Abstract
We earlier observed that treating rat proximal tubules with concentrations of angiotensin II (ANG II) that directly stimulate Na-K-ATPase activity changed how Na-K-ATPase subsequently eluted from an ouabain-affinity column. In this study we tested whether ANG II increases the rate of elution in response to ligands that trigger the decay of E2-P, which implies a change in functional properties of Na-K-ATPase, or by decreasing the amount subsequently eluted with SDS, which suggests a change in how Na-K-ATPase interacts with other proteins. We utilized a new digoxin-affinity column and novel lines of opossum kidney (OK) cells that coexpress the rat AT1a receptor and either the wild-type rat α1-isoform of Na-K-ATPase or a truncation mutant missing the first 32 amino acids of its NH2 terminus. We characterized how rat kidney microsomes bind to and elute from the digoxin-affinity column and demonstrated that they are heterogeneous in the rate at which they release digoxin in response to ligands that trigger the decay of E2-P. Incubating OK cells with ANG II stimulated the ensuing elution of wild-type rat α1-subunit by increasing the kinetic response to ligands that cause a decay of E2-P without affecting the amount later eluted with SDS. In contrast, ANG II had no effect on the kinetic response of the truncation mutant but decreased the amount eluted with SDS. These data suggest that ANG II regulates both the kinetic properties of Na-K-ATPase and its interaction with other proteins by a mechanism(s) involving its NH2 terminus.
Keywords: cardiac glycosides, ouabain, epithelial cells, sodium
the mechanisms by which angiotensin II (ANG II) directly stimulates the short-term activity of Na-K-ATPase in the proximal tubule (1, 2, 7) are incompletely understood, which limits our knowledge of how ANG II controls sodium reabsorption and how ANG II and angiotensin-converting enzyme (ACE) inhibitors affect the development of hypertension and the treatment of congestive heart failure (3, 29). When cells are exposed to ANG II, at least part of the stimulation that occurs during the first 15 min is due to the accumulation of Na-K-ATPase in the plasma membrane, which is triggered by an increase in the phosphorylation of Na-K-ATPase by PKC (4, 5).
ANG II may also stimulate Na-K-ATPase activity by other mechanisms. For example, briefly (≤2 min) treating rat proximal tubules with ANG II increased the amount of Na-K-ATPase eluted from an ouabain-affinity column with a solution containing 150 mM Na + ATP + EDTA and decreased the amount subsequently eluted with denaturing concentrations of SDS (39). Cardiac glycosides preferentially bind to the E2-P conformation of Na-K-ATPase (8), and the solution containing 150 mM Na + ATP + EDTA converts E2-P to E1 (8, 13). Therefore, the accelerated elution could have been due to an increase in the rate of conversion of E2-P to E1, the rate-limiting step in the reaction mechanism of Na-K-ATPase (14). The ratio of E2-P to E1 is one of the factors that control the apparent affinity for intracellular sodium and extracellular potassium (16, 31). Thus the effect of ANG II on increasing the amount eluted with 150 mM Na could be related to an earlier report that ANG II increases the affinity for intracellular sodium (1). On the other hand, the α1-isoform, which has a low affinity for ouabain (8, 26), accounts for 99.9% of Na-K-ATPase in rat kidney (20). Thus all of the bound Na-K-ATPase could have been eluted with the first solution without an increase in the rate of conversion of E2-P to E1. This idea led us to consider whether all of the Na-K-ATPase on the column was equally free to transition from E2-P to E1.
The plasma membranes of LLC-PK1 cells, a proximal tubule cell line from pig kidney, contain pools of pumping and nonpumping Na-K-ATPase molecules, and the movement of Na-K-ATPase molecules out of the nonpumping pool leads to an overall increase in Na-K-ATPase activity (18). Na-K-ATPase molecules that do not pump are located in caveolae (18), where they physically interact with other proteins as part of a signaling complex (36, 38). These proteins could in turn constrain the ability of Na-K-ATPase to change conformation and release ouabain and thereby account for Na-K-ATPase eluted with SDS. If this were the case, then our observation that ANG II decreased the amount of Na-K-ATPase eluted with SDS (39) could indicate that ANG II is stimulating Na-K-ATPase activity by causing a redistribution of Na-K-ATPase within the plasma membrane.
We hypothesized that the accelerated elution of Na-K-ATPase from the ouabain-affinity column was due to either a modification of the intrinsic kinetic properties of Na-K-ATPase or to a change in how the Na-K-ATPase interacts with proteins that constrain its ability to release ouabain. The purpose of the present study was to determine whether ANG II is stimulating the rate of release of Na-K-ATPase in direct response to ligands that trigger the decay of E2-P or by decreasing the amount of Na-K-ATPase that remains bound and is subsequently eluted with denaturing concentrations of SDS. Our experiments made use of new lines of opossum kidney (OK) cells that coexpress the rat AT1a receptor and either wild-type or mutant forms of the α1-isoform of the Na-K-ATPase. We also present and characterize a new digoxin-affinity column. Digoxin is a cardiac glycoside that binds to rat kidney Na-K-ATPase ∼10 times tighter than ouabain (24), which improves our ability to detect differences in the rate at which the rat α1-subunit is releasing cardiac glycoside.
MATERIALS AND METHODS
Preparation of affinity column
To prepare the digoxin-affinity medium, we hydrated and washed1gofdry epoxy-activated Sepharose (Amersham Biosciences) with distilled water on a sintered glass filter. The swollen beads were added to 65 ml of 4 mM digoxin dissolved in 77% ethanol. The suspension was incubated with mixing for ∼50 h at 45°C. Thereafter, the slurry was poured into a chromatography column, and the ethanol mixture was allowed to drain. The remaining beads were washed with 20 bed volumes of 0.1 M Na2CO3 at pH 11.0. Beads were then added to 50 ml of 1.0 M ethanolamine at pH 8.5, and the mixture was incubated with mixing at 45°C for ∼18 h. The slurry was drained through the column, and the beads were washed with 20 bed volumes of 0.1 M Na2CO3 at pH 11.0. The resulting affinity matrix was stored in the dark at 4°C in the final wash solution. Sham columns were made using the same procedure in the absence of digoxin.
