Abstract
Riboswitches are a novel class of genetic control elements that function through the direct interaction of small metabolite molecules with structured RNA elements. The ligand is bound with high specificity and affinity to its RNA target and induces conformational changes of the RNA's secondary and tertiary structure upon binding. To elucidate the molecular basis of the remarkable ligand selectivity and affinity of one of these riboswitches, extensive all-atom molecular dynamics simulations in explicit solvent (≈1 μs total simulation length) of the aptamer domain of the guanine sensing riboswitch are performed. The conformational dynamics is studied when the system is bound to its cognate ligand guanine as well as bound to the non-cognate ligand adenine and in its free form. The simulations indicate that residue U51 in the aptamer domain functions as a general docking platform for purine bases, whereas the interactions between C74 and the ligand are crucial for ligand selectivity. These findings either suggest a two-step ligand recognition process, including a general purine binding step and a subsequent selection of the cognate ligand, or hint at different initial interactions of cognate and noncognate ligands with residues of the ligand binding pocket. To explore possible pathways of complex dissociation, various nonequilibrium simulations are performed which account for the first steps of ligand unbinding. The results delineate the minimal set of conformational changes needed for ligand release, suggest two possible pathways for the dissociation reaction, and underline the importance of long-range tertiary contacts for locking the ligand in the complex.
INTRODUCTION
Riboswitches represent a novel class of genetic control elements found mainly in the 5′-untranslated regions of bacterial mRNAs, but also in 5′- and 3′-untranslated regions and introns in (pre)mRNAs of plants and fungi (1–3). They regulate gene expression through direct and specific interactions with small metabolite molecules. Riboswitches can recognize a wide variety of different small molecule targets including important coenzymes such as FMN, cobalamine, S-adenosylmethionine, S-adenosylhomocysteine, thiamine pyrophosphate, amino acids such as lysine and glycine, the purine bases guanine, adenine and preQ1 and even metal ions such as Mg2+(4–6). They normally bind their ligands with high affinity and often reject even close chemical relatives of their cognate ligands. The binding of the metabolite is thought to induce large-scale structural changes of the secondary and tertiary structure of the riboswitch RNA element. This may result in either the masking of the Shine–Dalgarno sequence of the mRNA and the blocking of ribosome binding, or in the formation of an terminator hairpin loop preventing the transcription of the mRNA. Thus, it is obvious that structural rearrangements and conformational dynamics are at the heart of the function of many riboswitches.
High-resolution structural information from X-ray crystallography is so far available only for the ligand-bound states of the aptamer domains of a number of riboswitches (7,8). Only in the case of the lysine sensing riboswitch, a complete structure is also available for the ligand-free aptamer domain (9,10). However, in this rather untypical case the free state is highly similar to the bound state and ligand recognition involves only moderate structural changes of the RNA. The conformation of the ligand-free state of riboswitches and the conformational transitions upon ligand binding have been investigated mostly by chemical and enzymatic probing, fluorescence spectroscopy and NMR spectroscopy (4,11–13). The most extensive set of experimental data is probably available for the two closely related purine-sensing riboswitches that bind to either guanine or adenine and strongly suggests that conformational dynamics plays an important role in ligand binding and recognition at multiple levels (14–16). The X-ray structures of the guanine and adenine bound aptamer domains reveal that the ligand is almost completely buried in the RNA with only 5 % of its surface still solvent accessible (14,15). The ligand is bound by nucleotides in the center of a three-way helical junction that form a novel intermolecular base triple with the ligand (Figure 1). The N3/N9-edge of the purine ligand is involved in three hydrogen bonds with U51 of the riboswitch, whereas its Watson–Crick edge forms a Watson–Crick base pair with C74. An additional hydrogen bond involves the 2′-OH group of U22 and the N7-nitrogen of the purine base. The compact RNA tertiary structure is further stabilized by long-range base pairing interactions between the loops L2 and L3 capping the helices II and III, respectively.
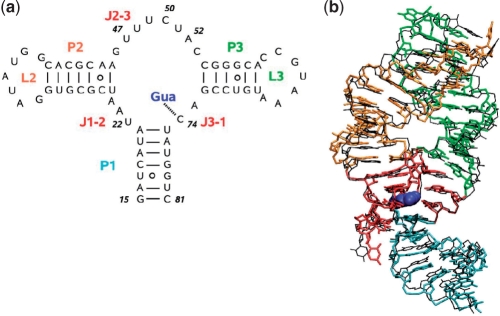
Figure 1.
Guanine sensing riboswitch aptamer domain represented by (a) its secondary structure including stem base pair hydrogen bonds and residue numbering and (b) a representative MD snapshot (color) overlaid with the X-ray structure (15) (black). The colors indicate various segments of the RNA: stems P1, P2 and P3 are in cyan, orange and green, respectively; loops L2 and L3 are in orange and green, respectively; the junction-connecting segments J1-2, J2-3 and J3-1 are in red; the ligand guanine is shown in blue. For clarity, hydrogen and oxygen atoms of the phosphate groups were omitted.
The deep burial of the ligand in the RNA-fold already suggests that the ligand can only access the binding pocket through an extensive induced-fit mechanism (14) where the binding pocket is not preformed. This idea is supported by chemical probing data and NMR experiments (16–18), which indicate that the ligand binding core of the aptamer domain is either in an open conformation not stabilized by base to base hydrogen bonding interactions or exists as a dynamic ensemble of rapidly interconverting conformations. Kinetic data from fluorescence experiments (11,12,19) and time-resolved NMR (20) show that ligand binding and dissociation is a very slow process occurring on a time scale of seconds, which is also indicative of substantial conformational rearrangements. Furthermore, time-resolved NMR (20) and fluorescence quenching experiments with site-specific labeled RNAs (11,12,19) indicate that ligand recognition is a multistep hierarchical process, where local folding of some of the structural elements is the prerequisite for the rearrangement of other structural elements.
In line with these findings, Batey and coworkers (19) suggested a two-step model for ligand binding to the aptamer domain. In this model, first the Watson–Crick interactions between the ligand and C74 of guanine riboswitch are established, while the ligand binding pocket is in an open conformation. Subsequently, the remainder of the binding pocket closes to establish interactions between the N3/N9-edge of the ligand and U51. In this case, the ligand should enter the binding site from the J2/J3-junction side of the binding pocket. On the other hand, NMR-data (20) indicate that the guanine riboswitch is capable of forming a transient complex with purine analogs such as adenine as long as they are capable of base pairing interactions through their N3/N9 edge. This either suggests a general unspecific docking step between U51 and the ligand followed by a selection step depending on the Watson–Crick hydrogen bonding potential of the ligand with C74 or points toward different modes of the initial interactions with the RNA for cognate and noncognate ligands.
However, conformational dynamics is apparently not only restricted to the ligand binding process. The bound state of the riboswitch aptamer domain in complex with its cognate ligand appears to be dynamic as well. Single molecule FRET studies of the adenine riboswitch bound to adenine indicated a conversion between a closed state with tertiary interactions between loops II and III and a more open global conformation, even in the presence of the ligand (11). O'Neill and coworkers (21) used fluorescence lifetime measurements of the fluorescent adenine analog 2-aminopurine to obtain evidence for at least three different conformations in the bound state as reported by three different ligand fluorescence lifetimes. However, atomic resolution information about the different intermediate stages of the ligand binding process, the pathways of their interconversion and the dynamics in the ligand binding pocket are not available from these methods.
Modern computer simulation techniques can supplement the information coming from these experimental techniques by realistic atomic resolution pictures of molecular motions (22,23). In particular, classical molecular dynamics (MD) simulations can provide a detailed description of RNA conformational dynamics in the condensed phase (24–32). They are still affected by imperfections of the atomistic force field (33,34) and typically limited to tenth or hundreds of nanoseconds. However, accurately performed simulations can provide qualitative information about the structural and dynamic aspects of RNA, which cannot be obtained by other techniques.
