Abstract
Archaeal splicing endonucleases (EndAs) are currently classified into three groups. Two groups require a single subunit protein to form a homodimer or homotetramer. The third group requires two nonidentical protein components for the activity. To elucidate the molecular architecture of the two-subunit EndA system, we studied a crenarchaeal splicing endonuclease from Pyrobaculum aerophilum. In the present study, we solved a crystal structure of the enzyme at 1.7-Å resolution. The enzyme adopts a heterotetrameric form composed of two catalytic and two structural subunits. By connecting the structural and the catalytic subunits of the heterotetrameric EndA, we could convert the enzyme to a homodimer that maintains the broad substrate specificity that is one of the characteristics of heterotetrameric EndA. Meanwhile, a deletion of six amino acids in a Crenarchaea-specific loop abolished the endonuclease activity even on a substrate with canonical BHB motif. These results indicate that the subunit architecture is not a major factor responsible for the difference of substrate specificity between single- and two-subunit EndA systems. Rather, the structural basis for the broad substrate specificity is built into the crenarchaeal splicing endonuclease itself.
INTRODUCTION
Most RNA primary transcripts undergo several processing steps to become mature and functional. In the case of tRNA maturation, primary transcripts are trimmed to have correct 5′- and 3′-termini, many bases at specific positions are modified according to the tRNA species, and -CCA-3′ terminal sequences are added if they are lacking (1). In addition, many tRNA genes encode introns that must be removed by a specific set of enzymes (2).
In yeast, the first step of tRNA intron removal, cleavage of phosphodiester bonds between an exon and intron, is performed by tRNA intron endonuclease (2). The enzyme is composed of four different polypeptides: Sen2p, Sen34p, Sen15p and Sen54p (3). Sen2p and Sen34p possess RNase activity, and Sen54p is considered to specify the position of introns by recognizing structural features present in the tRNA bodies (3,4). The same kind of enzyme is also present in human cells (5). However, the human counterpart is reported to be associated with protein factors essential for pre-mRNA 3′ end processing (5,6). One of the associated factors is hClp1, which has polynucleotide kinase activity toward 3′-exons of tRNAs and short-interfering RNAs (7). Recently, mutations in the human tRNA intron endonuclease subunit genes were shown to be associated with pontocerebellar hypoplasias, representing a group of neurodegenerative autosomal recessive disorders (8).
In Archaea, the cleavage of phosphodiester bonds between an exon and intron is processed by a splicing endonuclease (2,9), which is also involved in intron removal in rRNA (10). Furthermore, the enzyme is considered to be involved in removal of mRNA introns (11,12) and rRNA maturation (13–16) through recognition of a bulge–helix–bulge (BHB) motif composed of a couple of 3-nt bulges split by a 4-bp helix (2,9). Subunits in eucaryal tRNA intron endonuclease and archaeal splicing endonuclease have a common ancestor (17,18).
The archaeal splicing endonucleases (EndAs) can be classified into three types according to their subunit structures (9). Homotetramers (α4) consist of four identical polypeptides with a molecular mass of 18 000–20 000. Homodimers (α2) are composed of two identical polypeptides with a molecular mass of 30 000–45 000. The X-ray structures for these two types have been solved (19–21). A comparison of the structures reveals that the gene for the homodimeric enzyme appears to have been formed by duplication of the endA gene of the homotetramer type, and the duplicated genes fused to make one large subunit (20). Then, the N-terminal domain lost amino acid residues required for RNase activity. The third type, a heterotetramer (α2β2), was recently discovered by several groups (22–25). Enzymes of this type consist of two sets of two polypeptides, namely the catalytic (α) and structural (β) subunits. However, there was no conclusive evidence that they adopt a heterotetrameric subunit structure. In the present study, we elucidated an X-ray structure for a splicing endonuclease from crenarchaeon Pyrobaculum aerophilum (PAE-EndA), which requires two polypeptides for its activity. Based on the structure, we could convert the enzyme to a homodimer by connecting the structural and catalytic subunits without losing the broad substrate specificity that is one of the characteristics of the heterotetrameric splicing endonuclease. On the other hand, a deletion of six amino acid residues in a Crenarchaea-specific loop abolished the endonuclease activity even on a substrate with the canonical BHB motif.
MATERIALS AND METHODS
Construction of expression plasmids
Cells of P. aerophilum P1 (Japan Collection of Microorganisms accession number: 9630) were propagated according to the supplier's instructions. The cell lysate was used as a DNA source for the PCR template. DNA fragments encoding PAE2269 (catalytic subunit) protein and PAE0789 (structural subunit) protein were amplified by PCR with the primers (PAE2269-F + PAE2269-R, and PAE0789-F + PAE0789-R, respectively) listed in Table S1. The PAE0789 coding sequence was cloned into pET15b (Novagen) between the NdeI and BamHI sites in the vector to obtain pET15b-PAE0789.
To construct an expression plasmid that simultaneously expresses the two proteins in a single cell, we first cloned a DNA fragment encoding the catalytic subunit protein in between the NdeI and BglII restriction sites of pETDuet-1 (Novagen), called pETDuet-PAE2269. Next, the 6×His-tag coding region in pET15b followed by the PAE0789 open reading frame sequence was amplified from pET15b-PAE0789 with pETupstream and pETterminator primers (Table S1). The amplified product was digested with NcoI and BamHI and cloned in between the NcoI and BamHI sites in the multicloning-site-1 of the pETDuet-PAE2269 to form pETDuet-PAE-EndA.
To prepare plasmids for expressing linker-connected variants, PCR reactions (with primer sets in Table S1) were performed with the plasmid pETDuet-PAE-EndA as a template. The primer combinations for each variant are: 0789+None and None+2269 (for NoAA); 0789 + LP and None+2269 (for LP); 0789+LP and E+2269 (for LPE); and 0789+LP and EI+2269 (for LPEI). After the reaction, the amplified DNAs were self-ligated to obtain plasmids with linker-connected protein-coding sequences. To construct plasmids that express deletion variants, we performed PCR with the following primer sets (Table S1): PAE2269Del-Fw1 and PAE2269Del-RV (for Del-1); PAE2269Del-Fw1 and Del-Rev-1 (for Del-2); PAE2269Del-Fw1 and Del-Rev-2 (for Del-3). Then, the amplified DNAs were self-ligated.
Expression and purification of PAE-EndA subunits and complexes
The protein expression protocol was the same as for Sulfolobus tokodaii EndA subunits (23). First, Escherichia coli strain Rosetta-gami (DE3) (Novagen) was transformed with the expression plasmid described above. A colony was picked up, and the bacteria were grown in LB broth [1% (w/v) Bacto-tryptone, 0.5% (w/v) Bacto-yeast extract, 0.5% (w/v) NaCl] at 37°C supplemented with 100 μg/ml carbenicillin and 34 μg/ml chloramphenicol. When the optical density at 600 nm (OD600) reached 0.1, isopropyl β-d-thiogalactopyranoside (IPTG) was added to yield a final concentration of 10 μM. Then, the culture was incubated at 18°C for 20 h. After 20 h, a typical culture had an OD600 of ∼1.0. Cells were harvested by centrifugation at 8000 g for 10 min, washed and suspended in 1/25 of culture volume of Buffer A [20 mM Tris–HCl (pH 8.0), 50 mM NaCl]. The cell suspensions were stored at –20°C until use.