General procedure for running chromatography columns with rat kidney microsomes
Rat kidney microsomes (∼2.5 mg of total protein) were suspended in ∼1 ml of a loading buffer containing 30 mM NaCl, 3 mM ATP, 4 mM MgCl2, and 20 mM imidazole, pH 7.4, and combined with ∼1 ml of digoxin-affinity medium in a chromatography column at 4°C. The column was capped at both ends, laid horizontally, and rocked end to end for 60 min. The column was positioned vertically, the caps were removed, and the column was drained from the bottom until all the fluid reached the top of the chromatography matrix. More loading buffer was then added to wash off unbound protein. In one experiment (Fig. 1), ∼2.5 ml of loading buffer were added. In all other experiments, ∼5 ml were added. After all the loading buffer had entered the bed of the column, an eluting solution containing 150 mM NaCl, 3 mM ATPNa2, 4 mM EDTA, and 20 mM imidazole, pH 7.4, was added to the top of the column. Once the eluting solution had completely entered the bed of the column, the columns were put at room temperature, and a solution containing 2% SDS was added. In all experiments, the flow rate was relatively constant at ∼0.5 ml/min at both 4°C and room temperature. Each fraction was ∼0.6 ml; thus it took ∼1.2 min/fraction.
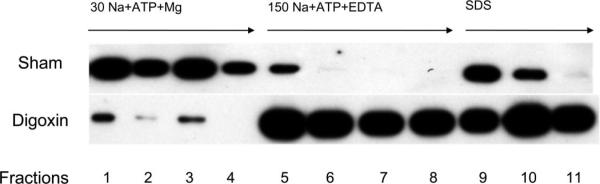
Fig. 1.
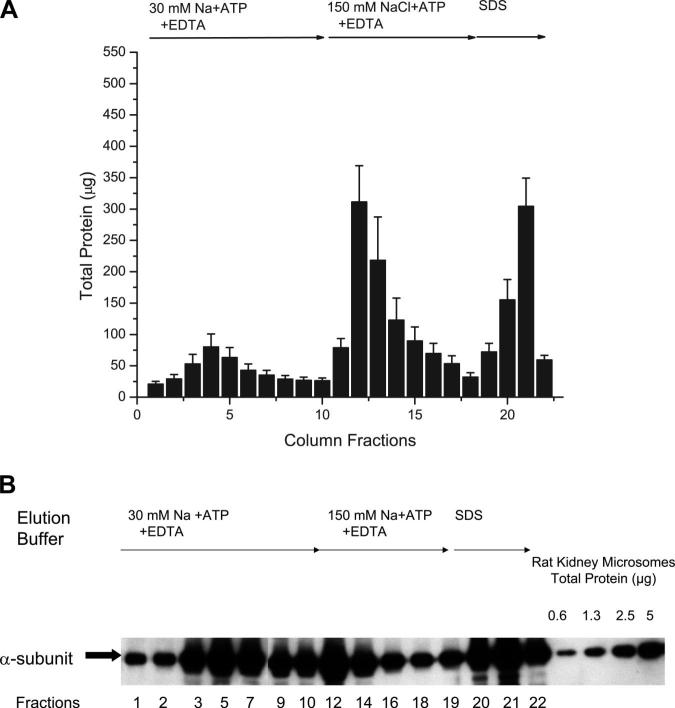
An immunoblot prepared using an antibody to the α-subunit of Na-K-ATPase that shows specific elution of rat kidney Na-K-ATPase from the digoxin-affinity column. After digoxin-affinity and sham columns of the same size were loaded with the same amount of protein in the presence of ligands (30 mM Na + ATP + Mg) that promote the E2-P conformation of Na-K-ATPase, much more Na-K-ATPase washed through the sham column than through the digoxin-affinity column (fractions 1−4). On the other hand, much more Na-K-ATPase was eluted from the digoxin-affinity column than from the sham column by a solution containing ligands (150 mM Na + ATP + EDTA) that trigger a decline in E2-P (fractions 5−8) and by a solution containing denaturing concentrations of SDS (fractions 9−11).
Preparation of rat kidney microsomes
Rat kidney microsomes were prepared as previously described (40). The kidneys were removed from the rat, minced whole on ice, and homogenized with a glass-Teflon Wheaton Potter-Elvehjem homogenizer (1,000 rpm) in 10 vols of buffer containing 50 mM imidazole (pH 7.4), 2 mM EDTA, and 250 mM sucrose. The homogenate was centrifuged at 6,000 g for 20 min, and the supernatant was spun again at 48,000 g for 60 min at 4°C. The subsequent pellet was suspended in the homogenization buffer at a final protein concentration of ∼5 mg/ml and was stored at −70°C. Before use, all microsomes were alternately frozen at −70°C and thawed six times to permeabilize them to ATP (21).