In this work, we employ classical MD simulations to study the structure and the conformational dynamics of the aptamer domain of the guanine-sensing riboswitch (G-riboswitch). Representing the riboswitch by the AMBER99 all-atom force field (35,36) in explicit water, in total ≈1 μs simulation time was collected. To investigate the structural basis of ligand affinity and selectivity, the structural features of G-riboswitch bound to either its cognate ligand guanine or to the noncognate ligand adenine are studied and compared with NMR and crystallographic data (15–17,37). Furthermore, simulations of the G-riboswitch in a hypothetical ligand-free conformation are performed to study the initial response of the system to unbinding. To probe possible pathways of ligand binding and unbinding, we employed additional nonequilibrium MD simulations (38). By pulling the ligand out from its binding pockets by an external force acting along two possible reaction coordinates, the unbinding process of guanine is triggered. Moreover, the first step of guanine binding is studied by simulating the prebinding state of the G-riboswitch. That is, we place the guanine ligand close to the binding pocket of the free G-riboswitch and monitor the penetration of guanine to the binding pocket (27). The simulations suggest a two-step ligand recognition process, involving a general purine binding step via U51 and a subsequent selection of the cognate ligand via C74.
METHODS
All MD simulations were performed using the GROMACS suite of programs (version 3.3) (39–41). The AMBER99 force field (35,36) was employed to describe the G-riboswitch and its ligands, adenine and guanine. Partial charges of the ligands were derived using the RESP (42) approach, which is in line with the force field parameterization. The RNA was placed in a rhombic dodecahedron box (edge length ∼8 nm), which was subsequently filled with about 11 000 TIP3P water molecules (43). To neutralize the system, 66 sodium or 33 magnesium ions were placed randomly in the simulation box. The starting structure of the guanine-bound G-riboswitch complex was taken from the Protein Data Bank (44,45) [PDB structure 1Y27 (15)]. The G-riboswitch with adenine as the bound ligand was modeled by fitting the adenine to the ligand position in the crystallographic structure. The guanine complex and the free G-riboswitch were simulated for 150 ns, the adenine complex for 100 ns.
A cut-off of 1.0 nm was used for the Lennard–Jones interactions. The interactions between atoms within 1.0 nm were evaluated at every time step. The particle mesh Ewald method (46) was employed to treat Coulomb interactions, using a switching distance of 1.0 nm, a grid of 0.12 nm and a beta value of 3.1 nm−1. Long-range dispersion corrections for energy and pressure were applied. Constant pressure p and temperature T were maintained by weakly coupling the system to an external bath at 1 bar and 298 K, using the Berendsen barostat and thermostat, respectively (47). The system was coupled to the temperature bath with a coupling time of 0.1 ps. The pressure coupling time was 0.5 ps and the isothermal compressibility 4.5×10−5 bar−1. The bond distances and the bond angle of the solvent water were constrained using the SETTLE algorithm (48). All other bond distances were constrained using the LINCS algorithm (49). A leap-frog integrator with a integration time step of 2 fs was used. Analysis of the trajectories was performed with tools from the GROMACS package and with modified versions of them. To define the presence of an hydrogen bond, an acceptor–donor distance <0.35 nm and a donor-hydrogen-acceptor angle >150° was requested. Figures showing molecular structures were generated using the graphical package VMD (50).
To study the unbinding of the ligand, a spring (i.e. a harmonic potential) was connected to the ligand and slowly retracted (51). This has the effect of pulling the ligand away from its initial location. Two pulling coordinates were used: the distance between the centers of mass of guanine and the base of residue C74 as well as the corresponding distance between guanine and U51. We employed a spring constant between 6000 and 10 000 kJ mol−1 nm−2 and a pulling rate of 0.0001 nm ps−1. For each choice of the pulling coordinate, we performed five independent 10 ns simulations of the guanine-bound G-riboswitch complex in magnesium solution (virtually the same results are obtained for sodium ions).
RESULTS
G-riboswitch bound to guanine
Structures of the G-riboswitch bound to its cognate ligand guanine were sampled from MD simulations with a total length of 300 ns at 298 K, using either Na+ or Mg2+ as counter-ions. We choose these limiting cases of the ion environment for several reasons. First, including Na+ ions only allows us to study if the structure is stable in the absence of Mg2+ ions. Including Mg2+ ions only, on the other hand, facilitates the study of Mg2+ binding sites of the G-riboswitch. Although the resulting Mg2+ concentration appears unrealistically high, it did not severely affect the structure and the dynamics of the riboswitch on the time scales under consideration (see below). Furthermore, it should be stressed that a ratio of 22 trivalent cobalt hexamine ions to 1 RNA molecule was required to successfully crystallize the G-riboswitch (14), and that NMR experiments showed no structural deformations of the RNA using Mg2+/RNA ratios as high as 20/1 (16). Finally, the true ion environment depends on the specific conditions of a given experiment and therefore will vary from case to case.
First, we will discuss the structural features observed for the guanine-bound G-riboswitch in our simulations in the presence of Na+ ions. Despite the absence of stabilizing Mg2+ ions, the structure of the complex was found to be quite stable throughout the simulation. That is, the average root mean square deviation (RMSD) with respect to the X-ray structure (15) was only 0.23 nm. Figure 1 shows the secondary structure as well as a representative MD snapshot of the guanine-bound G-riboswitch. The complex is characterized by two ‘kissing hairpins’ (labeled P2 and P3) and a stem (P1) connected by junction segments (J1-2, J2-3 and J3-1), which accommodate the ligand. Stem P1 consists of beginning and end segments of the RNA (residues 15–21 and 75–81, respectively), which are involved in base pairing interactions. Hairpins P2 and P3 are formed by residues 25–45 and 54–72, respectively, and are capped by the two seven-residue loops L2 and L3. These two hairpins are interacting with each other through the formation of five long-range base pairs. Two of these base pairs are aligned through standard Watson–Crick pairing, while the others form noncanonical base pairs in agreement with X-ray and NMR studies (14–16). As observed in experiment (15,17), the residues in the junction area are involved in intramolecular base triple (residues A21:U75:C50 and A23:G46:C53) and base pairing interactions (residues U22:A52 and U20:A76). Details on the base pair hydrogen bonding are given in Table 1.
Table 1.
