Abstract
ST1710, a member of the multiple antibiotic resistance regulator (MarR) family of regulatory proteins in bacteria and archaea, plays important roles in development of antibiotic resistance, a global health problem. Here, we present the crystal structure of ST1710 from Sulfolobus tokodaii strain 7 complexed with salicylate, a well-known inhibitor of MarR proteins and the ST1710 complex with its promoter DNA, refined to 1.8 and 2.10 Å resolutions, respectively. The ST1710–DNA complex shares the topology of apo-ST1710 and MarR proteins, with each subunit containing a winged helix-turn-helix (wHtH) DNA binding motif. Significantly large conformational changes occurred upon DNA binding and in each of the dimeric monomers in the asymmetric unit of the ST1710–DNA complex. Conserved wHtH loop residues interacting with the bound DNA and mutagenic analysis indicated that R89, R90 and K91 were important for DNA recognition. Significantly, the bound DNA exhibited a new binding mechanism.
INTRODUCTION
Microbial antibiotic resistance is a result of either inactivation or reduced accumulation of antibiotics within an organism. Proteins belonging to the multiple antibiotic resistance regulators (MarR) family reportedly regulate the expression of proteins conferring resistance to multiple antibiotics, organic solvents, household disinfectants, oxidative stress agents and pathogenic factors (1–3). For example, in the absence of the appropriate stimulus, Escherichia coli MarR proteins negatively regulate the marRAB operon, and repression of this operon is alleviated by exposure to a variety of phenolic compounds, most notably sodium salicylate (1). Similarly, MexR negatively regulates an operon in Pseudomonas aeruginosa that, when expressed, encodes a tri-partite multi-drug efflux system that results in an increased resistance to multiple antibiotics, including tetracycline, β-lactams, chloramphenicol, novobiocin, trimethoprim, sulfonamides and fluoroquinolones (4,5). Some members of the MarR family of DNA-binding proteins, for example hypothetical uricase regulator (HucR) and organic hydroperoxide resistance regulator (OhrR), mediate a cellular response to reactive oxidative stress (ROS) (6,7). The Deinococcus radiodurans HucR was shown to repress its own expression as well as that of an uricase. This repression is alleviated both in vivo and in vitro upon binding uric acid, the substrate for uricase. As uric acid is a potent scavenger of reactive oxygen species, and D. radiodurans is known for its remarkable resistance to DNA-damaging agents, these observations indicate a novel oxidative stress response mechanism (8–10). Similar to HucR, the OhrR protein of Bacillus subtilis also mediates a response to oxidative stress; however, for OhrR, it is oxidation of a lone cysteine residue by organic hydroperoxides that abrogates DNA binding (11,12).
We have reported two different crystal forms of ST1710 (13) and others (14). The structure showed the winged helix-turn-helix (wHtH) motif at the DNA binding site that obviously belonged to the MarR family of proteins. The crystal structures of proteins in the MarR family have also been determined from a number of organisms including MarR from E. coli (15), MexR from P. aeruginosa (16), SarR from Staphylococcus aureus (17), Slya-like protein from Enterococcus faecalis (18), OhrR from B. subtilis (19), HucR from D. radiodurans (20) and MTH313 from Methanobacterium thermoautotrophicum (21). Sequence comparisons of these proteins with ST1710 showed less than 25% identity. A homology search in the non-redundant protein database using Blastp revealed that ST1710 has about 51% identity to the Sulfolobus acidocaldarius (22) and Sulfolobus solfataricus (23) sequences and about 41% identity to the Metallosphaera sedula sequence (24). However, none of the proteins closely related to ST1710 have been biochemically or structurally characterized.
Sodium salicylate is well known to inhibit MarR activity both in vitro and in vivo, at millimolar concentration levels (25). Sodium salicylate is routinely used as a model inhibitor of MarR to induce MarA expression in E. coli and Salmonella typhimurium, thereby conferring a Mar phenotype (25–28). The structure of E. coli MarR was solved with two salicylate molecules per monomer and both of them are highly exposed to the solvent. It seems that salicylate may have stabilized the crystal packing since in the absence of salicylate, the crystals could not be used for structure determination in the case of E. coli MarR (15). Recently, the structure of MTH313, a MarR homolog from M. thermoautotrophicum, was solved in the free form and complexed with salicylate; these analyses revealed a large asymmetrical conformational change that is mediated by the binding of sodium salicylate to two distinct locations in the dimer (21).
The members of the MarR family of regulatory proteins recognize double stranded DNA by their wHtH motifs (15–19). Footprinting experiments revealed that MarR binds as a dimer at two different, but similar, sites in marO, protecting 21 bp of DNA on both strands at a single site without bending its target (29,30). One of the MarR families of proteins, the OhrR protein complexed with the ohrA operator with a 29-bp duplex was solved, which revealed the interactions between them. The protein–DNA contact region included the major groove of the −10 element, and indicated that OhrR, and probably MarR and MexR as well, repress transcription by blocking the access of RNA polymerase to this promoter element (19). In addition, the mutational analysis of the RNA polymerase binding site, the −10 element of the OhrR-, MarR- and MexR-regulated promoters, revealed the loss of DNA binding ability by ∼10-fold when this region was altered (11,16,30,31).
On the basis of the sequence of the ohrA promoter, we previously identified a putative promoter for ST1710 and showed binding ability by gel-mobility shift assays (13). This promoter is located immediately upstream of the first ATG of the ST1710 gene and downstream of the STS1709 gene. To understand the importance of MarR family members in antibiotic resistance and other biological processes, here, we solved the ST1710 in three different forms: (i) apo-form (native), (ii) complexed with its inhibitor, sodium salicylate (salicylate complex) and (iii) complexed with its promoter DNA. A slight conformational change on the side chains of protein residues’ was observed when bound to the salicylate ligand, compared to the apo-form. The DNA bound to the wHtH motif of one monomer on the dimeric ST1710, and specifically interacted with the residues R84, R89, R90 and K91. A significantly large conformational change was observed between the monomers of the dimeric protein bound to the DNA, and also with the apo/salicylate complex structures. Significantly, a distinct mode of DNA binding was observed with the bound DNA passing over the protein molecule rather than passing through the 2-fold related axis of the molecule as previously observed (19).
MATERIALS AND METHODS
Cloning, expression and purification of ST1710
The gene encoding the MarR family regulator protein (ST1710) from Sulfolobus tokodaii (S. tokodaii) strain 7 was amplified from genomic DNA by PCR, using the primers 5′-ggaattCATATGTTAGAAAGTAATGAAAACAGAATAC-3′ and 5′-ggaattGGATCCTTATTACTGACTAATTTCCTCAATTCTTTTC-3′. The PCR fragment was digested with NdeI and BamHI and cloned into the pET21a(+) expression vector. The plasmid was transformed into the E. coli BL21-CodonPlus (DE3)—RIL-X strain (Stratagene), and the selenomethionine-containing ST1710 proteins were over-expressed and purified as described in our earlier studies (13).
