Abstract
Estradiol (E2) regulates gene expression at the transcriptional level by functioning as a ligand for estrogen receptor alpha (ERα) and estrogen receptor beta (ERβ). E2-inducible proteins c-Myc and E2Fs are required for optimal ERα activity and secondary estrogen responses, respectively. We show that E2 induces 21 microRNAs and represses seven microRNAs in MCF-7 breast cancer cells; these microRNAs have the potential to control 420 E2-regulated and 757 non-E2-regulated mRNAs at the post-transcriptional level. The serine/threonine kinase, AKT, alters E2-regulated expression of microRNAs. E2 induced the expression of eight Let-7 family members, miR-98 and miR-21 microRNAs; these microRNAs reduced the levels of c-Myc and E2F2 proteins. Dicer, a ribonuclease III enzyme required for microRNA processing, is also an E2-inducible gene. Several E2-regulated microRNA genes are associated with ERα-binding sites or located in the intragenic region of estrogen-regulated genes. We propose that the clinical course of ERα-positive breast cancers is dependent on the balance between E2-regulated tumor-suppressor microRNAs and oncogenic microRNAs. Additionally, our studies reveal a negative-regulatory loop controlling E2 response through microRNAs as well as differences in E2-induced transcriptome and proteome.
INTRODUCTION
Estradiol (E2) controls several biological processes by functioning as a ligand for nuclear receptors estrogen receptor alpha (ERα) and beta (ERβ) (1). ERs may participate in the genomic (transcriptional) and non-genomic actions of E2 (1,2). The genomic action involves binding of ERα to the regulatory regions of target genes either directly or through protein–protein interaction. DNA-bound ERα then recruits various co-regulatory molecules to induce chromatin modifications that either increase or decrease the level of target gene transcription. Several extracellular signal activated kinases phosphorylate and modulate ERα activity; this may be responsible for altered E2 responses such as resistance to anti-estrogen therapy in breast cancer (2). The major kinases that modulate ERα activity include ERK1/2, AKT, RSK, PAK1, p38 kinase and SRC (2–4).
A significant number of studies so far have focused on E2:ERα-mediated transcriptome. The effect of E2 on gene expression at post-transcriptional level is yet to be elucidated. MicroRNAs are a class of evolutionarily conserved non-coding RNAs that control gene expression at the post-transcriptional level (5,6). They regulate gene expression through either direct cleavage of the target mRNAs or by translational inhibition (5,7–9). However, control of gene expression at the level of translation by microRNAs is cell cycle dependent (10). A highly conserved microRNA family is predicted to target 300 mRNA species and together affect ∼30% of the protein-coding genes (11,12). Cellular stress and RNA-binding proteins, which usually bind to sequences in between microRNA-recognition sites on mRNAs, determine the target specificity of microRNAs (13,14). Additional functions of microRNAs include transcriptional activation of genes with complementary promoter sequences (15) and chromatin modification (16,17).
RNA polymerase II enzyme transcribes the majority of microRNA genes (a minority by polymerase III) to produce a primary microRNA (18,19). Approximately 50% of microRNAs are transcribed from introns of protein-coding genes while the rest are intergenic with primary transcripts as long as 4 kb (19). MicroRNA genes usually appear in polycistronic clusters and more than 50% are located in cancer associated genomic regions at fragile sites (5,20). Thus, an alteration in the expression of a single microRNA can have a profound effect on cellular physiology because of their enormous influence on the expression of multiple genes.
Three recent reports describe microRNA-expression patterns in breast cancer; two of these have evaluated the expression pattern in relation to ERα, ErbB2 and intrinsic subtypes (21–23). Overall, microRNA levels were lower in poorly differentiated tumors compared to well-differentiated tumors and microRNA profile rather than mRNA-expression profile correlated more accurately with cell differentiation. ERα positive breast cancers displayed a distinct microRNA-expression profile including elevated expression of Let-7 family members of microRNAs and miR-21 compared to ER-negative breast cancers (21). Blenkiron et al. described microRNA-expression profiles in relation to five intrinsic subtypes (luminal A, luminal B, normal-like, HER2+ and basal) of breast cancer (22,24,25). Luminal subtypes and basal type breast cancers display distinct microRNA-expression patterns (22). ERα expression and activity in breast cancer may be regulated by a Estrogen-ERα-microRNA-regulatory loop as mir-206 inhibits ERα translation by binding to the 3′ UTR of ERα mRNA, whereas ERα agonists block miR-206 expression (26).
Despite enormous progress in understanding the expression pattern of microRNAs in breast cancer, regulation of their expression in different breast cancer subtypes is not known. We focused on the effect of E2 on microRNA expression in breast cancer cells with two goals: (i) to determine whether E2 regulates the expression of any of the microRNAs expressed at higher levels in luminal type A/ERα-positive breast cancer; and (ii) to determine whether these E2-regulated microRNAs subsequently control the expression of E2-regulated genes at the post-transcriptional level. We also examined whether extracellular signal activated kinases such as AKT modulate E2-regulated microRNA expression. Results from this study reveal a unique role for E2 in regulating the expression of luminal type A-enriched microRNAs with tumor suppressor and oncogenic functions and the ability of AKT to alter E2-regulated microRNA expression.
MATERIALS AND METHODS
Cell lines
MCF-7 cells containing the bicistronic vector control (MCF-7p) or the constitutively active AKT (MCF-7AKT) were generated by retrovirus-mediated gene transfer and have been described previously (27). Virus infected cells were selected using one microgram per ml puromycin and mass culture instead of selected clones were used to avoid clonal bias. The constitutively active AKT1 construct with deletion of N-terminal pleckstrin homology domain (amino acids 4–129) but addition of myristylation signal has been described previously (3). Cells were maintained in MEM plus 10% fetal calf serum and one microgram per ml of puromycin. Cells were switched to phenol red free MEM plus 5% dextran-charcoal treated serum (CCS) for at least 4 days prior to experiments.
MicroRNA-expression analysis
Cells were treated with the solvent ethanol or 10−8 M E2 for 4 h and RNA was prepared using the mirVana™ microRNA isolation kit (Ambion Inc., Austin, TX). MicroRNA microarray and hybridization to microRNA arrays have been described previously (28,29). Assays were done in duplicate with RNA from two independent clones. Since all the probes were printed twice on the microarray in two independent blocks, four measurements were available for every microRNA under each experimental condition. Signal intensity of tRNA-Thr was used for normalization between samples and relative differences in individual microRNA expression with or without E2 treatment was then calculated. Differentially expressed microRNAs were identified using a linear mixed effect model that describes the relationship among three factors, including cell type, E2 treatment, probe blocks and interactions between cell type and E2 treatment. The linear mixed effect model can be described as:
where i, j, k, l, n represent the the microRNA, the cell types (MCF-7p or MCF-7AKT), the conditions (with and without E2 treatment), the RNA samples and block on the array, respectively. αij and βik represent the effect of cell types and conditions, and γijk denotes the interaction of these two factors. The RNA sample and block are considered as random effects, and the RNA sample factor is nested in the cell type and condition factors.
E2-induced changes in Let-7f, miR-98 and miR-21 was verified in four independent RNA preparations by quantitative reverse transcription polymerase chain reaction (qRT-PCR) using The TaqMan® microRNA assays designed to detect and accurately quantify mature microRNAs (Applied Biosystems, Foster City, CA). Primers specific to small RNA, RNU66, were used for normalization (Applied Biosciences). Target genes of differentially expressed microRNAs were predicted using TargetScan (12).
