Abstract
The androgen receptor (AR) plays a pivotal role in the onset and progression of prostate cancer by promoting cellular proliferation. Recent studies suggest AR is a master regulator of G1-S progression and possibly a licensing factor for DNA replication yet the mechanisms remain poorly defined. Here we report that AR targets the human Cdc6 gene for transcriptional regulation. Cdc6 is an essential regulator of DNA replication in eukaryotic cells and its mRNA expression is inversely modulated by androgen or antiandrogen treatment in androgen-sensitive prostate cancer cells. AR binds at a distinct androgen-response element (ARE) in the Cdc6 promoter that is functionally required for androgen-dependent Cdc6 transcription. We found that peak AR occupancy at the novel ARE occurs during the G1/S phase concomitant with peak Cdc6 mRNA expression. We also identified several of the coactivators and corepressors involved in AR-dependent Cdc6 transcriptional regulation in vivo and further characterized ligand-induced alterations in histone acetylation and methylation at the Cdc6 promoter. Significantly, AR silencing in prostate cancer cells markedly decreases Cdc6 expression and androgen-dependent cellular proliferation. Collectively, our results suggest that Cdc6 is a key regulatory target for AR and provide new insights into the mechanisms of prostate cancer cell proliferation.
INTRODUCTION
Androgens are steroid hormones responsible for the development and functional maintenance of male reproductive and accessory sex tissues. They exert their physiologic actions by binding to the androgen receptor (AR), a 110-kDa member of the nuclear receptor family of ligand-activated transcription factors (1). AR mediates androgen action by binding to specific DNA sequences termed androgen response elements (ARE) found within promoter or enhancer regions of AR-target genes (2–5). When bound with androgen, AR can activate target gene transcription by recruiting distinct coregulatory factors including enzymes that covalently modify histones and remodel chromatin (6,7), as well as the Mediator complex that directly interfaces with the RNA polymerase II (RNA pol II) basal machinery (8,9). When bound with antiandrogenic compounds, AR can repress target gene transcription by recruiting negative coregulatory factors termed corepressors (6,7). Although numerous AR coactivators and corepressors have been reported and characterized, many of the genes directly bound and regulated by AR in vivo remain poorly defined.
Significantly, AR plays a pivotal role in the onset and progression of prostate cancer by promoting the growth and proliferation of prostate cancer cells (1,10,11). Mechanistic investigations have revealed that AR acts as a master regulator of G1-S phase progression in androgen-dependent prostate cancer cells (12) and that AR protein is degraded at mitosis during each cell cycle (13). These findings suggest that AR may be acting as a licensing factor for DNA replication in androgen-sensitive prostate cancer cells, and that mitotic AR degradation is required to license a new round of DNA replication. Treatment options for prostate cancer include androgen-ablation therapy that initially triggers apoptosis or cell-cycle arrest of prostate cancer cells (14–16). Paradoxically, nearly all invasive or metastatic prostate cancers eventually progress into a fatal androgen-independent disease, yet most of these cancers continue to express AR and remain dependent on AR for growth and survival (1,10,11). Therefore, identifying the specific genes regulated by AR will be critical for understanding the mechanisms of androgen-dependent and -independent prostate cancer cell growth and proliferation.
Cdc6 is an essential regulator of DNA replication in eukaryotic cells (17). Along with a subset of other key replication factors, Cdc6 helps form a pre-replication complex at the origins of DNA replication early in G1 phase thereby ‘licensing’ these sites to bind DNA polymerase and initiate DNA replication during S phase (18,19). The expression and functional activity of Cdc6 is tightly regulated in a cell-cycle-dependent manner thus ensuring that the entire genome is replicated only once in each cell division. Indeed, Cdc6 is considered an oncogene and its deregulated expression can lead to under- or over-replication, DNA damage and genetic instability (18). In mammalian cells, Cdc6 expression peaks during the G1/S transition and is transcriptionally regulated in a cell-cycle- and E2F-dependent manner (20–22). Given AR's presumptive role as a licensing factor for DNA replication, it has been proposed that Cdc6 and possibly other replication factors might be regulatory targets for AR-signaling pathways (13). Interestingly, when synchronized prostate cancer LNCaP cells are treated with the antiandrogenic compound bicalutamide (Casodex), the cells fail to enter S phase and concomitantly downregulate Cdc6 mRNA expression (23). Furthermore, AR binds at the human Cdc6 promoter in vivo and androgens were found to regulate Cdc6 gene expression in AR-positive prostate cancer cells and xenografts (24,25).
In this study, we investigated whether AR targets the human Cdc6 gene for transcriptional regulation in prostate cancer cells in a cell-cycle-dependent manner. Using androgen-sensitive LNCaP cells, we found that Cdc6 mRNA and protein expression is activated or repressed in the presence of androgen or antiandrogen, respectively. We identified a 15 bp palindromic ARE in the Cdc6 promoter (−734 bp upstream of the transcription start site) and show that AR occupies this site in vitro and in vivo. Mutagenesis of the ARE abolishes AR binding as well as androgen-dependent Cdc6 transcription. Intriguingly, and consistent with its presumptive role as a DNA replication licensing factor, we show that peak occupancy of AR at the Cdc6 promoter occurs during the G1/S phase of the cell cycle, concomitant with peak Cdc6 mRNA expression. Silencing AR expression markedly decreases both Cdc6 expression and androgen-dependent cellular proliferation. Significantly, we also identified several specific coactivators and corepressors involved in AR-dependent Cdc6 transcriptional regulation and further characterized androgen- and antiandrogen-induced alterations in histone H3 acetylation and methylation patterns at the Cdc6 promoter. Collectively, our results suggest that Cdc6 is a key regulatory target gene in androgen-responsive prostate cells and may have important implications for prostate cancer cell growth and proliferation.
MATERIALS AND METHODS
Cell culture
LNCaP, DU145 and PC3 cells were obtained from the American Type Culture Collection (Manassas, VA). 1532T-f:AR cells were generated and cultured as described previously (26). LNCaP cells were routinely maintained in RPMI 1640 (Invitrogen, Carlsbad, CA) with 10% fetal bovine serum (FBS; Gemini Bioproducts) along with penicillin and streptomycin (Invitrogen). DU145 and PC3 cells were grown in DMEM with 10% FBS and penicillin/streptomycin. In the androgen starvation experiments, cells were grown in phenol red-free medium containing charcoal/dextran-stripped FBS (CDS–FBS, Gemini Bioproducts). All cells were maintained in a humidified incubator at 37°C and 5% CO2.
Antibodies and reagents
Specific antibodies against Cdc6, AR, MED1, MED6, MED14, MED17, SRC1, SRC3, NCoR, SMRT, HDAC1, HDAC2, HDAC3 and α-tubulin were all from Santa Cruz Biotechnology (Santa Cruz, CA). Antibodies against H3R17-2me, H3K4-2me and unmodified Histone 3 were from Abcam (Cambridge, MA) and antibodies against acetylated-H3 and H3K9-3me were from Millipore/Upstate Biotech (Billerica, MA). Horseradish peroxidase-conjugated anti-rabbit IgG and anti-mouse IgG were from Cell Signaling (Beverly, MA). Enhanced Chemiluminescence (ECL) reagents were from Amersham Pharmacia/GE Healthcare (Piscataway, NJ). The Luciferase Assay kit was from Promega (Madison, WI) and the dihydrotestosterone (DHT), nocodazole and R1881 were all from Sigma (St Louis, MO). Casodex (Bicalutamide) was generously provided by AstraZeneca (United Kingdom).
RT–PCR analysis
Total RNA was extracted from prostate cancer cell lines using TRIzol reagent (Invitrogen). One microgram of total RNA was incubated with reverse transcriptase (Invitrogen) and 200 mM deoxynucleoside triphosphates (dNTPs) to generate first-strand cDNA and then amplified by PCR using Fast Start Taq polymerase (Roche) with primers specific for either Cdc6 (24) or β-actin (27) for 25 cycles. The PCR products were then analyzed on 1.5% agarose gel stained with ethidium bromide.
Real-time PCR
PCR was performed using the Opticon Continuous Fluorescence Detection System (MJ Research) using a SYBR® Green PCR kit (Invitrogen). Human β-actin was used as an internal control for real time RT–PCR analyses. Non-specific rabbit IgG was used as an internal control for real time ChIP.
