then future experiments to determine whether the expression of catalytically inactive topo 2 triggers a G2 delay, carried out in a conditional KD background, should allow any requirement for the topo 2 protein itself in generating a signal in response to excess catenation, to be elucidated. In this regard it is intriguing that budding yeast cells expressing only catalytically inactive top2 have been shown to undergo a stable G2 arrest, requiring an intact DNA damage checkpoint (44). Moreover, recent studies have revealed that for an ICRF-193-induced G2 checkpoint to be triggered in HeLa cells the presence of topo 2α phosphorylated at Ser1524, is required (56). Phospho-Ser1524 appears to act as a binding site for recruitment of the multi-functional Mediator of DNA damage Checkpoint protein-1 (MDC1) (57). Where cells expressed only non-phosphorylatible S1524A topo 2α, ICRF-193 failed to induce a G2 delay, despite cells being defective in decatenation. Hence, whatever triggers the ICRF-193-associated G2 delay in HeLa cells (catenation, DSBs, changes in chromatin topology, or something else) the signal is mediated through topo 2α Ser1524P. It is possible, therefore, that the accumulation of excess catenation arising from an absence of topo 2 protein would be unable to activate a G2 checkpoint response. Further work is required to elucidate whether excess DNA catenation can trigger a G2 arrest of the vertebrate cell cycle and if it can, the mechanism involved.
SUPPLEMENTARY DATA
Supplementary Data is available at NAR Online.
FUNDING
The Biotechnology and Biological Sciences Research Council (grant number BBS/B/04994); and by Cancer Research-UK (grant number C9609/A3527).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We would like to thank Dr Gary Felsenfeld for supplying the HS4 DNA, Dr Mahito Nakanishi for kindly providing the plasmid pRSVZeo and Andy Jessop for help with flow cytometry.
REFERENCES
- 1.Buerstedde JM, Takeda S. Reviews and protocols in DT40 research. Subcell. Biochem. 2006;40:1–477. [Google Scholar]
- 2.Hudson DF, Morrison C, Ruchaud S, Earnshaw WC. Reverse genetics of essential genes in tissue-culture cells: ‘dead cells talking'. Trends Cell Biol. 2002;12:281–287. doi: 10.1016/s0962-8924(02)02281-x. [DOI] [PubMed] [Google Scholar]
- 3.Martin SE, Caplen NJ. Applications of RNA interference in mammalian systems. Annu. Rev. Genomics Hum. Genet. 2007;8:81–108. doi: 10.1146/annurev.genom.8.080706.092424. [DOI] [PubMed] [Google Scholar]
- 4.Chung TD, Drake FH, Tan KB, Per SR, Crooke ST, Mirabelli CK. Characterization and immunological identification of cDNA clones encoding two human DNA topoisomerase II isozymes. Proc. Natl Acad. Sci. USA. 1989;86:9431–9435. doi: 10.1073/pnas.86.23.9431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Niimi A, Suka N, Harata M, Kikuchi A, Mizuno S. Co-localization of chicken DNA topoisomerase IIalpha, but not beta, with sites of DNA replication and possible involvement of a C-terminal region of alpha through its binding to PCNA. Chromosoma. 2001;110:102–114. doi: 10.1007/s004120100140. [DOI] [PubMed] [Google Scholar]
- 6.Akimitsu N, Adachi N, Hirai H, Hossain MS, Hamamoto H, Kobayashi M, Aratani Y, Koyama H, Sekimizu K. Enforced cytokinesis without complete nuclear division in embryonic cells depleting the activity of DNA topoisomerase IIalpha. Genes Cells. 2003;8:393–402. doi: 10.1046/j.1365-2443.2003.00643.x. [DOI] [PubMed] [Google Scholar]
- 7.Chaly N, Chen X, Dentry J, Brown DL. Organization of DNA topoisomerase II isotypes during the cell cycle of human lymphocytes and HeLa cells. Chromosome Res. 1996;4:457–466. doi: 10.1007/BF02265053. [DOI] [PubMed] [Google Scholar]