Locked nucleic acid (LNA) treatment and western blot analysis
LNA against Let-7f, miR-98 and miR-21 as well as control LNA were obtained from Exiqon, Inc (Woburn, MA). LNA (5 nM) was transfected into cells grown in 60 mm plates using Lipofectamine reagent (Invitrogen, Carlsbad, CA). After 2 days, cells were split into two 60-mm plates. E2 or ethanol was added to cells 24 h after splitting and harvested for western analysis 24 h post E2 treatment. All experiments were done three to five times with similar results. Antibodies against AIB1 (BD biosciences, San Jose, CA), E2F1, E2F2, Maspin (Santa Cruz Biotechnology, Santa Cruz, CA), c-Myc (Upstate Biotechnology, Lake Placid, NY), EZH2 (Cell Signaling, Danvers, MA) and PDCD4 (Abcom, Cambridge, MA) were used for western analysis as per instructions of manufacturers.
Total RNA isolation, northern blotting and quantitative reverse transcription polymerase chain reaction (qRT–PCR)
Total RNA from ethanol and E2-treated cells was isolated using an RNeasy kit from Qiagen (Valencia, CA). Northern blotting was performed as described previously (3). RNA was reverse transcribed using a single-stranded cDNA synthesis kit (Invitrogen, Carlsbad, CA) and subjected to quantitative polymerase chain reaction (Q-PCR) using Syber green (Applied Biosciences, Foster City, CA). Primers used were: Dicer forward primer TCACCTGCCTCACTTGACCTGAAA; Dicer reverse CGCTTTCAAACTGCTGCTCAT; beta-actin forward TGGATCAGCAAGCAG; and beta-actin reverse GCATTTGCGGTGGAC.
Chromatin immunoprecipitation assay
The entire ERα chromatin immunoprecipitation-microarray (ChIP-on-chip) data set of MCF-7p and MCF-7AKT cells as well as statistical analysis has been described (27). ERα ChIP assay was performed as described previously (30). ChIP DNA was subjected to Q-PCR using the primers 5′-TCTTTGACCCACTAGCCTTGCAGT-3′ and 5′-TCACTTCTCTCTGGGCCACAGTTT-3′ to determine E2-inducible changes in ERα binding to the central-binding site close to miR-21 identified in ChIP-on-chip. The chromosomal location of ERα binding was determined using UCSC genome browser (www.genome.ucsc.edu). E2-inducible changes in ERα binding were not detected with ChIP DNA obtained by immunoprecipitation with control IgG.
Transient transfection and luciferase assay
The 3′ untranslated regions of AIB1 (nucleotides 6872–7145 of NM_181659) or c-Myc (nucleotides 1861–2360 of NM_002467) were cloned into a cytomegalovirus (CMV) enhancer-promoter driven luciferase gene at the 3′-end of the coding sequence. MCF-7 cells maintained in MEM plus 5% CCS were transfected with the above plasmid and the internal control plasmid renila luciferase using lipofectamine (Invitrogen). E2 was added 24 h after transfection and luciferase assay was performed 16 h after E2 addition using dual luciferase assay kit from Promega (Madison, WI).
RESULTS
E2 regulates the expression of microRNAs
We recently reported the ERα-binding pattern in MCF-7 cells with a retrovirus vector control (MCF-7p) and MCF-7 cells overexpressing constitutively active AKT1 (MCF-7AKT) (27). We observed ERα-binding sites associated with 1667 and 1908 genes in one-hour E2-treated MCF-7p and MCF-7AKT cells, respectively. However, not all of the genes with ERα-binding sites had an altered transcript level after E2 treatment. Also, not all E2-regulated genes, as identified by microarray analysis of both cell types with or without E2 treatment for 4 h, contained ERα-binding sites suggesting that there are other mechanisms by which E2 regulates gene expression. In fact, among 833 E2-regulated genes in MCF-7p cells, only 299 genes contained ERα-binding sites (see Supplementary Data for select genes). Although secondary estrogen response through E2-regulated transcription factors such as E2F1/2 can explain the lack of correlation between ERα binding and target gene expression (31), control of gene expression by E2-regulated microRNAs has not been investigated in detail. To investigate this possibility, we determined microRNA-expression patterns in MCF-7p and MCF-7AKT cells with and without E2 treatment for 4 h. We observed 21 E2-inducible and 7 E2-repressible microRNAs in MCF-7p cells (statistical cutoff P-value <0.05 and fold change >1.5 or <0.7) (Table 1).
Table 1.
The effect of E2 on microRNA expression in MCF-7p and MCF-7AKT cells
| MCF-7p cells |
MCF-7AKT cells |
||
|---|---|---|---|
| Name | Fold change | Name | Fold change |
| E2-inducible microRNAs | |||
| miR-Let-7f | 3.2 | miR-520d | 4.8 |
| miR-Let-7a | 2.9 | ||
| miR-Let-7d | 2.7 | ||
| miR-Let-7c | 2.6 | ||
| mir-Let-7g | 2.6 | ||
| miR-203 | 2.4 | ||
| miR-Let-7b | 2.3 | ||
| miR-Let-7e | 2.2 | ||
| miR-98 | 2.1 | ||
| miR-21 | 2.0 | ||
| miR-200a | 2.0 | ||
| miR-103 | 1.8 | ||
| miR-200c | 1.8 | ||
| miR-107 | 1.7 | ||
| miR-17-5p | 1.7 | ||
| miR-23a | 1.6 | ||
| miR-200b | 1.6 | ||
| miR-30c | 1.6 | ||
| miR-30b | 1.5 | ||
| miR-424 | 1.5 | ||
| miR-let7i | 1.5 | ||
| E2-repressible microRNAs | |||
| miR-302b* | 0.4 | miR-524 | 0.3 |
| miR-506 | 0.5 | miR-518d | 0.3 |
| miR-524* | 0.5 | miR-518e | 0.3 |
| miR-27a | 0.6 | miR-506 | 0.3 |
| miR-27b | 0.6 | miR-409-5p | 0.3 |
| miR-143 | 0.6 | miR-216 | 0.3 |
| miR-9 | 0.7 | miR-518c* | 0.5 |
| miR-526b | 0.5 | ||
| miR-34b | 0.5 | ||
| miR-337 | 0.5 | ||
| miR-146 | 0.5 | ||
| miR-128b | 0.5 | ||
| miR-124a | 0.5 | ||
| miR-211 | 0.6 | ||
| miR-143 | 0.6 | ||
| miR-128a | 0.6 | ||
| miR-126* | 0.6 | ||
| miR-126 | 0.6 | ||
| miR-1 | 0.6 | ||
| miR-10b | 0.7 | ||
| MicroRNAs that display differences in basal expression between two cell typesa | |||
| miR-520d | 5.1 | miR-200a | 0.6 |
| miR-let7g | 0.5 | miR-182 | 0.6 |
| miR-337 | 0.5 | miR-17-5p | 0.6 |
| miR-20b | 0.5 | miR-1 | 0.6 |
| miR-Let-7i | 0.6 | miR-203 | 0.6 |
| miR-98 | 0.6 | miR-200c | 0.7 |
Cells were treated with E2 for 4 h and microRNA expression array analysis was performed. Only those microRNA whose expression differences with a P-value of <0.05 among untreated and E2-treated cells are presented. miR-98 was induced by E2 in both cell types (2.1-fold in MCF-7p cells, 1.4-fold in MCF-7AKT cells) whereas miR-143 (to same level) and miR-506 (0.50 in MCF-7p cells and 0.30 in MCF-7AKT cells) were repressed by E2 in both cell types.
aMicroRNA with fold change >1 is expressed at higher levels in MCF-7AKT cells, whereas fold change <1 represent microRNAs expressed at higher levels in MCF-7p cells.