The following procedures were performed at 4°C, unless otherwise noted. The frozen suspensions were thawed, and the cells were disrupted by the addition of hen egg-white lysozyme (final concentration, 200 μg/ml) followed by incubation on ice for 30 min. The viscosity of the lysates was reduced by sonication. Then, the lysates were incubated at 80°C for 20 min and clarified by centrifugation (20 000 g for 10 min at 4°C). To enrich the proteins with the 6×His-tag at their N-termini, the cleared lysate was mixed with TALON metal-affinity resin (Clontech, Co2+-chelated agarose) equilibrated with Buffer A (15-μl bed volume of the resin per 1 ml lysate). After gentle agitation for 20 min at 4°C, the mixture was separated into the resin and supernatant by centrifugation at 700 g for 10 min. The supernatant was removed, and the resin was washed once with 1 ml of Buffer B [50 mM sodium phosphate buffer (pH 8.0), 300 mM NaCl] per 15-μl resin and washed again with 0.5 ml of Buffer B + 10 mM imidazole per 15-μl resin. The protein bound to the TALON resin was eluted with 0.3 ml of Buffer B + 150 mM imidazole per 15-μl resin. The elution process was repeated twice. The eluate was dialyzed against Buffer C [20 mM Tris–HCl (pH 8.0), 750 mM KCl, 10% (v/v) glycerol] and used for further analyses. Protein concentrations of the fractions after dialysis and the following procedures were estimated by measuring the absorbance of the fractions at 280 nm. An absorbance of 1 was regarded as 1 mg/ml (26).
Gel filtration
The endonuclease protein in the 6×His-PAE-EndA fraction isolated through elution from TALON resin in Buffer C was concentrated with an Amicon Ultra 4 (Millipore, molecular weight cut-off: 10 000). After the protein concentration was adjusted to an ∼5 mg/ml with Buffer C, 0.5 ml of the protein solution was injected into Superdex 200 10/300 (GE Healthcare) pre-equilibrated with Buffer C at a flow rate of 0.5 ml/min. Fractionation (0.5 ml/fraction; 1 min/fraction) was initiated after 15 min of injection. Fractions were subjected to endonuclease assay and sodium dodecylsulfate (SDS) polyacrylamide gel electrophoresis (PAGE) analysis. Activity peak fractions were used for crystallization experiments.
Analytical ultracentrifugation
Wild-type PAE-EndA and the linker variants at a concentration of 1 mg/ml in Buffer C were subjected to sedimentation velocity experiments by using an Optima XL-A analytical ultracentrifuge (Beckman-Coulter) equipped with an eight-hole An50Ti rotor at 20°C. The analyses were conducted at Research Institute of Biological Science, Katakura Industry Co. Ltd. Buffer C was used as the reference solution. Sedimentation velocity data with a scanning light of 280-nm wavelength were acquired at a rotor speed of 50 000 r.p.m. The sedimentation coefficient distribution function, c(s), and the molecular mass distribution, c(M), were obtained with the SEDFIT program (version 2008) (27).
SDS–PAGE
Protein samples were resolved on SDS–PAGE with a 15% separation gel. To visualize protein bands on the gel, the gel was stained with GelCode-Blue (Pierce), according to the supplier's instructions.
Crystallization
Activity peak fractions from gel filtration were pooled and concentrated to ∼8 mg/ml by using an Amicon Ultra 4 (Millipore, molecular weight cut-off: 10 000).
Screening for crystallization conditions by the sitting-drop vapor-diffusion technique was performed by using 96-well CrystalClear P Strips (Douglas Instruments). One microliter of the concentrated 6×His-PAE-EndA protein solution was mixed with an equal volume of reservoir solution, and then the drop was allowed to reach equilibrium with 100 μl of the reservoir solution. Initial crystallization conditions for the reservoir solution were screened at 25°C by using Crystal Screen (28), Crystal Screen II (Hampton Research), and Wizard Screens I and II (Emerald BioStructures). Out of 194 conditions, the condition [Crystal Screen No. 50, 0.5 M lithium sulfate monohydrate, 15% (w/v) polyethylene glycol (PEG) 8000] that produced crystals with the best resolution was identified. For phasing by introduction of platinum, the crystals were soaked in the reservoir solution supplemented with 2 mM K2Pt(NO2)4 for 24 h before being scooped into a loop.
X-ray data collection and processing
For collecting diffraction data, a crystal was scooped into a nylon loop and soaked briefly in reservoir solution supplemented with 20% (v/v) glycerol and then frozen by rapid submersion in liquid nitrogen. Diffraction data sets for native crystals (λ = 1.000 Å) and single-wavelength anomalous dispersion (SAD) data sets for Pt-introduced crystals (λ = 1.0718 Å) were collected under liquid-nitrogen-cooled conditions of 100 K at NW12 of the Photon Factory Advanced Ring in Tsukuba, Japan, by using a Quantum-210 detector (Advanced Detector System Corporation). All data sets were indexed, integrated and scaled by using HKL2000 (29). Native crystal diffracted up to 1.7-Å resolution. The crystals belong to the orthorhombic space group with cell parameters a = 69.8, b = 70.7, c = 131.0 Å, α = β = γ = 90°. There is one heterotetramer per asymmetric unit. Statistics for data collection are summarized in Table S2.
Structure determination and refinement
Initial attempts to solve the crystal structure by using the molecular replacement method with known EndA were unsuccessful. Therefore, the crystal structure of PAE-EndA was determined by the SAD method. The 10 platinum sites of PAE-EndA were initially calculated with SHELEXC/D (30) with the data set collected at the peak wavelength. Phases were determined in the program autoSHARP (31) by using the platinum peak data set and were improved upon solvent flattening with SOLOMON (32). Automatic model building with ARP/wARP (33) allowed us to trace about 80% of the residues. Manual building of the remaining model and further crystallographic refinement were performed with the COOT program (34) and Refmac5 (35). The final model (with 443 water molecules) refined to a 50.0–1.7-Å resolution has an R-factor of 21.0% and an Rfree of 25.2%. The final model was checked by PROCHECK (36), revealing good stereochemical parameters with no residues outside of the allowed regions of the Ramachandran plot. Statistics for data collection and refinements are summarized in Table S2. Graphic representations developed by using the coordinates were prepared with a PyMOL graphics system (DeLano Scientific LLC, Palo Alto, CA, USA. http://www.pymol.org).
Splicing endonuclease assay
A typical endonuclease reaction in a total volume of 40 μl contained 2 μg (0.04 A260 unit) of S. tokodaii tRNATrp precursor (23) or its variant, 8 μl of 5×EndA Buffer [100 mM Tris–HCl (pH 8.5), 50 mM MgCl2] and 1 μl of the enzyme fraction in Buffer C. The reaction was incubated at 70°C for 30 min. Then, the RNA was isolated by phenol extraction and ethanol precipitation. The RNA was dissolved in FDE [90% (v/v) formamide, 10 mM ethylenediaminetetraacetic acid (pH 8.0), 0.02% (w/v) bromphenol blue, 0.02% (w/v) xylene cyanol] and resolved by 8.3-M urea/10% PAGE. The RNA bands were visualized by methylene blue staining.
