Abstract
In this article, a panel of ssDNA aptamers specific to Staphylococcus aureus was obtained by a whole bacterium-based SELEX procedure and applied to probing S. aureus. After several rounds of selection with S. aureus as the target and Streptococcus and S. epidermidis as counter targets, the highly enriched oligonucleic acid pool was sequenced and then grouped under different families on the basis of the homology of the primary sequence and the similarity of the secondary structure. Eleven sequences from different families were selected for further characterization by confocal imaging and flow cytometry analysis. Results showed that five aptamers demonstrated high specificity and affinity to S. aureus individually. The five aptamers recognize different molecular targets by competitive experiment. Combining these five aptamers had a much better effect than the individual aptamer in the recognition of different S. aureus strains. In addition, the combined aptamers can probe single S. aureus in pyogenic fluids. Our work demonstrates that a set of aptamers specific to one bacterium can be used in combination for the identification of the bacterium instead of a single aptamer.
INTRODUCTION
Staphylococcus aureus is among the most pathogenic bacteria causing a wide spectrum of diseases ranging from minor wound infections to life-threatening infections such as endocarditis, septicemia, pneumonia and toxic shock syndrome (1). Rapid and accurate identification of S. aureus from infected samples is critical for the subsequent treatment of infectious diseases caused by S. aureus. Currently, microbiological detection such as classified cultivation, dyeing observation and corresponding zymology experiments are the most important methods to identify S. aureus but remain time-consuming and are not highly sensitive.
Systematic Evolution of Ligands by Exponential Enrichment (SELEX) is a high-flux screening technique that involves the progressive selection of aptamers by repeated rounds of partitioning and amplification from a large random synthetic oligonucleic acid library (2–3). Aptamers evolved from SELEX have a wide variety of targets ranging from small organic molecules to complex proteins or even whole intact cells (4–10).
For single protein or small molecule recognition, a single aptamer may be very useful as the aptamer has high specificity and affinity. But for whole bacteria detection, a single aptamer might give a false negative result. It is well known that bacteria of different growth states may preferentially express different sets of molecules to allow their growth and virulence factors to be under precise controls (11,12). Bacteria even undergo antigenic variations to escape the hosts’ surveillance (13,14). Therefore, detection of bacteria based on a single molecular probe (antibody, aptamer) might omit the positive bacteria which happen to negatively express specific molecules or express mutated ones. In some sense, targeting several sites of the whole bacterial cell simultaneously may increase the sensitivity of detection. At the same time, combining multiple aptamers will not increase the probability of false positive results by synergistic addition of cross-reactivity of aptamers when each aptamer shows no cross-reactivity to specimens of closely related species, thus maintaining specificity of the detection.
In this article, we obtained a panel of ssDNA aptamers specific against S. aureus by the subtractive SELEX, and demonstrated a superior effect of the combined use of these aptamers compared to an individual aptamer in the recognition of different strains of S. aureus or cells in different growth states. We also demonstrated the potential of the aptamer mixture to differentiate pathogens in clinical infectious diseases by probing single S. aureus in pyogenic fluids smear. Our work may provide a novel method for fast and convenient detection of pathogens.
MATERIALS AND METHODS
Bacterial strains and clinical specimens
The standard laboratory strain of S. aureus (ATCC 8325-4) and the clinical isolation strains of S. aureus (196, 04018), Streptococcus (A5005) and S. epidermidis (26069) were preserved in our laboratory (15). All these gram positive coccus were incubated at 37°C in Trypicase Soy Broth medium (OXOID LTD.). A gram negative bacteria of our laboratory stock, Escherichia coli DH5α, were grown in Luria-Bertani medium (Oxoid Ltd) at 37°C. To prepare the targets for selection, the gram positive bacteria were cultured to early log growth phase with OD600 of about 0.2 nm. One hundred microliters of the bacterial culture was removed, diluted with medium, coated on the TSB agar plates and cultured at 37°C for 12 h to count colony forming units. The rest of the bacteria were then harvested by centrifugation at 8000g for 10 min at 4°C. The sediment of bacteria was washed with PBS for three times. Cells were then fixed with 90% (v/v) methanol at −20°C for 10 min followed by additive fixation in absolute methanol at −20°C overnight. All the bacteria were preserved in absolute methanol at −20°C, and an aliquot was removed and washed with PBS before use. For the characterization of aptamers, bacteria were also cultured until the indicated growth phase (early-log growth phase with OD600 of about 0.2 nm, mid-log growth phase with OD600 of about 0.5, late-log growth phase with OD600 of about 1.5 nm and stationary growth phase with OD600 of about 6.0 nm). Cells were harvested and treated the same as above, except that E. coli cells were coated and cultured on LB agar plates.