Crystallization and data collection
Native crystals of ST1710 were produced to medium size (0.1 × 0.1 × 0.1 mm) within the period of a week at 20°C by the sitting drop vapor diffusion method (32), by adding 1 μl of protein solution to 1 μl of well solution, containing 18% PEG 8 K, 0.2 M calcium acetate, and 0.1 M sodium cacodylate, pH 6.5. For the ST1710–salicylate complex, crystals were soaked in the mother liquor containing 0.2 M of sodium salicylate for 3 min. Twenty percent ethylene glycol was used as a cryo-protectant and the complete data set was collected for the native and salicylate complex with the in-house R-axis VII system (RIGAGU MSC). These crystals belonged to the tetragonal space group, P41212, and the processed data statistics are given in Table 1. The 30 bp synthetic oligonucleotide containing the putative promoter sequence (5′-AATAATGTCATTGTTAACAATAGCAAAAAT-3′) and its complementary oligonucleotide (5′-ATTTTTGCTATTGTTAACAATGACATTATT-3′) were annealed completely to form the DNA-duplex and complexed with the ST1710 protein at an equimolar ratio. Initial crystals of ST1710–DNA complex were produced at 20°C by the sitting drop vapor diffusion method (32), by adding 1 μl of protein–DNA complex solution to 1 μl of well solution, containing 30% (v/v) 2-Methyl-2,4-pentanediol (MPD), 0.02 M calcium chloride dehydrate and 0.1 M sodium acetate trihydrate, pH 3.8. Preliminary tiny crystals that were seeded in the equilibrated drops of the same condition grew up to a maximum dimension of 0.3 mm within a 2-week period. Complete Multiple Anomalous Dispersion (MAD) data sets were obtained at 100 K using a Jupiter210 CCD detector (RIGAGU MSC) on the RIKEN structural genomics beamline I (BL26B1) at SPring-8, Hyogo, Japan. These crystals belonged to the centered orthorhombic space group C2221, with cell dimensions a = 94.44, b = 106.73 and c = 82.26 Å. The native and Se-edge MAD data sets were processed up to 2.10 Å using the HKL2000 suite (33) (Table 1).
Table 1.
Data collection and refinement statistics of the ST1710 native, salicylate, DNA complex and their mutants
| Data collection | ST1710-salicylate | ST1710-native | ST1710–DNA complex |
R89A | R90A | K91A | ||
|---|---|---|---|---|---|---|---|---|
| Peak | Edge | Remote (Low energy) | ||||||
| Wavelength | 1.54178 | 1.54178 | 0.97896 | 0.97930 | 1.000 | 1.54178 | 1.54178 | 1.54178 |
| Space group | P41212 | P41212 | C2221 | P41212 | P41212 | P41212 | ||
| Cell dimensions (Å) | a = b = 46.14, c = 137.64 | a = b = 46.04, c = 138.45 | a = 94.44, b = 106.73, c = 82.26 | a = b = 45.86, c = 138.17 | a = b = 45.90, c = 137.42 | a = b = 45.96, c = 138.29 | ||
| No. of molecules/asu | 1 | 1 | 2 | 1 | 1 | 1 | ||
| Resolution Range (Å) | 40–1.80 (2.07–2.0)a | 40–2.0 (2.07–2.0) | 50–2.10 (2.18–2.10) | 40–2.20 (2.28–2.20) | 40–1.90 (1.97–1.90) | 50–2.10 (2.18–2.10) | ||
| Unique reflections | 13 381 | 10 380 | 24 349 | 24 204 | 24 231 | 8143 | 12 085 | 9163 |
| Redundancy | 13.4 (7.6) | 18.5 (19.1) | 9.6 (9.8) | 12.5 (13.0) | 13.0 (12.6) | 12.2 (12.9) | ||
| Completeness (%) | 91.3 (99.8) | 96.0 (94.4) | 99.1 (98.7) | 99.1 (98.8) | 99.0 (98.6) | 100 (100) | 97.7 (95.1) | 98.1 (100) |
| Rmergeb | 0.100 (0.264) | 0.104 (0.321) | 0.080 (0.294) | 0.072 (0.284) | 0.067 (0.281) | 0.091 (0.389) | 0.065 (0.326) | 0.095 (0.302) |
| Refinement statistics | ||||||||
| Resolution Range (Å) | 20–1.80 | 20–2.0 | 20.0–2.10 | 20–2.20 | 20–1.90 | 20.0–2.10 | ||
| Reflections used in the refinement | 13 138 | 10 244 | 24 202 | 7877 | 11 906 | 8997 | ||
| Total no. of reflections used for working set | 12 246 | 9423 | 22 476 | 7292 | 10 943 | 8258 | ||
| Rworkc | 0.233 | 0.211 | 0.237 | 0.201 | 0.205 | 0.218 | ||
| Total no. of reflections used for Rfree | 892 | 821 | 1726 | 585 | 963 | 739 | ||
| Rfreed | 0.265 | 0.252 | 0.287 | 0.245 | 0.239 | 0.270 | ||
| No. of protein atoms | 1142 | 1142 | 2305 | 1136 | 1136 | 1138 | ||
| No. of nucleic acid atoms | – | – | 466 | |||||
| No. of water molecules | 152 | 156 | 154 | 125 | 160 | 127 | ||
| No. ligand molecules | 1 | |||||||
| No. Ca2+ ions | – | 1 | – | 1 | 1 | 1 | ||
| Average B factor (Å2) | 29.6 | 31.8 | 51.2 | 28.1 | 33.5 | 36.9 | ||
| Ramachandran statistics | ||||||||
| Most favored regions (%) | 97.7 | 98.5 | 98.1 | 97.0 | 97.7 | 96.2 | ||
| Allowed regions (%) | 2.3 | 1.5 | 1.9 | 3.0 | 2.3 | 3.8 | ||
| PDB code | 3GEZ | 3GF2 | 3GFI | 3GFJ | 3GFL | 3GFM | ||
RMSD in bond angles and bond lengths are varied from 0.005 to 0.007 (Å) and 1.0–1.1 (°). Solvent content is about 55%.
aValues in parentheses are for the highest resolution shell.
bRmerge = ∑h ∑i | I(h, i)−<I(h)> | /∑h ∑i I(h, i), where I(h, i) is the intensity value of the ith measurement of h and <I(h)> is the corresponding mean value of I(h) for all i measurements.
cR factor = ∑ | |Fobs|−|Fcalc| |/|Fobs|, where |Fobs| and |Fcalc| are the observed and calculated structure factor amplitudes, respectively.
dRfree is the same as R factor, but for a 5–7% subset of all reflections.
Structure determination and refinement
The native and salicylate complex of ST1710 structures were determined by the molecular replacement method, using our previous ST1710 structure (PDB code, 2eb7), as a search model. The solution was found by automated-MOLREP, within the CCP4 program suite, and the refinement was carried out using CNS (34). The protein model was built using the programs Quanta (35) and Coot (36). The native and salicylate complexes were refined to resolutions 1.80 and 2.0 Å, respectively (Table 1). Since the molecular replacement method was not successful for phasing the ST1710–DNA complex, we collected and processed the Se-MAD data sets. The ST1710–DNA complex structure was successfully phased by the MAD method with the three different wavelength data sets collected at the Se-edge, using the program SOLVE (37). Solvent flattening and initial model building were performed by RESOLVE (37). Improvement of the partial model derived from RESOLVE was performed with the ARP/wARP program (38). We observed unambiguous density for the DNA bases and built the DNA-model using the program Quanta (35). The final model was refined and manually fitted using CNS (33), Coot (36) and Quanta (34). The final model with 285 protein residues and 23 nucleic acid bases, except for 4 and 3 residues in the N-terminal of A and B chains, respectively, was refined to a crystallographic R-factor of 0.237 (Rfree = 0.287) at 2.10 Å resolution, using synchrotron radiation X-ray data collected at cryo temperature (see Table 1). Figures were prepared with the program Pymol (39). The coordinates and structure factors for the native, salicylate, and ST1710–DNA complex have been deposited in the Protein Data Bank, under the accession codes 3GEZ, 3GF2 and 3GFI, respectively.