Measuring the amount of Na-K-ATPase
The amount of α-subunit in fractions off the columns was measured by immunoblotting (22) and converted to absolute amounts of Na-K-ATPase by using the amount of α-subunit in rat kidney microsomes as a standard. Immunoblot signals were quantified in arbitrary units using a Fuji LAS-1000 System and Image Gauge version 3.3 software. For each blot, we constructed a standard curve of arbitrary units compared with known amounts of α1-subunit, fit the data with the equation y = (a)·ln(x) + b by using least-squares, and used the resulting equation to calculate the amount of α-subunit in each fraction. The absolute amount of α-subunit in the microsomes was determined by measuring Na-K-ATPase activity under optimal conditions (6) and then calculating the amount of Na-K-ATPase that had to be present to account for the observed activity. For this calculation, we knew that the maximum rate of ATP hydrolysis per pump was 10,000 molecules of ATP per minute (15) and that the functional unit was an αβ complex (23) that would have a molecular mass of ∼140 kDa. In the rat kidney microsomes used for standards, the specific activity of the Na-K-ATPase was 2.8 ± 0.4 μmol Pi·mg−1·min−1. Assuming that all of the Na-K-ATPase was active, we determined that the α-subunit was ∼4% of the total protein.
Plasmid and OK cell lines
The coding sequence for the rat α1-subunit of Na-K-ATPase subcloned into the pRc/CMV plasmid was a generous gift from Dr. J. Lingrel (37). A plasmid encoding the rat α1-subunit of Na-K-ATPase in which the sequence encoding the first 32 amino acids was replaced by a Myc epitope tag was generously provided by Dr. Marie-Josee Duran in the laboratory of Dr. Thomas Pressley. OK cells that stably express the rat AT1a receptor were generously provided by Dr. T. Thekkumkara (35) and were routinely cultured in DMEM-F12 with 10% fetal calf serum, 50 IU/ml penicillin, and 50 μg/ml streptomycin at 37°C. For transfection, 0.5 ml of cells at 5 × 106 cells/ml in unsupplemented DMEM-F12 medium were mixed with 20 μg of plasmid DNA in a 0.4-cm gap cuvette and subjected to electroporation (260 V, 950 μF). The cells were then plated on 100-mm culture dishes with regular growth medium and allowed to attach overnight. The next day, the cells were split onto three 150-mm plates and cultured for 2 wk in growth medium further supplemented with either 10 or 100 μM ouabain for selection. A pooled population was obtained. Expression of the rat α1-subunit was confirmed by testing the ability of different concentrations of ouabain to inhibit 86Rb influx (25). Selection with 10 μM ouabain was maintained throughout subsequent culture of the stable cell lines.
Procedure to treat OK cells with or without ANG II, load equal amounts of protein on digoxin-affinity columns, and subsequently elute protein using three different solutions
Cells grown on 150-mm plastic culture dishes to ∼80% confluency were brought to room temperature, and either ANG II at a final concentration of 10 nM or an equal volume of buffer was added to each. Five minutes later, the plates were put on ice, the medium was removed, and the cells were rinsed three times with ice-cold Hanks’ solution without added calcium, magnesium, or phenol red. After the Hanks’ solution was removed, 1.5 ml of ice-cold lysis buffer containing 30 mM NaCl, 3 mM ATP, 4 mM Mg, 25 mM imidazole, 1 mM 4-(2-aminoethyl)benzene sulfonyl fluoride, 0.8 μM aprotinin, 20 μM leupeptin, 40 μM bestatin, 15 μM pepstatin A, 14 μM E-64, 1 μM okadaic acid, 1 μM microcystin, and 5 μM phenylarsine oxide, pH 7.4, was added to each of the plates. The cells were scraped off and placed into centrifuge tubes, which were put on ice and rocked end to end for 5 min. The samples were centrifuged at 4°C for 5 min at ∼800 relative centrifugal force, and the resulting supernatant was collected. In each experiment, ∼4.9 mg of protein from control and ANG II-treated cells were combined with 1 ml of digoxin-affinity matrix in a chromatography tube, which was mixed end to end for 60 min at 4°C. The column was mounted vertically, and the affinity matrix was washed at 4°C with 5 ml of a solution containing 30 mM NaCl, 3 mM ATP, 4 mM MgCl2, and 25 mM imidazole, pH 7.4. Next, 5 ml of a solution containing 30 mM NaCl, 3 mM ATP, 4 mM EDTA, and 25 mM imidazole, pH 7.4, were added to the top of the column, and nine fractions were collected. As the preceding solution drained to the top edge of the chromatography media, 5 ml of a solution containing 150 mM NaCl, 3 mM ATP, 4 mM EDTA, and 25 mM imidazole, pH 7.4, were added to the top of the column, and nine fractions were collected. The column was put at room temperature, 4 ml of 2% SDS were added to the top of the column, and four fractions of equal volume were collected. The average fraction size was ∼0.6 ml, and the flow rate was ∼0.5 ml/min at both 4°C and room temperature.
Experimental design and statistical analysis for testing the effect of ANG II
In each experiment, half of the cells were treated with ANG II and half were not. Protein from cells treated with ANG II was put on one digoxin-affinity column. Protein from the control cells was simultaneously put on a comparable column. Both columns were run side by side under the same conditions. At the end of the experiment, the amount of protein in each of the fractions was measured in duplicate in the same assay. All the data from all the experiments performed on a given cell line were analyzed at the same time using the General Linear Model in SPSS, which in this case is equal to a two-way ANOVA with repeated measures with two between-subject factors. This design was chosen to look for differences in the amount of protein eluted in each fraction between columns loaded with protein from control and ANG II-treated cells. The first between-subject factor accounts for the difference between individual experiments (n = 6). The second between-subject factor measures the difference between control cells and those treated with ANG II. If there was a significant difference between the second between-subject factor (P ≤ 0.05), a Bonferroni post hoc analysis was performed to determine whether there was a significant difference (P ≤ 0.05) in the amount of protein in individual fractions from control versus ANG II-treated cells. Fractions that were significantly different from each other with or without ANG II are indicated where appropriate.