ChIP analyses
LNCaP cells were cultured in CDS–FBS containing media for 3 days and then treated with DHT (10 nM) for 4 h. For the Casodex treatment, LNCaP cells were cultured in 10% FBS RPMI media and then treated with Casodex (30 μM) for 2 h. Antibodies specific for AR, coactivators, corepressors and covalently modified histones were then used to immunoprecipitate formaldehyde cross-linked chromatin–protein complexes as outlined in detail (28,29). The immunoprecipitated DNA was then analyzed via semi-quantitative and real time PCR using primers spanning the Cdc6 promoter region: Primer set A: forward, 5′-gaa acc cta gtg ttt cgc cat aaa ag-3′ (−825 bp), and reverse, 5′-ggt aaa gtt cta cac acc tat ata aag-3′(−604 bp); Primer set B: forward, 5′-gcc ttc acg aaa tgt act cca c-3′ (−1394 bp), and reverse, 5′-caa cta gta agt gga aga gct ag-3′ (−1254 bp); Primer set C: forward, 5′-gct ctc tca ttg gct gta act c-3′ (−149 bp), and reverse, 5′-cgc tcg cgc caa atc cga atg-3′ (+5 bp); Primer set D: forward, 5′-gag act ata act cta cag att g-3′ (+14976 bp), and reverse, 5′-ctt gaa aca agt ggc ttc atc-3′ (+15152 bp). All ChIP experiments were carried out at least three times. Image processing and quantitation of the semi-quantitative PCR data was performed using the Quantity One software (Bio-Rad) (Supplementary Figure S1).
Plasmids
The pGL3-Cdc6-1.7-kb promoter-Luc plasmid was provided by Dr Joseph Nevins (Duke) and has been described previously (20). The pSG5-AR construct was provided by Dr Frank Claessens (Katholieke Universiteit Leuven), the pRSV-TRβ vector was from Dr Herb Samuels (NYU) and the pCMV-GR construct was from Dr Michael Garabedian (NYU). The pGL2-Cdc6-ARE construct was generated by PCR by amplifying the Cdc6 promoter region containing the putative ARE and GATA sites (bps −781 to −575) using primers (forward) 5′-ggg gta ccc ata att ccc tcc cca tga tgt gtg g-3′ and (reverse) 5′-gaa gat ctc tcc tga tgg ctg aac tag tga ttt tta tgg-3′ and then subcloning the amplified fragment into pGL2-Basic (Promega, Madison, WI). The pGL2-Cdc6-AREmt construct was created via PCR-generated mutagenesis of the pGL2-Cdc6-ARE construct as described (30) using primers (forward) 5′-gaa gat gca caA GGc ata ata ttc tta ggt tg-3′ and (reverse) 5′-cct aag aat att atg CCT tgt gca tct tc-3′. The pGL2-Cdc6-GATA-mt.1 and pGL2-Cdc6-GATA-mt.2 constructs were generated via PCR-generated mutagenesis using primers (forward) 5′-gca caa atc aag act tac tat gat gaa gc-3 and (reverse) 5′-gct tca tca tag taa gtc ttg att tgt gc-3′ for mutant 1 and primers (forward) 5′-gca caa atc aag aac cac tat gat gaa gc-3′ and (reverse) 5′-gct tca tca tag tgg ttc ttg att tgt gc-3′ for mutant 2.
Luciferase reporter gene assays
LNCaP cells were cultured in CDS–FBS containing media for 3 days and then seeded (1 × 105 cells) in 12-well plates and transfected with 100 ng pGL3-Cdc6-1.7 kb or empty pGL3 control using Lipofectamine PLUS reagent (Invitrogen, Carlsbad, CA). In separate experiments, 500 ng of either pGL2-Cdc6-ARE, pGL2-Cdc6-AREmt, pGL2-Cdc6-GATA-mt.1, pGL2-Cdc6-GATA-mt.2, or empty pGL2 control were transfected via Lipofectamine PLUS reagent. Three hours post-transfection, the medium was replaced with fresh media containing or lacking R1881 (10 nM final) for an additional 24 h. For experiments involving addition of Casodex, cells were grown in normal FBS prior to transfection. DU145 cells grown in normal 10% FBS were seeded (1 × 105 cells) in 12-well plates and cotransfected with 500 ng of pGL2-Cdc6-ARE and 500 ng of either pSG5-AR, pCMV-GR, pRSV-TRβ, or their corresponding empty vectors via Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA). Three hours post-transfection, the medium was replaced with fresh media containing or lacking R1881 (10 nM), Dexamethasone (10 nM final), or T3 (100 nM) for an additional 24 h. Cells were harvested and equivalent amounts of protein were assayed for luciferase activity using an assay kit (Promega) and a luminometer. Luciferase values were normalized by using a β-galactosidase (pSV-ßgal) (Promega) expression vector as internal control.
Electromobility shift assay
The wild-type Cdc6 promoter ARE oligo 5′GAA GAT GCA CAG AAC ATA ATA TTC TTA GGT TG3′ and its complementary strand were annealed generating a double-stranded template with protruding BglII ends. Similarly, the mutant ARE oligo 5′GGT AAG AAG ATG CAC AAT ATT CTT AGG TTG3′ and its complement were annealed generating a double-stranded template with protruding BglII ends. As a positive control, a double-stranded MMTV long terminal repeat promoter ARE template was generated as described previously (26). The double-stranded AREs were labeled by filling in with [α-32P]dATP (50 µCi; Amersham Biosciences) and Klenow enzyme. To purify the full-length AR protein used for EMSA, FLAG-tagged human AR (f:AR) was immunopurified from a stable 1532T-f:AR cell line using anti-FLAG M2 antibodies coupled to agarose beads (Sigma) as described previously (26). Purified f:AR protein was incubated for 15 min at room temperature in binding buffer containing 10 mM Tris–Cl (pH 7.9), 50 mM KCl, 1 mM dithiothreitol, 10% glycerol, 1 µg/µl bovine serum albumin, 0.5 µg of poly (dI-dC), 1 mM EDTA, 0.1% Nonidet P-40 along with 2 ng of 32P-labeled double-stranded ARE probe. The reactions were electrophoresed in a prerun 5% polyacrylamide gel, 0.5× Tris borate–EDTA at 100 V for 3–4 h. The gel was then dried and autoradiographed.
RNA interference
AR siRNA was generated using the Silencer siRNA Construction Kit (Ambion) using the specific target sequence 5′-GAC CTA CCG AGG AGC TTT C-3′ as previously outlined (31). A scrambled non-specific siRNA smart pool was from Dharmacon Research, Inc., as previously described (27). The AR siRNA (200 nM final) was transfected into LNCaP cells using Lipofectamine with Plus reagent.
Cell proliferation assay
LNCaP cells were androgen-starved in RPMI media containing 10% CDS–FBS for 48 h and then seeded (1 × 105 cells) in 12-well plates and transfected with AR siRNA or a non-specific control siRNA (200 nM final). Forty-eight hours post-transfection, the cells were trypsinized and seeded (5 × 104) in 12-well plates and allowed to proliferate for an additional 48 h in CDS–FBS with or without 10 nM DHT or 30 µM Casodex. Cell proliferation was measured by manually counting cell numbers. Experiments were performed in quadruplicate.
LNCaP cell synchronization
LNCaP cells cultured in normal serum were seeded 5 × 105 cells per 6-well plate and then transfected with AR siRNA or a non-specific control siRNA (200 nM final). The cells were either arrested at the border of G1/S using the double-thymidine-block methodology as described previously (32) or arrested in G2/M by treating the cells with 100 ng/ml nocodazole for 18 h. Then cells were then harvested for RT–PCR, immunoblot or ChIP analyses as described above. Synchronization was verified by fixing and staining the cells with propidium iodide (20 µg/ml) and then analyzing them using a Cytomics FC500 flow cytometer (Beckman Coulter Inc.).