Differential scanning calorimetric (DSC) analysis of salicylate binding to ST1710
DSC experiments were carried out using a VP-capillary DSC platform (Microcal, USA). For the DSC measurements, the protein concentration was fixed at 0.5 mg/ml in 20 mM Tris–HCl buffer (pH 8.0) containing 150 mM NaCl. Dialyzed protein sample and the sodium salicylate were filtered through a 0.22 μM pore size membrane and complexed with different concentrations of sodium salicylate (0–300 mM) and were loaded on to the capillary system. The scan rate was 90°C/hr for all experiments. We calculated the binding constant (Kd) of sodium salicylate using the DSC curves analysis using the Origin software (Microcal, USA) and the Graphpad Prism 2.0, a non-linear curve-fitting algorithm (GraphPad software).
Site-directed mutagenesis of ST1710 and gel-mobility shift assays
Initially, the ST1710 plasmid was prepared with a Qiagen miniprep kit. The Quickchange site-directed mutagenesis kit (Stratagene) was used to create the DNA-binding site mutants (R89A, R90A and K91A), and the resultant plasmids were transformed into JM109 cells. N-terminal sequencing was carried out for all the mutants, which were expressed and purified in a manner similar to the native protein (13).
To evaluate the protein–DNA interactions in solution, a gel-mobility shift assay was used. The purified ST1710 was incubated in binding buffer (10 mM Tris–HCl, pH 8.0, 200 mM NaCl, 20 mM MgCl2 and 5 mM β-ME) at pre-determined concentrations, and the 30-mer DNA (100 nM) was added. The reaction mixture was incubated for 20 min at room temperature and mixed with 1 μl of 50% glycerol before loading onto the gel. The free DNA and ST1710–DNA complexes were resolved on a 10% polyacrylamide gel (running buffer, 1X TBE, constant voltage, 200 V; temperature, 4°C). Inhibition of ST1710 was analyzed in the presence of increasing concentrations of sodium salicylate by gel-mobility shift analysis. The free and complexed nucleic acids were stained by fluorescent SYBR Green (EMSA Kit, Invitrogen) and the bands visualized with a UV transilluminator (LAS-3000, FUJIFILM, Japan).
RESULTS AND DISCUSSION
Biophysical analysis of salicylate binding to ST1710 and inhibition assays
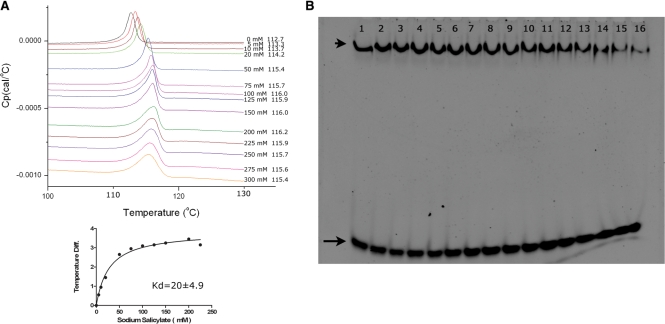
Previous in vitro and in vivo analyses of MarR family of proteins suggested that salicylate is a broad inhibitor for MarR activity at the millimolar concentration levels (1). To investigate whether salicylate binds to ST1710, we used a differential scanning calorimetric method for the binding analysis. Various concentrations of sodium salicylate (0–300 mM) were mixed with the protein and the heat capacities (Cp) measured with the scan rate of 90°C/hr (Figure 1A). The binding constant (Kd) was 20 ± 4.9 mM for sodium salicylate as calculated by the GraphPad Prism software, while the Origin program produced similar values. This analysis clearly suggests that salicylate binds to ST1710, and the binding constants were comparable with other members of MarR family (21).
Figure 1.
Salicylate binding and inhibition of ST1710–DNA complex assays. (A) DSC analysis of salicylate binding to ST1710. Typical excess heat capacity curves of ST1710 in the absence/presence of sodium salicylate ligand, at a scan rate of 90°C/hr. Salicylate concentration and peak temperature are noted on each curve. The binding constant was calculated using the non-linear regression fit, using the GraphPad Prism software. (B) Gel-mobility shift assay showing ST1710–DNA complex inhibition. All reactions were carried out in binding buffer containing 150 μM of protein and various amounts of sodium salicylate (to a final concentration of 0–250 mM; lanes 1–16 with 0, 1, 5, 10, 20, 30, 40, 50, 60, 70, 80, 90, 100, 150, 200 and 250 mM, respectively) were added and incubated at room temperature for 20 min. To this reaction mixture, 100 nM of 30-mer DNA added, and after for 20 min., the reactions were fractionated by 10% native PAGE and the DNA stained by SYBR Green (EMSA Kit, Invitrogen). The positions of the free and complex DNA are indicated by an arrow and an arrowhead, respectively.
Next, to visualize the protein–DNA inhibition by sodium salicylate, we used the gel-mobility shift assays. While the ST1710–DNA complex formed more compact than free DNA alone, the addition of salicylate inhibited the DNA–protein complex. At concentrations above 100 mM, the most of the DNA in the salicylate treated complex released from ST1710. This analysis clearly demonstrates that the ST1710–DNA complex formation was inhibited with increasing concentrations of salicylate (Figure 1B). Taken together, the data indicates that salicylate bound to the ST1710 in solution and inhibited the protein–DNA complex when it exceeded the intracellular concentration levels.