For comparing specific activities of the linker variants, 200 000 c.p.m. of [32P]-labeled S. tokodaii tRNATrp precursor was included to the reaction above, and incubated at 60°C. Then, 10 μl was withdrawn at each time point. The reaction was stopped by adding an equal volume of FDE and the product RNA were resolved by 8.3-M urea/10% PAGE.
For Mini-BHL (37), the substrates were prepared by transcription reactions by the use of T7 RNA polymerase and [α-32P]-CTP (MP Biomedicals, 111 TBq/mmole) (23). In a total reaction volume of 10 μl, the transcript (15 000 c.p.m.) was incubated with 0.3 μg of EndA protein at 65°C. After the incubation, an equal volume of FDE was added to the reaction and directly loaded onto an 8.3-M urea/15% polyacrylamide gel.
Radioactivity was detected by exposing the gel to ImagePlate (Fujifilm) and then scanning the exposed plate with BAS-2500 (Fujifilm). Radioactive signals on the scanned images were quantified by Multi Gauge software (Fujifilm).
RESULTS
Pyrobaculum aerophilum splicing endonuclease requires two proteins for its activity
The genome of P. aerophilum (38) encodes two genes homologous to the endA gene from Methanocaldococcus jannaschii (39). The open reading frames for the two proteins were designated as PAE2269 and PAE0789. We assumed that the two proteins are concurrently required for splicing endonuclease activity, because this requirement has been shown for S. tokodaii, Sulfolobus solfataricus and Nanoarchaeum equitans (22–25). The PAE2269 protein could be a catalytic subunit because it possesses conserved histidine that is required for RNase activity. The PAE0789 protein could be a structural subunit. To confirm our assumption, we expressed each protein with an N-terminal 6×His-tag in E. coli; then, we partially purified the proteins with a metal-affinity column and examined their splicing endonuclease activity. We found that the enzyme activity required both open reading frame products at the same time (Figure S1). In addition, we found that the two proteins interact with each other in solution in an unequivocal manner by attaching different epitope-tag sequences at the N-termini of each protein (Figure S2).
Crystal structure reveals PAE-EndA as a heterotetramer
To understand the nature of PAE-EndA, we conducted crystallization screens with the endonuclease fraction after the gel filtration (a typical fractionation pattern is shown in Figure S3). After the screening, we found that one reservoir solution [0.5 M lithium sulfate monohydrate, 15% (w/v) PEG 8000] gave us crystals with diffractions up to 1.7-Å resolution. The crystal structure of PAE-EndA was determined by the SAD method with Pt-induced crystal. The final refined model has an Rwork of 21.0% and an Rfree of 25.2% for the resolution range from 50 to 1.7 Å resolution, and it has good stereochemical quality. The refined structure contains all amino acid residues except for the three C-terminal residues (chain A) of the protein. Moreover, we could not fit the 20 N-terminal amino acid residues derived from the 6× His-tag sequences in chains A and C into the electron density map. The asymmetric unit contains a heterotetrameric subunit structure, α2β2, composed of two catalytic subunits (chains B and D) and two structural subunits (chains A and C) that are related by a noncrystallographic 2-fold axis (Figure 1A). Purified PAE-EndA appeared at the heterotetrameric size on the gel filtration column; this purified PAE-EndA had enzymatic activity and an apparent molecular mass of 56 kDa (Figures S3 and 3A). These results indicate that the heterotetrameric enzyme observed in the crystal structure is formed in solution.
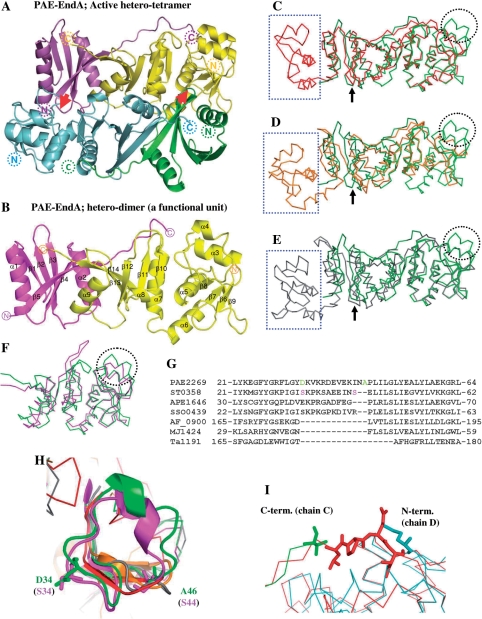
Figure 1.
Crystal structure of PAE-EndA. (A) Schematic representation of a crystal structure of PAE-EndA. The unit is composed of two catalytic subunits (cyan and yellow) and two structural subunits (green and magenta). Red arrows indicate L10-loop-like interactions between the structural and the catalytic subunits to form a hetero-tetramer. The positions of the N-termini and C-termini of the proteins are shown as dotted circles. (B) A functional hetero-dimer unit of PAE-EndA (chains A and B). The positions of the N-termini and C-termini of the proteins are shown as circles. (C–E) Superimposed structure of the PAE-EndA (PDB ID: 2ZYZ, chains A and B) on; (C) AFU-EndA (PDB ID: 1RLV, chain A); (D) TAC-EndA (PDB ID: 2OHC, chain A); and (E) MJA-EndA (PDB ID: 1A78, chains A and B). (F) Catalytic subunits of crenarchaeal origin, STO-EndA (PDB ID: 2CV8, chain B), and PAE-EndA (PDB ID: 2ZYZ, chain D) were superimposed. Through (C) to (F), color-assignments are as follows: dark green for PAE-EndA structural subunit; light green for PAE-EndA catalytic subunit; red for AFU-EndA; orange for TAC-EndA; pink for STO-EndA; gray for MJA-EndA. Positions for L10-like loop are indicated with black arrows. The EndA N-subdomain missing in the PAE-EndA structural subunit is enclosed by blue-dotted square. The extra loop missing in the euryarchaeal EndA catalytic subunits is circled with black dots. (G) Part of an amino acid sequence alignment for EndA catalytic domains or subunits. Amino acid sequences from catalytic subunits of crenarchaeal origin [PAE2269 (P. aerophilum, residues 1–183. GenBank accession number AAL64075), ST0358 (S. tokodaii, residues 1–180. Accession number BAB65337), APE1646 (Aeropyrum pernix, residues 1–187. Accession number BAA80647), and SSO0439 (S. solfataricus, residues 1–182. Accession number AAK40764)], catalytic domains of euryarchaeal homodimeric EndAs, [AF_0900 (A. fulgidus, residues 150–305. Accession number AAB90338), and Ta1191 (T. aciophilum, residues 156–289. Accession number CAC12316)], and a subunit of a euryarchaeal homotetrameric EndA, MJ1424 (M. jannaschii, residues 1–179. Accession number ABW02570) were aligned with CLUSTALW version 1.83 (43) on DNA Data Bank Japan (DDBJ) server (http://clustalw.ddbj.nig.ac.jp/top-j.html) with default parameters. (H) Magnified view of crenarchaeal EndA-specific extra loop. Residues for D34 and A46 of PAE-EndA (green) and S34 and S44 of STO-EndA catalytic subunits are presented as sticks. Color assignments for the structures are the same as in Figure 1C–F. (I) Magnified view of the superposed structure around the linker-loop region. Green, C-terminal region of the PAE-EndA structural subunit; cyan, N-terminal region of the PAE-EndA catalytic subunit; red, interdomain loop region of AF-EndA. The C-terminal residue of the PAE-EndA structural subunit (valine), the N-terminal residue of the PAE-EndA catalytic subunit (methionine), and the reference residues (-LPEI-) in the loop region for linker variants are shown as sticks.