E2 increased the expression of eight members of the Let-7 family microRNAs (≥2.2-fold increase), which previously have been shown to be overexpressed in luminal type A breast cancer (21,22). Similarly, E2 increased the expression of miR-21, which is also expressed at a higher level in luminal type-A breast cancer. AKT completely changed the pattern of E2 regulation of microRNA expression. In MCF-7AKT cells, E2 increased the expression of only one microRNA but reduced the expression of 20 microRNA species. Basal expression of 11 microRNAs was lower in MCF-7AKT cells compared to MCF-7p cells; whereas, one microRNA displayed elevated expression in MCF-7AKT cells. Seven of the microRNAs that displayed lower basal expression in MCF-7AKT cells were E2-inducible in MCF-7p cells. Only three microRNAs (miR-143, miR-506 and miR-98) showed similar E2-dependent regulation in both cell types.
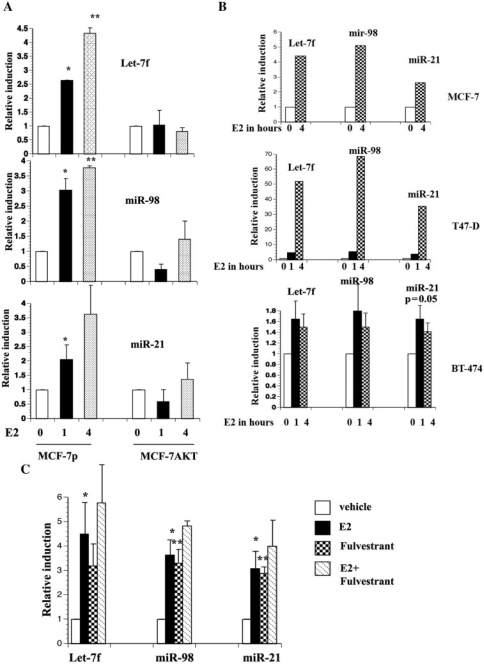
We performed microRNA qRT–PCR, which quantitatively measures only mature microRNAs, to confirm E2-inducible expression of Let7f, miR-21 and miR-98 (Figure 1A). Consistent with the microarray results, E2 increased the expression of these microRNAs in MCF-7p cells; none of the E2-induced changes in microRNA expression in MCF-7AKT cells was statistically significant. E2 increased the expression of all three of these microRNAs in parental MCF-7 cells that were used to generate the above cell lines (Figure 1B, top panel). E2 similarly increased Let-7f, miR-98 and miR-21 levels in T47-D cells, another ERα-positive cell line (Figure 1B, middle panel). Interestingly, in BT-474, an ERα-positive/HER2-positive cell line, E2-inducible expression of these microRNAs was modest (Figure 1B, bottom panel).
Figure 1.
(A) E2-inducible expression of Let-7f, miR98 and miR-21 in MCF-7p and MCF-7AKT cells. MCF-7p and MCF-7AKT cells were treated with E2 (10−8M) for indicated time and microRNA was subjected qRT–PCR. RNU66, small RNAs encoded in the intron of RPL5 gene (chr1:93 018 360–93 018 429) was used for normalization between samples. Mean plus standard error of the mean (SEM) are shown. *P < 0.03, ethanol versus 1-h E2-treated cells; **P < 0.001 ethanol versus 4 h E2-treated cells. (B) The effect of E2 on the expression of Let-7f, miR-98 and miR-21 in MCF-7 (top), T47-D (middle) and BT-474 (bottom) cells. (C) The effect of fulvestrant treatment on basal and E2-regulated expression of Let-7f, miR-98 and miR-21. Cells were pretreated with 100 nM fulvestrant for 24 h and then treated with E2 for 4 h. *P < 0.05, ethanol versus E2-treated; **P < 0.05, ethanol versus fulvestrant treated cells.
To determine the requirement of ERα for E2-regulated expression of the above microRNAs, we pretreated MCF-7 cells with fulvestrant for 24 h and then stimulated cells with E2 for 4 h. Fulvestrant inhibits ERα function through degradation. The basal levels of miR-98 and miR-21 microRNAs were elevated in fulvestrant-treated cells and were only marginally increased upon E2-treatment (Figure 1C). The basal level of Let-7f was marginally increased but it did not reach statistical significance. These results suggest that the unliganded ERα represses the expression of these microRNAs and E2 relieves this repression, which is consistent with a recent report on nuclear receptor-mediated microRNA regulation in Caenorhabditis elegans (32).
Putative targets of E2-regulated microRNAs
E2-inducible expression of Let-7 family members and reduced basal and/or E2-inducible expression of this family in MCF-7AKT cells is interesting because reduced expression of this family is linked to loss of differentiation and increased self-renewal of progenitor cells (33,34). Both Let-7 family members and miR-98 have the same seed sequence and target the same mRNAs. Ras family oncogenes and HMGA2 are the well-characterized targets of Let-7 (35,36). c-Myc is also a target of Let-7 (37) and miR-21 (38). Based on TargetScan and miRGen analyses, E2F1 and E2F2, E2-inducible transcription factors involved in secondary estrogen responses (31), are predicted targets of miR-205 and Let7/miR-98, respectively. E2F1 and E2F2 are also targets of miR20 (39); mir-20b is expressed at a higher level in MCF-7p cells compared to MCF-7AKT cells (Table 1). NCOA3/AIB1 is a target of miR-17-5p (40), which in our studies is E2-inducible microRNA (Table 1). EZH2 is a multifunctional protein that integrates Wnt and E2 signaling in breast cancer cells and the abnormal function of this protein is linked to several diseases (41). Based on TargetScan analysis, EZH2 is a likely target of mir-124a and mir-506, both of which are repressed by E2. The above genes thus constituted potential targets of E2-regulated microRNAs.
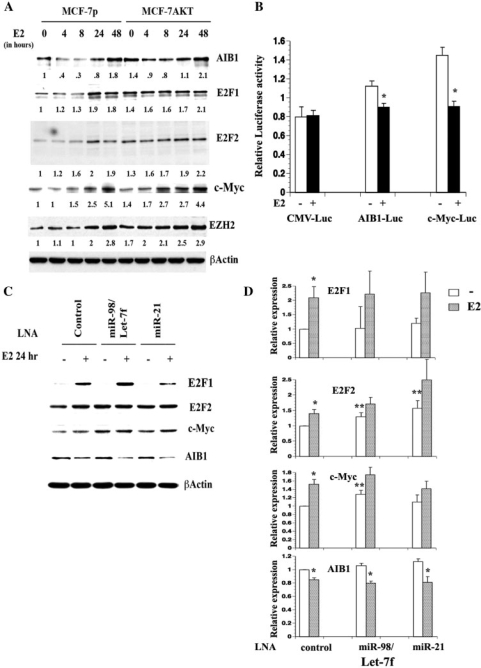
We focused our studies only to those genes that are involved in E2-signaling pathways and/or regulated by E2 at the transcriptional level and additionally controlled at the post-transcriptional level by E2-regulated microRNAs as part of an autoregulatory loop (Figure 2A). We observed repression of AIB1 and induction of EZH2 by E2 in both cell types. Additionally, while E2F1 and E2F2 displayed a delayed E2-inducible increase in protein levels in MCF-7p cells, basal levels of both of these proteins were elevated with a further E2-inducible increase in MCF-7AKT cells. Elevated basal levels of these two proteins in MCF-7AKT cells correlate with a lower level of miR20b in these cells compared to MCF-7p cells (Table 1). Previously, we reported little difference in the kinetics of E2-inducible expression of c-Myc mRNA in parental and CA-AKT overexpressing MCF-7 cells (3). However, at the protein level, E2-mediated increase in c-Myc is advanced in MCF-7AKT cells compared to MCF-7p cells (Figure 2A). None of the previous microarray profiling studies including our study observed an effect of E2 on EZH2 mRNA level and our ChIP-chip data did not identify ERα-binding sites associated with the EZH2 gene. Furthermore, RT–PCR analysis has confirmed the lack of an E2 effect on the mRNA levels of EZH2 (data not shown). Thus, E2 may control translation of AIB1 and EZH2 through microRNAs.