Structure of ST1710 complexed with salicylate
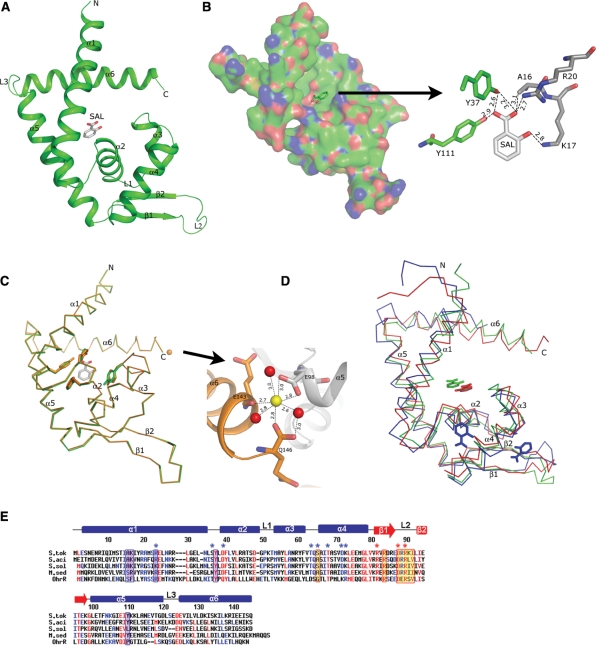
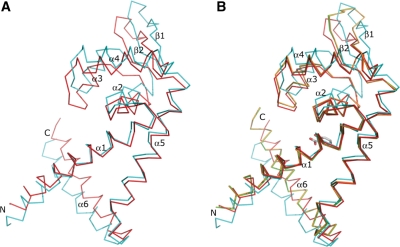
Our DSC and gel-mobility shift analyses confirmed that salicylate bound to the ST1710 and inhibited the protein–DNA complex formation. To see how salicylate binds to ST1710, we determined the ST1710–salicylate complex at a resolution of 1.80 Å, and refined to a final R value of 23.3% and an Rfree value of 26.5%. The overall structure of ST1710 within the complex was similar to our recently reported native structure, showing that it belongs to the α/β family of proteins and resembles those of the MarR family of proteins. It consists of six α-helices and two β-strands, arranged in the order of α1-α2-α3-α4-β1-β2-α5-α6 in the primary structure (Figure 2A). The asymmetric subunit contains one molecule, with overall dimensions of 60 × 39 × 26 Å. Two monomers are related by a crystallographic 2-fold axis to form the dimer (Supplementary Figure S1), and this is consistent with our gel-filtration analysis (13) as well as with studies of other MarR family proteins (15–19). The N- and C-terminal equivalent residues of each monomer, located at the α1, α5 and α6 helices, are closely intertwined and form a dimerization domain, which is stabilized by hydrophobic and hydrogen bonding interactions between the residues located within these regions. Apart from the dimerization domain, as observed in many DNA binding transcriptional regulators, the residues located at the α2-α3-α4-β1-β2 formed a wHtH DNA binding motif. In the dimer, the distances between the recognition helix (α4) to the recognition helix and the loop to loop (connecting the β-strands) of the wHtH domains are ∼30 and ∼70 Å, respectively.
Figure 2.
Structure of ST1710 native and salicylate complex and sequence comparison with closely related proteins. (A) A ribbon diagram of the ST1710–salicylate complex. The secondary structure assignments and the N-/C- termini are labeled on the structure. The bound salicylate is shown in the stick model. (B) An electrostatic representation of the ST1710 monomer. The basic regions are shown in blue, and the acidic regions are red. Close-up view of salicylate binding site interactions with protein residues. The hydrogen bonds are indicated by broken lines. (C) Structural comparison of the ST1710–salicylate complex with the native structure. The native structure is shown in orange color. The salicylate and key residues involved in interactions are shown in stick models and the bound Ca2+ ion in the native structure is represented by an orange sphere. The Ca2+ ion binding site is enlarged. (D) Superimposition of the ST1710–salicylate complex along with another known MarR family protein crystallized with salicylate. The E. coli MarR is represented by blue color and M. thermoautotrophicum MTH313 is shown in red. (E) Sequence analysis of ST1710 (S. tok) and its closely related proteins from different species: S. acidocaldarius (S. aci), S. solfataricus (S. sol), and M. sedula (M. sed) along with the OhrR protein. Conserved residues are indicated by red letters. The secondary structural elements in the primary sequences of ST1710 are indicated as α-helices (bars), β-strands (arrows) and loops (lines). The salicylate and DNA contacting residues in ST1710 are shown in blue-shaded and yellow-shaded boxes, respectively. DNA contacting residues of the wing in OhrR are indicated by red asterisks. Selected homologous/identical residues that interact with DNA in OhrR are indicated by blue asterisks.
The fine quality of the electron density map allowed us to identify unambiguously the specific salicylate binding site in the complex structure. The bound salicylate is located at the interface between the helical dimerization and wHtH DNA-binding domains (Figures 2A and B). The large portion of the DNA-binding and dimerization domains formed a salicylate-binding pocket. The salicylate ligand has many interactions with the protein residues (Figure 2B). The O2′ of salicylate is bonded to the side chain oxygens of Y37 and Y111; in addition, side chain oxygen of Y37 is also bonded to the O1′ of the ligand molecule. The ligand oxygen O1′ is hydrogen bonded to the side chain amino group (NH2) of residue R20, while the O2 of ligand molecule is hydrogen bonded to the side chain nitrogen of K17. The latter two interactions are from the symmetrically related molecule. Thus, the bound salicylate has many interactions. All of the residues which interact with the ligand are highly conserved among the closely related species (Figure 1E).
Next, to observe if any conformational change occurred in the salicylate liganded complex when compared to the native structure, we collected the new native data set from the crystal grown under the same conditions, solved at 2.0 Å and refined to a final R value of 21.1% and an Rfree value of 25.2%. The overall conformation of the complex is very similar to the native structure, with an rmsd of 0.11 Å for superposition of 141 Cα atoms (Figure 2C). However, a minor conformational change was observed in the side chain orientations of the ligand interacting residues and the DNA-binding wHtH motifs. We believe that allosteric changes might occur at the molecular level in solutions in the presence of inhibiting ligand, salicylate, which is not seen in the crystal structure. A divalent metal ion, Ca2+, was observed in the native structure, and it interacted with the C-terminal Q146 and E98 residues, which is closer to the DNA-binding loop of the symmetrically related molecule (Figure 2C).
It is interesting to compare the salicylate complexes within the MarR family of regulators. There are two complex structures available; MarR from E. coli and the more recently solved MTH313 from M. thermoautotrophicum. Although these proteins displayed sequence similarity ∼26% to ST1710, the superposition of the ST1710–salicylate complex with the two others revealed that the overall topology is similar, with an rmsd of 1.90 Å for 119 Cα atoms and 2.1 Å for 131 Cα atoms for E. coli MarR and M. thermoautotrophicum MTH313, respectively (Figure 2D). However, E. coli MarR was crystallized with two molecules of salicylate per dimer, both of which were highly exposed to the solvent (15), and these salicylate binding sites are not comparable to that with ST1710. The salicylate ligands in MarR hydrogen bonded to some of the amino acid residues (A70, T72, R77 and R86); however, the physiological relevance of either salicylate binding site could not be determined because the ligands were involved in interactions with protein molecules within the crystal which may stabilize the crystal lattice. On the other hand, the mode of salicylate binding between ST1710 and MTH313 is comparable, and the ligand adjusted up ∼2 Å and ∼3 Å towards the α5 helix upon binding. In contrast to the salicylate binding in ST1710, two direct and one water mediated protein residues were in contact with the ligand in MTH313. A comparison between the apo and salicylate complex of MTH313 revealed a significant asymmetrical conformational change that is mediated by the binding of sodium salicylate to two distinct locations in the dimer (21), whereas we did not observe such changes in the case of ST1710. The available in vivo and in vitro analyses suggest that the MarR family of regulators inhibits the activity in the presence of salicylate. Since we could see only the fixed side-chain orientation of the protein residues contacting the bound salicylate ligand, we believe possible dynamic or allosteric changes occurring upon salicylate binding to ST1710 to inhibit its activity would not be captured by crystallization.