Base pair hydrogen bonds in the kissing region (upper panel) and the junction region (lower panel) of G-riboswitch, as obtained from equilibrium MD simulation
| Base:base | Atom:atom | In Na+ solution |
In Mg2+ solution |
||||
|---|---|---|---|---|---|---|---|
| Free | G | A | Free | G | A | ||
| Kissing region | |||||||
| G37:C61 | O6:N4 | s | s | s | s | s | s |
| N1:N3 | s | s | s | s | s | s | |
| N2:O2 | s | s | s | s | s | s | |
| G37:U34 | N2:O2 | s | s | s | s | s | s |
| C61:A65 | O2:N6 | w | w | w | s | s | s |
| U34:A65 | O2:N6 | s | w | w | s | s | s |
| N3:N7 | s | s | s | s | s | s | |
| G38:C60 | O6:N4 | s | s | s | s | s | s |
| N1:N3 | s | s | s | s | s | s | |
| N2:O2 | s | s | s | s | s | s | |
| A33:A66 | N1:N6 | w | w | w | s | s | s |
| N6:N7 | w | w | w | s | s | s | |
| G38:A66 | N2:N1 | w | w | w | w | s | s |
| A35:A64 | N7:N6 | – | – | – | w | w | – |
| N6:N1 | w | w | – | w | w | – | |
| G62:U63 | N2:O4 | s | s | s | – | – | w |
| Junction region | |||||||
| U20:A76 | O4:N6 | s | s | s | s | s | s |
| N3:N1 | s | s | s | s | s | s | |
| A76:U49 | N3:N3 | – | w | – | – | – | – |
| A21:U75 | N6:O4 | s | s | s | s | s | s |
| N1:N3 | s | s | s | s | s | s | |
| U75:C50 | O2:N4 | – | s | – | – | w | – |
| U22:A52 | O4:N6 | s | s | s | s | s | s |
| N3:N1 | s | s | s | s | s | s | |
| A23:G46 | N1:N2 | s | s | s | s | s | s |
| N6:N3 | s | s | s | s | s | s | |
| G46:C53 | N2:O2 | s | s | s | s | s | s |
| N1:N3 | s | s | s | s | s | s | |
| O6:N4 | s | s | s | s | s | s | |
Compared are the free riboswitch and the guanine-bound (G) and the adenine-bound (A) states. Weak (w) and strong (s) hydrogen bonds correspond to hydrogen bond probabilities 0.4 ≤ PHB ≤ 0.8 and PHB > 0.8, respectively
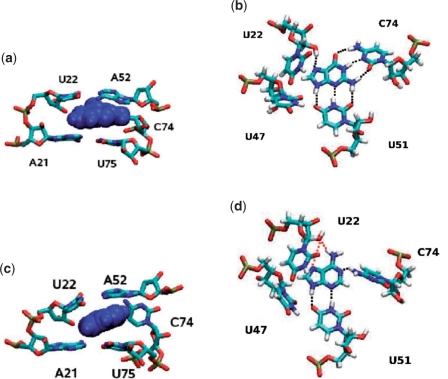
To illustrate the binding mode of the guanine ligand observed during the simulations, Figure 2 shows the details of the structural arrangement of guanine bound to the G-riboswitch. Similar to the findings in X-ray and NMR experiments (14,15,17), two base pair interactions are observed between the ligand and RNA. One is an intermolecular Watson–Crick G-C base pair between the ligand and residues C74, the other is between the N3/N9 edge of the guanine ligand and residue U51. Both interactions are mediated by three hydrogen bonds. Furthermore, in the Na+ simulations the C2 oxygen of U47 is in hydrogen bond distance [average O2(U47)–N9(ligand) distance: 0.34 nm], but the average donor-hydrogen-acceptor angle (O2(U47)-H9-N9) of 109° is unusually small for a ‘true’ hydrogen bond. Moreover, a hydrogen bond is also observed between the U22 2′-hydroxyl group and the N7 of the guanine, in agreement with crystallographic results (15).
Figure 2.
Details of the RNA–ligand binding interactions in (top) the guanine and (bottom) the adenine complex of the G-riboswitch. Left panels present the ligand in blue and residues A21-U22, A52 and C74-U75. Right panels present the ligand and residues U22, U47, U51, and C74. Hydrogen bonds between RNA and the ligand are indicated by black and red dotted lines. Red-dotted neighboring hydrogen bonds indicate that these bonds occur alternatively rather than simultaneously.
The effects of using either mono- or divalent ions on the overall structural features of the complex are quite small. When using Mg2+ instead of Na+ ions, the RMSD between X-ray and simulated structures decreases by only 0.03 nm to 0.21 nm—a difference to the RMSD of the Na+ simulation that is well within the variations expected when comparing multiple simulations (52). Table 1 lists the probabilities of hydrogen bonds in the kissing loop and junction region for the various ionic environments. Generally speaking, we find that the presence of divalent ions significantly strengthens the base pair interactions between the two kissing hairpins (53). On the other hand, sodium ions show the tendency to penetrate deeper than magnesium ions into the negatively charged region between the kissing hairpins (54). The latter might be related to the fact that hydrated sodium ions have a smaller van der Waals radius (0.25 nm) than hydrated magnesium ions (0.43 nm).
Despite the absence of major overall structural changes, the different ionic environment may well affect the local structure and the hydrogen bonding properties of the complex. For example, the Mg2+-simulations exhibit a higher (20%) probability for a hydrogen bond between the ligand N9 position and the O2 oxygen of U47. The average N9 (ligand)-O2(U47) distance of 0.29 (±0.02) nm is shorter than in the Na+ simulations and the value of the donor-hydrogen-acceptor angle 128° (±30°) is larger. Concerning the hydrogen bond between N9 (ligand) and U51, the probability of H9 to be hydrogen bonded to the oxygen (O4) of residue U51 decreases from 90% in Na+ environment to 60% in Mg+ environment. This is again related to the donor-hydrogen-acceptor angle, which decreases form 160° (±15°) in Na+ to 150° (±15°) in Mg+.
Interestingly, the simulations in Mg+ environment show some additional conformational fluctuations with regard to residue C50, which forms a base triple with the A21:U75 Watson–Crick base pair of helix 1 (P1) that is located right underneath the ligand. The distance between the center of mass of the ligand and the base C50 oscillates between 0.65 and 0.80 nm with a concomitant loss of hydrogen bonding between C50 and U75 and a destabilization of the base triple upon which the ligand stacks. The state at 0.65 nm is ∼ four time more populated than the state at 0.80 nm. The lifetimes of the two states are in the order of tens of nanoseconds. In addition, the distance between the ligand and the base A73 which interacts in a water-mediated manner with the U22-A52 base pair directly above of the ligand oscillates between 0.70 nm and 0.85 nm. There is a concomitant increase in the distance between A73 and the U22:A52 base pair from 0.40 to 0.66 nm when the ligand-A73 distance increases. The two different substates have life times in the order of 25 ns. In this case, both states are populated similarly. The two oscillations involving either the C50-ligand distance or the A73-ligand distance, respectively, and the associated conformational changes are not correlated, though.
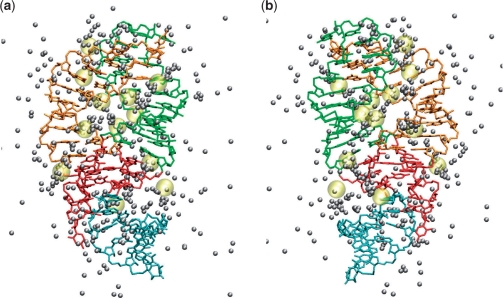
Figure 3 shows a representative MD snapshot of the G-riboswitch that reveals the highly populated magnesium sites of the system [i.e. where an ion can be found for at least two-thirds (100 ns) of the simulation time]. A total of seven highly occupied sites were found during the 150 ns simulation, four near the phosphate groups of the RNA and three near the oxygen of the cystidine and guanosine bases. Also shown in Figure 3 are the positions of cobalt hexamine ions found in the structure of the hypoxanthine-riboswitch complex as observed in X-ray experiments (14). In addition to the binding sites found in the simulation, the X-ray data indicate the presence of divalent ions also at the kissing region around A33 and along the junction between the two hairpins (residues 45–47). We note that, as a consequence of the microsecond magnesium–water residence times, only a part of the existing binding sites can be found within the sampling time of 150 ns. Table 2 lists the locations of magnesium binding sites as observed in the simulation and in the X-ray structure. The magnesium ions are mainly found along the stem of hairpins P2 and P3, in the junction regions J1-2 and J3-1, and in the kissing region around residue 37.
Figure 3.
Magnesium sites of the G-riboswitch as indicated by the positions of Mg2+ ions (gray balls) extracted every 10 ns from the MD trajectory. For comparison, the positions of cobalt hexamine ions found in the X-ray structure of hypoxanthine complex (14) are shown in yellow. The ligand is omitted. Using the same perspective and color code as in Figure 1, a representative MD snapshot of the G-riboswitch is shown as a front (a) and a back (b) view.
Table 2.
Magnesium sites of guanine-bound G-riboswitch as obtained from equilibrium MD simulation (see text for definition), compared with cobalt hexamine ion binding sites of hypoxanthine-riboswitch complexes as obtained from X-ray structure (14)
| U22 | A23 | U25 | C26 | C28 | G31 | A33 | G37 | G38 | |
|---|---|---|---|---|---|---|---|---|---|
| MD | Op | Op | O2 | Op | |||||
| X-Ray | Op | Op | O6 | Op | O6 | ||||
| A45 | C54 | G56 | C58 | U67 | C71 | A73 | |||
| MD | O6 | O2 | Op | ||||||
| X-Ray | Op | Op | O6 | Op | Op | Op | Op | ||
The location of the ion is given with respect to its closest residue and labeled according to its closest atom: phosphate oxygen (Op), carbonyl oxygen (O2, O4 and O6), and imino group (NH).