The suppurative fluids specimens were sampled from patients of infectious diseases at Department of Burn, 304th Clinical Division of PLA General Hospital, China. The pyogenic fluids smear on glass slides were fixed with absolute methanol at −20°C for 10 min before use.
Random library and primers
The synthetic ssDNA library and a 5′ FITC labeled library consisted of a random sequence of 45 nt flanked by two primer hybridization sites (5′-GCAATGGTACGGTACTTCC-N45-CAAAAGTGCACGCTACTTTGCTAA-3′, termed GP45). A 5′ (+) strand primer (5′-GCAATGGTACGGTACTTCC-3′, termed Plong-1) and a 3′ (−) strand primer (5′-TTAGCAAAGTAGCGTGCACTTTTG-3′, termed P11) were used to synthesize double stranded DNA (dsDNA) with standard PCR procedure. The primer Plong-1 and a structured 3′ (−) strand primer (5′-GCTAAGCGGGTGGGACTTCCTAGTCCCACCCGCTTAGCAAAGTAGCGTGCACTTTTG-3′, termed Pstemloop-3) were used to synthesize single stranded DNA (ssDNA) by unequal length strand PCR. The random library and primers as well as the FITC-labeled library and aptamers were synthesized by Invitrogen Co. Ltd., Beijing, China.
Preparation of ssDNA by unequal length strand PCR
Using the primers Plong-1 and Pstemloop-3, the unequal length of ssDNA molecules were produced by PCR as follows: A 100 μl of PCR mixture contains 10 μl of 10 × PCR buffer, 0.2 mM dNTPs, 0.5 μM each primer, 10 nM template and 2.5 U Taq DNA polymerase. The mixture was thermally cycled 30 times through 95°C for 1 min, 37°C for 30 s and 58°C for 40 s, and followed by 5 min extension at 58°C. PCR products of unequal length strand were analyzed by electrophoresis in a 10% polyacrylamide-7 M urea gel and the lower band of interest was purified from the gel for the next round of selection.
SELEX procedure
The subtractive SELEX strategy as described previously (9) was performed with a few modifications. Briefly, the synthetic ssDNA library was denatured by heating at 100°C for 5 min and cooled immediately on ice for 10 min before selection. The denatured ssDNA library (1500 pmol for initial round; 25 pmol for subsequent rounds) was incubated with 107 S. aureus 8325-4 cells at 37°C for 1 h in 200 μl screening buffer (0.1 µg/µl salmon sperm DNA (0.1 µg/µl tRNA, 1%BSA, 1 × PBS, 0.05% (v/v) Tween-20). Unbound ssDNAs were washed with 1 ml washing buffer (1 × PBS, 0.05% (v/v) Tween-20) by centrifugation at 8000g for 10 min, and the sediment of the bacteria–aptamers complex were used as a template for the amplification of bound aptamers by the two step unequal length PCR procedure as follows: the 100 μl PCR mixture underwent five thermal cycles. Then, 10 μl of the PCR product aliquot was removed and served as the template which underwent appropriate cycles until the expected product was visible on a 10% denatured polyacrylamide-7 M urea gel by EB staining. The remaining PCR products were amplified as described above. The amplified ssDNAs in PCR products were purified by 10% denatured PAGE. Then the recovered ssDNAs were quantitated by OD260/280 ratio and ready for the next round of SELEX. The counter-selection was introduced from the third round of selection as follows: The ssDNA pool was first incubated with 107 counter target bacterial cells (Streptococcus A5005 for the third, fourth and seventh round and S. epidermidis 26 069 for the fifth and sixth round) at 37°C for 1 h in 200 μl screening buffer, and then the unbound ssDNAs in supernatant were withdrawn by centrifugation and incubated with target bacteria. To acquire aptamers with high affinity and specificity, the washing strength was enhanced gradually by increasing the frequencies of washes (from three to five). After several rounds of selection, the enrichment of aptamer candidates was monitored by radiolabeled ssDNA pool-binding assays. The highest affinity pool was amplified into dsDNA with primer Plong-1 and P11, and then cloned into E. coli DH5α using the pUC19-T vector system (TAKARA Biotechnology Co. Ltd.). Separated colonies were picked out randomly and their sequences were determined by Autolab Biotechnology Co. Ltd., Beijing, China.