- 8.Christensen MO, Larsen MK, Barthelmes HU, Hock R, Andersen CL, Kjeldsen E, Knudsen BR, Westergaard O, Boege F, Mielke C. Dynamics of human DNA topoisomerases IIalpha and IIbeta in living cells. J. Cell Biol. 2002;157:31–44. doi: 10.1083/jcb.200112023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grue P, Grasser A, Sehested M, Jensen PB, Uhse A, Straub T, Ness W, Boege F. Essential mitotic functions of DNA topoisomerase IIalpha are not adopted by topoisomerase IIbeta in human H69 cells. J. Biol. Chem. 1998;273:33660–33666. doi: 10.1074/jbc.273.50.33660. [DOI] [PubMed] [Google Scholar]
- 10.Heck MM, Hittelman WN, Earnshaw WC. Differential expression of DNA topoisomerases I and II during the eukaryotic cell cycle. Proc. Natl Acad. Sci. USA. 1988;85:1086–1090. doi: 10.1073/pnas.85.4.1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meyer KN, Kjeldsen E, Straub T, Knudsen BR, Hickson ID, Kikuchi A, Kreipe H, Boege F. Cell cycle-coupled relocation of types I and II topoisomerases and modulation of catalytic enzyme activities. J. Cell Biol. 1997;136:775–788. doi: 10.1083/jcb.136.4.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Null AP, Hudson J, Gorbsky GJ. Both alpha and beta isoforms of mammalian DNA topoisomerase II associate with chromosomes in mitosis. Cell Growth Differ. 2002;13:325–333. [PubMed] [Google Scholar]
- 13.Woessner RD, Mattern MR, Mirabelli CK, Johnson RK, Drake FH. Proliferation- and cell cycle-dependent differences in expression of the 170 kilodalton and 180 kilodalton forms of topoisomerase II in NIH-3T3 cells. Cell Growth Differ. 1991;2:209–214. [PubMed] [Google Scholar]
- 14.Yang X, Li W, Prescott ED, Burden SJ, Wang JC. DNA topoisomerase IIbeta and neural development. Science. 2000;287:131–134. doi: 10.1126/science.287.5450.131. [DOI] [PubMed] [Google Scholar]
- 15.Linka RM, Porter AC, Volkov A, Mielke C, Boege F, Christensen MO. C-terminal regions of topoisomerase IIalpha and IIbeta determine isoform-specific functioning of the enzymes in vivo. Nucleic Acids Res. 2007;35:3810–3822. doi: 10.1093/nar/gkm102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Porter AC, Farr CJ. Topoisomerase II: untangling its contribution at the centromere. Chromosome Res. 2004;12:569–583. doi: 10.1023/B:CHRO.0000036608.91085.d1. [DOI] [PubMed] [Google Scholar]
- 17.Wang JC. Cellular roles of DNA topoisomerases: a molecular perspective. Nat. Rev. Mol. Cell Biol. 2002;3:430–440. doi: 10.1038/nrm831. [DOI] [PubMed] [Google Scholar]
- 18.Carpenter AJ, Porter AC. Construction, characterization, and complementation of a conditional-lethal DNA topoisomerase IIalpha mutant human cell line. Mol. Biol. Cell. 2004;15:5700–5711. doi: 10.1091/mbc.E04-08-0732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paddison PJ, Caudy AA, Sachidanandam R, Hannon GJ. Short hairpin activated gene silencing in mammalian cells. Methods Mol. Biol. 2004;265:85–100. doi: 10.1385/1-59259-775-0:085. [DOI] [PubMed] [Google Scholar]
- 20.Toyoda E, Kagaya S, Cowell IG, Kurosawa A, Kamoshita K, Nishikawa K, Iiizumi S, Koyama H, Austin CA, Adachi N. NK314, a topoisomerase II inhibitor that specifically targets the alpha isoform. J. Biol. Chem. 2008;283:23711–23720. doi: 10.1074/jbc.M803936200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sakaguchi A, Kikuchi A. Functional compatibility between isoform alpha and beta of type II DNA topoisomerase. J. Cell Sci. 2004;117:1047–1054. doi: 10.1242/jcs.00977. [DOI] [PubMed] [Google Scholar]
- 22.Toyoda Y, Yanagida M. Coordinated requirements of human topo II and cohesin for metaphase centromere alignment under Mad2-dependent spindle checkpoint surveillance. Mol. Biol. Cell. 2006;17:2287–2302. doi: 10.1091/mbc.E05-11-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Deming PB, Cistulli CA, Zhao H, Graves PR, Piwnica-Worms H, Paules RS, Downes CS, Kaufmann WK. The human decatenation checkpoint. Proc. Natl Acad. Sci. USA. 2001;98:12044–12049. doi: 10.1073/pnas.221430898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Downes CS, Clarke DJ, Mullinger AM, Gimenez-Abian JF, Creighton AM, Johnson RT. A topoisomerase II-dependent G2 cycle checkpoint in mammalian cells. Nature. 1994;372:467–470. doi: 10.1038/372467a0. [DOI] [PubMed] [Google Scholar]