Figure 2.
Putative targets of E2-induced microRNAs. (A) MCF-7p and MCF-7AKT cells were treated with E2 for indicated time and western blotting was performed. (B) 3′-UTR of AIB1 or c-Myc reduces E2-inducible expression of a luciferase reporter under the control of CMV enhancer-promoter. MCF-7 cells were transfected with indicated reporters along with the internal control Renila-luciferase under TK promoter-enhancer and luciferase activity was measured in untreated and E2-treated cells. *P < 0.05, ethanol treated versus E2-treated cells. (C) LNA against Let-7f/miR-98 and miR-21 differentially affect basal and E2-inducible levels of c-Myc and E2F2 proteins in MCF-7p cells. Cells were treated with control or specific LNA and treated with E2 for 24 h. Western blot analysis was done with indicated antibodies. (D) Expression levels of E2F-1, E2F-2, c-Myc and AIB1 from three to five independent experiments, as measured by densitometric scanning (Mean plus SEM), are indicated. *P < 0.05, ethanol versus E2-treated cells; **P < 0.05, control LNA treated versus Let-7f/mir-98 or miR-21 LNA-treated cells. Note the effects of LNA against Let-7f/miR98 and miR-21 on E2F2 and c-Myc but not on E2F1 and AIB1 protein levels.
The 3′ UTRs of mRNAs are considered to be the primary targets of microRNAs although recent studies indicate the coding sequences are also targeted (8,9). We cloned the predicted microRNA target sequences in the 3′ UTR of AIB1 and c-Myc into a CMV-driven luciferase-reporter vector and investigated the effects of E2 on luciferase levels (Figure 2B). E2 did not have an effect on luciferase expression in cells transfected with parental vector. In contrast, E2-reduced luciferase expression in cells transfected with vectors containing either the AIB1 or c-Myc 3′ UTR. These results suggest that the 3′ UTR of AIB-1 and c-Myc genes contain sequences that are targeted by E2-regulated microRNAs. Note that based on ChIP-on-chip data, cloned 3′-UTR sequences of both AIB1 and c-Myc lack ERα-binding sites (27).
Let-7f/miR-98 and miR-21 may be part of negative-regulatory loop that control E2-induced levels of c-Myc, E2F1 and E2F2 proteins
Our previous studies have shown E2-mediated increase in mRNA levels of c-Myc, E2F1 and E2F2 in MCF-7p and MCF-7AKT cells (3,27). We used LNA-mediated knockdown of Let-7f/miR98 to determine whether reducing the levels of these microRNAs lead to changes in E2-inducible expression of c-Myc, E2F1 and E2F2 at the protein level. Neither LNA against Let-7f/miR-98 nor LNA against miR-21 had any effect on E2-inducible expression of E2F1 (Figure 2C and D). Consistent with TargetScan prediction, LNA against Let7f/miR-98 increased E2-inducible levels of E2F2. Surprisingly, basal level of E2F2 was also increased in miR-21 LNA treated MCF-7p cells compared to control LNA-treated cells. LNA against Let-7f/miR-98 but not miR-21 increased the basal levels of c-Myc proteins. LNA against Let-7f or miR-98 individually was ineffective, possibly due to their redundant targets (data not shown). E2 mediated reduction of AIB1 protein levels was unaffected by LNA against Let-7f/miR-98 or miR-21 (Figure 2C and D). Thus, E2-induced Let7f/miR-98 controls the expression levels of expected targets (E2F2 and c-Myc), whereas miR-21 unexpectedly controls the expression of E2F2. Also, the overall protein level changes (1.5–2-fold) that we observed in cells transfected with LNA-targeting specific microRNAs are within the range predicted and verified by proteomic approaches (11,42).
Bioinformatics analysis of potential targets of E2-regulated microRNAs identify additional E2-responsive genes
We then investigated whether any luminal type A specific genes expressed in primary breast cancers are putative targets of E2-regulated microRNAs using available bioinformatics tools. We used TargetScan among several available search engines because this program is more robust with regards to its prediction algorithm and several of its predictions have been verified by proteomics (11,42). Indeed, major genes that define a luminal type A phenotype or are overexpressed in luminal type A breast cancers including FOXA1, GATA-3, ERα and MyB are putative targets of E2-regulated microRNAs (Supplementary Table S1).
We next determined whether any of the E2-regulated mRNAs are the targets of E2-regulated microRNAs. The 21 E2-induced microRNAs in MCF7p cells belong to 10 distinct microRNA families. We analyzed whether these microRNAs-target genes that are induced or repressed by E2 in MCF-7 cells. Results of microarray analysis of untreated and 4 h E2-treated MCF-7p and MCF-7AKT cells were used for this purpose (27). E2-induced microRNAs are predicted to target 94 E2-inducible, 142 E2-repressible and 541 non-E2-regulated mRNAs in MCF-7 cells. Detailed statistical analysis is presented in Table 2. Supplementary Table S2 contains name of these genes, E2-dependent changes in their mRNA levels after 4 h of E2 treatment in both cell types, and microRNAs that may target them. MicroRNAs repressed by E2, which belongs to four families (hsa-miR-302b* and hsa-miR-524* are excluded from this analysis due to the lack of prediction from TargetScan), are predicted to target 67 E2-inducible, 116 repressed and 407 non-E2-regulated genes. Supplementary Table S2 contains names and E2-regulated expression of these genes. Taken together, E2-regulated microRNAs potentially target 1021 genes in MCF-7p cells. Similar analysis in MCF-7AKT cells showed that microRNAs upregulated by E2 in these cells may target 20 E2-inducible genes, 20 E2-repressed genes and 68 non-E2-regulated genes. MicroRNAs repressed by E2 in these cells may target 148 E2-inducible genes, 111 E2-repressed genes and 554 non-E2 target genes (Table 2 and Supplementary Table S3). Overall, E2-regulated microRNAs potentially control 850 genes in MCF-7AKT cells.
Table 2.