Radiolabeled pool binding assays
The original naive library and enriched pools of each round were radiolabeled through phosphorylation reaction by T4 polynucleotide kinase (PNK) and [γ-32P] ATP (Furui Biology Engineer Co. Ltd., Beijing, China) using a DNA 5′ terminal labeling kit (MEGALABEL Kit, TAKARA Biotechnology Co. Ltd) as follows: 3 μl of ssDNA pools (5−10 pmol), 1 μl of 10× Phosphorylation buffer, 5 μl of γ-32 P ATP (10 mCi/ml) and 1 μl of T4 PNK were mixed and incubated at 37°C for 40 min. The enzymatic activity was then inactivated by heating at 70°C for 10 min. Radiolabeled pools were ethanol precipitated and incubated with 107 bacteria at 37°C for 45 min in 100 μl of screening buffer. After centrifugation, the bacterial sediment was washed by 1× PBS until there is no radioactivity in the supernatant. The bacterial sediment was dried by air. The amount of radiolabeled ssDNA in each sample was monitored by scintillation counting.
Confocal imaging of bacteria bound with aptamers
The FITC-labeled naive library and aptamers (240 nM each) were incubated with 107 bacteria at 37°C for 45 min in 500 μl of screening buffer, and then centrifuged to discard the unbound aptamers in supernatant. The bacterium–ssDNA complex sediment was washed with PBS three times, resuspended in PBS and dropped on a glass slide to make a thin smear of the bacteria sample. Bacteria were then fixed to the slide by passing the slide three or four times through the flame of a Bunsen burner. Imaging of the bacteria was performed with a confocal microscope (ZEISS, LSM 510META) under 488 nm exciting light and visible light. For the detection of pathogens in a clinical setting, the FITC labeled aptamers were incubated with pyogenic fluids smear on glass slides from patients of severe burn wound infection at 37°C for 45 min. The pathogens of infection were determined by a bacteria culture test by a clinical laboratory. The bacteria in the clinical specimen were then imaged with confocal microscopy.
Flow cytometry analysis
The FITC-labeled aptamers (240 nM each) were incubated with 107 bacteria in 500 μl of screening buffer at 37°C for 45 min. Bacteria were subsequently washed with 1 ml of washing buffer containing 0.05% (v/v) Tween-20 and resuspended in 300 μl PBS. The fluorescence intensity of bacteria was monitored with a FACSCalibur cytometer (B-D, USA) by counting 30 000 events. The FITC-labeled naive library served as a negative control. The results were analyzed by FCS express V3 software.
Characterization of the binding parameters of aptamers to bateria
Binding affinities of the individual aptamer to S. aureus (ATCC 8325-4) was obtained through monitoring the mean fluorescence intensity of target bacteria binding with FITC-labeled aptamers by a FACSCalibur cytometer as described (4). The concrete steps were the same as Flow cytometry analysis. The equation Y = BmaxX/(Kd+X) (Origin Pro 7.5) was used to calculate the median effective concentration, EC50.
RESULTS
Preparation of the unequal length single-stranded products using the special primer with stem-loop structure
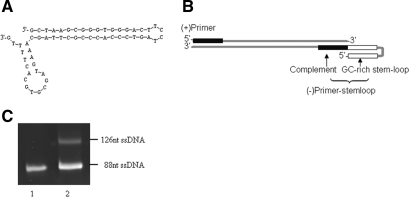
The 3′(−) primer Pstemloop-3 consists of two compositions, the 5′-GC rich reverse repeat sequence with the ability to form stem-loop structures and the 3′-complement sequence of the template (Figure 1A). Due to the high Tm of the primer, the advanced structures still remain when the Taq DNA polymerase function at the temperature range of 55–65°C and thus prevent the elongation of the (+) strand (Figure 1B). Therefore, the PCR products exhibited two bands with different migration rates in denatured gel electrophoresis (Figure 1C); the lower band is at the same location level of the single strand template and the higher band is the extension product of Pstemloop-3 with reverse repeat sequence of 38 nt longer than the template.