Binding of G-riboswitch to adenine
To explore the structural and dynamic basis for the binding specificity of the G-riboswitch, we also performed simulations where the guanine ligand was replaced by an adenine molecule. The overall structural features of the resulting complex are quite similar to the case of the guanine-bound G-riboswitch. For example, these simulations showed an average RMSD to the X-ray structure of the guanine complex of 0.24 and 0.26 nm in the presence of sodium and magnesium ions, respectively. As detailed in Table 1, the kissing interactions between the two hairpins are only little affected and the residues in the junction region are still involved in intramolecular base pairing and base triple interactions.
Naturally, the main changes upon replacement of the ligand occur in the binding pocket. Figure 2 compares the arrangements of RNA residues around the two purines as found in the end of the MD trajectory. On average, adenine is bound to the RNA through four hydrogen bonds, that is, three less than in the case of guanine. Two of the four remaining ligand–RNA hydrogen bonds are between the N3/N9 edge of adenine and residue U51. An hydrogen bond is also observed between the 2′-hydroxyl group of residue U22 and the N6/N7 edge (O2′-H2′:N7 or O2′:H6-N6). That renders this part of the intermolecular hydrogen-bonding network similar to the one found in the guanine complex. In addition, a novel hydrogen bond between the ligand and the RNA connects the adenine (N1) and the amino group of residue C74 (N4). The hydrogen bond results in a changed orientation of residue C74 in order to form this hydrogen bond to the ligand. This distortion causes the loss of the stacking interactions between C74 and A73 as well as between C74 and U75, while the other stacking and hydrogen bond interaction between the residues of the ligand binding core are mostly preserved.
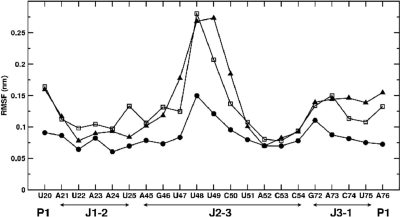
The missing RNA–ligand interactions are also reflected in the atomic root mean square fluctuations of the two systems with respect to their average structures. Figure 4 shows that the main difference of the atomic fluctuations are observed in the junction region, i.e. the adenine bound G-riboswitch RNA is significantly more flexible than the guanine complex. In particular, enhanced conformational fluctuations are observed in the J2–J3 junction region and to a lesser extent in the J3–J1 junction region. Residues U48 and U49 of the J2–J3 junction show the most pronounced deviations from the average structure. This is mainly related to unfavorable interactions between the ligand and residue C74, which hamper the stacking of the adenine ligand on top of the base pair A21:U75. In the guanine complex, on the other hand, favorable stacking interactions between the ligand and A21:U75 are observed (Figure 2). Subsequently, the adenine-induced C74 reorientation causes residue C50 to move away from the base of U75 and to break the base triple with the U75:A21 base pair. As a consequence, the backbone segment of residues U49 and U48 is released, as shown by the fluctuation values in Figure 4. Interestingly, the rearrangements in the binding pockets due to the presence of the noncognate ligand adenine described above do not affect the presence of the other base pairs and base triple interactions in the junction region.
Figure 4.
Root mean square fluctuations (in nm) of various residues in the junction region, as obtained from equilibrium MD simulations of guanine-bound (balls), adenine-bound (squares), and free (triangles) G-riboswitch.
The observed missing hydrogen bonds and stacking interactions in the adenine complex are expected to explain the high binding specificity of the G-riboswitch for guanine. The difference in the ligand–RNA interaction energy for adenine and guanine is 55 kJ/mol, i.e. the guanine interacts stronger with the RNA molecule than adenine does. Assuming that entropic effects and interactions with the solvent do not play a major role, our results are in qualitative agreement with the difference in the observed binding free energies, which was reported (4,19) as −50 and −20 kJ/mol for guanine and adenine, respectively.
Free G-riboswitch
As a further modification of the binding pocket, we performed simulations of the G-riboswitch where the guanine ligand was completely removed prior to the start of the simulation. Although this rather artificial system can certainly not describe the experimentally observed free state of the riboswitch, it nevertheless indicates the first response of the riboswitch upon unbinding. On the relatively short time scale (300 ns) of the simulation investigation, the overall structural features of the free G-riboswitch were again quite similar to the case of the ligand bound G-riboswitch. The simulated RNA shows an average RMSD of 0.28 and 0.30 nm to the X-ray structure of the guanine complex in presence of sodium and magnesium ions, respectively. The absence of the ligand did not affect the kissing interactions between the two hairpins, see Table 1. This agrees with the results of high-resolution NMR spectroscopy in this region (16,37) which also show the same base pairing pattern in the free and bound state of G-riboswitch.
The absence of the ligand, however, does severely affect the junction region. The absence of the RNA–ligand base pair interactions results in the loss of the base triple A21:U75:C50 and subsequently destabilizes the ligand binding core. As shown in Figure 4, these missing interactions induce large conformational fluctuations of residues U48–U49 at the junction. Furthermore, the fluctuations of the junction segment G46-C50 create an opening between nucleotides of the ligand binding core where the residue U49 is free to move away from the base A76 and to point to the solvent. Something similar can be observed for residue C50 which moves away from the base of U75 to point to the solvent and break the base triple with the U75:A21 base pair. Residues U47 and U48 stack on top of each other, which creates an opening between regions P1 and J2–J3. It is interesting to note that the base solvent accessible surface of U49 and C50 tend to fluctuate between 1.0 and 1.5 nm2 as compared with 1.0 and 1.1 nm2 in the guanine bound riboswitch. For U47 and U48, on the other hand, the solvent accessible surface shows a tendency to decrease during the simulation time due to the developing stacking interaction between the two residues which is not observable in the ligand bound state. The other base triple interactions in the ligand binding core domain are conserved also in the absence of the ligand, at least within the simulated time. The presence of the base triple interaction A23:G46:C53 in the junction area was also deduced from NMR experiments on the free RNA in presence of magnesium ions (37).
Nonequilibrium simulations of the prebinding state
To investigate how the ligand may access the binding core and which interactions will be stabilized during this process, we performed 10 independent nonequilibrium MD simulations (10 ns each) of the prebinding state of the G-riboswitch. That is, we positioned the ligand within a distance of 1.1 nm from the binding pocket (calculated from the center of mass of residues 51 and 74 and the center of mass of the guanine ligand) at a conformation found in the end of the MD trajectory of the free G-riboswitch, and followed the time evolution of this nonstationary initial state of the system. In particular, the ligand was positioned near the opening created by the flexible residues U48 and U49 in the junction area (see Free G-riboswitch section), at a distance ≳1.1 nm from U51 and C74.
In five out of 10 simulations, the ligand actually did enter the binding pocket within the simulation time. As detailed in Table 3, in three of these cases the ligand came close to (only) residue U51, while it came close to (both) U51 and C74 in the two other cases. To facilitate the access of the binding pocket near residues U51 and C74, the ligand was found to destabilize the stacking interactions of these residues with C50 and A73, respectively. Moreover, we observed the absence of hydrogen bonding between residues C50 and U75 throughout the simulation as well as a weakening of the interactions of base pair A21:U75. All other base pair interactions did not change during the simulation, i.e. they are the same as found for the free riboswitch, see Table 1.
Table 3.