RESULTS
Cdc6 expression in LNCaP cells is modulated in an androgen- and antiandrogen-dependent fashion
Several previous studies indirectly link AR-signaling pathways with Cdc6 expression in androgen-sensitive prostate cancer cells. For example, when AR-expressing prostate cancer LNCaP cells are synchronized in the G0/G1 phase of the cell cycle and then treated with the antiandrogenic compound Casodex, the cells fail to enter S phase and concomitantly downregulate Cdc6 mRNA expression (23). Similarly, a significant loss of transcription efficiency of a human Cdc6 promoter-reporter gene was noted in AR-negative prostate cancer cells as compared with that observed in AR-positive prostate cells (24). Moreover, previous LNCaP gene array studies have shown that Cdc6 mRNA expression increases in the presence of androgen (33) and decreases in the presence of the antiandrogen (23). Indeed, we performed human gene microarray analyses with androgen-sensitive LNCaP cells and also identified Cdc6 as an androgen-responsive gene (data not shown).
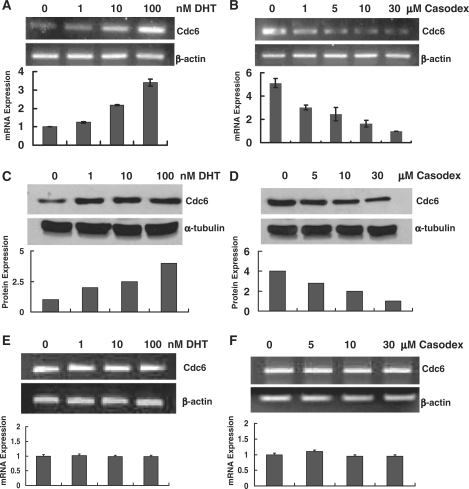
In an effort to validate our microarray data linking androgen stimulation with Cdc6 expression, RNA was extracted from LNCaP cells treated with either androgen (dihydrotestosterone or DHT) or antiandrogen (Casodex) and then analyzed by RT–PCR using primers specific for the Cdc6 gene. As shown in Figure 1A and B, and in agreement with the previous microarray studies, Cdc6 mRNA increased in a DHT dose-dependent manner as much as 3-fold, whereas Cdc6 mRNA decreased in the presence of increasing concentrations of Casodex as much as 5-fold. Immunoblot analyses showed that DHT- or Casodex-induced increases or decreases in Cdc6 mRNA were accompanied by a corresponding increase or decrease in protein expression (Figure 1C and D). By contrast, when AR-negative DU145 prostate cancer cells were treated with either DHT or Casodex, no significant increase or decrease in Cdc6 mRNA levels was detected (Figure 1E and F). Taken together, our findings show that androgens and antiandrogens modulate Cdc6 mRNA and protein expression in androgen-sensitive prostate cancer cells, but not in prostate cancer cells lacking AR expression.
Figure 1.
Cdc6 expression in androgen-sensitive prostate cancer cells is modulated in an androgen- and antiandrogen-dependent manner. (A and C) LNCaP cells were androgen-starved for 72 h and then treated with different concentrations of DHT 8 h. (B and D) Androgen-starved LNCaP cells were first cultured in CDS–FBS media with 10 nM R1881 for 24 h to elevate basal Cdc6 expression, and then were treated with different concentrations of Casodex for 8 h. Total RNA was analyzed by RT–PCR using primers specific for Cdc6 and β-actin (A and B). Bar graph represents results as the mean ± S.E. of triplicate reactions. Twenty-five micrograms of whole cell extract was probed by immunoblot with anti-Cdc6 and anti-tubulin antibodies (C and D). (E and F) DU145 cells were androgen-starved for 3 days and then treated with different concentrations of DHT and Casodex for 8 h. Total RNA was analyzed by RT–PCR using primers specific for Cdc6 and β-actin as outlined above.
Identification of an AR-binding site in the Cdc6 promoter
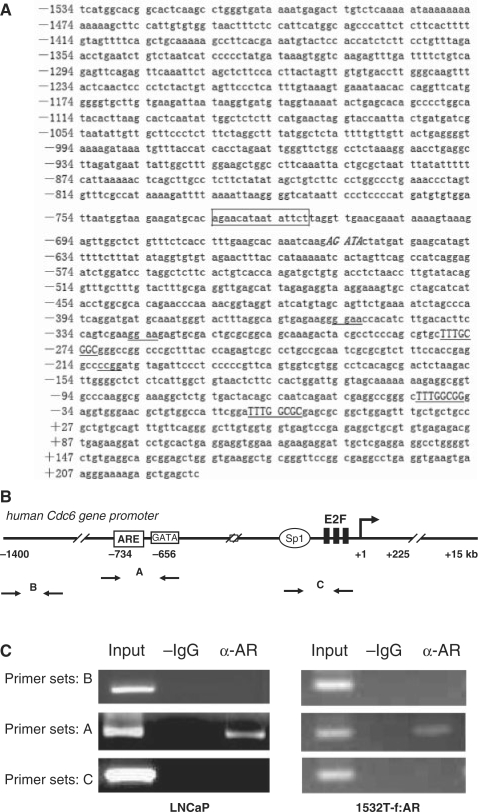
In view of the importance of Cdc6 in regulating eukaryotic DNA replication, we were interested in determining whether AR directly targets the Cdc6 gene for transcriptional regulation. Interestingly, a search of the Cdc6 promoter region for potential AR-binding sites revealed the presence of a canonical class I steroid hormone response element (AGAACAnnnTATTCT) (34) spanning positions −734 to −720-bp upstream of the transcription start site (Figure 2A). It has been reported that binding sites for the GATA and ETS families of transcription factors are enriched proximal to cognate AR-binding sites in the genome and may play cooperative roles in mediating an androgen response (3,5,35). Notably, a GATA element was detected in the Cdc6 gene promoter just downstream of the putative ARE at position −656 bp and three ETS-binding sites were found at positions −355, −326 and −211 bp (Figure 2A). To determine whether endogenously expressed AR in LNCaP cells directly associates with the Cdc6 promoter at or near to the putative ARE region in vivo, we performed chromatin-immunoprecipitation (ChIP) assays using an anti-AR antibody and PCR primer sets corresponding to different regions of the Cdc6 promoter (Figure 2B). Utilizing LNCaP cells cultured in normal serum, we detected AR occupancy at the Cdc6 gene-promoter region containing the putative ARE, but not at an upstream-promoter region, nor at a downstream region containing the E2F-binding elements (Figure 2C). Identical results were observed when ChIP was performed using prostate cancer 1532T cells stably transfected with ectopic FLAG-tagged AR (1532T-f:AR) (26) (Figure 2C).
Figure 2.
AR binds at the Cdc6 gene in vivo in a promoter region containing a consensus ARE. (A) DNA sequence of the human Cdc6 promoter region. The ARE is boxed, E2F sites are capitalized and underlined, the GATA site is capitalized and in italics, and the ETS sites are underlined. (B) Schematic representation of the human Cdc6 promoter showing relative location of three PCR primer sets (A, B and C) used for ChIP analyses. (C) Chromatin prepared from LNCaP and 1532T-f:AR cells cultured in normal serum was used for ChIP along with anti-AR antibodies. Semi-quantitative PCR was performed using the three primer sets shown in (B).
We next tested whether recombinant AR can specifically bind to the Cdc6 ARE in vitro by performing DNA electromobility shift assays. Accordingly, full-length human FLAG-AR (f:AR) was purified from 1532T-f:AR cells as described (26) and then incubated with a radiolabeled Cdc6 ARE. A mutated Cdc6 ARE and a MMTV long terminal repeat promoter ARE were used as negative and positive controls, respectively. As shown in Figure 3A and B, f:AR purified from DHT-treated 1532T-f:AR cells bound efficiently with the wild-type Cdc6 ARE but not with mutated Cdc6 ARE, whereas f:AR purified from androgen-starved cells bound only weakly to the wild-type Cdc6 ARE. Addition of an anti-AR antibody confirmed the presence of AR in the protein–DNA complex and as previously reported, dramatically enhanced the AR:ARE interaction (36) (Figure 3C). The binding specificity of f:AR for the Cdc6 ARE was confirmed by the ability of a molar excess of unlabeled wild-type ARE to compete for and inhibit binding with the radiolabeled ARE (Figure 3D). To confirm that endogenously expressed AR from prostate cancer cells directly binds to the Cdc6 ARE in vitro, we performed DNA electromobility assays using nuclear extract prepared from either AR-positive LNCaP cells or AR-negative PC3 cells. As expected, LNCaP nuclear extract exhibited strong Cdc6 ARE-binding activity, whereas no binding activity was observed with the PC3 nuclear extract (Figure 3E). In sum, these results show that AR directly binds to ARE in the Cdc6 gene promoter.