Overall structure of the ST1710–DNA operator complex
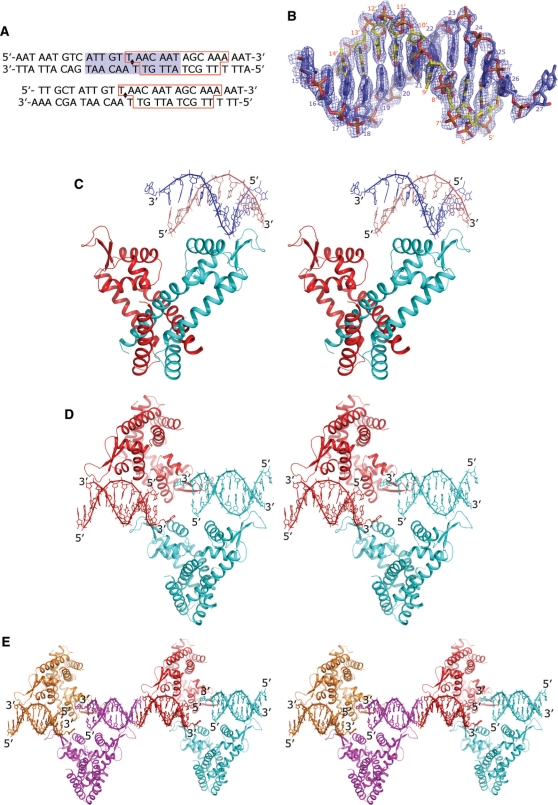
In our earlier studies, we identified the putative promoter for ST1710, which is located upstream of the st1710 gene (13). The gel-mobility shift analysis suggested that protein–DNA interactions were competitive and concentration dependent (Supplementary Figure S2). Here, we complexed the ST1710 and 30-mer duplexed DNA (Figure 3A), crystallized, and collected the data set up to a resolution of 2.1 Å, under the space group C2221. Initially, we were unsuccessful in solving the structure by molecular replacement using the coordinates of our native structure. Later, we solved the ST1710–DNA complex by the MAD method using three different data sets collected at the Se-edge, and the structure was refined to a final R value of 23.7% and an Rfree value of 28.7%. A high quality electron density map enabled us to build the nucleic acids unambiguously (Figure 3B). The overall structure of ST1710–DNA complex is shown in Figures 3C–E. The protein crystallized as a dimer in the asymmetric unit (asu) with the DNA fragment bound to one of the dimeric molecules (Figure 3C). Part of the packing diagram with two and four asymmetric unit of the molecules related by the crystallographic 2-fold axis is also given (Figures 3D and E). The duplex-DNA fragments observed in two adjacent asu of the dimeric molecules shown in magenta and red (Figure 3E), probably form one double-stranded DNA molecule of the full-length promoter DNA. The full-length (30-mer) of the DNA-duplex in the crystals were also confirmed by dissolving the complex crystals (ST1710–DNA) in water, after washing them a few times in reservoir solution, and then analyzed DNA-duplex, using agarose gel electrophoresis. These analyses indicated that the DNA-duplex present in the crystal was, indeed, the full-length DNA (Supplementary Figure S3). To clarify further the DNA binding to ST1710, we prepared an additional DNA duplex with exactly 2-fold related sequence based on the observed fragments in the present crystal (5′-TTGCTATTGTTAACAATAGCAAAAAT-3′) (Figure 3A, bottom sequence), crystallized, and solved the structure. The overall structure of this new complex resembles the present one, and it recognizes the DNA fragment irrespective of the sequence heterogeneity (our unpublished data). As shown in Figure 3E, one full-length duplex-DNA was interacted with four molecules of the ST1710, which was coming from four different asu of the dimeric molecules. Thus, the analysis of the ST1710–DNA complex in the present study suggests that the protein:DNA ratio is 2:1. Interestingly, the bound DNA passes over the wHtH motif, making contacts only at the loop regions.
Figure 3.
Structure of ST1710–DNA complex. (A) DNA sequence used for crystallization (30-mer and 26-mer duplex). The blue background shows the 2-fold related sequence. The red boxed residues were observed in the crystal structure. (B) The final 2Fo-Fc omit electron density map with the nucleic acids contoured at 1σ level is shown in blue mesh. The template strand and its complementary strands are shown in blue and yellow stick models. (C) Stereo view of the asymmetric unit of ST1710–DNA complex. The secondary structural assignments, N- and C-terminal ends are labeled in one of the dimeric monomers. The bound nucleic acids are shown as stick representations. (D) Stereo view of the two dimers of the asymmetric unit related by 2-fold axis to form the tetramer for recognition of one promoter DNA. (E) Part of the crystal packing is shown (stereo view). Each asymmetric unit of the complex is shown in a unique color. The 5′- and 3′- ends of each nucleotide chain is labeled.
Interactions between the ST1710 and promoter DNA
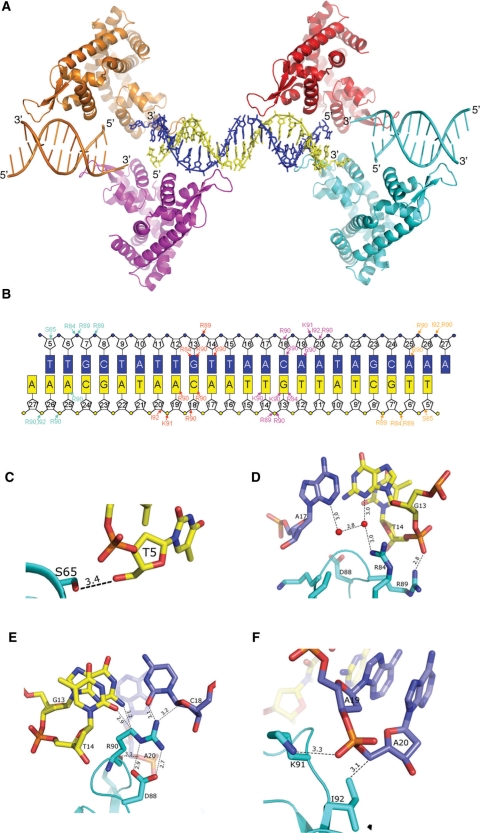
As explained above, Figure 4A represents the full length duplex-DNA bound to four monomers of the symmetry-related dimeric molecules. Although we used only a 30-mer duplexed DNA for our crystallization studies, we could see the duplexed-DNA consisting of T5 to A27 and T5′ to A27′ of the bases bound to the protein (Figures 4A and B). The 4 and 3 bases at the 5′- and 3′-end, respectively, were highly disordered in both of the DNA-strands and hence not modeled. The bound DNA adapted a B-form right handed structure, passing over the protein molecule by only contacting at the wHtH loop regions. The protein–DNA interactions seen in the asu of the complex (Figures 3B and C) were essentially same in all of the four symmetry-related molecules. Interestingly, as observed in the OhrR-ohrA operator complex, the –10 region (TAACAAT) of the promoter DNA (15–21) was recognized by the wHtH domains (Figures 4B–F). Of the bound 54 nucleotides, only 22 nucleotides make 36 contacts with six protein residues (Figures 4B–F). The side chain oxygen of S65 was bonded to the O5′ of Thy5′. Interestingly, the side chain (NH1) of residue R84 formed water-mediated hydrogen bonds to the N3 of bases G13′ and Ade17. In addition, side chain (NH1) of R89 hydrogen bonded to the backbone phosphate oxygen (O2P) of Thy14′. The residue R90 hydrogen bonded to the O4′ and O2 of base Cyt18 and the same residue made two salt-bridge contacts with D88. This salt bridge may assist in fixing the conformation of residue R90 in order to make contact with the nucleic acid base, Cyt18. Besides, the side chain atom (CD) bonded to the bases of Gua13′ (N2) and Thy14′ (O2). The side chain of K91 interacted with backbone phosphate of Ade19 and Ile91 to C5 of Ade20. Thus, the following residues S65, R84, D88, R89, R90, K91 and I92 interacted with the bound promoter DNA. To evaluate the protein–DNA interactions at the loop region, we prepared three mutant proteins (R89A, R90A and K91A) and analyzed the binding ability by gel-mobility shift assays. All three mutants failed to bind to DNA, suggesting that these three residues are important for protein–DNA interactions (Supplementary Figure S4). We also crystallized all three mutant proteins under the native protein conditions and solved their structures by molecular replacement method as explained previously (Table 1). All three mutant structures resembled the native ST1710, except very little changes were observed in the wHtH loop regions (Supplementary Figure S5). Furthermore, DNA-binding residues in ST1710 were highly conserved among the closely related proteins and OhrR (Figure 2E). The winged loop region connecting the strands β1 and β2 apparently plays a major role in modulating their conformation for binding to the DNA molecule, and this mode of recognition is anticipated for the proteins closely related to ST1710 as well as the family of MarR regulators. We observed Ca2+ ions in all of the mutant and native structures, but not in the salicylate and DNA-complex structures. Superimposition of the native, salicylate and DNA-complex structures suggested that the C-terminal helix (α6) in the DNA complex deviated greatly from the metal ion binding site and in the salicylate complex slight changes in side chain orientations were observed (Supplementary Figure S6). However, the functions of this metal ion observed in the native and mutant structures of ST1710 will need to be further investigated.