Figure 1.
Unequal strand length PCR using a primer with stem-loop structure. (A) The secondary structure of the (−) primer Pstemloop-3. (B) Schematic of PCR product. The (−) primer has a GC-rich reverse repeat sequence in the 5′ terminus, which can form the stem-loop structures and prevent the (+) strand elongation. (C) PCR products were analyzed by denaturing gel electrophoresis. Lane 1, the ssDNA of the naive library GP45; lane 2, the unequal length ssDNA products.
The whole bacteria SELEX for enrichment of candidate aptamers
The subtractive selection to intact biological entities does not require a prior knowledge of the complex, providing a new idea for development of molecular probes in detection of pathogens. Our subtractive selection strategy against S. aureus was similar to previous studies with a few modifications.
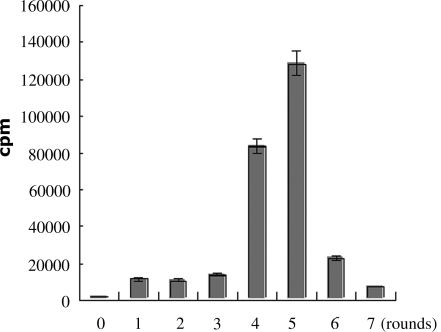
The enrichment of selection pools were monitored by the binding assays of enriched pools. The results showed directly that fifth round pool was enriched best with radioactivity of about 100-fold excess of the naive ssDNA library (Figure 2). The sixth and seventh round showed a gradually decreased enrichment, which may be a result of loss of structured specific aptamers through PCR amplification.
Figure 2.
The monitoring of enrichment of selection process by binding assay of radiolabeld pools with S. aureus. The number 0 represents the naive library and the numbers 1–7 represents the first to seventh round selected pool, respectively. Twenty-five picomoles of each radiolabeled pool were incubated with 107 S. aureus 8325-4 for 45 min. The binding capacity was detected by scintillation counting.
Identification of a panel of aptamers specific to S. aureus
The enriched DNA pool of the fifth round was cloned into E. coli DH5α using the pUC-19 T vector system. Fifty-five positive clones were picked out at random out of about 500 clones and their sequences were determined by Invitrogen Co. Ltd, Beijing, China. The homology and secondary structure of these sequences were analyzed by the software of MEME on line (http://meme.sdsc.edu/meme/intro.html) (16) and RNA STRUCTURE 4.5, respectively. The fifty-five sequences were then divided into nine families based on homology of DNA sequence and the similarity of secondary structure (data not shown).
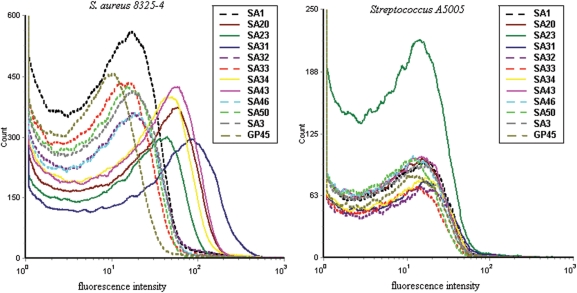
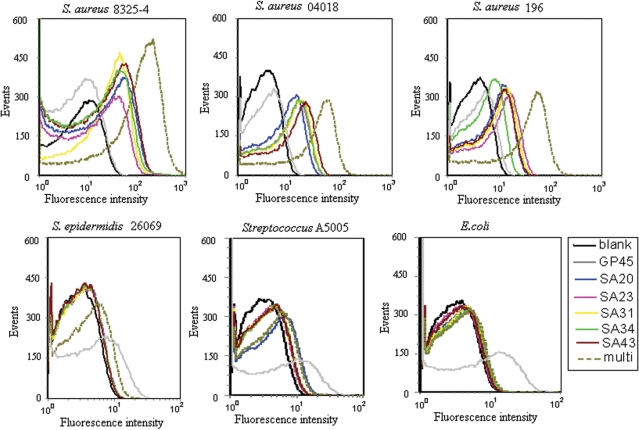
Eleven candidate sequences with stable structures and representative motifs out of eight families were chosen for further characterization, with one to two sequences from each family. One family was excluded from characterization because of the least numbers and unstable structure of the members. The binding assays were performed with flow cytometry to screen the specific aptamers for S. aureus 8325-4. Results showed that most of the aptamers could discriminate S. aureus from streptococcus. Five aptamers (SA20, SA23, SA31, SA34 and SA43) with high ranked specificity and sensitivity were selected for further investigation (Figure 3). The five aptamers were determined to be specific to S. aureus with a similar calculated median effective concentration of 8325-4 binding in the nanomolar (nM) range (Table 1) and were not cross-reactive to Streptococcus A5005, S. epidermidis 26069 and E. coli DH5α (Figure 4A and B). The sequences and the representative structures of the strong binders were shown as Supplementary Data (Supplementary Figure 1).