The heterotetrameric PAE-EndA has a rectangular parallelepiped-like structure, with approximate dimensions of 60 Å × 50 Å × 35 Å (Figure 1A). The two dimers (chains A, B and chains C, D) do not show any significant differences [the root mean square deviations (RMSDs) of the Cα 0.697 Å] except for the N- and C-terminal regions of the structural subunits (chains A and C, Figure S4). The structural subunit consists of five β strands (β1–β5) and two α helices (α1 and α2, Figure 1B). The five β strands form a mixed antiparallel/parallel β sheet (β1-β2-β3-β4-β5; underlines denote parallel strands) that is surrounded by two α helices (α1 and α2). The catalytic subunit consists of seven α helices (α3–α9) and nine β strands (β6–β14, Figure 1B). In the N-terminal catalytic subunit, four β strands form a mixed antiparallel/parallel β sheet (order: β8-β7-β6-β9) that is surrounded by four α helices (α3–α6). In the C-terminal catalytic subunit, five β strands form a mixed antiparallel/parallel β sheet (order: β10-β11-β12-β13-β14) that is surrounded by three α helices (α7–α9). Between the structural and the catalytic subunits, an antiparallel intermolecular β sheet that is important for dimerization (β5–β14 interaction) is formed by β5 (structural subunit) and β14 (catalytic subunit). The C-terminal long loop of the structural subunit extends to the N-terminal of the catalytic subunit (Figure 1A and 1I).
Comparison with other EndAs
The archaeal splicing endonucleases can be distinctly grouped into three families: homodimers (α2), hom-tetramers (α4) and heterotetramers, or dimers of heterodimers (α2β2) (9). Crystal structures of the homodimeric splicing endonucleases from Archaeoglobus fulgidus (AFU-EndA; PDB accession number 1RLV) (20) and Thermoplasma acidophilum (TAC-EndA; PDB accession numbers 2OHC and 2HOE) (21) have been reported. In addition, the crystal structure of AFU-EndA in complex with an RNA substrate has been revealed (PDB accession number 2GJW) (40). One crystal structure for a homotetrameric splicing endonuclease, the M. jannaschii splicing endonuclease (MJA-EndA; accession number 1A79), has been reported (19). For heterotetramers or dimers of heterodimers, two crystal structures for nonfunctional dimers of the catalytic subunit have been reported: the splicing endonuclease from S. tokodaii (STO-EndA; accession number 2CV8) and the splicing endonuclease from S. solfataricus (SSO-EndA, no structural data is deposited on PDB) (24).
The splicing endonuclease from P. aerophilum (PAE-EndA, this work) belongs to the dimers of heterodimers family (α2β2). The subunit interaction is mainly due to a hydrophobic interaction and hydrogen bonds. Calculations for subunit interactions are summarized in Table S3. The interface between the structural and catalytic subunits (between chains A and B, C and D) covers ∼20% of the total accessible surface of the two proteins (20% for between A and B, 23% for between C and D). These values are higher than the values for MJA-EndA (14% for between A and B as well as for C and D, respectively), probably due to the small size of the structural subunit in PAE-EndA and the C-terminal extension of the structural subunit towards the N-terminus of the catalytic subunit (Figure 1A and 1I). The interface between the functional units (A + B and C + D) covers 17% of the total accessible area. This value is comparable to values for MJA-EndA (19%), AFU-EndA (24%), and TAC-EndA (15%).
For the functional unit, the crystal structure of PAE-EndA is similar to the structures of other EndAs (Figure 1C–E). The structure of the PAE-EndA functional unit (dimer of the structural and catalytic subunits, chains A and B) aligns well with the monomer structure of AFU-EndA (chain A of 1RLV, RMSD = 1.12 Å for the 141 Cα atoms of the catalytic subunit). Superposition of the Cα atoms of the PAE-EndA functional unit and TAC-EndA monomer (chain A of 2OHC) gives an RMSD of 1.99 Å for 120 atoms of the catalytic subunits. When the functional unit of homotetrameric MJA-EndA (chains A and B of 1A78) was compared with the PAE-EndA functional unit, the RMSD of the Cα atoms (157 atoms of the catalytic subunit) was 1.49 Å. Similar to other EndAs, the functional unit of PAE-EndA is formed by a tight interaction between C-terminal regions of the structural and catalytic subunits, including the β9–β′9-like interaction (for PAE-EndA; β5–β14 interaction) (19,20). Interactions between the functional units are also conserved, including the structural subunit L10-loop (for PAE-EndA; L5-loop, between β4 and β5) interaction with the other catalytic subunit (19,20), but the loop conformations are different from each other (Figure 1C–E, black allows). There are some structural differences between PAE-EndA and other EndAs. PAE-EndA has a long loop insertion containing a short α4 helix (the loop between α3 and β8). However, the euryarchaeal EndAs (AFU-, TAC-, and MJA-EndAs) lack this region (Figure 1C–E, circled in dotted black). This loop is maintained in a catalytic subunit of STO-EndA of crenarchaeal origin (dotted black circle in Figure 1F) when it is superposed on the catalytic subunit of PAE EndA (RMSD = 1.64 Å for the 153 Cα atoms of the chain D of the PAE-EndA and chain B for 2CV8). When amino acid sequence alignment was conducted among MJA-EndA, catalytic domains of AFU-EndA (residues 150–305) and TAC_EndA (residues 156–289), and catalytic subunits of PAE-EndA, STO-EndA, SSO-EndA and Aeropyrum penix EndA, the amino acid sequence of the extra loop aligned well among crenarchaeal EndA catalytic subunits. However, this part is missing in euryarchaeal EndAs and makes a large gap in the alignment. Thus, it is possible that the existence of the extra loop is common to crenarchaeal EndAs. Interestingly, when the amino acid residues of the extra loops at the border of the gap [Figure 1G, D34 and A46 for PAE-EndA (highlighted in green), S34 and S44 for STO-EndA (highlighted in pink)] were mapped on the superimposed crystal structures, the positions of the insertion seemed almost identical (Figure 1H). The other difference is that the first EndA-N subdomain at the N-terminus of the structural domain of the functional unit is missing in PAE-EndA (Figure 1C–E, square in dotted blue).