Figure 4.
ST1710–promoter DNA interactions. (A) A ribbon diagram of ST1710–DNA complex colored as in Figure 4E. The full-length promoter DNA is shown in stick model. The T5-A27 and T5′-A27′ strands are in blue and yellow, respectively. (B) Schematic representation showing all ST1710–DNA contacts. The bases are shown in rectangles. The protein–DNA contacts in each of the ST1710 monomer are represented by the same color in (A). (C–F) The close-up view of the critical protein–DNA interactions in the complex is shown in A–D. The residues of nucleic acids and protein are shown in stick models. The hydrogen bonds are shown in broken lines.
Conformational changes
In our recent report, we noticed a small change only at the loop region connecting strands β1 and β2 in the protein conformers crystallized in two different space groups and the overall structure was identical with an rmsd of 0.519 Å for 141 Cα atoms (13). In a similar way, when we compared the present ST1710–salicylate complex and native structure crystallized under the same conditions, a similar structural conformation was revealed with an rmsd of 0.11 Å for superposition of 141 Cα atoms. Additionally, the subunits in the dimer were identical. In contrast, the superimposition of the ST1710–DNA complex subunits (A and B chains) on one another revealed a large local conformational change all along the structure, excluding the helices α1 and α5 with an rmsd of 2.85 Å for 142 Cα atoms; however, the overall structural topology was similar (Figure 5A). A displacement of ∼3.5–5.5 Å was seen all along the winged HtH motif region and the C-terminal helix showed the displacement of around 2–3 Å. The winged HtH motif of A-chain where the DNA is recognized was elevated up compared to the B-chain, while the C-terminal helix α6 was shifted down.
Figure 5.
Structural comparison of ST1710–DNA complex monomers with its native structure and salicylate complex. (A) Superimposition of ST1710–DNA complex monomers. As shown in Figure 2C one monomer of the complex is shown in cyan and the other monomer is in red. (B) The ST1710–DNA complex monomers superimposed with the ST1710 native structure and salicylate complex. The native structure and salicylate complex are represented by orange and green, respectively.
As seen in Figure 5B, significant changes were also observed between the subunits of the DNA-complex and the native and salicylate complex. Superposition of the native and salicylate complexes on to the A-chain showed greater differences than on the B-chain: an rmsd of 2.6 and 2.9 Å with the A-chain; and 0.9 and 1.0 Å with the B-chain, for the native structure and salicylate complex, respectively. One of the wHtH motifs may have been relocated to establish contact with the DNA, and by this mode of recognition, the wHtH loop regions were stabilized. The temperature factor for this loop region in the B-chain increased significantly, although it formed a dimer related by a non-crystallographic 2-fold axis, observed in the asymmetric unit of the cell (Supplementary Figure S7). It is noteworthy to mention that the temperature factor was higher for the DNA-complex B-chain when not only compared to the DNA-complex A-chain, but also to the native, salicylate complex and all three of the mutant structures. Apparently, when protein binds to the DNA, the wHtH loop becomes stabilized and the temperature factor of that region is lowered; this observation was similar to that of the salicylate-complex which was solved at the highest resolution. It is also interesting to note that in the dimer, the distances between the loop to loop (connecting the β-strands) of the wHtH domains were reduced by ∼10 Å for the ST1710–DNA complex, compared to the native and salicylate complexes.
Mode of nucleic-acid binding
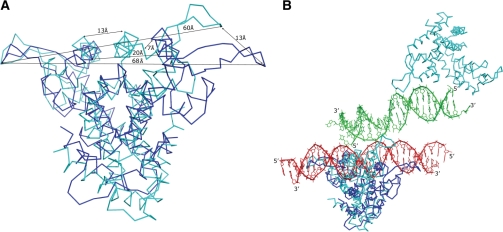
There are several MarR families of regulators reported to date from different organisms including E. coli (15), P. aeruginosa (16), S. aureus (17), E. faecalis (18), B. subtilis (19), D. radiodurans (20) and M. thermoautotrophicum (21). However, only the structure of OhrR from B. subtilis is available with its promoter and revealed their interactions. It is intriguing to compare our ST1710–DNA complex with OhrR-ohrA operator binding to clarify the binding mechanism. The superimposition of the ST1710–DNA complex on the OhrR-ohrA operator revealed large conformational changes, with an rmsd of 4.4 Å for 215 Cα atoms when compared to ST1710 before binding to its cognate promoter (rmsd 2.80 for 238 Cα atoms) (Figures 6A and B). Compared to the ST1710–DNA complex with its native structure and salicylate complex in the previous section, we found a similar change in the protein and unexpectedly found conformational changes in the mode of DNA recognition also. In the OhrR-ohrA complex, the distance between the subunits loop-to-loop was around 67 Å and the recognition helix (α4)−helix was about 20 Å (Figure 6A), although the wings of the subunits translocated about 16 Å compared to the structure of reduced OhrR (19). The bound 2-fold related promoter sequence recognized the protein wHtH loop-to-loop, with substantial widening and deepening of the major groove that resulted from insertion of the recognition helix (α4) of the HtH motif. In contrast to this mode of binding, the bound DNA in ST1710 passed over the wHTH motif without deepening the structure through the 2-fold axis, even though the protein contacting residues are highly conserved between these two proteins and among the MarR family of regulators (Figures 2E, 4A and 6B). This unexpected mode of DNA-binding originated due to the translocation of one of the subunits around 13 Å towards the 2-fold axis, reducing the distance between the recognition helix of the subunits to 13 Å (Figures 6A and B). Thus, the DNA passing through the 2-fold axis deepening the recognition helices as observed in OhrR-ohrA operator complex would be impossible for that of ST1710.