Figure 3.
The binding capacity of candidate aptamers to S. aureus by flow cytometry analysis. The FITC-labeled individual aptamer was incubated with 107 target bacteria (S. aureus 8325-4) or non-target bacteria (Streptococcus A5005) at 37°C for 45 min. The selected aptamers with higher ranked specificity and sensitivity were depicted with solid lines, and the GP45 naive library control and candidates with lower ranked specificity and sensitivity were depicted with dot lines. Representative results of three separate experiments.
Table 1.
EC50 of individual aptamers or aptamer combinations to S. aureus 8325-4
| Aptamer name | Calculated EC50 (nM) |
|---|---|
| SA20 | 70.86 ± 39.22 |
| SA23 | 61.50 ± 22.43 |
| SA31 | 82.86 ± 33.20 |
| SA34 | 72.42 ± 35.23 |
| SA43 | 210.70 ± 135.91 |
| Multiple aptamers | 479.98 ± 209.94 |
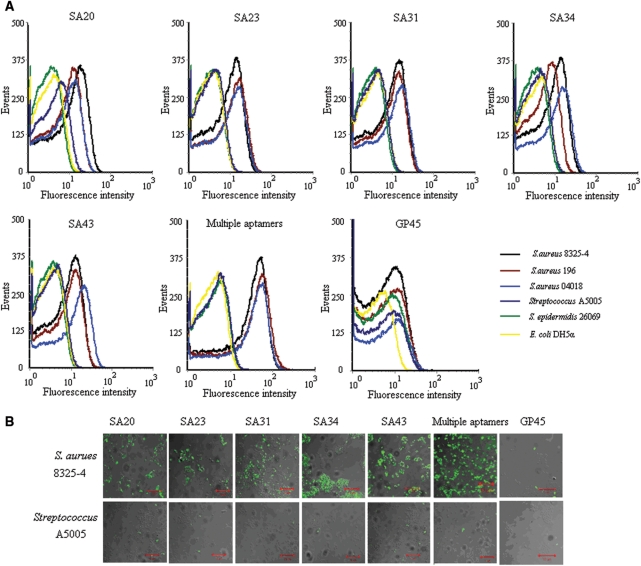
Figure 4.
The characterization on specificity of individual aptamer and combined aptamers for S. aureus. The FITC-labeled individual aptamer or aptamer combination were incubated with 107 bacteria at 37°C for 45 min. The concentration of aptamers in the binding buffer was 240 nM each. Representative results of three separate experiments. (A) Flow cytometry assay for the binding of aptamers with bacteria. The different color curves represent different strains of bacteria. (B) Overlaid confocal images of S. aureus 8325-4 and Sreptolococcus A5005 stained by aptamers from fluorescence and optical images. A 10 μm bar was shown.