Summary of the five runs of the prebinding simulations, in which the ligand was initially positioned near the binding pocket of free G-riboswitch
| 1 | 2 | 3 | 4 | 5 | |
|---|---|---|---|---|---|
| GUA-U51 | →0.50 | →0.48 | →0.65 | →0.65 | →0.35 |
| GUA-C74 | →0.60 | →0.86 | →0.58 | →1.15 | →0.79 |
| U49-C50 | 0.50 | 0.54 | →0.5 | 0.50 | →1.0 |
| C50-U51 | →0.60 | →0.75 | →0.75 | →0.85 | →0.85 |
| U51-A52 | →0.95 | →0.45 | →0.60 | →0.40 | →0.45 |
| A52-C53 | 0.36 | 0.35 | 0.36 | 0.37 | 0.39 |
| A73-C74 | 0.40 | 0.40 | 0.37 | →0.45 | →0.49 |
| C74-U75 | 0.44 | 0.43 | 0.44 | 0.40 | →0.45 |
Shown are various ligand–base and base–base distances (in nm) in the binding pocket. Distances are calculated between the centers of mass. When a distance changed significantly during the simulation time, the final value is reported in bold face, otherwise the average value is given.
As expected, the simulation time is too short to allow for the complete relaxation of the ligand in the binding pocket and to form the base pair interactions observed in the equilibrium simulations of the guanine complex. Nevertheless, it is interesting to note that the loss of the interaction of C50 with the A21:U75 base pair and the resulting flexibility around nucleotides U48 and U49 as observed in the simulations of the free RNA may already create an opening that is large enough to allow the ligand access the binding pocket.
Nonequilibrium simulations of guanine unbinding
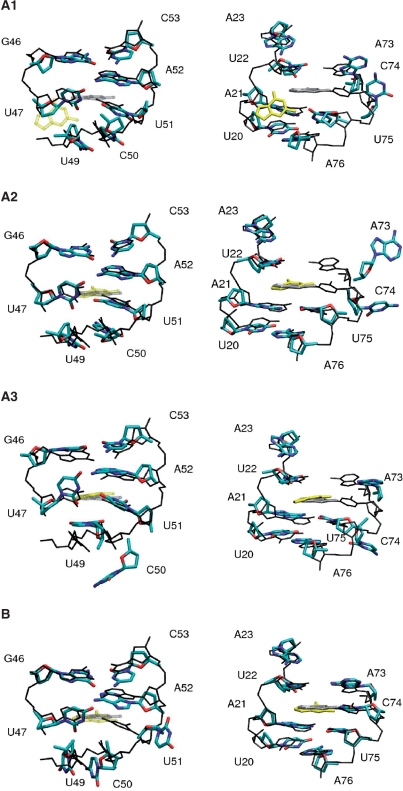
To investigate the changes in hydrogen bonding and base stacking that are required for the unbinding of the ligand, the guanine ligand was artificially pulled out of its binding pocket (see Methods section). As the unbinding of the ligand requires to break the hydrogen bonds between the ligand and residues C74 and U51, we separately used two reaction coordinates: (i) the distance between the ligand and the riboswitch residue C74 and (ii) the distance between the ligand and the riboswitch residue U51. For each choice of the pulling coordinate, we performed five independent simulations (10 ns each) of the guanine-bound G-riboswitch complex. We observed four different types of conformational rearrangements, which are discussed using the examples of trajectories A1, A2, A3, and B. For each case, Table 4 summarizes the changes observed in the binding pocket, and Figure 5 shows structural details of the conformational rearrangements.
Table 4.
Summary of the unbinding simulations of the guanine-bound G-riboswitch in magnesium ion containing solution
| EQ | A1 | A2 | A3 | B | |
|---|---|---|---|---|---|
| GUA-U22 | 0.46 | →0.7 | 0.46 | 0.42 | 0.44 |
| GUA-C50 | 0.70 | 0.78 | 0.70 | →1.2 | →0.9 |
| GUA-U51 | 0.54 | →1.0 | 0.54 | 0.56 | pc |
| GUA-A52 | 0.49 | →0.9 | 0.50 | 0.49 | 0.53 |
| GUA-C74 | 0.55 | pc | pc | pc | 0.56 |
| U49-C50 | 0.44 | 0.42 | 0.42 | →0.8 | 0.38 |
| C50-U51 | 0.39 | 0.40 | 0.40 | →1.0 | →1.2 |
| U51-A52 | 0.41 | 0.42 | 0.40 | 0.42 | →1.2 |
| A73-C74 | 0.38 | →0.6 | →0.9 | →0.9 | 0.37 |
| C74-U75 | 0.40 | →0.9 | →1.1 | →1.0 | 0.38 |
As pulling coordinate (pc) the distance between the ligand and riboswitch base C74 (cases A1, A2, A3) and U51 (case B) are employed, respectively. Shown are various ligand–base and base–base distances (in nm) in the binding pocket. Distance are calculated between the centers of mass. When a distance changes significantly during the simulation time, the final value is reported in bold face, otherwise the average value is given. As a comparison, the corresponding results from the equilibrium simulation (EQ) of the complex are listed.
Figure 5.
Conformational rearrangements observed in various unbinding simulations of the guanine-bound G-riboswitch, using the distance between the ligand and riboswitch residue C74 (cases A1, A2, A3) and U51 (case B) as the pulling coordinate. The nonequilibrium structures (in color, bases and sugars only) are compared with equilibrium structures (in black and gray, hydrogen atoms and oxygen atoms of the phosphate groups are omitted). For clarity, left panels show only residues 20–23 and 73–76, while right panels show only residues 46–47 and 49–53.
In A1-type simulations (two out of five trajectories), the ligand moves initially (t ≲ 4 ns) towards residues U48 and U49 of the junction, which causes the breaking of all seven hydrogen bonds between the ligand and the RNA in the first 4 ns of the simulation. That takes place in the following order: first C74:ligand, then U22:ligand and at last U51:ligand and U47:ligand. During the following 2 ns, the stacking interactions between binding pocket residues A73-C74 and C74-U75 disappear, moreover, the long range hydrogen bonding interaction between residues C50 and U75 is no longer observed. Nucleotide U49 reorients itself towards the solvent and its solvent accessible surface increases. All other base pair and stacking interactions in the junction region remain stable. Thus, overall in these trajectories, we find a behavior similar to that observed in the simulations of the free guanine riboswitch where the nucleotides U47-C50 are released from their stabilizing interactions and their reorientation creates an opening through which the ligand could leave the binding pocket. In the course of these two simulations, the ligand becomes more accessible to the solvent, as indicated by the increase of its solvent accessible surface from 1.0 to 1.4 nm2.
The main effect observed in A2-type simulations (two out of five) is an early (t ≲ 2 ns) flipping of residue C74 towards the solvent. As a consequence, the stacking interactions of C74 with A73 and U75 are not observed anymore, while the remaining hydrogen network of the junction region is not affected. In contrast to A1-type simulations, the ligand stays in the binding pocket and remains hydrogen bonded to residues U51, U22 and U47.
In A3-type simulations (one out of five), we also observe an initial flipping of residue C74 towards the solvent. In this case, however, not only the stacking interactions involving C74 but also the one between U49-C50 and C50-U51 disappear. The solvent accessible surface for bases C50 and U49 increases, thus indicating a reorientation towards the solvent, whereas it remains constant for U48. The base pairing interaction of C50 with U75 of the A21:U75 base pair is broken. In this respect, the A3-type simulations resemble the A1-type trajectories. In addition, the water-mediated hydrogen bond between A73 and U22 is broken and U49 which is involved in a base triple with the A76:U20 in the ligand bound state moves away from A76. As observed in A2-type trajectories, the remaining hydrogen network of the junction region is not affected and the ligand stays in the binding pocket.
When the distance between the ligand and residue U51 is used as pulling coordinate, all five simulations (type B) exhibit qualitatively the same behavior. The main effect is a flipping of residue U51 towards the solvent, which causes the loss of the stacking interactions of residues C50-U51 and U51-A52 and of the C50-U75 base pairing interaction. All other interactions in the binding pocket region are preserved and the ligand stays in the binding pocket and remains hydrogen bonded to residues C74, U22 and U47. Accordingly, in the simulations with the pulling coordinate between U51 and the ligand, the system stays very close to its starting structure. In contrast to the A-type simulations, no opening of the binding pocket in the area of nucleotides U47 and U48 is observed. Thus, the loss of three hydrogen bonds between U51 and the ligand does not appear to trigger the subsequent loss of additional stabilizing interactions in the direct vicinity of the ligand. This is in contrast to what happens upon loss of the C74:ligand intermolecular hydrogen bonds during the unbinding process, while residue C74 turns toward the solvent (Figure 5).