ENGINEERING PAE-ENDA TO CONVERT TO A HOMODIMERIC FORM
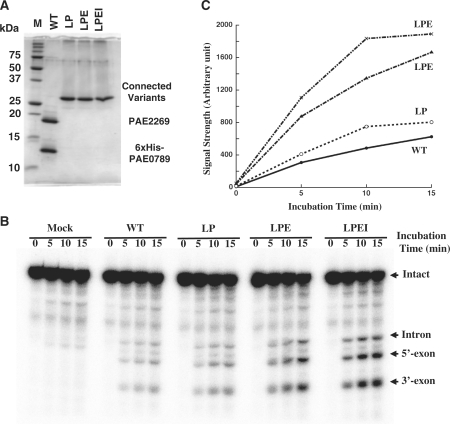
Figure 1I shows the superimposed X-ray structures for the interdomain loop region of AFU-EndA, the C-terminal residues of the structural subunit and the N-terminal residue of the catalytic subunit of PAE-EndA. An interesting observation about the crystal structure of PAE-EndA is that the C-terminal residues of the structural subunit form an extended arm toward the N-terminal residue of the catalytic subunit. This extension is not observed in MJA-EndA, and the extension seems to overlap with the interdomain loop connecting the N-terminal and C-terminal domain of AFU-EndA. We expected that the introduction of approximately three amino acid residues between the C-terminus of the structural subunit (PAE0789 protein) and the N-terminus of the catalytic subunit (PAE2269 protein) could connect the two subunits without a large change in the mutual organization of the subunits and could make the enzyme a homodimer. Therefore, we constructed plasmids that express variant PAE-EndAs in which the C-terminus of the structural subunit with 6×His-tag (6×His-PAE0789) was fused to the N-terminus of the catalytic subunit (PAE2269 protein) with the introduction of zero (variant NoAA), two (-Leu-Pro-, variant LP), three (-Leu-Pro-Glu-, variant LPE) or four (-Leu-Pro-Glu-Ile-, variant LPEI) amino acids between the subunits. We selected the introduced amino acid residues by comparing the missing part in PAE-EndA with the interdomain loop of AFU-EndA (Figure 1I). All of the variants could be expressed in E. coli. However, the variant NoAA could not be recovered as a soluble protein after incubation at 80°C (data not shown). The other variants (LP, LPE and LPEI) could be expressed and recovered as soluble proteins after the purification steps used for the co-expression system proteins (Figure 2A). Variants that remained soluble were metal-affinity purified and subjected to a 15-min time course endonuclease assay. All variants tested possessed splicing endonuclease activity toward a substrate with the canonical BHB motif, S. tokodaii tRNATrp precursor (Figure 2B). Then, the amount of accumulation for the 5′-exon was measured as shown in Figure 4C. Each variant exhibited accumulation of the 5′-exon with good linearity up to 10 min. Some differences could be observed for their activities. At the 5-min time point, each variant accumulated more 5′-exon as compared to the amount of 5′-exon accumulated by the wild-type endonuclease (LP; 1.3-fold, LPE; 2.9-fold, LPEI; 3.6-fold, Figure 2C).
Figure 2.
Engineered homodimeric PAE-EndAs retain splicing endonuclease activity. (A) GelCode-Blue-stained 15% SDS–PAGE gel. In each lane, 2 μg of the fraction eluted from TALON resin was loaded. (B) Splicing endonuclease assay. Sulfolobus tokodaii tRNATrp precursor was used as the substrate. Lane assignments are; M, molecular mass marker in (A); mock, mock incubation with enzyme solvent; WT, wild-type 6xHis-PAE-EndA; LP, variant LP; LPE, variant LPE; LPEI, variant LPEI. (C) Accumulation of 5′-exon on the time course.
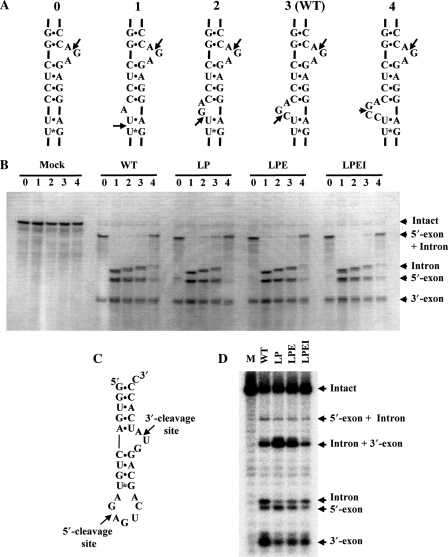
Figure 4.
Engineered homodimeric PAE-EndAs cleave substrates with noncanonical BHB motif. (A) Secondary structural representation of bulge number variants. The molecules were designed by deleting (variants 0, 1 and 2) or adding (variant 4) nucleotides at the 5′-exon–intron boundary of the S. tokodaii tRNATrp precursor. Arrows indicate cleavage sites of PAE-EndA and its variants. (B) Methylene blue-stained gel for the splicing endonuclease assay. The amount of protein used for the assay was 0.5 μg. (C) Secondary structural representation of a Mini-BHL molecule. Arrows in the figure indicate expected cleavage sites for the heterotetrameric splicing endonuclease. (D) Autoradiogram of a splicing assay gel. The reaction was carried out for 20 min. Abbreviations for the variants are the same as in Figure 2.
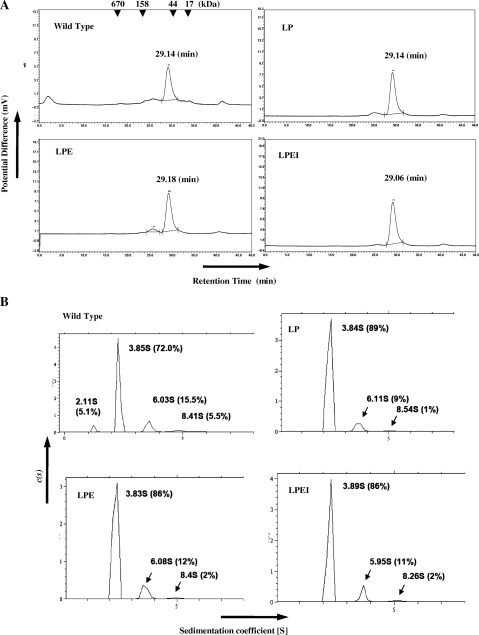
To investigate subunit assembly, the wild-type PAE-EndA and the variants (fractions after the metal-affinity fractionation) were subjected to gel filtration analysis through a Superdex 200 10/300 (GE Healthcare) column. Retention times of the variants through the gel filtration column were almost the same as that of the wild-type endonuclease (29.14 min for wild type, 29.14 min for LP, 29.18 min for LPE and 29.06 min for LPEI, respectively, Figure 3A), indicating the formation of the heterotetrameric subunit structure for the wild-type endonuclease and the homodimeric structure for the variants in solution. Formation of the heterotetramer for the wild type and the homodimer for the variants was further confirmed by analytical ultracentrifugation (Figure 3B). The main peak of the wild-type endonuclease has a sedimentation coefficient of 3.85 S with a calculated molecular mass of 55.0 kDa. Meanwhile, the sedimentation coefficient values of the main peaks for the wild-type endonuclease and the variants were almost identical (3.84 S with a calculated molecular mass of 60.4 kDa for LP, 3.85 S with a calculated molecular mass of 62.0 kDa for LPE, and 3.89S with a calculated molecular mass of 59.3 kDa for LPEI, respectively). These data also suggest that the engineered linker variant proteins formed homodimers.
Figure 3.