The combined use of five aptamers in recognition of S. aureus
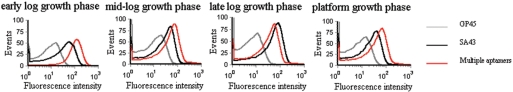
Because of a space-temporal antigenic expression model and high antigenic variation by bacteria, the detection of bacteria based on a single molecular probe may be prone to a false negative result. Therefore, combined detection of multiple molecular characteristics on bacteria with a panel of aptamers targeting different sites will improve the test sensitivity theoretically. The five aptamers were verified to recognize different molecular targets on S. aureus by a competition experiment (Supplementary Figure 2). The feasibility of using mixture of these five aptamers for recognizing S. aureus was then tested by flow cytometry and confocal microscopy in this study. Because we evaluate individual aptamer under concentration of 240 nM that is far higher than the concentration to induce maximum binding for anyone aptamer (Supplementary Figure 3), i.e. the corresponding target is saturated with each aptamer, we also use equimolarly mixed multiple aptamers with 240 nM each to gain a similar saturation state to evaluate the effect of mixture. The results indicated that mixed aptamer probes kept the specific property of an individual aptamer, i.e. distinguish S. aureus including 8325-4, 196 and 04018 from S. epidermidis, streptococcus and E. coli. (Figure 4A and B). The individual aptamer showed various degrees of recognition to different strains of S. aureus, which demonstrated a different expression profile by different strains (Figure 4A). Importantly, the combined application of aptamers showed enhanced fluorescence intensities, and thus a higher positive rate or binding capacity, against different strains of S. aureus in early growth state (Figure 5) and against 8325-4 of different growth states compared to the single probe (Figure 6), suggesting a more sensitive and effective strategy by multiple aptamers in bacteria identification. Because different aptamers recognize different targets and do not interfere with each other (Supplementary Figure 2); the EC50 value of each aptamer in the mixture will be the same as it is in the individual binding experiment. Therefore, the EC50 of the ssDNA in the mixture will be approximately the summation of EC50 of each aptamer theoretically. Accordingly, a mixture of aptamers with individual EC50s amongst 61.50–210.70 nM is characterized by an EC50 of 479.98 nM (Table 1).
Figure 5.
The comparison of sensitivity of combined aptamers to single aptamer for different S. aureus strains. Three different S. aureus strains in early log growth state were incubated with individual aptamer or aptamer combinations. The concentration of aptamer in the mixture was 240 nM each. Representative results of three separate experiments.
Figure 6.
The comparison of sensitivity of combined aptamers to single aptamer for S. aureus in different growth states. The S. aureus 8325-4 in different growth states were incubated with a single aptamer SA34 or aptamer combinations before flow cytometry analysis. The concentration of aptamer in the mixture was 240 nM each. Representative results of three separate experiments.
The specific aptamer mixture was then incubated with suppurative fluids smear on glass slides from patients of severe burn wound infection to investigate the potential of aptamers in the clinical setting. The aptamer mixture can recognize most of bacteria in one S. aureus infection specimen, and recognize a small portion of bacteria in another infection specimen that reported to be of mixed infection of Streptococcus and S. aureus (Figure 7).
Figure 7.
The recognition of S. aureus in specimens of infectious diseases by aptamer mixture. The FITC labeled aptamer mixture was incubated with suppurative fluids smear on glass slides from patients of severe burn wound infection. The fluorescence patterns were observed with a confocal microscope. Upper row: S. aureus infection specimen; lower row: the specimen of mixed infection of Streptococcus and S. aureus.
DISCUSSION
The aptamer pool rather than an individual aptamer is recommendable in revealing a comprehensive list of biomarkers due to the cumulative effect of multiple aptamers (17). But until now, there is no study focusing on using a panel of aptamers to detect pathogenic bacterium. Here, we demonstrated for the first time that simultaneously probing several targets by a set of specific aptamers proved to be more efficient than probing by an individual aptamer in detection of S. aureus. Unlike single target SELEX which generates an enriched pool of aptamers to the single target, the complex targets SELEX ends up with an enriched pool of aptamers to various targets (9,10). The different sets of aptamers to a single target usually have competitive effect as a result of competitive binding for the same site or a stereospecific blockade. But the aptamers to different targets trend to have synergistic effect on the recognition of the complex target. Therefore, even though the binding of individual aptamers usually exceeds that of the enriched pool in the case of single target SELEX procedure, it is expected that the binding efficiencies of individual aptamers to S. aureus is much lower than that of the enriched pool in this study, which demonstrated about 10 times the binding of naive library of the individual aptamer versus a 100-fold increase of binding of the enriched pool over the naive library. These results are also supportive for the combining use of multiple aptamers for increased sensitivity. As a matter of fact, the mixture of the five aptamers proved to be superior to individual aptamer in the detection of different S. aureus strains and different growth states. Besides the detection of different S. aureus strains, the detection of pathogen in different growth phases also is of great importance to clinical samples, as the bacteria in clinical specimens of different phases of the infectious disease (latency, active stage, quiescent stage, incipient stage or advanced stage, etc.), may be in a different growth cycle. Anti-bacteria drugs also will interfere the growth cycles of the pathogen. The mixture of five aptamers has been validated to recognize S. aureus well in the clinical specimens in a further experiment. It is suggested that the combination of a group of specific aptamers could also be used for a fast and convenient detection of other pathogens.