Number of genes potentially targeted by E2-regulated microRNAs in MCF-7p and MCF-7AKT cells
| mRNAs induced by E2 (FC > 1.5) | mRNAs repressed by E2 (FC ≤ 1.5) | Non-E2-regulated mRNAs (−1.5 < FC < 1.5) | |
|---|---|---|---|
| E2 upregulated microRNA targets in MCF-7p cells | 94 | 142 | 541 |
| E2 downregulated microRNA targets in MCF-7p cells | 67 | 116 | 407 |
| E2 upregulated microRNA targets in MCF-7AKT cells | 20 | 20 | 68 |
| E2 downregulated microRNA targets in MCF-7AKT cells | 148 | 111 | 554 |
Cellular mRNAs are subclassified into E2-induced, E2-repressed and unaffected by E2 but expressed in MCF-7p and MCF-7AKT cells based microarray analysis of ethanol or E2 (4-h) treated cells (27). FC = Fold change.
miR-21-regulatory region is associated with ERα-binding sites
We used our ChIP-on-chip data set (27) and other bioinformatics tools to investigate how E2 regulates the expression of microRNAs. MicroRNAs can be transcribed as independent units or as parts of intronic sequences in large transcribed genes (19). MicroRNAs that reside in the intergenic sequence are transcribed as 3–5-kb transcripts with clearly defined 5′ and 3′ boundaries. The promoter/enhancer regions of such microRNA genes are enriched for five transcription factor-binding sites: MSX-1, TLX2 (Hox11L1), CDC5, SRF and ZNF238 (19). Table 3 provides summary of E2-regulated microRNAs that are associated with ERα-binding sites in their regulatory region or located in the intronic region of known E2-regulated genes. An ERα-binding site was assigned to a microRNA if it is located within 20 kb of the 5′ transcription start site or within 20-kb of the 3′ transcription termination site (27). Additional details of ERα-binding site distribution around microRNAs that are regulated by E2 in MCF-7p and MCF-7AKT cells or ERα-binding sites along with E2-regulated expression of host genes that harbor these microRNAs are provided as supplementary files (Supplementary Table S4 and Supplementary Table S5).
Table 3.
E2-regulated microRNAs associated with ERα-binding sites or located in the intragenic region of E2-regulated genes
| MicroRNA | Fold change in microRNA expression after E2 treatment in MCF-7p cells | ERα-binding sites after E2 treatment |
|---|---|---|
| MicroRNAs with ERα-binding sites | ||
| miR-21 | 1.98 | 1 |
| miR-23a | 1.62 | 4 |
| miR27a | 0.64 | 4 |
| miR27b | 0.60 | 5 |
| MicroRNA | Fold change in microRNA levels after E2 treatment of MCF-7p cells | Host gene | E2 effect on the host gene |
|---|---|---|---|
| MicroRNAs located in the intragenic regions of E2-regulated genes | |||
| miR30c | 1.58 | NF-YC | −1.2 |
| miR-9 | 0.654 | C1orf61 | 1.16 |
| Let-7c | 2.6 | C21orf34 | −1.9 |
| Let-7g | 2.6 | TMEM113 | 1.3 |
miR-27b is located within aminopeptidase gene, which is an E2-inducible gene and displayed five and eight ERα-binding sites in E2-treated MCF-7p and MCF-7AKT cells, respectively.
We further evaluated ERα binding to miR-21-regulatory regions by ChIP assay. Location of the miR-21 transcribed region in relation to ERα-binding sites and results of the ChIP assay confirming ERα binding are shown in Figure 3. Unliganded ERα bound to three regions in MCF-7p cells (Figure 3A). E2 reduced the number of binding sites to one. ChIP assay was used to confirm ERα binding to the region II (central region). Unliganded ERα binding to this region was observed in MCF-7p cells (10-fold higher levels of PCR product compared to IgG control), which was unaffected upon E2 treatment (Figure 3B). However, ERα binding to this region was enhanced by E2 in MCF-7AKT cells. Thus, the miR-21 gene is associated with an authentic ERα-binding site. The fact that E2 reduced the number of miR-21 associated ERα-binding sites in MCF-7p cells and that this reduction in binding correlates with induction of miR-21 suggests that unliganded ERα suppresses the expression of this microRNA and E2 relieves that repression. This observation is consistent with data presented in Figure 1C and recent observations in C. elegans (32). In summation, our studies reveal a complex mechanism of E2-mediated changes in microRNA expression and the consequences of these changes on post-transcriptional control of gene expression.
Figure 3.
The regulatory regions of miR-21 contain ERα-binding sites. (A) ERα DNA-binding patterns to the genomic region harboring miR-21 gene on chromosome 17. Black bars represent ERα-binding sites in two cell types observed under ethanol and E2-treated condition. (B) ChIP analysis of ERα binding to the genomic region (central bar) shown in A. ChIP DNA obtained with ERα antibody was subjected to semi-quantitative PCR (top) or Q-PCR (bottom) with specific primers and relative enrichment of ERα binding is shown after normalizing ERα binding in ethanol MCF-7p or MCF-7AKT cells to one unit. The level of ERα binding in E2-treated MCF-7AKT cells was significantly higher than in untreated cells.
Dicer is an E2-inducible gene
To determine whether E2 controls microRNA processing, we searched our ChIP-on-chip datasets and microarray datasets of untreated and E2-treated MCF-7p and MCF-7AKT for ERα binding and E2-regulated expression of genes associated with microRNA processing (27). The only gene associated with an ERα-binding site is Dicer (Figure 4A) and Dicer mRNA was induced by E2 as measured by northern analysis and qRT–PCR (Figure 4B). AKT only delayed E2-mediated induction of Dicer. These results suggest that E2 regulates microRNA machinery at the level of transcription as well as processing.
Figure 4.
Dicer is an E2-inducible gene. (A) ERα DNA-binding patterns to the genomic region harboring Dicer. Black bars represent ERα-binding sites in two cell types observed under ethanol-treated or E2-treated condition. (B) E2-inducible expression of Dicer in MCF-7p and MCF-7AKT cells was measured by northern blotting (top panel). The same blot was reprobed with 36B4 to ensure integrity of RNA. qRT–PCR confirming E2-inducible expression of Dicer is also shown (mean plus SEM, bottom panel). *P < 0.04 ethanol-treated versus E2-treated cells.
DISCUSSION
Deregulation of microRNA expression is observed in various diseases including cancer, cardiovascular diseases and neurodegenerative disorders (43,44). While biological functions of microRNAs are being studied extensively, signaling pathways that control their expression are just beginning to be explored. Cytokines and hypoxia are known modulators of microRNA expression (45,46). In this study, we investigated the role of E2 in modulating microRNA expression in breast cancer cells. Unlike the effect of E2 on mRNA-encoding genes where the majority of them are repressed (47), 21 of the 28 E2-regulated microRNAs are induced. Our results on E2-regulated expression of miR-21 differ from a publication that appeared in this journal during revision of our manuscript (48). Wickramasinghe et al. (48) found lower miR-21 expression in MCF-7 but not T47-D cells treated with E2 for 6 h compared to untreated cells and this E2-mediated downregulation of miR-21 correlated with elevated expression of several miR-21 target genes. Reasons for this discrepancy are unknown. However, we note that there are several variants of MCF-7 and we have previously shown that these variants express different levels of co-activators involved in ERα-mediated gene expression (49). The second possibility is that E2 regulates miR-21 in a biphasic manner (induction followed by repression), which helps in fine-tuning of the E2 response. We also note that E2-mediated upregulation of miR-21 observed in our study correlates with reported higher levels of miR-21 expression in primary ERα+ breast cancers (21).
Possible mechanisms of E2-regulated expression of microRNAs
Based on the results of our ChIP-on-chip study and microarray expression analysis (27), at least three distinct mechanisms appear to be involved in E2-mediated upregulation of microRNAs. First is the direct binding of ERα to the regulatory regions of microRNAs. The regulatory regions of miR-21 and miR-23a contain ERα-binding sites. With respect to miR-21, unliganded ERα binds to three regions in MCF-7p cells but only one region in MCF-7AKT cells. E2 either enhances or decreases interaction of ERα to the region shared by both cell types, depending on AKT activity. How this unusual interaction pattern of ERα to the miR-21-regulatory region impacts the expression of this microRNA is unknown.