Other procedures and sources of antibodies
Protein was determined by bicinchoninic acid assay (Pierce Biotechnology) following the manufacturer's suggestions. The antibody to the α1-isoform of Na-K-ATPase was obtained from Sigma (clone M8-Pl-A3).
RESULTS AND DISCUSSION
We first characterized how the rat kidney α1-isoform in rat kidney microsomes interacts with the digoxin-affinity column as a function of ligands that change Na-K-ATPase conformation. We then used this information and the new lines of OK cells to test whether ANG II accelerates the rate of elution of the rat kidney Na-K-ATPase as a function of time in response to solutions that trigger the decay of E2-P.
Binding and recovery of rat kidney microsomes requires digoxin
To characterize our new digoxin-affinity column, we first tested whether the binding of plasma membranes to the column required the presence of digoxin. These and the following experiments were carried out at 4°C to slow the rate at which digoxin is released from Na-K-ATPase and, in the absence of detergents, to maintain protein-protein interactions that could affect the rate at which Na-K-ATPase changes conformation and mimic our earlier experimental conditions (39). Equal amounts of rat kidney microsomes were applied to sham and digoxin-affinity columns in the presence of a solution containing 30 mM Na + ATP + Mg, which promotes E2-P and the binding of Na-K-ATPase to cardiac glycosides (8). In the presence of this solution, a large amount of Na-K-ATPase washed through the sham column without binding (Fig. 1, fractions 1−4). In contrast, most of the Na-K-ATPase applied to the digoxin-affinity column bound, and very little appeared in the subsequent wash of the column with the solution containing 30 mM Na + ATP + Mg (Fig. 1, fractions 1−4). Adding a solution containing 150 mM Na + ATP + EDTA, which has been shown to reverse the binding of ouabain to the human red cell Na-K-ATPase (13), removed large amounts of rat renal microsomal Na-K-ATPase from the digoxin-affinity column (Fig. 1, fractions 5−8) and almost no Na-K-ATPase from the sham column (Fig. 1, fractions 6 −8). The small amount of Na-K-ATPase in fraction 5 (Fig. 1) likely represents unbound Na-K-ATPase that was still washing off the column in the presence of the buffer containing 30 mM Na + ATP + Mg. The subsequent addition of denaturing concentrations of SDS eluted more Na-K-ATPase from the digoxin-affinity than it did from the sham column (Fig. 1, fractions 9−11).
It was difficult to tell from the experiment shown in Fig. 1 how much Na-K-ATPase was being eluted from the digoxin-affinity column in response to the SDS, because Na-K-ATPase was still coming off the digoxin-affinity column in response to the buffer containing 150 mM Na + ATP + EDTA (Fig. 1, fractions 7 and 8). Therefore, we repeated the experiment using a much larger volume of the solution containing 150 mM Na + ATP + EDTA. The digoxin and sham columns were loaded with rat kidney microsomes, and all the unbound protein was washed off the columns with the solution containing 30 mM Na + ATP + Mg (data not shown). The subsequent addition of the solution containing 150 mM Na + ATP + EDTA eluted a distinct peak from the digoxin-affinity column (Fig. 2A, fractions 1−9, closed symbols) and little Na-K-ATPase from the sham column (Fig. 2A, fractions 1−9, open symbols). After this peak, little additional Na-K-ATPase was eluted (Fig. 2A, fractions 9 −16, closed symbols). Nevertheless, a distinct peak of Na-K-ATPase was eluted from the digoxin-affinity column by the subsequent addition of SDS (Fig. 2A, fractions 18−20, closed symbols). In contrast, a much smaller amount of Na-K-ATPase was eluted with SDS from the sham column (Fig. 2A, fractions 18−20, open symbols). The data in Fig. 2A show that the presence of digoxin on the affinity column greatly increased the recovery of rat kidney microsomes in response to both the solution containing 150 mM Na + ATP + EDTA and the solution containing SDS. These data support the hypothesis that plasma membranes bind to the column via the digoxin, and not to the column matrix. Likewise, these results are consistent with the conclusion that rat kidney microsomes contain a population of Na-K-ATPase molecules that are readily eluted from digoxin by ligands that trigger the decay of E2-P and a population that is resistant to elution.
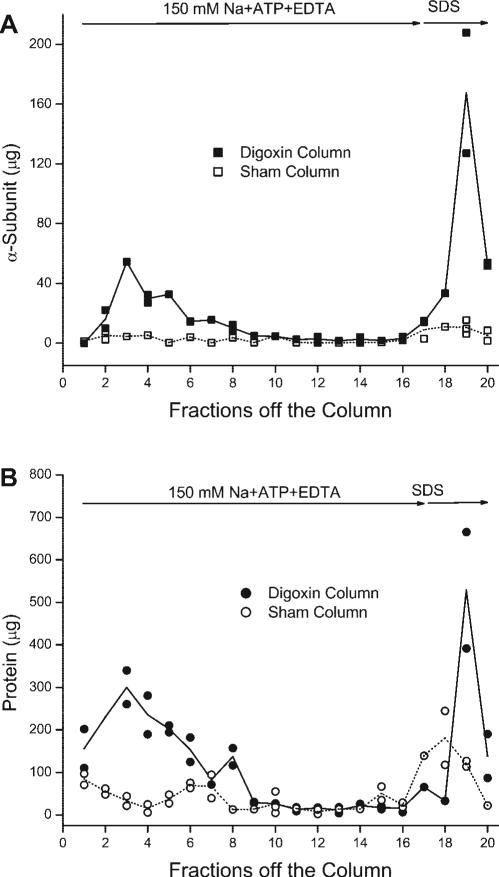
Fig. 2.