A key procedure of the SELEX is the preparation of the ssDNA, retaining the positive (+) strand with binding or catalytic activities by removal of the negative (−) strand. The commonly used method is the asymmetric PCR or specific biotinylation of the (−) strand followed by streptavidin separation. The asymmetric PCR has the disadvantages of low yield, (−) strand interference, and the common occurrence of smeared bands. Biotin based ssDNA preparation is timesaving but relatively expensive and undesirable if another biotin based separation is planned during a different step of the selection protocol. Here we improved the method of ssDNA preparation by unequal length PCR with a structured (−) strand primer as a substitute for the special modified (−) strand primer as described by Williams et al. (18). Our work is motivated by the characteristic that the DNA template with advanced structures can prevent the extension of Taq DNA polymerase. A peculiar design of a structured (−) primer allowing strand separation following PCR was used to prepare ssDNA. The sequence of structured primer may be randomized but obeys the rules that the 3′ of the primer is complementary to the template, and that the 5′ of the primer forms a hairpin structure. The (−) primer or (−) strand should keep its hairpin structure during annealing and elongation process of PCR procedure so as to prevent the extension of (+) strand. Therefore, the stability of the hairpin is critical, and we use a long reverse repeat sequences with high GC content to form a long stem and a few bases in the media to form a small loop in this study. This improved method is cheap, and not restricted to the difficult synthesis of the special (−) primer for most companies. The structured primer in this study, Pstemloop-3, formed a stable stem-loop structure in the 5′ position and may interfere in the annealing process of the 3′-complement sequence to the template, thus causing less efficient synthesis of the (−) strand. Therefore, the longer strand is much less intense than the lower band in Figure 1C. But the synthesis efficiency of the interest (+) strand is not influenced and thus recommendable to prepare ssDNA in a SELEX procedure.
To gain highly efficient selection, we made several modifications to the SELEX procedure in this article. First, we performed PCR directly with the bacteria–ssDNA complex as the template without the step of the ssDNA partition from the targets, as we found that no specific products appeared when using the bacteria itself as the PCR template and an improved recovering efficacy of the ssDNA of this method compared to a partition approach (Supplementary Figure 4). Secondly, the two closely related bacteria to S. aureus in evolution, Streptococcus (A5005) and S. epidermidis (26 069) were used as a counter target successively to subtract the common molecules that are not specific for S. aureus. All these strategies could be used for efficient selection of aptamers against other pathogens in the future.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Chinese National High-Tech Research and Development Program (2006AA02Z408). Funding for open access charge: 2006AA02Z408.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Heath Acuff Elrod and Dr Shiyong Sun at Emory University School of Medicine, USA for linguistic revision on the manuscript. We thank all the members in our laboratory for their help.