- 25.Damelin M, Sun YE, Sodja VB, Bestor TH. Decatenation checkpoint deficiency in stem and progenitor cells. Cancer Cell. 2005;8:479–484. doi: 10.1016/j.ccr.2005.11.004. [DOI] [PubMed] [Google Scholar]
- 26.Kaufmann WK. Dangerous entanglements. Trends Mol. Med. 2006;12:235–237. doi: 10.1016/j.molmed.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 27.Sauer B, Henderson N. Targeted insertion of exogenous DNA into the eukaryotic genome by the Cre recombinase. New Biol. 1990;2:441–449. [PubMed] [Google Scholar]
- 28.Petruti-Mot AS, Earnshaw WC. Two differentially spliced forms of topoisomerase IIalpha and beta mRNAs are conserved between birds and humans. Gene. 2000;258:183–192. doi: 10.1016/s0378-1119(00)00465-0. [DOI] [PubMed] [Google Scholar]
- 29.Bell AC, West AG, Felsenfeld G. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell. 1999;98:387–396. doi: 10.1016/s0092-8674(00)81967-4. [DOI] [PubMed] [Google Scholar]
- 30.Arakawa H, Buerstedde JM. DT40 gene disruptions: a how-to for the design and the construction of targeting vectors. Subcell. Biochem. 2006;40:1–9. doi: 10.1007/978-1-4020-4896-8_1. [DOI] [PubMed] [Google Scholar]
- 31.Eguchi A, Kondoh T, Kosaka H, Suzuki T, Momota H, Masago A, Yoshida T, Taira H, Ishii-Watabe A, Okabe J, et al. Identification and characterization of cell lines with a defect in a post-adsorption stage of Sendai virus-mediated membrane fusion. J. Biol. Chem. 2000;275:17549–17555. doi: 10.1074/jbc.M910004199. [DOI] [PubMed] [Google Scholar]
- 32.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gillespie DA, Walker M. Mitotic index determination by flow cytometry. Subcell. Biochem. 2006;40:355–358. doi: 10.1007/978-1-4020-4896-8_25. [DOI] [PubMed] [Google Scholar]
- 34.Franklin R, Sale JE. 2D cell cycle analysis. Subcell. Biochem. 2006;40:405–408. doi: 10.1007/978-1-4020-4896-8_35. [DOI] [PubMed] [Google Scholar]
- 35.Szuts D, Sale JE. Subnuclear immunofluorescence. Subcell. Biochem. 2006;40:395–398. doi: 10.1007/978-1-4020-4896-8_33. [DOI] [PubMed] [Google Scholar]
- 36.Yao F, Svensjo T, Winkler T, Lu M, Eriksson C, Eriksson E. Tetracycline repressor, tetR, rather than the tetR-mammalian cell transcription factor fusion derivatives, regulates inducible gene expression in mammalian cells. Hum. Gene Ther. 1998;9:1939–1950. doi: 10.1089/hum.1998.9.13-1939. [DOI] [PubMed] [Google Scholar]
- 37.Bell AC, West AG, Felsenfeld G. Insulators and boundaries: versatile regulatory elements in the eukaryotic. Science. 2001;291:447–450. doi: 10.1126/science.291.5503.447. [DOI] [PubMed] [Google Scholar]
- 38.Chung JH, Whiteley M, Felsenfeld G. A 5′ element of the chicken beta-globin domain serves as an insulator in human erythroid cells and protects against position effect in Drosophila. Cell. 1993;74:505–514. doi: 10.1016/0092-8674(93)80052-g. [DOI] [PubMed] [Google Scholar]
- 39.Pikaart MJ, Recillas-Targa F, Felsenfeld G. Loss of transcriptional activity of a transgene is accompanied by DNA methylation and histone deacetylation and is prevented by insulators. Genes Dev. 1998;12:2852–2862. doi: 10.1101/gad.12.18.2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Spence JM, Phua HH, Mills W, Carpenter AJ, Porter AC, Farr CJ. Depletion of topoisomerase IIalpha leads to shortening of the metaphase interkinetochore distance and abnormal persistence of PICH-coated anaphase threads. J. Cell Sci. 2007;120:3952–3964. doi: 10.1242/jcs.013730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Andoh T, Ishida R. Catalytic inhibitors of DNA topoisomerase II. Biochim. Biophys. Acta. 1998;1400:155–171. doi: 10.1016/s0167-4781(98)00133-x. [DOI] [PubMed] [Google Scholar]