Subunit assembly of the wild-type and engineered variants of PAE-EndA. (A) Gel filtration profiles. Arrowheads at the top of the figure indicate the elution position of the marker with corresponding molecular weights. Horizontal axis; retention time (min), vertical axis; Potential difference proportional to A280. (B) Analytical centrifugation profiles. The analysis were conducted at Research Institute of Biological Science, Katakura Industry Co. Ltd. Horizontal Axis; sedimentation coefficient, [S]; vertical axis, sedimentation distribution function, c(s). Abbreviations for the variants are the same as Figure 2.
Engineered homodimeric PAE-EndAs retain the substrate specificity of the heterotetrameric splicing endonuclease
Heteromeric splicing endonucleases can cut substrates with a noncanonical BHB motif with altered bulge nucleotide numbers (24,25). The two splicing endonucleases from S. solfataricus and N. quitans used in those experiments are predicted to possess an EndA-N subdomain in their structural subunits as judged by the number of amino acid residues in the subunits. Furthermore, although AFU-EndA can cleave the substrate with altered bulge nucleotide numbers within the tRNA body with less efficiency, an AFU-EndA deletion mutant of the ‘mature domain’, which is equivalent to the EndA-N subdomain that is lacking in PAE-EndA, does not cleave the bulge–helix–loop (BHL) motif with or without mature tRNA bodies (37). Therefore, to see if PAE-EndA, which is composed of structural subunits without the EndA-N subdomain, can exhibit splicing endonuclease activity on a noncanonical BHB-motif within a mature tRNA context, we tested substrates of S. tokodaii tRNATrp variants with altered numbers of bulge nucleotides at the 5′-exon–intron boundary (Figure 4A). As shown in Figure 4B, the wild-type PAE-EndA could cleave only the intron–3′-exon border for the variant without bulge moiety at the 5′-exon–intron boundary (lane 0 in WT), resulting in the appearance of RNA bands for only the 3′-exon and 5′-exon + intron. Furthermore, the enzyme did not favor the 4-nt bulge at the 5′-exon–intron boundary (lane 4 in WT) on this substrate, as shown by the remaining 5′-exon + intron band and the less intense bands for the 5′-exon and intron. Other substrates could be efficiently cleaved, resulting in three RNA bands representing the 5′-exon, intron and 3′-exon. One interesting observation is that when the lengths of the products were compared, the 5′-exon and 3′-exon remained the same, and the introns differ in length by one base. This observation indicates that the endonuclease seemed to measure the 2-nt distance from the 4-bp helix in the modified BHB motifs. For linker variant EndAs, the band pattern of the RNAs was almost identical to that of WT. Thus, the substrate specificity is not altered by the change in subunit organization from heterotetramer to homodimer.
The heteromeric splicing endonucleases possess broader substrate specificity than their euryarchaeal homomeric counterparts (12,24,25,41). An example is the activity toward Mini-BHL (Figure 4C) shown by Tocchini-Valentini et al. (37). They found that the splicing endonuclease from S. solfataricus could cleave the Mini-BHL molecule without a tRNA body, whereas homodimeric AFU-EndA and homotetrameric MJA-EndA failed to cleave the molecule (37). We used this molecule to assess the substrate specificity of the variants. The wild-type PAE-EndA cleaved the Mini-BHL molecule in an expected manner (Figure 4D, lane WT). Interestingly, the homodimeric variants also exhibited the splicing endonuclease activity toward the Mini-BHL molecule, and the cutting manner was almost identical to that of wild-type endonuclease (Figure 4D, lanes LP, LPE and LPEI]. These results indicate that the subunit structure of the crenarchaeal enzyme is not responsible for its substrate specificity.
Engineered PAE-EndA without the extra-loop lose the splicing endonuclease activity
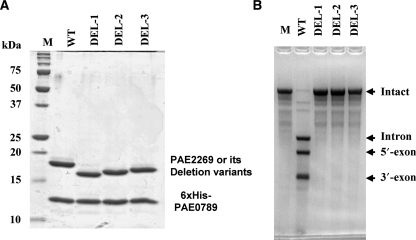
As shown in Figure 1F–H, there is an extra loop that seemed to be specific for crenarchaeal EndA catalytic subunits. In the case of PAE-EndA, the residue numbers of the loop are D34–A46. To examine the role of the loop in the splicing endonuclease function, we made PAE-EndA variants in which parts of the loop are deleted. The residues deleted were R29-D39 (DEL-1), L31-D39 (DEL-2) and D34-D39 (DEL-3). The variants were expressed in E. coli and purified by metal-affinity chromatography. The eluted fractions contained two protein components (Figure 5A), meaning that the two proteins formed a complex in solution. Further gel filtration analysis of these fractions also supported the complex formation (Figure S5). However, when the splicing endonuclease assay was conducted, we found that all of the deletion variants lost their activity (Figure 5B). An assay with five times more variants did not change the results (data not shown). It is surprising since that the variants were not deleted the residues that are needed for RNase activity. They also lost activity toward substrates with noncanonical BHB motifs (data not shown).
Figure 5.
Extra-loop deletion variants lose the splicing endonuclease activity. (A) GelCode-Blue-stained 15% SDS–PAGE gel. In each lane, 2 μg of the fraction eluted from TALON resin was loaded. (B) Splicing endonuclease assay. Sulfolobus tokodaii tRNATrp precursor was used as the substrate. Lane assignments are; M, molecular mass marker for (A); WT, wild-type 6×His-PAE-EndA; DEL-1, variant DEL-1; DEL-2, variant DEL-2; DEL-3, variant DEL-3. The amount of the protein used for the assay was 0.5 μg.
DISCUSSION
Pyrobaculum aerophilum splicing endonuclease forms a heterotetramer
Previous studies have proved that EndAs from S. solfataricus, S. tokodaii, and N. equitans require two proteins for their enzymatic activity (22–25). We used fractions of E. coli-expressed PAE-EndA collected after metal-affinity chromatography and gel filtration to obtain the crystal structure of the enzyme. The crystal structure unequivocally revealed for the first time that the enzyme forms a heterotetramer.
To date, all of the completed crenarchaeal genomes encode two distinct genes for homologs of the MJA-EndA subunit, suggesting that all of the crenarchaeal species possess a heterotetrameric type of splicing endonuclease (42). The reason why the enzyme in Crenarchaea has a distinctive subunit structure from the enzyme in its euryarchaeal counterpart has not yet been elucidated. One possibility is that the enzyme activity is regulated through the separate regulation of each subunit. As shown in EndAs for S. solfataricus (24) and S. tokodaii (PDB accession number: 2CV8), the catalytic subunit can form a homodimer. However, the homodimers are inactive (23,24 and Figure S1) and require the integration of the structural subunit into the complex to form active enzyme. Therefore, by regulating the expression of the structural subunit, it is possible to regulate enzymatic activity in the cell. However, we must study the Crenarchaea gene expression profile of the two proteins to uncover the biological significance of the existence of the structural subunit.