Figure 3.
AR directly binds to the Cdc6 ARE in vitro in the presence of ligand. (A–D) Full-length human FLAG-AR (f:AR) purified from DHT-cultured prostate cancer 1532T cells stably expressing ectopic f:AR (26) was incubated with a 32P-radiolabeled Cdc6 ARE and then assayed by EMSA (see Methods section). A mutated Cdc6 ARE and a MMTV long terminal repeat promoter ARE were used as negative and positive controls, respectively. In (B), equal amounts of f:AR purified from either androgen-starved (−DHT) or DHT-treated (+DHT) 1532T-f:AR cells was used. In (C), anti-AR antibodies were added to the binding reaction as indicated. In (D), a molar excess of cold unlabeled wild-type ARE or mutated ARE was added to binding reactions as indicated. (E) Nuclear extract prepared from DHT-treated LNCaP or PC3 cells was incubated with the wild-type 32P-radiolabeled Cdc6 ARE and assayed by EMSA as described above.
The Cdc6 promoter ARE confers androgen-dependent transcriptional activation
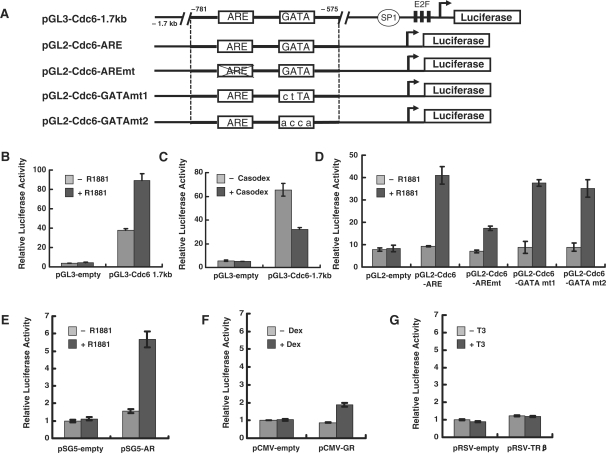
We next sought to determine whether the ARE in the Cdc6 gene promoter is responsible for the androgen-induced increase in Cdc6 mRNA observed in androgen-sensitive prostate cancer cells (Figure 1). To address this question, we first measured transcription from a Cdc6 promoter-luciferase reporter gene (pGL3-Cdc6-1.7 kb) containing 1.7 kb of the native human Cdc6 promoter region (−1700 to +7) (20). Androgen-starved LNCaP cells were transfected with either pGL3-Cdc6-1.7 kb or an empty pGL3 control and then cultured with or without the synthetic androgen R1881 (Figure 4B). Basal expression from the pGL3-Cdc6-1.7 kb construct (minus R1881) was considerably higher than that observed with the pGL3 control, presumably due to the presence of Sp1 and E2F sites in the Cdc6 promoter construct near the transcription start site. Consistent with the idea that the ARE region is required for androgen-dependent Cdc6 transcription, addition of R1881 stimulated transcription from pGL3-Cdc6-1.7 kb greater than 2-fold (Figure 4B), whereas addition of the antiandrogen Casodex reduced transcription by nearly 2-fold (Figure 4C). Moreover, androgen-dependent transcription from a Cdc6 promoter reporter construct (pGL2-Cdc6-ARE) containing only the ARE and GATA region (bps −781 to −575) was stimulated nearly 4-fold in LNCaP cells (Figure 4D) and in prostate cancer DU145 cells transiently transfected with AR (Figure 4E).
Figure 4.
The ARE in Cdc6 promoter confers androgen-dependent transcriptional activation. (A) Schematic representation of human Cdc6 promoter-luciferase constructs. (B) Seventy-two hours post-androgen starvation, LNCaP cells were transfected with the pGL3-Cdc6-1.7 kb reporter construct and then treated with or without 10 nM R1881 for 24 h. (C) LNCaP cells cultured in 10% FBS were transfected with pGL3-Cdc6-1.7 kb and then treated with 30 µM Casodex for 10 h. (D) Seventy-two hours post-androgen starvation, LNCaP cells were transfected with the indicated reporter constructs and then treated with or without 10 nM R1881 for 24 h. (E–G) DU145 cells cultured in 10% FBS were cotransfected with pGL2-Cdc6-ARE and the indicated nuclear receptor expression constructs and then treated with or without 10 nM R1881 (panel E) 10 nM dexamethasone (panel F) or 100 nM T3 (panel G) for 24 h. Whole cell extract were prepared and assayed for luciferase activity. Luciferase values are presented as the mean ± SE of triplicate transfections.
Given that the Cdc6 ARE comprises a consensus class I steroid hormone response element, we next asked whether the element might confer other steroid hormone responses. Consistent with this notion, transient overexpression of the glucocorticoid receptor in DU145 cells activated transcription from the pGL2-Cdc6-ARE reporter greater than 2-fold (Figure 4F), yet overexpression of the class II thyroid hormone receptor elicited no response in the presence of its cognate ligand (Figure 4G). To further examine whether the Cdc6 ARE region is directly responsible for the observed androgen-induced transcription, we carried out site-directed mutagenesis of the putative ARE (AGAACA→AAGCCA) within the context of the pGL2-Cdc6-ARE reporter construct (see ‘Materials and Methods’ section). As expected, mutation of the Cdc6 ARE significantly decreased R1881-induced transcription (Figure 4D). In contrast, two different site-directed mutations of the putative GATA site (mutant 1: AGATA→ACTTA and mutant 2: AGATA→AACCA) within the context of the pGL2-Cdc6-ARE reporter had only a modest inhibitory effect on R1881-stimulated transcription (Figure 4D). In summary, our data demonstrate that the novel ARE located within Cdc6 promoter (−734 bp to −720 bp) is functionally required for androgen-dependent Cdc6 gene expression, whereas a functional role for the downstream GATA site remains unclear (see ‘Discussion’ section).
Androgen- and antiandrogen-induced coregulator recruitment and covalent histone modifications at the Cdc6 gene promoter in prostate cancer cells
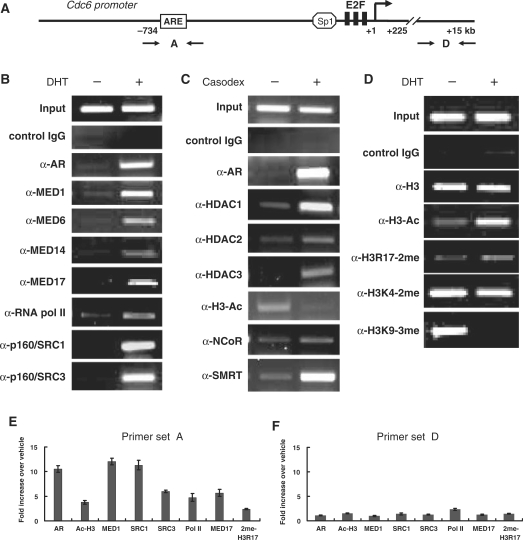
Our findings showing that androgens and antiandrogens modulate Cdc6 transcription, and that AR directly binds at a novel ARE in the Cdc6 gene promoter, strongly suggest that AR targets the Cdc6 gene for transcriptional regulation. Importantly, AR transcriptional activity in prostate cells is critically dependent on its interaction with accessory coregulatory factors and enzymes, and its subsequent recruitment of these cofactors to its respective target genes (6,7). To better understand how AR regulates Cdc6 gene expression, we performed ChIP assays with LNCaP cells treated with androgen (DHT) or antiandrogen (Casodex) using both semi-quantitative and real time PCR with primers spanning the ARE in Cdc6 promoter (Figure 5A). Of note, anti-AR antibodies revealed strong ligand-dependent AR occupancy at the Cdc6 ARE in the presence of both DHT and Casodex (Figure 5B, C and E). In order to identify specific cofactors recruited by AR to the Cdc6 promoter, antibodies against known NR transcriptional coactivators and corepressors were utilized. Furthermore, antibodies specific for acetylated or methylated histone H3 were employed to identify androgen- and antiandrogen-induced alterations in histone modifications at the Cdc6 promoter.