The second mechanism of E2-regulated microRNA expression may involve E2-inducible expression of mRNA-encoding genes that harbor microRNA genes in their intronic regions. NF-YC and C21orf34 are repressed by E2 (5) yet microRNAs within their introns are induced by E2. It remains to be determined whether these microRNAs are transcribed from an intronic promoter independent of the host gene. The third possibility is that E2 regulates the expression of transcription factors that control the expression of microRNAs. In this respect, TRANSFAC database analysis revealed selective enrichment of binding sites for MRF-2, PBX-1, RUNX1, RUNX2 and GATA-3-transcription factors in microRNA genes regulated by E2. Microarray and subsequent validation by qRT–PCR revealed MSX1, RUNX1 and RUNX2 as E2-inducible genes and MRF-2 and PBX-1 as E2-repressible genes in MCF-7 cells (data not shown). Previous studies have shown E2-regulated expression of GATA-3 in breast cancer cells (50). Which of these transcription factors contribute to E2-dependent microRNA expression remains to be addressed.
The fact that eight members of the Let-7 family are induced by E2 is interesting and suggests that a common pathway is involved in the expression and/or processing of Let-7 family microRNAs. This pathway is under the control of E2. In this respect, LIN28 has been shown to suppress the processing of all Let-7 family microRNAs without interfering with processing of other microRNAs (51). However, neither LIN28a nor LIN28b has ERα-binding sites and our microarray studies and RT–PCR analysis indicate lack of E2-regulated LIN28 expression. We cannot rule out a post-translational activation mechanism of LIN28 by E2-regulated signaling pathways, which may induce enhanced processing of Let-7 family members. Among the several genes studied that are known to be involved in the processing of microRNAs, only Dicer is associated with ERα-binding sites and is induced by E2 (Figure 4). Oncomine database analysis revealed overexpression of Dicer in ERα-positive breast cancers compared to ERα-negative breast cancers, which is consistent with our results of E2-regulated expression of this gene. Thus, E2 not only regulates the expression of specific microRNAs, but also may have global effects on microRNA-regulated gene expression by altering their rate of processing.
Among the microRNAs repressed by E2, only miR-27a and mir-27b contain ERα-binding sites. E2 may indirectly repress the expression of other microRNAs through a transcriptional repressor. In this respect, c-Myc has been shown to be a global repressor of microRNA expression (52). c-Myc is an E2-inducible gene, which is further negatively regulated by E2-induced Let-7 (Figure 2). Let-7 directly represses c-Myc translation and also destabilizes c-Myc transcript levels by inhibiting translation of IMP-1, a RNA-binding protein that stabilizes c-Myc transcripts (37,53). Thus, pathways that disrupt the finely balanced positive and negative effects of E2 on c-Myc expression may play a significant role in breast cancer progression.
The global effect of AKT on E2-regulated microRNA expression is puzzling and may involve AKT-dependent changes in the activity of transcription factors. The failure of E2 to induce the expression of miR-200a, miR-17-5p, miR-200a, miR-200c and miR-203 in MCF-7AKT cells could be due to AKT-mediated downregulation of the basal expression of these microRNAs and the effect of E2 on them being below the detection level in our assay (Table 1). miR-520d appears to be an exception to the general phenomena of AKT-dependent decrease in microRNA expression as AKT increased the basal levels of this microRNA by ∼5-fold and this microRNA was further induced by E2 in AKT overexpressing cells. miR-520d may be E2-inducible in parental cells also but its expression is below the level for detection. The significance of AKT-mediated induction of miR-520d could not be predicted as targets of this microRNA are yet to be identified.
Potential role of E2-regulated microRNAs in breast cancer
The majority of E2-regulated microRNAs appear to have a tumor suppressor role by participating in a negative feedback loop to restrict E2 action. For example, E2-inducible expression of Let-7 family members may limit the expression of Ras and c-Myc oncogene levels and promote differentiation of cancer cells. Control of c-Myc levels by Let-7 is particularly relevant to E2 signaling because c-Myc cooperates with ERα in the expression of select target genes and Let-7 may play a role in attenuating such cooperative gene expression (54). E2-induced microRNAs may control secondary estrogen responses by limiting the expression levels of E2F family members (31). Additionally, E2-induced microRNA miR-17-5p may control ERα primary responses by reducing the levels of the AIB1 coactivator (40). Figure 5 schematically represents how E2-regulated microRNAs may control E2 responses by targeting c-Myc and E2F2.
Figure 5.
A model depicting the possible effects of E2-regulated microRNAs on E2 response and differentiated phenotype of ERα-positive breast cancer cells.
E2-regulated miR-21 may have dual roles in cancer. It may protect cancer cells against cell death and promote metastasis: this effect of miR-21 is likely due to translational repression of pro-apoptotic PDCD4 and metastasis suppressor Maspin (55). In this respect, overexpression of miR-21 in MCF-7 cells leads to enhanced tumor growth in nude mice (56). On the other hand, miR-21 may reduce self-renewal of cancer cells with stem-cell-like properties by inhibiting the translation of stem cell renewal genes Oct-4, c-Myc, Nanog and Sox2 (38). For unknown reasons, we did not observe an effect of LNA against miR-21 on PDCD4 expression in E2-treated MCF-7 cells and these cells do not express Maspin (data not shown). Our results differ from that of Wickramasinghe et al. (48) with respect to PDCD4 possibly reflecting differences in the variants of MCF-7 cells used in the two studies. In our MCF-7 cells, PDCD4 is an E2-inducible gene and is associated with ERα-binding sites. Additionally, based on TargetScan prediction, PDCD4 is a target of E2-inducible miR-200a (Table 1 and Supplementary Table S1). Therefore, these multiple levels of E2 action may have obscured the effects of miR-21 LNA on PDCD4 protein levels in E2-treated cells.
AKT is a major signaling molecule in breast cancer. MMTV-AKT1 transgenic mice develop ERα-positive breast cancer when exposed to chemical carcinogens suggesting a significant crosstalk between AKT and ERα in breast cancer (57). The extent to which this crosstalk relies on modulation of microRNA expression is not known and may potentially provide important clues into the initiation of ERα-positive breast cancers. AKT decreased E2-inducible expression of both tumor suppressor and oncogenic microRNAs. E2-dependent reduction of miR-126 in only AKT overexpressing cells is relevant to breast cancer as this microRNA is considered both a tumor suppressor and metastasis inhibitor (58).
In summary, we show the effects of E2 on modulating the microRNA-expression pattern as well as the potential for extracellular signal activated kinases to modulate this action of E2. With respect to breast cancer, our observations may have implications on responses to endocrine therapy as signaling pathways that modulate ERα function may change the balance between tumor suppressor and oncogenic microRNAs induced by E2 and thus alter the response to anti-estrogen therapy.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Department of Defense BC053295 concept award; National Institutes of Health (R01CA89153); IU Simon Cancer Center breast cancer pilot grant to HN; R01DK074967, DF/HCC Breast Cancer Spore from NCI, and the DFCI Women's; Cancers Program to MB. Funding for open access charge: Institutional funds.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank other members of Nakshatri Lab for various reagents and discussion. We also thank Dr Jay Patel for critical reading of the manuscript, IU Simon Cancer Center Flow Cytometry Resource Facility and Translational Genomic Core facilities for assistance. HN is Marian J. Morrison Professor of Breast Cancer Research.