Elution of Na-K-ATPase from the digoxin-affinity column (A, ■) was closely correlated with the elution of total protein (B, •), whereas little Na-K-ATPase (A, □) or total protein (B, ○) was eluted from the sham column. After unbound protein was washed off (data not shown), ligands that trigger a decline in the amount of E2-P (150 mM Na + ATP + EDTA) eluted much more Na-K-ATPase α-subunit (A, fractions 1−16) and protein (B, fractions 1−16) from a digoxin-affinity column than from a sham column. The remainder was removed by denaturing concentrations of SDS (fractions 17−21). In A, the amount of α-subunit in some fractions was measured up to 3 times, and data points represent individual measurements. In B, duplicate measurements were made in each fraction. In both A and B, the line indicates the mean value. This experiment was repeated a total of 3 times with similar results.
The pattern of elution seen by measuring the total protein (Fig. 2B, fractions 1−20, closed symbols) is consistent with the pattern observed with the α-subunit (Fig. 2A, fractions 1−20, closed symbols) and is more straightforward to quantify. When equal volumes of each fraction are measured by immunoblotting, one must run each fraction at multiple dilutions to obtain a signal that is in the responsive range of the antibody, which is a nonlinear function of the amount of Na-K-ATPase. Therefore, in subsequent experiments with rat kidney microsomes, we measured the amount of protein to estimate the amount of eluted Na-K-ATPase.
Capacity of the digoxin-affinity column
The total protein that is specifically recovered from the digoxin-affinity column is the sum of the protein that is eluted by ligands causing the decay of E2-P and that eluted with SDS (Fig. 2). The amount of protein eluted from the digoxin-affinity column increased in a linear fashion as the amount of loaded protein increased from 0.5 to 2.5 mg (Fig. 3). Over this same range there was little protein recovered from the sham columns (Fig. 3). When the amount of protein loaded onto the columns was increased to 5 mg, the amount of protein recovered from the digoxin-affinity column began to saturate and significantly more protein eluted with SDS from the sham columns, which indicates nonspecific binding.
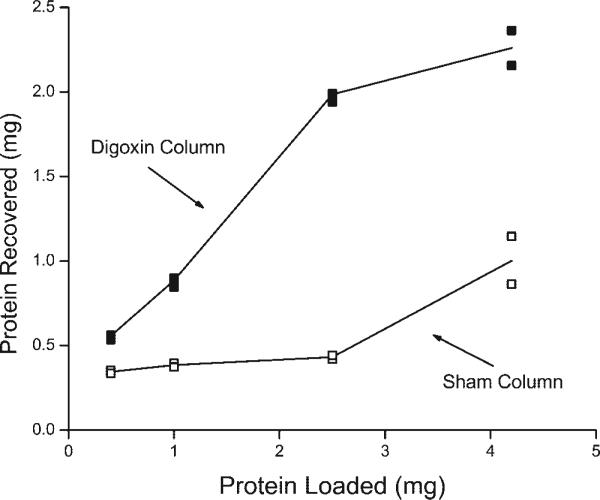
Fig. 3.
Total amount of protein recovered from digoxin-affinity and sham columns with a bed volume of 1 ml as a function of the amount of protein initially loaded onto the columns. The total protein is the sum of the amount eluted with the solution containing 150 mM Na + ATP + EDTA plus the amount eluted with SDS. Values are means of the protein eluted from 2 digoxin-affinity and 2 sham columns run at each of the indicated amounts of protein.
Over the linear range of binding (0.5 to 2.5 mg), we recovered 30−80% of the applied protein (Fig. 3), consistent with approximately one-half of the applied protein being in apical membranes devoid of Na-K-ATPase. The maximum amount of rat kidney microsomes that specifically bound to the digoxin-affinity column was ∼1.5 mg protein/ml bed volume (Fig. 3), which contained ∼400 μg of α-subunit (Fig. 2). Under the experimental conditions used in Fig. 3, the binding capacity of the ouabain-affinity column was ∼4 μg rat kidney α-subunit/ml bed volume (data not shown). This value is very close to our previous report that the ouabain-affinity column bound ∼5 μg rat kidney α-subunit/ml bed volume (40). Thus the digoxin-affinity column binds ∼100 times more rat kidney α-subunit than the ouabain-affinity column, which is more effective in isolating low-abundant Na-K-ATPase with a high affinity for ouabain (11).
Evaluating how the α1-subunit in rat kidney microsomes releases digoxin
Based on the work of Huang and Askari (13), we earlier assumed that solution containing 150 mM Na + ATP + EDTA eluted Na-K-ATPase (39) by stimulating the decay of E2-P to E1 via the accepted steps in the reaction mechanism of Na-K-ATPase: E2-P → E2 → E1 → E1·ATP → Na·E1·ATP. Under normal physiological conditions, ouabain dissociates after phosphate is lost (8, 30, 32), and for most forms of Na-K-ATPase, the rate at which ouabain is subsequently released is very slow in the absence of ligands that promote the formation of E1 (8, 13). The rat α1-subunit, however, releases ouabain at a much faster rate (26). To slow the rate of dissociation and to increase the amount of Na-K-ATPase that bound to our ouabain-affinity column, we conducted our experiments at 4°C (39, 40). At this colder temperature, it is possible that the solution containing 150 mM Na + ATP + EDTA was eluting Na-K-ATPase by the Hofmeister effect, in which sudden jumps in salt concentration convert E2-P to E1-P (27), a conformation to which cardiac glycosides do not bind (8).