Figure 5.
Androgen- and antiandrogen-induced recruitment of coregulatory factors and covalent histone modifications at the Cdc6 promoter in vivo. (A) Schematic representation of the human Cdc6 promoter showing PCR primers used for ChIP. LNCaP cells were androgen-starved for 72 h and then treated with 10 nM DHT for 4 h (B and D), or cultured in normal serum for 72 h and then treated with 30 μM Casodex for 2 h (C). ChIP was then performed using the specific antibodies indicated on the left-hand side of the panels followed by semi-quantitative PCR. Anti-acetylated-histone H3 is indicated by α-H3-Ac; anti-dimethylated-histone H3-Arg17 is indicated by α-H3R17-2me; anti-dimethylated-histone H3-Lys4 is indicated by α-H3K4-2me; and anti-trimethylated-histone H3-Lys9 is indicated by α-H3K9-3me. Quantitative image processing of the PCR data can be found in Supplementary Figure S1. (E and F) ChIP was carried out as described in (B)–(D) followed by real-time PCR using the indicated PCR primer sets. Bar graph shows results as the mean ± SE of triplicate assays.
Mediator is a conserved transcriptional coregulatory complex that plays a vital role in nuclear hormone receptor (NR)-regulated gene expression (8,9). The complex binds to DNA-bound NRs and promotes the assembly and activation of RNA pol II and its associated factors at core promoters and may also play a role in facilitating the recruitment of histone-modifying enzymes. A single subunit of the complex termed MED1 targets Mediator to AR and other NRs in the presence of their cognate ligand. A recent study found that MED1 was indispensable for androgen-dependent transcription of the prostate specific antigen (PSA) gene in LNCaP cells and that RNAi silencing of MED1 was accompanied by a significant reduction in RNA pol II recruitment at the PSA gene promoter (37). Consistent with these findings, our lab recently found that MED1 silencing in LNCaP cells markedly inhibited androgen-dependent cellular proliferation and progression into the G2/M phase of the cell cycle (38).
Given the importance of MED1-Mediator in prostate cancer cell proliferation and cell-cycle progression, we first used anti-MED1 antibodies to precipitate formaldehyde cross-linked chromatin from DHT-stimulated LNCaP cells. Similar to previous LNCaP ChIP studies at the PSA gene promoter (28,37), we detected robust androgen-dependent recruitment of MED1 at the Cdc6 promoter (Figure 5B and E). Moreover, the androgen-dependent binding of other Mediator subunits (MED6, MED14 and MED17) support the notion that AR recruits the entire Mediator complex to the Cdc6 gene promoter. Significantly, and consistent with Mediator's role in facilitating the recruitment of RNA pol II (9), we also detected DHT-induced binding of RNA pol II at the Cdc6 promoter (Figure 5B and E).
Histone acetylation, a process dynamically controlled by histone acetyltransferases (HATs) and histone deacetylases (HDACs), has a profound impact on chromatin structure and consequently gene transcription (39). In general, acetylation of lysine residues on histones H3 and H4 facilitates transcriptional activation whereas deacetylation reverses this effect. The p160/SRC family of coactivators interact with NRs in a ligand-dependent manner and enhance their transcriptional transactivation (40). Two members of this family, p160/SRC-1 and p160/SRC-3, possess intrinsic HAT activity and bind and work together with other more potent HATs (CBP, p300 and PCAF) to acetylate histones. Interestingly, we detected strong DHT-dependent recruitment of both p160/SRC-1 and p160/SRC-3 at the Cdc6 promoter that was concomitantly associated with an increase in histone H3 acetylation (Figure 5B, D and E). By contrast, treatment of LNCaP cells with the antiandrogen Casodex triggered a pronounced decrease in histone H3 acetylation and was accompanied by the recruitment of HDACs-1, -2 and -3 (Figure 5C). The NR corepressors NCoR and SMRT, which directly bind to AR in the presence of antiandrogens, are thought to recruit HDACs to AR (6,7). Notably in this regard, we observed strong Casodex-dependent recruitment of SMRT, but not NCoR, to the Cdc6 promoter (Figure 5C). Although the recruitment of SMRT is consistent with transcriptional repression (6), the data contrasts earlier studies showing strong Casodex-dependent recruitment of both NCoR and SMRT at the PSA gene (41–43) and suggest that structural differences between the PSA and Cdc6 promoter/enhancer regions differentially influence AR–corepressor complex recruitment and binding.
Histone methylation has emerged as an equally important modification linked to both transcriptional activation and repression (39). In particular, methylation of histone H3-lysine 4 (H3K4) is associated with active transcription, while methylation of histone H3-lysine 9 (H3K9) is associated with gene silencing and heterochromatin formation (44). Furthermore, methylation of histone H3 arginine residues (R2, R17 and R26) are covalent modifications associated with ligand-dependent transcriptional activation by NRs (45). Strikingly, we observed a marked DHT-dependent decrease in H3K9 tri-methylation at the Cdc6 promoter as well as a modest increase in H3R17 di-methylation (Figure 5D and E), both marks consistent with Cdc6 transcriptional activation. By contrast, we failed to observe any significant increase in H3K4 methylation following addition of DHT. Collectively, these findings suggest that in addition to Mediator and HAT-containing coactivators, androgen-bound AR may also recruit H3K9-demethylase and H3-arginine-methyltransferase activities to the Cdc6 promoter (see ‘Discussion’ section). Conversely, antiandrogen-bound AR recruits SMRT–HDAC complexes that deacetylate histones at the Cdc6 promoter and facilitate gene silencing.
AR promotes cell-cycle-dependent Cdc6 mRNA expression in prostate cancer cells
AR is thought to act as a master regulator of G1-S phase progression in androgen-dependent prostate cancer cells (12). In most of the immortalized mammalian cell lines tested thus far, Cdc6 mRNA expression peaks during the G1/S phase (20–22). In view of our data showing that AR binds at Cdc6 promoter and regulates transcription in a ligand-dependent manner, we investigated whether AR is involved in regulating cell-cycle-dependent Cdc6 mRNA expression in prostate cancer cells. LNCaP cells grown in normal serum were first transfected with AR siRNA or a non-specific control siRNA and then synchronized in G1/S via thymidine block or in G2/M via nocodazole treatment (see ‘Materials and Methods’ section). Synchronization was confirmed by flow cytometry and untreated (unsynchronized) cells were used as controls (Figure 6A).
Figure 6.
AR promotes cell-cycle-dependent Cdc6 mRNA expression in prostate cancer cells. LNCaP cells cultured in normal serum were first transfected with AR siRNA or a non-specific control siRNA (200 nM final) and then synchronized in the G1/S phase via thymidine block or in the G2/M phase via nocodazole treatment. Untreated (unsynchronized) cells were used as controls. Cell-cycle distribution of the synchronized LNCaP cells was determined by flow cytometry. (A) The mean average of the percentage of cells in the sub-G0, G1/S and G2/M phases of the cell cycle from three separate synchronization experiments is shown. (B) Whole-cell extract was prepared and equivalent amounts of protein were probed by immunoblot using anti-AR or anti-α-tubulin antibodies. (C) Total RNA was analyzed by RT–PCR using primers specific for Cdc6 and β-actin. (D) RNA samples from (C) were analyzed using real-time PCR. Bar graph represents results as the mean ± SE of triplicate assays. (E) ChIP was were carried out as described in Figure 6 using PCR primer set A. Non-specific rabbit IgG was used as a negative control.
Similar to previous findings, we observed that Cdc6 mRNA levels in LNCaP cells are higher at G1/S than at G2/M (Figure 6C and D, left panels). AR protein levels also peak at G1/S relative to G2/M (Figure 6B, left panels) and when ChIP assays were performed using synchronized LNCaP cells, we detected a markedly higher occupancy of AR at the Cdc6 gene promoter at G1/S relative to G2/M (Figure 6E, left panels). Interestingly, when LNCaP cells were transfected with AR siRNA, there was a notable decrease in Cdc6 mRNA levels at G1/S (Figure 6C and D, right panels) that was accompanied by a sharp loss of AR occupancy at the Cdc6 promoter as determined by ChIP (Figure 6E, right panels). These findings correlate elevated AR protein expression and occupancy at the Cdc6 gene at G1/S phase with elevated levels of Cdc6 mRNA at G1/S, and consistent with its presumptive role as a licensing factor for the onset of S phase, suggest that AR promotes cell-cycle-dependent expression of Cdc6 mRNA.