Lack of EndA-N subdomain in the structural subunit of PAE-EndA
One large structural difference of PAE-EndA from the other known EndA structures resides in the structural subunit. The structural subunit of the PAE-EndA lacks the EndA-N subdomain, which is suggested to possess an electrostatic interaction with the mature tRNA body domain in AFU-EndA (37). Lack of the subdomain in PAE-EndA without loss of its enzymatic activity toward substrates with noncanonical BHB motifs clearly shows that the subdomain is not involved in substrate binding and recognition by the crenarchaeal splicing endonuclease. This observation is in agreement with the fact that some Crenarchaea are predicted to have structural subunits significantly smaller (90–110 amino acid residues, molecular mass of ∼11 000) than a subunit of MJA-EndA (∼20 000). The enzymes classified into this category probably do not require the EndA-N subdomain for their function. On the other hand, some other crenarchaeal splicing endonucleases are predicted to have a structural subunit with a similar size (∼20 000) as their catalytic counterparts. These large structural subunits can be considered to retain the EndA-N subdomains. It is of great interest to examine if the subdomain is required for the functions of the crenarchaeal enzymes with a large structural subunit.
Subunit structure is not a determinant of substrate specificity for the crenarchaeal EndA
It is interesting that the substrate specificity of the enzyme toward the bulge-number variants with a tRNA body and towards the mini BHL without a tRNA body was maintained after the conversion of the subunit structure from a heterotetramer to a homodimer. These results suggest that the substrate specificity is not determined by a flexible subunit co-ordination due to the heterotetramer. Rather, the broad substrate specificity is already built into the whole architecture of the protein itself, as discussed below.
Crenarchaea-specific extra loop is important for splicing endonuclease activity
We are currently examining the structural basis of the broad specificity of the enzyme toward noncanonical BHB motifs. One strong candidate site is the inserted extra loop in the catalytic subunit that is not found in homodimeric or homotetrameric enzymes of euryarchaeal origin (circled in black dot in Figures 1C–F). Unfortunately, the deletion variants around the loop lost splicing endonuclease activity even toward a substrate with a canonical BHB motif (Figure 5B). However, this result means that important information for the enzyme is included in D34–D39 (see DEL-3 variant in Figure 5). Through the characterization of physical and biochemical properties of the variants, we are approaching the structural basis of the broad specificity of the enzyme.
ACCESSION NUMBER
The structure coordinate of the PAE-EndA has been submitted to RCSB database (PDB ID code 2ZYZ).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We are grateful to Dr Shin-ichi Yokobori of Tokyo University of Pharmacy and Life Sciences for providing materials for the first screening of EndAs to be crystallized. We also thank Ms Wang Ludan, Mr Sunao Fujioka, Mr Kenji Kawahara, Mrs Nimisha Vandan and Ms Maho Okuda for helping to prepare materials for the study. Funding for open access charge: Grant-in-Aid for Creative Scientific Research of Japan Society for the Promotion of Science.
Conflict of interest statement. None declared.
REFERENCES
- 1.Li H. Complexes of tRNA and maturation enzymes: shaping up for translation. Curr. Opin. Struct. Biol. 2007;17:293–301. doi: 10.1016/j.sbi.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 2.Abelson J, Trotta CR, Li H. tRNA splicing. J. Biol. Chem. 1998;273:12685–12688. doi: 10.1074/jbc.273.21.12685. [DOI] [PubMed] [Google Scholar]
- 3.Trotta CR, Miao F, Arn EA, Stevens SW, Ho CK, Rauhut R, Abelson JN. The yeast tRNA splicing endonuclease: a tetrameric enzyme with two active site subunits homologous to the archaeal tRNA endonucleases. Cell. 1997;89:849–858. doi: 10.1016/s0092-8674(00)80270-6. [DOI] [PubMed] [Google Scholar]
- 4.Di Nicola Negri E, Fabbri S, Bufardeci E, Baldi MI, Gandini Attardi D, Mattoccia E, Tocchini-Valentini GP. The eucaryal tRNA splicing endonuclease recognizes a tripartite set of RNA elements. Cell. 1997;89:859–866. doi: 10.1016/s0092-8674(00)80271-8. [DOI] [PubMed] [Google Scholar]
- 5.Paushkin SV, Patel M, Furia BS, Peltz SW, Trotta CR. Identification of a human endonuclease complex reveals a link between tRNA splicing and pre-mRNA 3′ end formation. Cell. 2004;117:311–321. doi: 10.1016/s0092-8674(04)00342-3. [DOI] [PubMed] [Google Scholar]
- 6.de Vries H, Ruegsegger U, Hubner W, Friedlein A, Langen H, Keller W. Human pre-mRNA cleavage factor II(m) contains homologs of yeast proteins and bridges two other cleavage factors. EMBO J. 2000;19:5895–5904. doi: 10.1093/emboj/19.21.5895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weitzer S, Martinez J. The human RNA kinase hClp1 is active on 3′ transfer RNA exons and short interfering RNAs. Nature. 2007;447:222–226. doi: 10.1038/nature05777. [DOI] [PubMed] [Google Scholar]
- 8.Budde BS, Namavar Y, Barth PG, Poll-The BT, Nurnberg G, Becker C, van Ruissen F, Weterman MA, Fluiter K, te Beek ET, et al. tRNA splicing endonuclease mutations cause pontocerebellar hypoplasia. Nat. Genet. 2008;40:1113–1118. doi: 10.1038/ng.204. [DOI] [PubMed] [Google Scholar]
- 9.Calvin K, Li H. RNA-splicing endonuclease structure and function. Cell Mol. Life Sci. 2008;65:1176–1185. doi: 10.1007/s00018-008-7393-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burggraf S, Larsen N, Woese CR, Stetter KO. An intron within the 16S ribosomal RNA gene of the archaeon Pyrobaculum aerophilum. Proc. Natl Acad. Sci. USA. 1993;90:2547–2550. doi: 10.1073/pnas.90.6.2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Watanabe Y, Yokobori S, Inaba T, Yamagishi A, Oshima T, Kawarabayasi Y, Kikuchi H, Kita K. Introns in protein-coding genes in Archaea. FEBS Lett. 2002;510:27–30. doi: 10.1016/s0014-5793(01)03219-7. [DOI] [PubMed] [Google Scholar]
- 12.Yoshinari S, Itoh T, Hallam SJ, DeLong EF, Yokobori S, Yamagishi A, Oshima T, Kita K, Watanabe Y. Archaeal pre-mRNA splicing: a connection to hetero-oligomeric splicing endonuclease. Biochem. Biophys. Res. Commun. 2006;346:1024–1032. doi: 10.1016/j.bbrc.2006.06.011. [DOI] [PubMed] [Google Scholar]