REFERENCES
- 1.Lowy FD. Staphylococcus aureus infections. New Engl. J. Med. 1998;339:520–532. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- 2.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 3.Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 4.Shangguan DH, Li Y, Tang ZW, Cao ZC, Chen HW, Mallikaratchy P, Sefah K, Yang CJ, Tan WH. Aptamers evolved from live cells as effective molecular probes for cancer study. Proc. Natl Acad. Sci. USA. 2006;103:11838–11843. doi: 10.1073/pnas.0602615103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ulrich H, Magdesian MH, Alves MJ, Colli W. In vitro selection of RNA aptamers that bind to cell adhesion receptors of Trypanosoma cruzi and inhibit cell invasion. J. Biol. Chem. 2002;277:20756–20762. doi: 10.1074/jbc.M111859200. [DOI] [PubMed] [Google Scholar]
- 6.Gopinath SC, Misono TS, Kawasaki K, Mizuno T, Imai M, Odagiri T, Kumar PK. An RNA aptamer that distinguishes between closely related human influenza viruses and inhibits haemagglutinin-mediated membrane fusion. J. Gen. Virol. 2006;87:479–487. doi: 10.1099/vir.0.81508-0. [DOI] [PubMed] [Google Scholar]
- 7.Chen F, Zhou J, Fengling L, Mohammed AB, Zhang XL. Aptamer from whole-bacterium SELEX as new therapeutic reagent against virulent Mycobacterium tuberculosis. Biochem. Biophys. Res. Commun. 2007;357:743–748. doi: 10.1016/j.bbrc.2007.04.007. [DOI] [PubMed] [Google Scholar]
- 8.Bruno JG, Kiel JL. In vitro selection of DNA aptamers to anthrax spores with electrochemiluminescence detection. Biosens. Bioelectron. 1999;14:457–464. doi: 10.1016/s0956-5663(99)00028-7. [DOI] [PubMed] [Google Scholar]
- 9.Wang CL, Zhang M, Yang G, Zhang DJ, Ding HM, Wang HX, Fan M, Shen BF, Shao NS. Single-stranded DNA aptamers that bind differentiated but not parental cells: subtractive systematic evolution of ligands by exponential enrichment. J. Biotechnol. 2003;102:15–22. doi: 10.1016/s0168-1656(02)00360-7. [DOI] [PubMed] [Google Scholar]
- 10.Shangguan DH, Cao ZC, Li Y, Tan WH. Aptamers evolved from cultured cancer cells reveal molecular differences of cancer cells in patient samples. Clin. Chem. 2007;53:1153–1155. doi: 10.1373/clinchem.2006.083246. [DOI] [PubMed] [Google Scholar]
- 11.Beier D, Gross R. Regulation of bacterial virulence by two-component systems. Curr. Opin. Microbiol. 2006;9:143–152. doi: 10.1016/j.mib.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 12.Bronner S, Monteil H, Prévost G. Regulation of virulence determinants in Staphylococcus aureus: complexity and applications. FEMS Microbiol. Rev. 2004;28:183–200. doi: 10.1016/j.femsre.2003.09.003. [DOI] [PubMed] [Google Scholar]
- 13.Loughman A, Sweeney T, Keane FM, Pietrocola G, Speziale P, Foster TJ. Sequence diversity in the A domain of Staphylococcus aureus fibronectin-binding protein A. BMC Microbiol. 2008;8:74. doi: 10.1186/1471-2180-8-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van der Woude MW, Bäumler AJ. Phase and antigenic variation in bacteria. Clin. Microbiol. Rev. 2004;17:581–611. doi: 10.1128/CMR.17.3.581-611.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang G, Gao Y, Dong J, Liu C, Xue Y, Fan M, Shen B, Shao N. A novel peptide screened by phage display can mimic TRAP antigen epitope against Staphylococcus aureus infections. J. Biol. Chem. 2005;280:27431–27435. doi: 10.1074/jbc.M501127200. [DOI] [PubMed] [Google Scholar]
- 16.Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. In: Altman RB, Brutlag DL, Karp PD, Lathrop RH, Searls DB, editors. Proceedings of the Second International Conference on Intelligent Systems for Molecular Biology. Menlo Park, CA: AAAI Press; 1994. pp. 28–36. [PubMed] [Google Scholar]
- 17.Berezovski MV, Lechmann M, Musheev MU, Mak TW, Krylov SN. Aptamer-facilitated biomarker discovery (AptaBiD) J. Am. Chem. Soc. 2008;130:9137–9143. doi: 10.1021/ja801951p. [DOI] [PubMed] [Google Scholar]
- 18.Williams KP, Bartel DP. PCR product with strands of unequal length. Nucleic Acids Res. 1995;23:4220–4221. doi: 10.1093/nar/23.20.4220. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.