To determine whether the solution containing 150 mM Na + ATP + EDTA was eluting Na-K-ATPase by pulling Na-K-ATPase toward E1 or by means of the Hofmeister effect, we tested how the presence and absence of Mg affected the elution of Na-K-ATPase. As before, rat kidney microsomes were bound in the presence of 30 mM Na + ATP + Mg. Under these conditions, we would expect that removing the free Mg in the continued presence of 30 mM Na and ATP would trigger the spontaneous decay of E2-P to E1 and lead to the release of digoxin. The rate constant for the spontaneous decay of E2-P at 0°C is 0.06 s−1 (28). Therefore, it should take ∼17 s for the original amount of E2-P to spontaneously decay to 37% of its original value at 4°C. Instead, we found that replacing the Mg with EDTA eluted only ∼10% of the bound protein during the first ∼6 min at 4°C (Fig. 4A, fractions 1−5, hatched bars). The peak response was in fraction 3, which occurred ∼4 min after the free Mg was reduced (Fig. 4A), much slower than the expected rate of spontaneous decay. After the column was eluted for 20 min with the Mg-free solution, 75% of the protein that originally bound to the column was eluted with SDS (Fig. 4A, fractions 17−20, hatched bars). In contrast, no protein was eluted when the Mg was maintained in the presence of 30 mM Na and ATP (Fig. 4A, fractions 1−16, solid bars), and more protein was eluted with denaturing concentrations of SDS (Fig. 4A, fractions 17−20, solid bars).
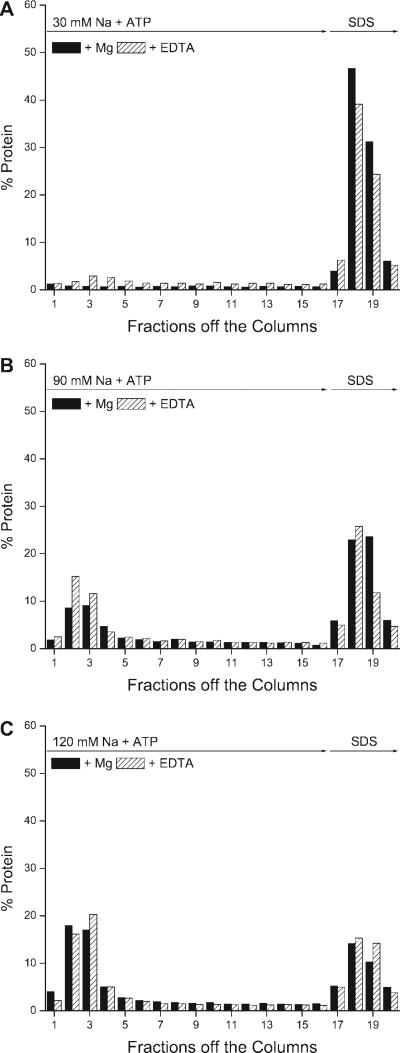
Fig. 4.
Effect of free Mg on the elution of protein from the digoxin-affinity column. A: an eluting solution containing 30 mM Na, 3 mM ATP, 4 mM EDTA, and 20 mM imidazole, pH 7.4, which reduces the free Mg, eluted more protein than a similar solution containing 30 mM Na, 3 mM ATP, and 4 mM Mg (fractions 1−16). The subsequent addition of SDS (fractions 17−20) eluted more protein from the columns previously eluted in the presence of Mg. B: the presence of Mg in the eluting buffer containing 90 mM Na + ATP reduced the amount of protein eluted from the column in fractions 1−5. Nevertheless, there was a substantial amount of protein eluted by the combination of 90 mM Na + ATP + Mg. The subsequent addition of SDS eluted more protein from columns eluted with 90 mM Na + ATP in the presence of Mg compared with those eluted in the absence of Mg (fractions 17−20). The eluting solution added to one of the columns consisted of 90 mM Na, 3 mM ATP, 4 mM EDTA, and 20 mM imidazole, pH 7.4; the other column was eluted with a similar solution in which 4 mM Mg replaced the EDTA. C: the presence of Mg had no effect on the amount of protein eluted in the first 5 fractions in the presence of 120 mM Na + ATP (fractions 2−5) or on the amount of protein eluted with SDS (fractions 17−20). The eluting solution containing 120 mM Na, 3 mM ATP, 4 mM EDTA, and 20 mM imidazole, pH 7.4, was added to one of the columns, and the same solution with 4 mM Mg in place of EDTA was added to the other column. Values are means of duplicates from representative experiments. The total amount of protein in each fraction was measured in duplicate and expressed as a percentage of the total protein eluted from each of the columns. All the data shown are from 1 experiment in which 6 separate columns were run at the same time under the same conditions. The entire experiment was done twice with similar results.
To determine how Na-K-ATPase was being eluted when the ionic strength was increased in the presence of ATP + EDTA (Fig. 1 and 2), we tested how the presence and absence of Mg affected the elution of protein when the NaCl was increased to the intermediate values of 90 and 120 mM Na in the presence of ATP. We reasoned that if Na + ATP was eluting Na-K-ATPase by pulling E2-P to E1, then we would expect that Mg should prevent elution by maintaining Na-K-ATPase in the E2-P conformation. In these experiments, rat kidney microsomes were bound to the digoxin-affinity column in the presence of 30 mM Na + ATP + Mg, and unbound protein was removed. We then added elution buffers containing either 90 mM + ATP + EDTA or 90 mM Na + ATP + Mg. Both solutions eluted much more protein (Fig. 4B, fractions 1−5, hatched and solid bars) than the solution containing 30 mM Na + ATP + EDTA (Fig. 4A, fractions 1−5, hatched bars). Slightly less protein was eluted with the solution containing 90 mM Na + ATP + Mg (Fig. 4B, fractions 1−5, solid bars) compared with the absence of Mg (Fig. 4B, fractions 1−5, hatched bars). Thus, at 90 mM Na, it appears that most of the protein was being eluted by the increase in ionic strength. When the columns were eluted with solutions containing 120 mM, there was a further increase in the amount of protein initially eluted compared with 90 mM Na (Fig. 4B, fractions 1−5) and no difference with or without Mg (Fig. 4C, fractions 1−5, solid vs. hatched bars). We conclude that increasing the NaCl from 30 to 150 mM ATP + EDTA was primarily eluting protein by means of the Hofmeister effect.