Figure 6.
Structural comparison of MarR family of protein–DNA complexes. Superposition of the OhrR-ohrA operator complex (PDB id, 1z9c) on the ST1710–DNA complex is shown without (A) and with nucleic acids (B). The protein and nucleic acids are shown in ST1710–DNA complex in cyan and green; while those in the OhrR-ohrA operator complex are shown in blue and red, respectively. The distances between the loop-to-loop and recognition helices are marked in (A). The 5′- and 3′- ends of each nucleotide chain are labeled.
In conclusion, this report describes the crystal structure of ST1710 in three different forms: apo-form, ST1710–salicylate and ST1710–DNA complex. We showed that the salicylate binding does not affect the overall structure. In addition we found that all the residues interacting with salicylate ligand are highly conserved among the closely related proteins. The ST1710 conformation changed dramatically upon binding to the DNA, and these changes might be necessary for its recognition. Structural analyses of the MarR family of regulators along with the bacterial transcription regulators such as BmrR (40) and CRP (41) and the eukaryotic transcription regulators like RFX1 (42) and histone H5 (43) suggested that orientations of the wHTH motifs were similar despite having little differences in loop lengths and its orientations (19). We believe most of the regulators containing wHTH motifs are prone to bind DNA through their acidic/basic residues located in their wings. However, the mode of DNA recognition depends on the subunit organization of the regulators as observed in the MarR family of proteins (ST1710, MarR).
ACCESSION NUMBERS
3GEZ, 3GF2, 3GFI, 3GFJ, 3GFL, 3GFM.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Proteomics Initiative (RSGI); the National Project on Protein Structural and Functional Analyses, Ministry of Education, Culture, Sports, Science and Technology of Japan. Funding for open access charge: RIKEN.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Dr K. Yutani and Ms. M. Takehira for DSC measurements and Dr T. Ishikawa for his moral support and encouragement.
REFERENCES
- 1.Alekshun MM, Levy SB. Regulation of chromosomally mediated multiple antibiotic resistance: the mar regulon. Antimicrob. Agents Chemother. 1999;41:2067–2075. doi: 10.1128/aac.41.10.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Miller PF, Sulavik MC. Overlaps and parallels in the regulation of intrinsic multiple-antibiotic resistance in Escherichia coli. Mol. Microbiol. 1996;21:441–448. doi: 10.1111/j.1365-2958.1996.tb02553.x. [DOI] [PubMed] [Google Scholar]
- 3.Aravind L, Anantharaman V, Balaji S, Mohan Babu M, Iyer LM. The many faces of the helix-turn-helix domain: transcription regulation and beyond. FEMS Microbiol. Rev. 2005;29:231–262. doi: 10.1016/j.femsre.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 4.Li XZ, Poole K. Organic solvent-tolerant mutants of Pseudomonas aeruginosa display multiple antibiotic resistance. Can. J. Microbiol. 1999;45:18–22. doi: 10.1139/cjm-45-1-18. [DOI] [PubMed] [Google Scholar]
- 5.Srikumar R, Paul CJ, Poole K. Influence of mutants in the mexR repressor gene on expression of the MexA-MexB-oprM multidrug efflux system of Pseudomonas aeruginosa. J. Bacteriol. 2000;182:1410–1414. doi: 10.1128/jb.182.5.1410-1414.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wilkinson SP, Grove A. HucR, a novel uric acid-responsive member of the MarR family of transcriptional regulators from Deinococcus radiodurans. J. Biol. Chem. 2004;279:51442–51450. doi: 10.1074/jbc.M405586200. [DOI] [PubMed] [Google Scholar]
- 7.Wilkinson SP, Grove A. Negative cooperativity of uric acid binding to the transcriptional regulator HucR from Deinococcus radiodurans. J. Mol. Biol. 2005;350:617–630. doi: 10.1016/j.jmb.2005.05.027. [DOI] [PubMed] [Google Scholar]
- 8.Hooper DC, Spitsin S, Kean RB, Champion JM, Dickson GM, Chaudhry I, Koprowski H. Uric acid, a natural scavenger of peroxynitrite, in experimental allergic encephalomyelitis and multiple sclerosis. Proc. Natl Acad. Sci. USA. 1998;95:675–680. doi: 10.1073/pnas.95.2.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kean RB, Spitsin SV, Mikheeva T, Scott GS, Hooper DC. The peroxynitrite scavenger uric acid prevents inflammatory cell invasion into the central nervous system in experimental allergic encephalomyelitis through maintenance of blood-central nervous system barrier integrity. J. Immunol. 2000;165:6511–6518. doi: 10.4049/jimmunol.165.11.6511. [DOI] [PubMed] [Google Scholar]
- 10.Ames BN, Cathcart R, Schwiers E, Hochstein P. Uric acid provides an antioxidant defense in humans against oxidant- and radical-caused aging and cancer: a hypothesis. Proc. Natl Acad. Sci. USA. 1981;78:6858–6862. doi: 10.1073/pnas.78.11.6858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fuangthong M, Helmann JD. The OhrR repressor senses organic hydroperoxides by reversible formation of a cysteine-sulfenic acid derivative. Proc. Natl Acad. Sci. USA. 2002;99:6690–6695. doi: 10.1073/pnas.102483199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fuangthong M, Atichartpongkul S, Mongkolsuk S, Helmann JD. OhrR is a repressor of ohrA, a key organic hydroperoxide resistance determinant in Bacillus subtilis. J. Bacteriol. 2001;183:4134–4141. doi: 10.1128/JB.183.14.4134-4141.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kumarevel TS, Tanaka T, Nishio M, Gopinath SCB, Takio K, Shinkai A, Kumar PKR, Yokoyama S. Crystal structure of the MarR family regulatory protein, ST1710, from Sulfolobus tokodaii strain 7. J. Struct. Biol. 2008;16:9–17. doi: 10.1016/j.jsb.2007.08.017. [DOI] [PubMed] [Google Scholar]
- 14.Miyazono K, Tsujimura M, Kawarabayasi Y, Tanokura M. Crystal structure of an archaeal homolog of multidrug resistance repressor protein, EmrR, from hyperthermophilic archaea Sulfolobus tokodaii strain 7. Proteins. 2007;67:1138–1146. doi: 10.1002/prot.21327. [DOI] [PubMed] [Google Scholar]
- 15.Alekshun MN, Levy SB, Mealy TR, Seaton BA, Head JF. The crystal structure of MarR, a regulator of multiple antibiotic resistance, at 2.3 Å resolution. Nat. Struct. Biol. 2001;8:710–714. doi: 10.1038/90429. [DOI] [PubMed] [Google Scholar]
- 16.Lim D, Poole K, Strynadka NCJ. Crystal structure of the MexR repressor of the mexRAB-oprM multidrug efflux operon of Pseudomonas aeruginosa. J. Biol. Chem. 2002;277:29253–29259. doi: 10.1074/jbc.M111381200. [DOI] [PubMed] [Google Scholar]