- 13.Tang TH, Rozhdestvensky TS, d'Orval BC, Bortolin ML, Huber H, Charpentier B, Branlant C, Bachellerie JP, Brosius J, Huttenhofer A. RNomics in Archaea reveals a further link between splicing of archaeal introns and rRNA processing. Nucleic Acids Res. 2002;30:921–930. doi: 10.1093/nar/30.4.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dennis PP, Ziesche S, Mylvaganam S. Transcription analysis of two disparate rRNA operons in the halophilic archaeon Haloarcula marismortui. J. Bacteriol. 1998;180:4804–4813. doi: 10.1128/jb.180.18.4804-4813.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chant J, Dennis P. Archaebacteria: transcription and processing of ribosomal RNA sequences in Halobacterium cutirubrum. EMBO J. 1986;5:1091–1097. doi: 10.1002/j.1460-2075.1986.tb04327.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Russell AG, Ebhardt H, Dennis PP. Substrate requirements for a novel archaeal endonuclease that cleaves within the 5′ external transcribed spacer of Sulfolobus acidocaldarius precursor rRNA. Genetics. 1999;152:1373–1385. doi: 10.1093/genetics/152.4.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kleman-Leyer K, Armbruster DW, Daniels CJ. Properties of H. volcanii tRNA intron endonuclease reveal a relationship between the archaeal and eucaryal tRNA intron processing systems. Cell. 1997;89:839–847. doi: 10.1016/s0092-8674(00)80269-x. [DOI] [PubMed] [Google Scholar]
- 18.Bujnicki JM, Rychlewski L. Prediction of a common fold for all four subunits of the yeast tRNA splicing endonuclease: implications for the evolution of the EndA/Sen family. FEBS Lett. 2000;486:328–329. doi: 10.1016/s0014-5793(00)02322-x. [DOI] [PubMed] [Google Scholar]
- 19.Li H, Trotta CR, Abelson J. Crystal structure and evolution of a transfer RNA splicing enzyme. Science. 1998;280:279–284. doi: 10.1126/science.280.5361.279. [DOI] [PubMed] [Google Scholar]
- 20.Li H, Abelson J. Crystal structure of a dimeric archaeal splicing endonuclease. J. Mol. Biol. 2000;302:639–648. doi: 10.1006/jmbi.2000.3941. [DOI] [PubMed] [Google Scholar]
- 21.Kim YK, Mizutani K, Rhee KH, Nam KH, Lee WH, Lee EH, Kim EE, Park SY, Hwang KY. Structural and mutational analysis of tRNA intron-splicing endonuclease from Thermoplasma acidophilum DSM 1728: catalytic mechanism of tRNA intron-splicing endonucleases. J. Bacteriol. 2007;189:8339–8346. doi: 10.1128/JB.00713-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tocchini-Valentini GD, Fruscoloni P, Tocchini-Valentini GP. Structure, function, and evolution of the tRNA endonucleases of Archaea: An example of subfunctionalization. Proc. Natl Acad. Sci. USA. 2005;102:8933–8938. doi: 10.1073/pnas.0502350102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yoshinari S, Fujita S, Masui R, Kuramitsu S, Yokobori S, Kita K, Watanabe Y. Functional reconstitution of a crenarchaeal splicing endonuclease in vitro. Biochem. Biophys. Res. Commun. 2005;334:1254–1259. doi: 10.1016/j.bbrc.2005.07.023. [DOI] [PubMed] [Google Scholar]
- 24.Calvin K, Hall MD, Xu F, Xue S, Li H. Structural characterization of the catalytic subunit of a novel RNA splicing endonuclease. J. Mol. Biol. 2005;353:952–960. doi: 10.1016/j.jmb.2005.09.035. [DOI] [PubMed] [Google Scholar]
- 25.Randau L, Calvin K, Hall M, Yuan J, Podar M, Li H, Soll D. The heteromeric Nanoarchaeum equitans splicing endonuclease cleaves noncanonical bulge-helix-bulge motifs of joined tRNA halves. Proc. Natl Acad. Sci. USA. 2005;102:17934–17939. doi: 10.1073/pnas.0509197102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stoscheck CM. Quantitation of protein. Methods Enzymol. 1990;182:50–68. doi: 10.1016/0076-6879(90)82008-p. [DOI] [PubMed] [Google Scholar]
- 27.Brown PH, Schuck P. A new adaptive grid-size algorithm for the simulation of sedimentation velocity profiles in analytical ultracentrifugation. Comput. Phys. Commun. 2008;178:105–120. doi: 10.1016/j.cpc.2007.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jancarik J, Scott WG, Milligan DL, Koshland DE, Jr., Kim SH. Crystallization and preliminary X-ray diffraction study of the ligand-binding domain of the bacterial chemotaxis-mediating aspartate receptor of Salmonella typhimurium. J. Mol. Biol. 1991;221:31–34. doi: 10.1016/0022-2836(91)80198-4. [DOI] [PubMed] [Google Scholar]
- 29.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 30.Sheldrick GM. A short history of SHELX. Acta Crystallogr., A. 2008;64:112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- 31.Vonrhein C, Blanc E, Roversi P, Bricogne G. Automated structure solution with autoSHARP. Methods Mol. Biol. 2007;364:215–230. doi: 10.1385/1-59745-266-1:215. [DOI] [PubMed] [Google Scholar]
- 32.Abrahams JP, Leslie AG. Methods used in the structure determination of bovine mitochondrial F1 ATPase. Acta Crystallogr., D, Biol. Crystallogr. 1996;52:30–42. doi: 10.1107/S0907444995008754. [DOI] [PubMed] [Google Scholar]
- 33.Perrakis A, Morris R, Lamzin VS. Automated protein model building combined with iterative structure refinement. Nat. Struct. Biol. 1999;6:458–463. doi: 10.1038/8263. [DOI] [PubMed] [Google Scholar]
- 34.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr., D, Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 35.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr., D, Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 36.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Cryst. 1993;26:283–291. [Google Scholar]
- 37.Tocchini-Valentini GD, Fruscoloni P, Tocchini-Valentini GP. The dawn of dominance by the mature domain in tRNA splicing. Proc. Natl Acad. Sci. USA. 2007;104:12300–12305. doi: 10.1073/pnas.0705537104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fitz-Gibbon ST, Ladner H, Kim UJ, Stetter KO, Simon MI, Miller JH. Genome sequence of the hyperthermophilic crenarchaeon Pyrobaculum aerophilum. Proc. Natl Acad. Sci. USA. 2002;99:984–989. doi: 10.1073/pnas.241636498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lykke-Andersen J, Garrett RA. RNA-protein interactions of an archaeal homotetrameric splicing endoribonuclease with an exceptional evolutionary history. EMBO J. 1997;16:6290–6300. doi: 10.1093/emboj/16.20.6290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xue S, Calvin K, Li H. RNA recognition and cleavage by a splicing endonuclease. Science. 2006;312:906–910. doi: 10.1126/science.1126629. [DOI] [PubMed] [Google Scholar]
- 41.Tocchini-Valentini GD, Fruscoloni P, Tocchini-Valentini GP. Coevolution of tRNA intron motifs and tRNA endonuclease architecture in Archaea. Proc. Natl Acad. Sci. USA. 2005;102:15418–15422. doi: 10.1073/pnas.0506750102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sugahara J, Kikuta K, Fujishima K, Yachie N, Tomita M, Kanai A. Comprehensive analysis of archaeal tRNA genes reveals rapid increase of tRNA introns in the order thermoproteales. Mol. Biol. Evol. 2008;25:2709–2716. doi: 10.1093/molbev/msn216. [DOI] [PubMed] [Google Scholar]
- 43.Thompson JD, Gibson TJ, Higgins DG. Multiple sequence alignment using ClustalW and ClustalX. Curr. Protoc. Bioinformatics. 2002 doi: 10.1002/0471250953.bi0203s00. Chapter 2, Unit 23. [DOI] [PubMed] [Google Scholar]