Heterogeneity in the elution of Na-K-ATPase
How does one account for the observed heterogeneity with which the Na-K-ATPase (Figs. 1, 2, and 4) eluted from our affinity columns (39)? First, it is important to appreciate that Na-K-ATPase that is eluted with SDS does not just represent nonspecific binding, because little Na-K-ATPase was eluted from sham columns (Fig. 2). Second, in the absence of detergents, distinct populations of Na-K-ATPase likely reside in separate membrane structures, likely to be in either closed or open vesicles.
We estimated the amount of Na-K-ATPase in closed versus open vesicles by doing Na-K-ATPase measurements. The original rat kidney microsomes had a Na-K-ATPase activity of 0.27 ± 0.05 μmol Pi·mg protein−1·min−1, as measured at optimal concentrations of Na, K, Mg, and ATP. Freezing and thawing these microsomes, which was always done before they were applied to the column, increased Na-K-ATPase activity to 0.53 ± 0.09 μmol Pi·mg protein−1·min−1. The increase in activity demonstrates that the freeze-thaw procedure opened closed vesicles. Nevertheless, treating frozen-thawed membranes with SDS in the presence of BSA, which is known to permeabilize membranes much more effectively than freezing and thawing (6), further increased the specific activity to 1.3 ± 0.22 μmol Pi·mg protein−1·min−1. Thus approximately half of the Na-K-ATPase in our frozen-thawed membranes was in closed vesicles. Theoretically, closed vesicles could be in either the right side-out or inside-out configuration. However, only vesicles in the right side-out configuration should bind to the column. Inside-out vesicles should pass through without binding, because the binding site for cardiac glycosides is on the inside of the vesicle.
On the basis of the above considerations, we conclude that half of the protein in basolateral membranes that bound to the column would have been in structures permeable to Mg and ATP, and the other half would have been in closed right side-out vesicles. Therefore, it is possible that some of the observed heterogeneity in the elution of Na-K-ATPase from the column could be due to the presence of trapped ligands in the right side-out vesicles. On the other hand, half of the bound protein would have been in permeable structures. Therefore, we expected that at least half of the bound protein would be rapidly eluted in response to ligands that cause a decay of E2-P. Instead, we found that only 10% or less was eluted over the expected time course when the free Mg was reduced (Fig. 4A, fractions 1−5, hatched bars). Therefore, a significant proportion of Na-K-ATPase that was in permeable structures must have stayed bound to the column for a much longer period of time for reasons other than the occlusion of pump ligands. Thus vesicle heterogeneity can account for some, but not all, of Na-K-ATPase that is eluted with SDS.
There is certainly evidence for functional heterogeneity in the reaction mechanism of Na-K-ATPase in animal cell membranes (10, 34). For example, the turnover of the phosphointermediate in dog kidney demonstrates both fast and slow components (34), and there are rapid and slow components to the release of occluded Rb (10). Heterogeneity in the rate at which Na-K-ATPase releases cardiac glycosides might also be explained by the extent to which Na-K-ATPase in the plasma membrane interacts with other proteins. Part of Na-K-ATPase in the kidney is localized in caveolae (38), and the binding of ouabain to Na-K-ATPase alters how it interacts with other proteins (36). We would expect that the binding of Na-K-ATPase to other proteins could stabilize its binding to cardiac glycosides and theoretically help account for a population of Na-K-ATPase that was slow to release bound digoxin. Thus our data and subsequent analysis support the hypothesis that there could be two populations of Na-K-ATPase molecules in rat kidney microsomes that vary in the rate at which they release cardiac glycosides, which leaves open the option that ANG II increased the rate of elution from the ouabain-affinity column by decreasing the amount of Na-K-ATPase that was eluted with SDS (39).
Binding and elution of rat kidney Na-K-ATPase from the lysates of OK cells
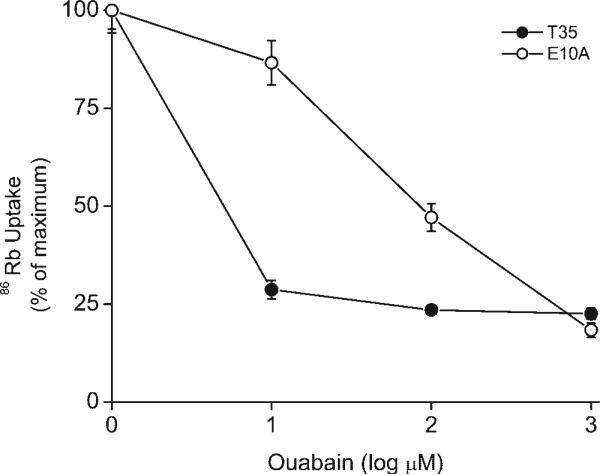
To study how ANG II regulates Na-K-ATPase activity, the wild-type rat α1-subunit was expressed in OK cells that stably express the rat AT1a receptor (T35 cells) and selected for growth by maintaining the cells in medium that contained 10 μM ouabain (materials and methods). In cells transfected with the wild-type rat α1-subunit and selected for growth (E10A cells), the rat α1-subunit accounts for 75% of the total Na-K-ATPase and the endogenous OK Na-K-ATPase makes up the other 25% (Fig. 5). The absolute pumping rate for the rat α1-subunit in E10A cells, measured as the difference in 86Rb influx in the presence of 1 μM and 6 mM ouabain, was the equivalent of 1.54 ± 0.34 nmol K+·mg−1·min−1 (n = 3).