Only in the A1-type simulations which use the C74-ligand distance as reaction coordinate, we observe a partial release of the ligand from the binding pocket as indicated by an increase in the ligand's solvent accessible surface. Interestingly, in these trajectories not only a breaking of all seven RNA–ligand hydrogen bonds is observed, but also the release of stabilizing tertiary interactions involving nucleotides U47-C50 that create an opening of the ligand binding pocket qualitatively similar to what was observed in the simulations of the free RNA or when the ligand is replaced by adenine (Figure 4). Furthermore, it is interesting to note that all 10 pulling simulations destabilize the hydrogen bonding interactions of the triple base A21:C50:U75 directly below the ligand, while the hydrogen bonding between the bases A52 and U22 above the ligand is not affected. As shown in Figure 2, the ligand is sandwiched between planes formed by the triple base A21:C50:U75 and the base pair A52:U22. In none of the pulling simulations we were able to observe a complete release of the ligand. Besides limitations due to short simulation times, this may be caused by our simple one-dimensional models of the pulling coordinate. Nevertheless, the A1-type simulations showed the breaking of up to seven hydrogen bonds and two stacking interactions.
DISCUSSION AND CONCLUSIONS
As outlined in the Introduction section, recent experiments have indicated that ligand recognition in purine riboswitches represents a multistep hierarchical process that requires different levels of conformational dynamics. To augment these experimental results with theoretical arguments, in the following we employ our MD simulations of the G-riboswitch domain in different functional states in order to gain further insight into these dynamic processes.
Equilibrium simulations of the G-riboswitch aptamer domain bound to its cognate ligand guanine have revealed that the AMBER99 force field faithfully reproduces the experimental X-ray structure on a 100 ns time scale. Changing ionic conditions from monovalent sodium ions to divalent magnesium ions resulted in essentially the same overall structures, which is in agreement with experimental findings where the structure was investigated either in the presence or the absence of magnesium (14,17). Also in agreement with NMR experiments is the finding that the tertiary interaction between loops L2 and L3 is stabilized by the presence of magnesium ions and that magnesium ions interact directly with the RNA in the region of the L2/L3 interactions (16,18). Minor variations of the guanine bound structure were observed with regard to fluctuations in the stacking interactions between the ligand and nucleotide C50, which participates in a base triple with the A21:U75 base pair as well as in the stacking interaction between the ligand and A73. For each of these two stacking interactions, two equally populated states with different distances between the center of mass of the ligand and the respective base were observed.
Population of such locally different conformational states might be an alternative explanation for the observation of O'Neill and coworkers (21), who reported several ligand bound states with different fluorescence properties and small apparent differences in their respective dissociation constants KD. These authors ascribed the occurrence of the different conformational states to a transient opening of the L2–L3-loop interaction and the resulting differences in interhelical stacking. However, experiments by Batey and coworker (13) have indicated that the loss of the L2–L3 interaction should result in a significant decrease of the KD, that is, by at least two orders of magnitude for optimal ligands and affecting the total loss of binding for suboptimal ligands, as the ones used in the experiments of O'Neill and coworkers. Furthermore, in line and chemical probing (13,14) as well as NMR experiment (16) have shown unambiguously that the loop–loop interaction is stable even at elevated temperatures and especially in the presence of magnesium. On the other hand, local structural variations, which lead to small changes in the stacking patterns around the ligand or in the hydrogen bonding patterns, may affect distinguishable conformational substates as observed in our simulations. These states might therefore provide an alternative explanation for the observations of several ligand-bound states of the purine riboswitch with different fluorescence properties.
The comparison of the equilibrium simulations describing the binding of guanine and adenine to the G-riboswitch (Binding of G-riboswitch to adenine section) reveals that adenine is bound to the RNA through only four hydrogen bonds, that is, three less than in the case of guanine. This leads to a significant difference in the calculated ligand–RNA interaction energy, which may explain the high binding specificity of the G-riboswitch found in experiment (19). While two hydrogen bonds of adenine and residue U51 remain intact, only one (compared with three in the case of bound guanine) hydrogen bond to residue C74 is formed. These changes of the hydrogen bonding pattern induce a geometry of the adenine:C74 base pair different from a canonical Watson–Crick base pair. As a consequence of this unfavorable geometry, the stacking of adenine to base pair A21:U75 is hampered and nucleotide C50 is released from the base triple with the A21:U75 base pair, which in turn causes enhanced fluctuations in the junction region, in particular, of residues U48 and U49 (Figure 4). Consequently, in the presence of the noncognate ligand adenine the ligand binding pocket never closes completely. These observations are in complete agreement with the experimental findings that C74 is the origin of the high binding specificity of G-riboswitch (20). Moreover, they are supported by the fact that the base pairing interactions between U51 and the ligand can possibly be formed for many different purine derivatives (20).
The removal of the ligand from the binding pocket was found to lead to structural consequences similar to those of the replacement of the cognate with a noncognate ligand, but on a larger scale. Most importantly, the absence of the ligand also releases C50 from the base triple with the A21:U75 base pair, which forms the base of the ligand binding pocket and leads to large conformational fluctuations in the backbone segment U47-C50. On the other hand, the U22:A52 base pair, which forms the roof of the ligand binding pocket, stays in place as does U51. It is interesting to note that the final conformation obtained in our equilibrium simulations of the free G-riboswitch shows some features that are in agreement with experimental results aimed at a characterization of the free RNA. A unique feature is the observed stacking between U48 and U49, while both nucleotides point toward the solvent. This geometry decreases the solvent accessible surface of U48 in the simulation of free RNA, whereas U48 is completely solvent exposed in the ligand-bound state. In agreement with this finding, fluorescence quenching experiment (12) using the fluorescent analog 2-aminopurine in position 48 show a low fluorescent state for the free RNA and a highly fluorescent state (i.e. absence of stacking) in the ligand bound state for this nucleotide. Furthermore, 2-aminopurine in position A21 and A24 shows no or only a small change of its fluorescence between the ligand bound and the free state. Accordingly, no changes in the solvent accessible surface for these two nucleotides are observed in our simulations.
The high backbone flexibility predicted by our simulations of the free G-riboswitch for nucleotides U47-C50 is also in agreement with a high reactivity of these nucleotides in chemical probing experiments (13). Thus, the simulated local unfolding shows important conformational features in agreement with experimental data. However, the chemical-probing experiment (13) and the NMR results of Noeske et al. (16) suggest that also nucleotides C51 and U22 are flexible in the free state, while the NMR results of Ottink et al. (37) suggest that the U22:A52 base pair is also present in the free RNA, in agreement with our simulations. Most importantly, we have found that the backbone flexibility of the U47-C50 segment observed in our simulations creates an opening of the binding pocket, which is in principle large enough to allow the ligand to enter or leave the binding pocket, as shown in our simulations of the prebinding state. Hence, even though our simulations of the free G-riboswitch might not reach the ‘true’ free state of the RNA as indicated by the discrepancies with the experimental results mentioned above, they nevertheless provide a picture of the minimal conformational rearrangements that are necessary to create an opening in the binding pocket, which allows for the binding or the dissociation of the ligand.