AR silencing inhibits Cdc6 expression and cell-cycle progression
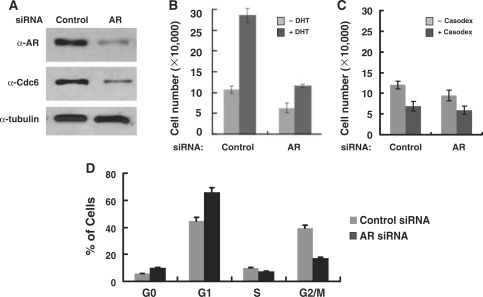
Given the importance of Cdc6 in regulating DNA replication, we asked whether loss of AR-mediated Cdc6 expression negatively effects androgen-dependent prostate cancer cell proliferation. Toward that end, equal numbers of androgen-starved LNCaP cells were transfected with AR siRNA or a non-specific control siRNA and then cultured with or without DHT for 2 days. In agreement with our earlier results, AR silencing concomitantly inhibits Cdc6 expression (Figure 7A) and in turn, dramatically decreases androgen-dependent cellular proliferation (Figure 7B). Indeed, loss of AR decreased the average DHT-induced LNCaP cell number by 61%, and consistent with a critical requirement for Cdc6 at the onset of S phase, loss of AR triggers cell-cycle arrest in the G0 and G1 phases (Figure 7D). Interestingly, we have also found that silencing of MED1, a key AR coactivator for Cdc6 transcription, likewise inhibited Cdc6 expression, decreased androgen-dependent proliferation, and triggered cell-cycle arrest in G1 phase (38). These results support the notion that AR and its associated coregulatory factors play an important role in licensing DNA replication by regulating the expression of Cdc6 and possibly other key replication factors.
Figure 7.
AR silencing inhibits Cdc6 expression and cell-cycle progression. (A) Whole cell extract prepared from LNCaP cells transfected with AR siRNA or a non-specific control siRNA (200 nM final) was probed by immunoblot with antibodies against AR, Cdc6 or α-tubulin. Equal numbers of androgen-starved LNCaP cells were transfected with AR siRNA or a non-specific control siRNA (200 nM final) and then cultured with or without DHT (10 nM) for 2 days (B), with or without Casodex (30 µM) for 2 days (C), after which time cell number was determined. Bar graph represents results as the mean ± SE of quadruplicate assays. (D) Androgen-starved LNCaP cells were transfected with AR siRNA or a non-specific control siRNA (200 nM final) and then cultured in DHT (10 nM) for 2 days after which time cell flow cytometry was performed.
DISCUSSION
In eukaryotic cells, Cdc6 plays a crucial functional role late in G1 phase by regulating the formation of pre-replication complexes that allow for the initiation of DNA synthesis at S phase. Indeed, whether or not Cdc6 is loaded on chromatin at the origins of DNA replication is believed to distinguish between cells that will continue through G1 to S phase versus those that will enter quiescence (17). Given its functional importance, Cdc6 expression is tightly regulated at the level of transcription, protein phosphorylation, and protein turnover (18,19). In mammalian cells, Cdc6 gene expression peaks at G1/S and is dependent on the presence of two E2F-binding sites in the proximal promoter which have been shown to confer both positive and negative transcriptional responses (20,21). G1 cyclins and cyclin dependent kinases have been proposed to activate promoter-bound E2F in mid to late G1 via the phosphorylation and displacement of inhibitory retinoblastoma (Rb) proteins (22).
Mallik et al. (25) first demonstrated the importance of an ARE in the Cdc6 promoter using luciferase assays, mutagenesis and ChIP in prostate cancer xenografts and cell lines. The study presented here extends this previous work with two important findings. First, we show for the first time that Cdc6 expression is under the regulatory control of AR in a cell-cycle dependent manner. Given the functional importance of Cdc6 in DNA replication, our results implicate the Cdc6 gene as a key regulatory target in AR-signaling pathways and provide new insights into AR's presumptive role as a master regulator of G1-S phase progression. Indeed, given that AR-signaling pathways become aberrantly hyperactivated in neoplastic prostate cells (10), it is conceivable that deregulated Cdc6 gene expression may promote prostate cancer cell growth and progression. Secondly, we have identified several specific coactivators and corepressors involved in AR-dependent Cdc6 transcriptional regulation and have further characterized androgen- and antiandrogen-induced alterations in histone H3 acetylation and methylation patterns at the Cdc6 promoter. Thus, the findings here have important implications for the fundamental molecular mechanisms used by AR to regulate transcription.
Considering the fact that other steroid NRs (e.g. the progesterone and glucocorticoid receptors) are potent regulators of cellular proliferation and G1-S phase progression (46), it is interesting to note that the ARE found in the Cdc6 promoter (AGAACAnnnTATTCT) constitutes a near perfect class I steroid hormone response element (34). Thus, other NRs in unrelated steroid hormone-responsive tissues might also target Cdc6 for transcriptional regulation. Consistent with this notion, we observed a modest yet reproducible ligand-dependent activation of a Cdc6-ARE reporter gene via the glucocorticoid receptor (Figure 4F). Binding sites for the GATA and ETS families of transcription factors have been identified proximal to AREs in the genome (35) and both types of factors have been proposed to facilitate AR binding to the ARE (3). Interestingly, both GATA and ETS-binding sites were detected in the Cdc6 gene promoter just downstream of the ARE (Figure 2A). Although we were unable to discern a functional requirement for either factor in the studies here, it remains plausible that GATA and/or ETS factors cooperate with AR in mediating the Cdc6 androgen response in vivo.
Significantly, we identified several of the transcriptional coregulatory factors involved in AR-dependent transcriptional regulation of the Cdc6 gene and further characterized androgen- and antiandrogen-induced alterations in histone H3 acetylation and methylation patterns at the Cdc6 promoter. Our findings show that in the presence of androgens, AR recruits Mediator and p160/SRC-HAT complexes to the Cdc6 promoter, which in turn facilitate histone acetylation at the promoter region and the recruitment of RNA pol II. Contrarily, in the presence of antiandrogens, AR recruits SMRT–HDAC complexes to the Cdc6 promoter that in turn facilitate histone deacetylation. We also observed a pronounced androgen-dependent decrease in H3K9 tri-methylation at the Cdc6 promoter as well as a modest increase in H3R17 di-methylation, both marks consistent with transcriptional activation (39). These findings suggest that in addition to the aforementioned coactivators, androgen-bound AR also recruits the histone H3-arginine methyltransferase CARM1 (which specifically facilitates H3R17 methylation) (47) and the histone H3-lysine demethylase JMJD2C (which specifically demethylates H3K9-3me) (48). Importantly, both CARM1 and JMJD2C have been previously shown to associate with AR and enhance AR-dependent transcription (41,47,49).
In agreement with our findings showing that AR silencing in LNCaP cells decreases Cdc6 expression, inhibits androgen-induced cell growth and triggers G0 and G1 cell-cycle arrest (Figure 7), we have likewise found that MED1 silencing in LNCaP cells also inhibits Cdc6 expression and decreases androgen-dependent progression into G2/M phase (38). These data, together with the ChIP findings discussed above, suggest that MED1-Mediator plays a particularly important coactivator role in AR-dependent transcriptional activation of Cdc6. Indeed, MED1 is indispensable for AR-dependent transcription of the PSA gene in LNCaP cells and MED1 silencing was accompanied by a significant reduction in RNA pol II recruitment at the PSA gene promoter (37). Thus in addition to the covalent histone modifications and chromatin remodeling events fulfilled by other types of AR coactivators, MED1-Mediator likely plays a rate-limiting role in facilitating the assembly and activation of RNA pol II and its associated factors at Cdc6 and other AR target genes.