REFERENCES
- 1.Jordan VC, O'Malley BW. Selective estrogen-receptor modulators and antihormonal resistance in breast cancer. J. Clin. Oncol. 2007;25:5815–5824. doi: 10.1200/JCO.2007.11.3886. [DOI] [PubMed] [Google Scholar]
- 2.Ali S, Coombes RC. Endocrine-responsive breast cancer and strategies for combating resistance. Nat. Rev. Cancer. 2002;2:101–112. doi: 10.1038/nrc721. [DOI] [PubMed] [Google Scholar]
- 3.Campbell RA, Bhat-Nakshatri P, Patel NM, Constantinidou D, Ali S, Nakshatri H. Phosphatidylinositol 3-kinase/AKT-mediated activation of estrogen receptor alpha: a new model for anti-estrogen resistance. J. Biol. Chem. 2001;276:9817–9824. doi: 10.1074/jbc.M010840200. [DOI] [PubMed] [Google Scholar]
- 4.Gururaj AE, Rayala SK, Vadlamudi RK, Kumar R. Novel mechanisms of resistance to endocrine therapy: genomic and nongenomic considerations. Clin. Cancer Res. 2006;12:1001s–1007s. doi: 10.1158/1078-0432.CCR-05-2110. [DOI] [PubMed] [Google Scholar]
- 5.Calin GA, Croce CM. Chromosomal rearrangements and microRNAs: a new cancer link with clinical implications. J. Clin. Invest. 2007;117:2059–2066. doi: 10.1172/JCI32577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hammond SM. MicroRNAs as tumor suppressors. Nat. Genet. 2007;39:582–583. doi: 10.1038/ng0507-582. [DOI] [PubMed] [Google Scholar]
- 7.Chendrimada TP, Finn KJ, Ji X, Baillat D, Gregory RI, Liebhaber SA, Pasquinelli AE, Shiekhattar R. MicroRNA silencing through RISC recruitment of eIF6. Nature. 2007;447:823–828. doi: 10.1038/nature05841. [DOI] [PubMed] [Google Scholar]
- 8.Tay Y, Zhang J, Thomson AM, Lim B, Rigoutsos I. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature. 2008;455:1124–1128. doi: 10.1038/nature07299. [DOI] [PubMed] [Google Scholar]
- 9.Forman JJ, Legesse-Miller A, Coller HA. A search for conserved sequences in coding regions reveals that the let-7 microRNA targets Dicer within its coding sequence. Proc. Natl Acad. Sci. USA. 2008;105:14879–14884. doi: 10.1073/pnas.0803230105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318:1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- 11.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 13.Kedde M, Strasser MJ, Boldajipour B, Vrielink JA, Slanchev K, le Sage C, Nagel R, Voorhoeve PM, van Duijse J, Orom UA, et al. RNA-binding protein Dnd1 inhibits microRNA access to target mRNA. Cell. 2007;131:1273–1286. doi: 10.1016/j.cell.2007.11.034. [DOI] [PubMed] [Google Scholar]
- 14.Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell. 2006;125:1111–1124. doi: 10.1016/j.cell.2006.04.031. [DOI] [PubMed] [Google Scholar]
- 15.Place RF, Li LC, Pookot D, Noonan EJ, Dahiya R. MicroRNA-373 induces expression of genes with complementary promoter sequences. Proc. Natl Acad. Sci. USA. 2008;105:1608–1613. doi: 10.1073/pnas.0707594105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Amaral PP, Dinger ME, Mercer TR, Mattick JS. The eukaryotic genome as an RNA machine. Science. 2008;319:1787–1789. doi: 10.1126/science.1155472. [DOI] [PubMed] [Google Scholar]
- 17.Lippman Z, Martienssen R. The role of RNA interference in heterochromatic silencing. Nature. 2004;431:364–370. doi: 10.1038/nature02875. [DOI] [PubMed] [Google Scholar]
- 18.Borchert GM, Lanier W, Davidson BL. RNA polymerase III transcribes human microRNAs. Nat. Struct. Mol. Biol. 2006;13:1097–1101. doi: 10.1038/nsmb1167. [DOI] [PubMed] [Google Scholar]
- 19.Saini HK, Griffiths-Jones S, Enright AJ. Genomic analysis of human microRNA transcripts. Proc. Natl Acad. Sci. USA. 2007;104:17719–17724. doi: 10.1073/pnas.0703890104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc. Natl Acad. Sci. USA. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mattie MD, Benz CC, Bowers J, Sensinger K, Wong L, Scott GK, Fedele V, Ginzinger D, Getts R, Haqq C. Optimized high-throughput microRNA expression profiling provides novel biomarker assessment of clinical prostate and breast cancer biopsies. Mol. Cancer. 2006;5:24. doi: 10.1186/1476-4598-5-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blenkiron C, Goldstein LD, Thorne NP, Spiteri I, Chin SF, Dunning MJ, Barbosa-Morais NL, Teschendorff AE, Green AR, Ellis IO, et al. MicroRNA expression profiling of human breast cancer identifies new markers of tumor subtype. Genome Biol. 2007;8:R214. doi: 10.1186/gb-2007-8-10-r214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl Acad. Sci. USA. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 25.Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey SS, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl Acad. Sci. USA. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Adams BD, Furneaux H, White BA. The micro-ribonucleic acid (miRNA) miR-206 targets the human estrogen receptor-alpha (ERalpha) and represses ERalpha messenger RNA and protein expression in breast cancer cell lines. Mol. Endocrinol. 2007;21:1132–1147. doi: 10.1210/me.2007-0022. [DOI] [PubMed] [Google Scholar]
- 27.Bhat-Nakshatri P, Wang G, Appaiah H, Luktuke N, Carroll JS, Geistlinger TR, Brown M, Badve S, Liu Y, Nakshatri H. AKT Alters genome-wide estrogen receptor alpha binding and impacts estrogen signaling in breast cancer. Mol. Cell Biol. 2008;28:7487–7503. doi: 10.1128/MCB.00799-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thomson JM, Parker JS, Hammond SM. Microarray analysis of miRNA gene expression. Methods Enzymol. 2007;427:107–122. doi: 10.1016/S0076-6879(07)27006-5. [DOI] [PubMed] [Google Scholar]
- 29.Thomson JM, Parker J, Perou CM, Hammond SM. A custom microarray platform for analysis of microRNA gene expression. Nat. Methods. 2004;1:47–53. doi: 10.1038/nmeth704. [DOI] [PubMed] [Google Scholar]
- 30.Carroll JS, Liu XS, Brodsky AS, Li W, Meyer CA, Szary AJ, Eeckhoute J, Shao W, Hestermann EV, Geistlinger TR, et al. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell. 2005;122:33–43. doi: 10.1016/j.cell.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 31.Bourdeau V, Deschenes J, Laperriere D, Aid M, White JH, Mader S. Mechanisms of primary and secondary estrogen target gene regulation in breast cancer cells. Nucleic Acids Res. 2008;36:76–93. doi: 10.1093/nar/gkm945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bethke A, Fielenbach N, Wang Z, Mangelsdorf DJ, Antebi A. Nuclear hormone receptor regulation of microRNAs controls developmental progression. Science. 2009;324:95–98. doi: 10.1126/science.1164899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Johnson CD, Esquela-Kerscher A, Stefani G, Byrom M, Kelnar K, Ovcharenko D, Wilson M, Wang X, Shelton J, Shingara J, et al. The let-7 microRNA represses cell proliferation pathways in human cells. Cancer Res. 2007;67:7713–7722. doi: 10.1158/0008-5472.CAN-07-1083. [DOI] [PubMed] [Google Scholar]