- 17.Liu Y, Manna A, Li R, Martin WE, Murphy RC, Cheung AL, Zhang G. Crystal structure of the SarR protein from Staphylococcus aureus. Proc. Natl Acad. Sci. USA. 2001;98:6877–6882. doi: 10.1073/pnas.121013398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu RY, Zhang RG, Zagnitko O, Dementieva I, Maltzev N, Watson JD, Laskowski R, Gornicki P, Joachimiak A. Crystal structure of Enterococcus faecalis Syla-like transcriptional factor. J. Biol. Chem. 2003;278:20240–20244. doi: 10.1074/jbc.M300292200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hong M, Fuangthong M, Helmann JD, Brennan RG. Structure of an OhrR-ohrA operator complex reveals the DNA binding mechanism of the MarR family. Mol. Cell. 2005;20:131–141. doi: 10.1016/j.molcel.2005.09.013. [DOI] [PubMed] [Google Scholar]
- 20.Bordelon T, Wilkinson SP, Grove A, Newcomer ME. The crystal structure of the transcriptional regulator HucR from Deinococcus radiodurans reveals a repressor preconfigured for DNA binding. J. Mol. Biol. 2006;360:168–177. doi: 10.1016/j.jmb.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 21.Saridakis V, Shahinas D, Xu X, Christendat D. Structural insight on the mechanism of regulation of the MarR family of proteins: high resolution crystal structure of a transcriptional repressor from Mathanobacterium thermoautotrophicum. J. Mol. Biol. 2008;377:655–667. doi: 10.1016/j.jmb.2008.01.001. [DOI] [PubMed] [Google Scholar]
- 22.Chen L, Bruegger K, Skovgaard M, Redder P, She Q, Torarinsson E, Greve B, Awayez M, Zibat A, Klenk HP, et al. The genome of Sulfolobus acidocaldarius, a model organism of the Crenarchaeota. J. Bacteriol. 2005;187:4992–4999. doi: 10.1128/JB.187.14.4992-4999.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.She Q, Singh RK, Confalonieri F, Zivanovic Y, Allard G, Awayez MJ, Chan-Weiher CCY, Clausen IG, Curtis BA, et al. The complete genome of the crenarchaeon Sulfolobus solfataricus P2. Proc. Natl Acad. Sci. USA. 2001;98:7835–7840. doi: 10.1073/pnas.141222098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Auernik KS, Maezato Y, Blum PH, Kelly RM. The genome of the metal mobilizing, extremely thermoacidophilic archaeon Metallosphaera sedula provides insights into bioleaching-associated metabolism. Appl. Environ. Microbiol. 2008;74:682–692. doi: 10.1128/AEM.02019-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alekshun MM, Levy SB. Alteration of the repressor activity of MarR, the negative regulator of the Escherichia coli marRAB locus, by multiple chemicals in vitro. J. Bacteriol. 1999;181:4669–4672. doi: 10.1128/jb.181.15.4669-4672.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alekshun MM, Levy SB. The mar regulon: multiple resistance to antibiotics and other toxic chemicals. Trends Microbiol. 1999;7:410–413. doi: 10.1016/s0966-842x(99)01589-9. [DOI] [PubMed] [Google Scholar]
- 27.Cohen SP, Levy SB, Foulds J, Rosner JL. Salicylate induction of antibiotic resistance in Escherichia coli: activation of the mar operon and a mar-independent pathway. J. Bacteriol. 1993;175:7856–7862. doi: 10.1128/jb.175.24.7856-7862.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sulavik MC, Dazer M, Miller PF. The Salmonella typhimurium mar locus: molecular and genetic analyses and assessment of its role in virulence. J. Bacteriol. 1997;179:1857–1866. doi: 10.1128/jb.179.6.1857-1866.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martin RG, Jair KW, Wolf RE, Jr., Rosner JL. Autoactivation of the marRAB multiple antibiotic resistance operon by the MarA transcriptional activator in Escherichia coli. J. Bacteriol. 1996;178:2216–2223. doi: 10.1128/jb.178.8.2216-2223.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martin RG, Rosner JL. Binding of purified multiple antibiotic-resistance repressor protein (MarR) to mar operator sequences. Proc. Natl Acad. Sci. USA. 1995;92:5456–5460. doi: 10.1073/pnas.92.12.5456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Evans K, Adewoye L, Poole K. MexR repressor of the MexAB-oprM multidrug efflux operon of Pseudomonas aeruginosa: Identification of MexR binding sites in the mexA-MexR intergenic region. J. Bacteriol. 2001;183:807–812. doi: 10.1128/JB.183.3.807-812.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McPherson A. Current approaches to macromolecular crystallization. Eur. J. Biochem. 1990;189:1–23. doi: 10.1111/j.1432-1033.1990.tb15454.x. [DOI] [PubMed] [Google Scholar]
- 33.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Meth. Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 34.Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. 1998;D54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 35.Oldfield TJ. A number of real-space torsion-angle refinement techniques for proteins, nucleic acids, ligands and solvent. Acta Crystallogr. 2001;D57:82–94. doi: 10.1107/s0907444900014098. [DOI] [PubMed] [Google Scholar]
- 36.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. 2004;D60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 37.Terwilliger TC, Berendzen J. Automated MAD and MIR structure solution. Acta Crystallogr. 1999;D55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Perrakis A, Morris R, Lamzin VS. Automated protein model building combined with iterative structure refinement. Nat. Struct. Biol. 1999;6:458–463. doi: 10.1038/8263. [DOI] [PubMed] [Google Scholar]
- 39.DeLano WL. The PyMOL Molecular Graphics System. DeLano Scientific. CA, USA: San Carlos; 2002. [Google Scholar]
- 40.Zheleznova EE, Markham PN, Neyfakh AA, Brennan RG. Structural basis of multidrug recognition by BmrR, a transcription activator of a multidrug transporter. Cell. 1999;96:353–362. doi: 10.1016/s0092-8674(00)80548-6. [DOI] [PubMed] [Google Scholar]
- 41.Schultz SC, Shields GC, Steitz TA. Crystal structure of a CAP-DNA complex: the DNA is bent by 90 degrees. Science. 1991;253:1001–1007. doi: 10.1126/science.1653449. [DOI] [PubMed] [Google Scholar]
- 42.Gajiwala KS, Chen H, Cornille F, Roques BP, Reith W, Mach B, Burley SK. Structure of the winged-helix protein hRFX1 reveals a new mode of DNA binding. Nature. 2000;403:916–921. doi: 10.1038/35002634. [DOI] [PubMed] [Google Scholar]
- 43.Ramakrishnan V, Finch JT, Graziano V, Lee PL, Sweet RM. Crystal structure of globular domain of histone H5 and its implications for nucleosome binding. Nature. 1993;362:219–223. doi: 10.1038/362219a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.