A flow cytometry approach was recently utilized to show that AR is degraded at the M phase of each cell cycle in different prostate cancer cell lines thus suggesting that mitotic AR degradation may be a requirement to license a new round of DNA replication (13). Consistent with a presumptive role as a DNA replication-licensing factor, we found that AR levels in LNCaP cells were significantly higher at G1/S than at G2/M (Figure 6). Furthermore, we found that peak occupancy of AR at the Cdc6 promoter occurs during the G1/S phase of the cell cycle, concomitant with peak Cdc6 mRNA expression, and when LNCaP cells were transfected with AR siRNA, there was notable decrease in Cdc6 mRNA levels at G1/S. These findings suggest that AR cooperates with E2F to promote cell-cycle-dependent Cdc6 gene expression. Interestingly, it has been reported that AR can directly interact with Rb proteins (50,51). Thus, in addition to the transactivation mechanisms outlined above, AR might also form complexes with Rb at the Cdc6 promoter that alleviate inhibitory constraints on promoter-bound E2F factors (25). In sum, our results suggest that Cdc6 is a key-regulatory target gene in androgen-sensitive prostate cells and help clarify the molecular role of AR during G1-S phase progression of the mammalian cell cycle.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health grant (DK054030 to J.D.F.). Funding for open access charge: National Institutes of Health.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors wish to thank Drs Joseph Nevins, Frank Claessens, Herb Samuels and Michael Garabedian for plasmids; Theresa Hyejeong Choi for assistance with the flow cytometry analyses; Drs Paul Yen, Madesh Belakavadi and Ravi Vijavargia for critically reading the manuscript; and AstraZeneca for kindly providing the Casodex used in this work.
REFERENCES
- 1.Dehm SM, Tindall DJ. Androgen receptor structural and functional elements: role and regulation in prostate cancer. Mol. Endocrinol. 2007;21:2855–2863. doi: 10.1210/me.2007-0223. [DOI] [PubMed] [Google Scholar]
- 2.Claessens F, Verrijdt G, Schoenmakers E, Haelens A, Peeters B, Verhoeven G, Rombauts W. Selective DNA binding by the androgen receptor as a mechanism for hormone-specific gene regulation. J. Steroid Biochem. Mol. Biol. 2001;76:23–30. doi: 10.1016/s0960-0760(00)00154-0. [DOI] [PubMed] [Google Scholar]
- 3.Wang Q, Li W, Liu XS, Carroll JS, Janne OA, Keeton EK, Chinnaiyan AM, Pienta KJ, Brown M. A hierarchical network of transcription factors governs androgen receptor-dependent prostate cancer growth. Mol. Cell. 2007;27:380–392. doi: 10.1016/j.molcel.2007.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bolton EC, So AY, Chaivorapol C, Haqq CM, Li H, Yamamoto KR. Cell- and gene-specific regulation of primary target genes by the androgen receptor. Genes Dev. 2007;21:2005–2017. doi: 10.1101/gad.1564207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Massie CE, Adryan B, Barbosa-Morais NL, Lynch AG, Tran MG, Neal DE, Mills IG. New androgen receptor genomic targets show an interaction with the ETS1 transcription factor. EMBO Rep. 2007;8:871–878. doi: 10.1038/sj.embor.7401046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heinlein CA, Chang C. Androgen receptor (AR) coregulators: an overview. Endocr. Rev. 2002;23:175–200. doi: 10.1210/edrv.23.2.0460. [DOI] [PubMed] [Google Scholar]
- 7.Chmelar R, Buchanan G, Need EF, Tilley W, Greenberg NM. Androgen receptor coregulators and their involvement in the development and progression of prostate cancer. Int. J. Cancer. 2007;120:719–733. doi: 10.1002/ijc.22365. [DOI] [PubMed] [Google Scholar]
- 8.Malik S, Roeder RG. Dynamic regulation of pol II transcription by the mammalian Mediator complex. Trends Biochem. Sci. 2005;30:256–263. doi: 10.1016/j.tibs.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 9.Belakavadi M, Fondell J. Role of the mediator complex in nuclear hormone receptor signaling. Rev. Physiol. Biochem. Pharmacol. 2006;156:23–43. doi: 10.1007/s10254-005-0002-0. [DOI] [PubMed] [Google Scholar]
- 10.Feldman BJ, Feldman D. The development of androgen-independent prostate cancer. Nat. Rev. Cancer. 2001;1:34–45. doi: 10.1038/35094009. [DOI] [PubMed] [Google Scholar]
- 11.Abate-Shen C, Shen MM. Molecular genetics of prostate cancer. Genes Dev. 2000;14:2410–2434. doi: 10.1101/gad.819500. [DOI] [PubMed] [Google Scholar]
- 12.Balk SP, Knudsen KE. AR, the cell cycle, and prostate cancer. Nucleic Recept. Signal. 2008;6:1–12. doi: 10.1621/nrs.06001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Litvinov IV, Vander Griend DJ, Antony L, Dalrymple S, De Marzo AM, Drake CG, Isaacs JT. Androgen receptor as a licensing factor for DNA replication in androgen-sensitive prostate cancer cells. Proc. Natl Acad. Sci. USA. 2006;103:15085–15090. doi: 10.1073/pnas.0603057103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huggins C, Hodges CV. Studies on prostatic cancer: I. The effect of castration, of estrogen and of androgen injection on serum phosphatases in metastatic carcinoma of the prostate. CA Cancer J. Clin. 1972;22:232–240. doi: 10.3322/canjclin.22.4.232. [DOI] [PubMed] [Google Scholar]
- 15.Kyprianou N, Isaacs JT. Activation of programmed cell death in the rat ventral prostate after castration. Endocrinology. 1988;122:552–562. doi: 10.1210/endo-122-2-552. [DOI] [PubMed] [Google Scholar]
- 16.Knudsen KE, Arden KC, Cavenee WK. Multiple G1 regulatory elements control the androgen-dependent proliferation of prostatic carcinoma cells. J. Biol. Chem. 1998;273:20213–20222. doi: 10.1074/jbc.273.32.20213. [DOI] [PubMed] [Google Scholar]
- 17.Coller HA. What's taking so long? S-phase entry from quiescence versus proliferation. Nat. Rev. Mol. Cell Biol. 2007;8:667–670. doi: 10.1038/nrm2223. [DOI] [PubMed] [Google Scholar]
- 18.Blow JJ, Gillespie PJ. Replication licensing and cancer – a fatal entanglement? Nat. Rev. Cancer. 2008;8:799–806. doi: 10.1038/nrc2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ayad NG. CDKs give cdc6 a license to drive into S phase. Cell. 2005;122:825–827. doi: 10.1016/j.cell.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 20.Yan Z, DeGregori J, Shohet R, Leone G, Stillman B, Nevins JR, Williams RS. Cdc6 is regulated by E2F and is essential for DNA replication in mammalian cells. Proc. Natl Acad. Sci. USA. 1998;95:3603–3608. doi: 10.1073/pnas.95.7.3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hateboer G, Wobst A, Petersen BO, Le Cam L, Vigo E, Sardet C, Helin K. Cell cycle-regulated expression of mammalian CDC6 Is dependent on E2F. Mol. Cell Biol. 1998;18:6679–6697. doi: 10.1128/mcb.18.11.6679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ohtani K, Tsujimoto A, Ikeda M, Nakamura M. Regulation of cell growth-dependent expression of mammalian CDC6 gene by the cell cycle transcription factor E2F. Oncogene. 1998;17:1777–1785. doi: 10.1038/sj.onc.1202105. [DOI] [PubMed] [Google Scholar]
- 23.Bai VU, Cifuentes E, Menon M, Barrack ER, Reddy GP-V. Androgen receptor regulates Cdc6 in synchronized LNCaP cells progressing from G1 to S phase. J. Cell Physiol. 2005;204:381–387. doi: 10.1002/jcp.20422. [DOI] [PubMed] [Google Scholar]