- 34.Ibarra I, Erlich Y, Muthuswamy SK, Sachidanandam R, Hannon GJ. A role for microRNAs in maintenance of mouse mammary epithelial progenitor cells. Genes Dev. 2007;21:3238–3243. doi: 10.1101/gad.1616307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mayr C, Hemann MT, Bartel DP. Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation. Science. 2007;315:1576–1579. doi: 10.1126/science.1137999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D, Slack FJ. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 37.Sampson VB, Rong NH, Han J, Yang Q, Aris V, Soteropoulos P, Petrelli NJ, Dunn SP, Krueger LJ. MicroRNA let-7a down-regulates MYC and reverts MYC-induced growth in Burkitt lymphoma cells. Cancer Res. 2007;67:9762–9770. doi: 10.1158/0008-5472.CAN-07-2462. [DOI] [PubMed] [Google Scholar]
- 38.Singh SK, Kagalwala MN, Parker-Thornburg J, Adams H, Majumder S. REST maintains self-renewal and pluripotency of embryonic stem cells. Nature. 2008;453:223–227. doi: 10.1038/nature06863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sylvestre Y, De Guire V, Querido E, Mukhopadhyay UK, Bourdeau V, Major F, Ferbeyre G, Chartrand P. An E2F/miR-20a autoregulatory feedback loop. J. Biol. Chem. 2007;282:2135–2143. doi: 10.1074/jbc.M608939200. [DOI] [PubMed] [Google Scholar]
- 40.Hossain A, Kuo MT, Saunders GF. Mir-17-5p regulates breast cancer cell proliferation by inhibiting translation of AIB1 mRNA. Mol. Cell Biol. 2006;26:8191–8201. doi: 10.1128/MCB.00242-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shi B, Liang J, Yang X, Wang Y, Zhao Y, Wu H, Sun L, Zhang Y, Chen Y, Li R, et al. Integration of estrogen and Wnt signaling circuits by the polycomb group protein EZH2 in breast cancer cells. Mol. Cell Biol. 2007;27:5105–5119. doi: 10.1128/MCB.00162-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Couzin J. MicroRNAs make big impression in disease after disease. Science. 2008;319:1782–1784. doi: 10.1126/science.319.5871.1782. [DOI] [PubMed] [Google Scholar]
- 44.Wang WX, Rajeev BW, Stromberg AJ, Ren N, Tang G, Huang Q, Rigoutsos I, Nelson PT. The expression of microRNA miR-107 decreases early in Alzheimer's disease and may accelerate disease progression through regulation of beta-site amyloid precursor protein-cleaving enzyme 1. J. Neurosci. 2008;28:1213–1223. doi: 10.1523/JNEUROSCI.5065-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kulshreshtha R, Ferracin M, Wojcik SE, Garzon R, Alder H, Agosto-Perez FJ, Davuluri R, Liu CG, Croce CM, Negrini M, et al. A microRNA signature of hypoxia. Mol. Cell Biol. 2007;27:1859–1867. doi: 10.1128/MCB.01395-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc. Natl Acad. Sci. USA. 2006;103:12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Frasor J, Danes JM, Komm B, Chang KC, Lyttle CR, Katzenellenbogen BS. Profiling of estrogen up- and down-regulated gene expression in human breast cancer cells: insights into gene networks and pathways underlying estrogenic control of proliferation and cell phenotype. Endocrinology. 2003;144:4562–4574. doi: 10.1210/en.2003-0567. [DOI] [PubMed] [Google Scholar]
- 48.Wickramasinghe NS, Manavalan TT, Dougherty SM, Riggs KA, Li Y, Klinge CM. Estradiol downregulates miR-21 expression and increases miR-21 target gene expression in MCF-7 breast cancer cells. Nucleic Acids Res. 2009;37:2584–2595. doi: 10.1093/nar/gkp117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kishimoto H, Wang Z, Bhat-Nakshatri P, Chang D, Clarke R, Nakshatri H. The p160 family co-activators regulate breast cancer cell proliferation and invasion through autocrine/paracrine activity of SDF-1{alpha}/CXCL12. Carcinogenesis. 2005;26:1706–1715. doi: 10.1093/carcin/bgi137. [DOI] [PubMed] [Google Scholar]
- 50.Eeckhoute J, Keeton EK, Lupien M, Krum SA, Carroll JS, Brown M. Positive cross-regulatory loop ties GATA-3 to estrogen receptor alpha expression in breast cancer. Cancer Res. 2007;67:6477–6483. doi: 10.1158/0008-5472.CAN-07-0746. [DOI] [PubMed] [Google Scholar]
- 51.Viswanathan SR, Daley GQ, Gregory RI. Selective blockade of microRNA processing by Lin28. Science. 2008;320:97–100. doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chang TC, Yu D, Lee YS, Wentzel EA, Arking DE, West KM, Dang CV, Thomas-Tikhonenko A, Mendell JT. Widespread microRNA repression by Myc contributes to tumorigenesis. Nat. Genet. 2008;40:43–50. doi: 10.1038/ng.2007.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Boyerinas B, Park SM, Shomron N, Hedegaard MM, Vinther J, Andersen JS, Feig C, Xu J, Burge CB, Peter ME. Identification of let-7-regulated oncofetal genes. Cancer Res. 2008;68:2587–2591. doi: 10.1158/0008-5472.CAN-08-0264. [DOI] [PubMed] [Google Scholar]
- 54.Cheng AS, Jin VX, Fan M, Smith LT, Liyanarachchi S, Yan PS, Leu YW, Chan MW, Plass C, Nephew KP, et al. Combinatorial analysis of transcription factor partners reveals recruitment of c-MYC to estrogen receptor-alpha responsive promoters. Mol. Cell. 2006;21:393–404. doi: 10.1016/j.molcel.2005.12.016. [DOI] [PubMed] [Google Scholar]
- 55.Zhu S, Wu H, Wu F, Nie D, Sheng S, Mo YY. MicroRNA-21 targets tumor suppressor genes in invasion and metastasis. Cell Res. 2008;18:350–359. doi: 10.1038/cr.2008.24. [DOI] [PubMed] [Google Scholar]
- 56.Si ML, Zhu S, Wu H, Lu Z, Wu F, Mo YY. miR-21-mediated tumor growth. Oncogene. 2007;26:2799–2803. doi: 10.1038/sj.onc.1210083. [DOI] [PubMed] [Google Scholar]
- 57.Blanco-Aparicio C, Perez-Gallego L, Pequeno B, Leal JF, Renner O, Carnero A. Mice expressing myrAKT1 in the mammary gland develop carcinogen-induced ER-positive mammary tumors that mimic human breast cancer. Carcinogenesis. 2007;28:584–594. doi: 10.1093/carcin/bgl190. [DOI] [PubMed] [Google Scholar]
- 58.Tavazoie SF, Alarcon C, Oskarsson T, Padua D, Wang Q, Bos PD, Gerald WL, Massague J. Endogenous human microRNAs that suppress breast cancer metastasis. Nature. 2008;451:147–152. doi: 10.1038/nature06487. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.