These findings are further supported by our unbinding simulations, especially by the A1-type trajectories that use the C74-ligand distance as the reaction coordinate. The calculations revealed that the loss of the C74-ligand hydrogen bonds trigger similar conformational changes as induced by the total absence of the ligand. For example, we also find the loss of the base triple interaction including C50 and the opening of the binding pocket in the vicinity of U48 and U49, which allowed at least a partial release of the ligand. Interestingly, the breaking of the U51-ligand hydrogen bonds did not result in subsequent conformational changes of the binding pocket that go further than the loss of the C50:U75 interaction. This suggest that, in contrast to C74, residue U51 is not involved in a long-range network of tertiary interactions through which conformational changes can be communicated. Considering the opposite process of ligand binding, this means that, even though the binding of a ligand could proceed first through interaction with U51, only the interaction with C74, which is specific for the cognate ligand, would trigger further conformational changes that leads to the actual closing of the binding pocket. In the case of a noncognate ligand the binding pocket would not close but remain partially open instead, thus facilitating rapid ligand release. In our simulations of the free riboswitch, U51 is slightly less mobile than U74 and much more rigid than, e.g. U47 and U48. This indicates that U51 might indeed be part of a rigid docking platform for initial ligand binding according to the model of Buck et al. (20). Alternatively, it might be possible that cognate and noncognate ligands interact in different ways with the RNA during the initial encounter. Cognate ligands might form the Watson–Crick base pair with C74 immediately as suggested by Batey and coworkers (19) inducing the closing of the binding pocket, whereas noncognate ligands can only interact productively with U51. This interaction would then not be sufficient to induce closure of the binding pocket and the noncognate ligand is dissociating again. However, to finally settle these question, longer simulations will be necessary.
In summary, we have performed extensive all-atom MD simulations to study the binding of purine bases to G-riboswitch. In agreement with previous experimental results (20), the simulations suggest a two-step ligand recognition process. In the first step, residue U51 in the aptamer domain functions as a general docking platform for purine bases, in the second step the interactions between C74 and the ligand are responsible for the observed ligand selectivity. Elucidating the possible role of residues in the binding core of riboswitches maybe helpful to improve the design of new ligands and aptamers, which can be employed in the biomedical field [e.g. in antibacterial strateg (55)] as well as in nanotechnology, e.g. to build synthetic circuits or nanomechanical devices (56).
FUNDING
Frankfurt Center for Scientific Computing; the Fonds der Chemischen Industrie; the Deutsche Forschungsgemeinschaft (in the SFB 579 ‘RNA-Ligand Interactions’). Funding for open access charge: Deutsche Forschungsgemeinschaft.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank Harald Schwalbe for numerous inspiring and helpful discussions.
REFERENCES
- 1.Nudler E, Mironov AS. The riboswitch control of bacteria metabolism. Trends Biochem. Sci. 2004;29:11–16. doi: 10.1016/j.tibs.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 2.Winkler WC. Riboswitches and the role of noncoding RNAs in bacterial metabolic control. Curr. Opin. Chem. Biol. 2005;9:594–602. doi: 10.1016/j.cbpa.2005.09.016. [DOI] [PubMed] [Google Scholar]
- 3.Edwards TE, Klein DJ, Ferre-d'Amare AR. Riboswitches: small molecule recognition by gene regulatory RNAs. Curr. Opin. Struct. Biol. 2007;17:273–279. doi: 10.1016/j.sbi.2007.05.004. [DOI] [PubMed] [Google Scholar]
- 4.Mandal M, Boese B, Barrick JE, Winkler WC, Breaker RR. Riboswitches control fundamental biochemical pathways inBacillus subtilisand other bacteria. Cell. 2003;113:577–586. doi: 10.1016/s0092-8674(03)00391-x. [DOI] [PubMed] [Google Scholar]
- 5.Winkler WC, Breaker RR. Regulation of bacterial gene expression by riboswitches. Ann. Rev. Microbiol. 2005;59:487–517. doi: 10.1146/annurev.micro.59.030804.121336. [DOI] [PubMed] [Google Scholar]
- 6.Miranda-Ríos J. The THI-box riboswitch, or how RNA binds thiamin pyrophosphate. Structure. 2007;15:259–265. doi: 10.1016/j.str.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 7.Schwalbe H, Buck J, Fürtig B, Noeske J, Wöhnert J. Structures of RNA switches: insight into molecular recognition and tertiary structure. Angew. Chem. Int. Ed. Engl. 2007;46:1212–1219. doi: 10.1002/anie.200604163. [DOI] [PubMed] [Google Scholar]
- 8.Montange RK, Batey RT. Riboswitches: emerging themes in RNA structure and function. Annu. Rev. Biochem. 2008;37:117–133. doi: 10.1146/annurev.biophys.37.032807.130000. [DOI] [PubMed] [Google Scholar]
- 9.Serganov A, Huang A, Patel DJ. Structural insights into amino acid binding and gene regulation by a lysine riboswitch. Nature. 2008;455:1263–1267. doi: 10.1038/nature07326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garst AD, Heroux A, Rambo RP, Batey RT. Crystal structure of the lysine riboswitch regulatory RNA element. J. Biol. Chem. 2008;283:22347–22351. doi: 10.1074/jbc.C800120200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lemay J-F, Penedo JC, Tremblay R, Lilley DMJ, Lafontaine DA. Folding of the adenine riboswitch. Chem. Biol. 2006;13:857–868. doi: 10.1016/j.chembiol.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 12.Rieder R, Lang K, Graber D, Micura R. Ligand-induced folding of the adenosine deaminase A-riboswitch and implications on riboswitch translational control. ChemBioChem. 2007;8:896–902. doi: 10.1002/cbic.200700057. [DOI] [PubMed] [Google Scholar]
- 13.Stoddard CD, Gilbert SD, Batey RT. Ligand-dependent folding of the three-way junction in the purine riboswitch. RNA. 2008;14:675–684. doi: 10.1261/rna.736908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Batey RT, Gilbert SD, Montange RK. Structure of a natural guanine-responsive riboswitch complexed with the metabolite hypoxanthine. Nature. 2004;432:411–415. doi: 10.1038/nature03037. [DOI] [PubMed] [Google Scholar]
- 15.Serganov A, Yuan Y, Pikovskaya O, Polonskaia A, Malinina L, Phan AT, Hobartner C, Micura R, Breaker RR, Patel DJ. Structural basis for discriminative regulation of gene expression by adenine- and guanine-sensing mRNAs. Chem. Biol. 2004;11:1729–1741. doi: 10.1016/j.chembiol.2004.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Noeske J, Buck J, Fürtig HRNB, Schwalbe H, Wöhnert J. Interplay of ‘induced fit' and preorganization in the ligand induced folding of the aptamer domain of the guanine binding riboswitch. Nucleic Acids Res. 2007;35:572–582. doi: 10.1093/nar/gkl1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Noeske J, Richter C, Grundl MA, Nasiri HR, Schwalbe H, Wöhnert J. An intermolecular base triple as the basis of ligand specificity and affinity in the guanine- and adenine sensing riboswitch RNAs. Proc. Natl Acad. Sci. USA. 2005;102:1372–1377. doi: 10.1073/pnas.0406347102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Noeske J, Schwalbe H, Wöhnert J. Metal-ion binding and metal-ion induced folding of the adenine-sensing riboswitch aptamer domain. Nucleic Acids Res. 2007;35:5262–5273. doi: 10.1093/nar/gkm565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gilbert SD, Stoddard CD, Wise SJ, Batey RT. Thermodynamic and kinetic characterization of ligand binding to the purine riboswitch aptamer domain. J. Mol. Biol. 2006;359:754–768. doi: 10.1016/j.jmb.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 20.Buck J, Fürtig B, Noeske J, Wöhnert J, Schwalbe H. Time-resolved NMR methods resolving ligand-induced RNA folding at atomic resolution. Proc. Natl Acad. Sci. USA. 2007;104:15699–15704. doi: 10.1073/pnas.0703182104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eskandari S, Prychyna O, Leung J, Avdic D, O'Neill MA. Ligand-directed dynamics of adenine riboswitch conformers. J. Am. Chem. Soc. 2007;129:11308–11309. doi: 10.1021/ja073159l. [DOI] [PubMed] [Google Scholar]