- 24.Robles LD, Frost AR, Davila M, Hutson AD, Grizzle WE, Chakrabarti R. Down-regulation of Cdc6, a cell cycle regulatory gene, in prostate cancer. J. Biol. Chem. 2002;277:25431–25438. doi: 10.1074/jbc.M201199200. [DOI] [PubMed] [Google Scholar]
- 25.Mallik I, Davila M, Tapia T, Schanen B, Chakrabarti R. Androgen regulates Cdc6 transcription through interactions between androgen receptor and E2F transcription factor in prostate cancer cells. Biochim. Biophys. Acta. 2008;1783:1737–1744. doi: 10.1016/j.bbamcr.2008.05.006. [DOI] [PubMed] [Google Scholar]
- 26.Wang Q, Udayakumar TS, Vasaitis TS, Brodie AM, Fondell JD. Mechanistic relationship between androgen receptor polyglutamine tract truncation and androgen-dependent transcriptional hyperactivity in prostate cancer cells. J. Biol. Chem. 2004;279:17319–17328. doi: 10.1074/jbc.M400970200. [DOI] [PubMed] [Google Scholar]
- 27.Pandey PK, Udayakumar TS, Lin X, Sharma D, Shapiro PS, Fondell JD. Activation of TRAP/Mediator subunit TRAP220/Med1 is regulated by mitogen-activated protein kinase-dependent phosphorylation. Mol. Cell Biol. 2005;25:10695–10710. doi: 10.1128/MCB.25.24.10695-10710.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Q, Sharma D, Ren Y, Fondell JD. A coregulatory role for the TRAP-Mediator complex in androgen receptor-mediated gene expression. J. Biol. Chem. 2002;277:42852–42858. doi: 10.1074/jbc.M206061200. [DOI] [PubMed] [Google Scholar]
- 29.Sharma D, Fondell JD. Ordered recruitment of histone acetyltransferases and the TRAP/Mediator complex to thyroid hormone-responsive promoters in vivo. Proc. Natl Acad. Sci. USA. 2002;99:7934–7939. doi: 10.1073/pnas.122004799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang YSF. Molecular mechanisms underlying KVS-1-MPS-1 complex assembly. Biophys. J. 2007;93:3083–3091. doi: 10.1529/biophysj.107.110320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Agoulnik IU, Vaid A, Bingman WE, III, Erdeme H, Frolov A, Smith CL, Ayala G, Ittmann MM, Weigel NL. Role of SRC-1 in the promotion of prostate pancer cell growth and tumor progression. Cancer Res. 2005;65:7959–7967. doi: 10.1158/0008-5472.CAN-04-3541. [DOI] [PubMed] [Google Scholar]
- 32.Whitfield ML, Sherlock G, Saldanha AJ, Murray JI, Ball CA, Alexander KE, Matese JC, Perou CM, Hurt MM, Brown PO, et al. Identification of genes periodically expressed in the human cell cycle and their expression in tumors. Mol. Biol. Cell. 2002;13:1977–2000. doi: 10.1091/mbc.02-02-0030.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao H, Whitfield ML, Xu T, Botstein D, Brooks JD. Diverse effects of methylseleninic acid on the transcriptional program of human prostate cancer cells. Mol. Biol. Cell. 2004;15:506–519. doi: 10.1091/mbc.E03-07-0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Verrijdt G, Haelens A, Claessens F. Selective DNA recognition by the androgen receptor as a mechanism for hormone-specific regulation of gene expression. Mol. Genet. Metab. 2003;78:175–185. doi: 10.1016/s1096-7192(03)00003-9. [DOI] [PubMed] [Google Scholar]
- 35.Masuda K, Werner T, Maheshwari S, Frisch M, Oh S, Petrovics G, May K, Srikantan V, Srivastava S, Dobi A. Androgen receptor binding sites identified by a GREF_GATA Model. J. Mol. Biol. 2005;353:763–771. doi: 10.1016/j.jmb.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 36.Ikonen T, Palvimo J, Kallio P, Reinikainen P, Janne O. Stimulation of androgen-regulated transactivation by modulators of protein phosphorylation. Endocrinology. 1994;135:1359–1366. doi: 10.1210/endo.135.4.7925097. [DOI] [PubMed] [Google Scholar]
- 37.Wang Q, Carroll JS, Brown M. Spatial and temporal recruitment of androgen receptor and its coactivators involves chromosomal looping and polymerase tracking. Mol. Cell. 2005;19:631–642. doi: 10.1016/j.molcel.2005.07.018. [DOI] [PubMed] [Google Scholar]
- 38.Vijayvargia R, May MS, Fondell JD. A coregulatory role for the mediator complex in prostate cancer cell proliferation and gene expression. Cancer Res. 2007;67:4034–4041. doi: 10.1158/0008-5472.CAN-06-3039. [DOI] [PubMed] [Google Scholar]
- 39.Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–412. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- 40.McKenna NJ, Lanz RB, O'Malley BW. Nuclear receptor coregulators: cellular and molecular biology. Endocr. Rev. 1999;20:321–344. doi: 10.1210/edrv.20.3.0366. [DOI] [PubMed] [Google Scholar]
- 41.Kang Z, Janne OA, Palvimo JJ. Coregulator recruitment and histone modifications in transcriptional regulation by the androgen receptor. Mol. Endocrinol. 2004;18:2633–2648. doi: 10.1210/me.2004-0245. [DOI] [PubMed] [Google Scholar]
- 42.Yoon HG, Wong J. The corepressors silencing mediator of retinoid and thyroid hormone receptor and nuclear receptor corepressor are involved in agonist- and antagonist-regulated transcription by androgen receptor. Mol. Endocrinol. 2006;20:1048–1060. doi: 10.1210/me.2005-0324. [DOI] [PubMed] [Google Scholar]
- 43.Shang Y, Myers M, Brown M. Formation of the androgen receptor transcription complex. Mol. Cell. 2002;9:601–610. doi: 10.1016/s1097-2765(02)00471-9. [DOI] [PubMed] [Google Scholar]
- 44.Klose RJ, Zhang Y. Regulation of histone methylation by demethylimination and demethylation. Nat. Rev. Mol. Cell Biol. 2007;8:307–318. doi: 10.1038/nrm2143. [DOI] [PubMed] [Google Scholar]
- 45.Stallcup MR. Role of protein methylation in chromatin remodeling and transcriptional regulation. Oncogene. 2001;20:3014–3020. doi: 10.1038/sj.onc.1204325. [DOI] [PubMed] [Google Scholar]
- 46.Boonyaratanakornkit V, Bi Y, Rudd M, Edwards DP. The role and mechanism of progesterone receptor activation of extra-nuclear signaling pathways in regulating gene transcription and cell cycle progression. Steroids. 2008;73:922–928. doi: 10.1016/j.steroids.2008.01.010. [DOI] [PubMed] [Google Scholar]
- 47.Koh SS, Li H, Lee Y-H, Widelitz RB, Chuong C-M, Stallcup MR. Synergistic coactivator function by coactivator-associated arginine methyltransferase (CARM) 1 and beta-Catenin with two different classes of DNA-binding transcriptional activators. J. Biol. Chem. 2002;277:26031–26035. doi: 10.1074/jbc.M110865200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wissmann M, Yin N, Muller JM, Greschik H, Fodor BD, Jenuwein T, Vogler C, Schneider R, Gunther T, Buettner R, et al. Cooperative demethylation by JMJD2C and LSD1 promotes androgen receptor-dependent gene expression. Nat. Cell Biol. 2007;9:347–353. doi: 10.1038/ncb1546. [DOI] [PubMed] [Google Scholar]
- 49.Majumder S, Liu Y, Ford OH, III, Mohler JL, Whang YE. Involvement of arginine methyltransferase CARM1 in androgen receptor function and prostate cancer cell viability. Prostate. 2006;66:1292–1301. doi: 10.1002/pros.20438. [DOI] [PubMed] [Google Scholar]
- 50.Hofman K, Swinnen J, Claessens F, Verhoeven G, Heyns W. The retinoblastoma protein-associated transcription repressor RBaK interacts with the androgen receptor and enhances its transcriptional activity. J. Mol. Endocrinol. 2003;31:583–596. doi: 10.1677/jme.0.0310583. [DOI] [PubMed] [Google Scholar]
- 51.Lu J, Danielsen M. Differential regulation of androgen and glucocorticoid receptors by retinoblastoma protein. J. Biol. Chem. 1998;273:31528–31533. doi: 10.1074/jbc.273.47.31528. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.