- 42.Grauslund M, Thougaard AV, Fuchtbauer A, Hofland KF, Hjorth PH, Jensen PB, Sehested M, Fuchtbauer EM, Jensen LH. A mouse model for studying the interaction of bisdioxopiperazines with topoisomerase IIalpha in vivo. Mol. Pharmacol. 2007;72:1003–1014. doi: 10.1124/mol.107.036970. [DOI] [PubMed] [Google Scholar]
- 43.McPherson JP, Goldenberg GJ. Induction of apoptosis by deregulated expression of DNA topoisomerase IIalpha. Cancer Res. 1998;58:4519–4524. [PubMed] [Google Scholar]
- 44.Baxter J, Diffley JF. Topoisomerase II inactivation prevents the completion of DNA replication in budding yeast. Mol. Cell. 2008;30:790–802. doi: 10.1016/j.molcel.2008.04.019. [DOI] [PubMed] [Google Scholar]
- 45.Classen S, Olland S, Berger JM. Structure of the topoisomerase II ATPase region and its mechanism of inhibition by the chemotherapeutic agent ICRF-187. Proc. Natl Acad. Sci. USA. 2003;100:10629–10634. doi: 10.1073/pnas.1832879100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Oestergaard VH, Knudsen BR, Andersen AH. Dissecting the cell-killing mechanism of the topoisomerase II-targeting drug ICRF-193. J. Biol. Chem. 2004;279:28100–28105. doi: 10.1074/jbc.M402119200. [DOI] [PubMed] [Google Scholar]
- 47.Roca J, Ishida R, Berger JM, Andoh T, Wang JC. Antitumor bisdioxopiperazines inhibit yeast DNA topoisomerase II by trapping the enzyme in the form of a closed protein clamp. Proc. Natl Acad. Sci. USA. 1994;91:1781–1785. doi: 10.1073/pnas.91.5.1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Germe T, Hyrien O. Topoisomerase II-DNA complexes trapped by ICRF-193 perturb chromatin structure. EMBO Rep. 2005;6:729–735. doi: 10.1038/sj.embor.7400465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hajji N, Pastor N, Mateos S, Dominguez I, Cortes F. DNA strand breaks induced by the anti-topoisomerase II bis-dioxopiperazine ICRF-193. Mutat. Res. 2003;530:35–46. doi: 10.1016/s0027-5107(03)00135-0. [DOI] [PubMed] [Google Scholar]
- 50.Huang KC, Gao H, Yamasaki EF, Grabowski DR, Liu S, Shen LL, Chan KK, Ganapathi R, Snapka RM. Topoisomerase II poisoning by ICRF-193. J. Biol. Chem. 2001;276:44488–44494. doi: 10.1074/jbc.M104383200. [DOI] [PubMed] [Google Scholar]
- 51.Wang L, Eastmond DA. Catalytic inhibitors of topoisomerase II are DNA-damaging agents: induction of chromosomal damage by merbarone and ICRF-187. Environ. Mol. Mutagen. 2002;39:348–356. doi: 10.1002/em.10072. [DOI] [PubMed] [Google Scholar]
- 52.Baumann C, Korner R, Hofmann K, Nigg EA. PICH, a centromere-associated SNF2 family ATPase, is regulated by Plk1 and required for the spindle checkpoint. Cell. 2007;128:101–114. doi: 10.1016/j.cell.2006.11.041. [DOI] [PubMed] [Google Scholar]
- 53.Chan KL, North PS, Hickson ID. BLM is required for faithful chromosome segregation and its localization defines a class of ultrafine anaphase bridges. EMBO J. 2007;26:3397–3409. doi: 10.1038/sj.emboj.7601777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang LH, Schwarzbraun T, Speicher MR, Nigg EA. Persistence of DNA threads in human anaphase cells suggests late completion of sister chromatid decatenation. Chromosoma. 2008;117:123–135. doi: 10.1007/s00412-007-0131-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Banaszynski LA, Chen LC, Maynard-Smith LA, Ooi AG, Wandless TJ. A rapid, reversible, and tunable method to regulate protein function in living cells using synthetic small molecules. Cell. 2006;126:995–1004. doi: 10.1016/j.cell.2006.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Luo K, Yuan J, Chen J, Lou Z. Topoisomerase IIalpha controls the decatenation checkpoint. Nat. Cell Biol. 2008;11:204–210. doi: 10.1038/ncb1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stucki M, Clapperton JA, Mohammad D, Yaffe MB, Smerdon SJ, Jackson SP. MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell. 2005;123:1213–1226. doi: 10.1016/j.cell.2005.09.038. [DOI] [PubMed] [Google Scholar]