- 22.Frenkel D, Smit B. Understanding Molecular Simulations. San Diego: Academic; 2002. [Google Scholar]
- 23.van Gunsteren WF, Bakowies D, Baron R, Chandrasekhar I, Christen M, Daura X, Gee P, Geerke DP, Glättli A, Hünenberger PH, et al. Biomolecular modeling: goals, problems, perspectives. Angew. Chem. Int. Ed. Engl. 2006;45:4064–4092. doi: 10.1002/anie.200502655. [DOI] [PubMed] [Google Scholar]
- 24.Zacharias M. Simulation of the structure and dynamics of nonhelical RNA motifs. Curr. Opin. Struct. Biol. 2000;10:311–317. doi: 10.1016/s0959-440x(00)00089-0. [DOI] [PubMed] [Google Scholar]
- 25.Rázga F, Zacharias M, Réblová K, Koča J, Šponer J. RNA kink-turns as molecular elbows: hydration, cation binding, and large-scale dynamics. Structure. 2006;14:825–835. doi: 10.1016/j.str.2006.02.012. [DOI] [PubMed] [Google Scholar]
- 26.Villa A, Widjajakusuma E, Stock G. Molecular dynamics simulation of the structure, dynamics, and thermostability of the RNA hairpins uCACGg and cUUCGg. J. Phys. Chem. B. 2008;112:134–142. doi: 10.1021/jp0764337. [DOI] [PubMed] [Google Scholar]
- 27.Mu Y, Stock G. Conformational dynamics in RNA-peptide binding: a molecular dynamics simulation study. Biophys. J. 2006;90:391–399. doi: 10.1529/biophysj.105.069559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kormos BL, Baranger AM, Beveridge DL. Do collective atomic fluctuations account for cooperative effects? molecular dynamics studies of the U1A-RNA complex. J. Am. Chem. Soc. 2006;128:8992–8993. doi: 10.1021/ja0606071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Auffinger P, Hashem Y. Nucleic acid solvation: from outside to insight. Curr. Opin. Struct. Biol. 2007;17:325–333. doi: 10.1016/j.sbi.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 30.Hashem Y, Westhof E, Auffinger P. Milestones in molecular dynamics simulations of RNA systems. In: Schwede T, Peitsch MC, editors. Computational Structural Biology. New York: World Scientific; 2008. [Google Scholar]
- 31.Fulle S, Gohlke H. Analyzing the flexibility of RNA structures by constraint counting. Biophys. J. 2008;94:4202–4219. doi: 10.1529/biophysj.107.113415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen S-J. RNA folding: conformational statistics, folding kinetics, and ion electrostatics. Annu. Rev. Biophys. 2008;37:197–214. doi: 10.1146/annurev.biophys.37.032807.125957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fadrná E, Špačková N, Štefl R, Koča J, Cheatham TR, III, Šponer J. Molecular dynamics simulations of guanine quadruplex loops: advance and force field limitations. Biophys. J. 2004;87:227–242. doi: 10.1529/biophysj.103.034751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pérez A, Marchán I, Svozil D, Šponer J, Cheatham TE, III, Laughton CA, Orozco M. Refinement of the AMBER force field for nucleic acids: improving the description of α/γ conformers. Biophys. J. 2007;92:3817–3829. doi: 10.1529/biophysj.106.097782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cornell WD, Cieplak P, Bayly CI, Gould IR, Merz KM, Ferguson DM, Spellmeyer DC, Fox T, Caldwell JW, Kollman PA. A second generation force field for the simulation of proteins, nucleic acids, and organic molecules. J. Am. Chem. Soc. 1995;117:5179–5197. [Google Scholar]
- 36.Wang J, Cieplak P, Kollman P. How does a restrained electrostatic potential (RESP) model perform in calculating conformational energies of organic and biological molecules? J. Comput. Chem. 2000;21:1049–1074. [Google Scholar]
- 37.Ottink OM, Rampersad S, Tessari M, Zaman GJ, Heus HA, Wijmenga SS. Ligand induced folding of the guanine sensing riboswitch is controlled by a combined predetermined-induced fit mechanism. RNA. 2007;13:2202–2212. doi: 10.1261/rna.635307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stock G, Wachtveitl J, Grubmüller H. Special issue on “Nonequilibrium Dynamics in Biomolecules”. Chem. Phys. 2006;323:1. [Google Scholar]
- 39.Berendsen HJC, van der Spoel D, van Drunen R. GROMACS: A message-passing parallel molecular dynamics implementation. Comput. Phys. Comm. 1995;91:43–56. [Google Scholar]
- 40.Lindahl E, Hess B, van der Spoel D. Gromacs 3.0: A package for molecular simulation and trajectory analysis. J. Mol. Mod. 2001;7:306–317. [Google Scholar]
- 41.van der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJC. GROMACS: fast, flexible and free. J. Comput. Chem. 2005;26:1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 42.Bayly CI, Cieplak P, Cornell WD, Kollman PA. A well-behaved electrostatic potential based method using charge restraints for deriving atomic charges: the RESP model. J. Phys. Chem. 1993;97:10269–10280. [Google Scholar]
- 43.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein M. Comparison of simple potential functioned for simulating liquid water. J. Chem. Phys. 1983;79:926. [Google Scholar]
- 44.Berman H, Henrick K, Nakamura H. Announcing the worldwide Protein Data Bank. Nature Struct. Biol. 2003;10:980. doi: 10.1038/nsb1203-980. [DOI] [PubMed] [Google Scholar]
- 45.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Darden T, York D, Petersen L. Particle mesh Ewald: An Nlog(N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98:10089. [Google Scholar]
- 47.Berendsen HJC, Postma JPM, van Gunsteren WF, Dinola A, Haak JR. Molecular dynamics with couplin to an external bath. J. Chem. Phys. 1984;81:3684. [Google Scholar]
- 48.Miyamoto S, Kollman PA. SETTLE: an analytical version of the SHAKE and RATTLE algorithms for rigid water models. J. Comput. Chem. 1992;13:952–962. [Google Scholar]
- 49.Hess B, Bekker H, Berendsen HJC, Fraaije JGEM. LINCS: a linear constraint solver for molecular simulations. J. Comput. Chem. 1997;18:1463–1472. [Google Scholar]
- 50.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1990;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 51.Grübmuller H, Heymann B, Tavan P. Ligand binding: molecular mechanics calculation of the streptavidin biotin rupture force. Science. 1996;271:997–999. doi: 10.1126/science.271.5251.997. [DOI] [PubMed] [Google Scholar]
- 52.Villa A, Fan H, Wassenaar T, Mark AE. How sensitive are nanosecond molecular dynamics simulations of proteins to changes in the force field? J. Phys. Chem. B. 2007;111:6015–6025. doi: 10.1021/jp068580v. [DOI] [PubMed] [Google Scholar]
- 53.Réblová K, Špačková N, Štefl R, Csaszar K, Koča J, Leontis NB, Šponer J. Non-Watson-Crick basepairing and hydration in RNA motifs: molecular dynamics of 5S rRNA Loop E. Biophys. J. 2003;84:3564–3582. doi: 10.1016/S0006-3495(03)75089-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Réblová K, Špačková N, Šponer JE, Koča J, Šponer J. Molecular dynamics simulations of RNA kissing-loop motifs reveal structural dynamics and formation of cation-binding. Nucleic Acids Res. 2003;31:6942–6952. doi: 10.1093/nar/gkg880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Blount KF, Wang JX, Lim J, Sudarsan N, Breaker RR. Antibacterial lysine analogs that target lysine riboswitches. Nat. Chem. Biol. 2007;3:44–49. doi: 10.1038/nchembio842. [DOI] [PubMed] [Google Scholar]
- 56.Davidson EA, Ellington AE. Synthetic RNA circuits. Nat. Chem. Biol. 2007;3:23–28. doi: 10.1038/nchembio846. [DOI] [PubMed] [Google Scholar]