Fig. 5.
86Rb influx is much more sensitive to inhibition by ouabain in parental opossum kidney (OK) cells that stably express the AT1a receptor (T35 cells) compared with T35 cells subsequently transfected with the wild-type rat α1 cDNA and selected for growth in medium with 10 μM ouabain (E10A cells). The rat α1-subunit is known to be much less sensitive to inhibition by ouabain. Therefore, these data show that E10A cells expressed the rat α1-subunit and that this isoform of Na-K-ATPase accounted for ∼75% of the ouabain-sensitive uptake of 86Rb uptake. Values are means ± SE (n = 6).
As the first step in determining how ANG II affects the elution of Na-K-ATPase, we tested the relationship between the amount of Na-K-ATPase (Fig. 6A) and the amount of protein (Fig. 6B) eluted from the digoxin-affinity column. Na-K-ATPase in the lysates of OK cells was bound to the digoxin-affinity column in the presence of a solution containing 30 mM NaCl + ATP + Mg that favors the E2-P conformation of Na-K-ATPase, and the unbound Na-K-ATPase was removed by washing the column with the loading buffer (materials and methods) as was done with the rat kidney microsomes. The pattern by which protein was subsequently eluted from the digoxin-affinity column in response to reducing the free Mg (Fig. 6A, fractions 1−10), increasing the NaCl concentration to 150 mM (Fig. 6A, fractions 11−18), and adding SDS (Fig. 6A, fractions 19−22) is very similar to the pattern observed with rat kidney microsomes. For instance, reducing the free Mg while maintaining the 30 mM Na and ATP only eluted a small peak of protein in columns loaded with either rat kidney microsomes (Fig. 4A, fractions 1−16)or the lysates of OK cells (Fig. 6A, fractions, 1−10). Likewise, increasing the NaCl concentration from 30 to 150 mM eluted a much larger amount of protein from columns loaded with either rat kidney microsomes (Figs. 2B, fractions 1−16)or protein from OK cells (Fig. 6A, fractions 11−18). Finally, the addition of SDS eluted a comparable amount of protein from columns loaded with either rat kidney microsomes (Fig. 2B, fractions 19 and 20) or protein from OK cells (Fig. 6A, fractions 19−21). As was the case with rat kidney microsomes (Fig. 2), protein was eluted from the column in the same pattern as the α-subunit of the Na-K-ATPase (Fig. 6B). Note that the data in Fig. 6B were obtained by taking equal aliquots from each fraction, and there was no attempt to quantify the antibody response as was done in Fig. 2A. The anti-α antibody used in this study detects both rat α1 and the endogenous OK Na-K-ATPase (data not shown). Nevertheless, the pattern by which protein from OK cells (Fig. 6A) and rat kidney micro-somes (Figs. 2B and 4) are eluted is very similar. Therefore, there are no data to suggest that the elution pattern is significantly altered by the small amount of endogenous Na-K-ATPase that is present in E10A cells. Thus we conclude that the Na-K-ATPase is eluting from the digoxin affinity column in a similar fashion whether it is present in rat kidney micro-somes or in the membranes of OK cells.
Fig. 6.
Total protein (A) and α-subunit of Na-K-ATPase (B) from the lysates of E10A cells were partially eluted from digoxin-affinity columns in response to 2 solutions that trigger the decay of E2-P; the remainder was eluted with SDS. The data in A are from 6 separate experiments; values are mean ± SE from all 6 experiments. The data in B are from 1 of the 6 experiments. An equal volume of sample was taken from each fraction for immunoblotting.
Effect of ANG II on the elution of wild-type rat α1-subunit from the lysates of OK cells
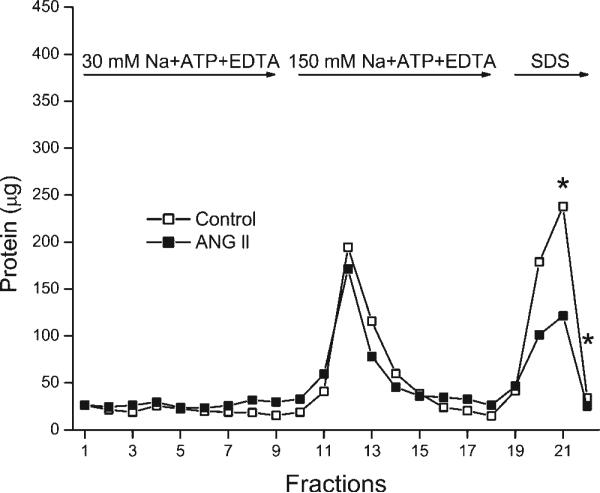
To test how ANG II is altering the interaction of the rat kidney Na-K-ATPase with digoxin, OK cells coexpressing the AT1a receptor and the wild-type rat α1-isoform of Na-K-ATPase were incubated with or without 10 nM ANG II for 5 min at room temperature. The cells were lysed in the presence of protease and phosphatase inhibitors and ligands (30 mM NaCl + ATP + Mg) that favor the E2-P conformation of Na-K-ATPase, and Na-K-ATPase was bound to the digoxin-affinity column (materials and methods). On digoxin-affinity columns loaded with protein from control cells, adding an eluting buffer (30 mM Na + ATP + EDTA) that maintained the original concentration of sodium and ATP but removed the free Mg resulted in the elution of protein with an average response that peaked in fraction 4 (Fig. 7, fractions 1−9, open symbols). On columns loaded with identical amounts of protein from cells that had been previously incubated with ANG II, the elution of protein in response to reducing the free Mg was slower, with an average response that reached a maximum in fraction 9 (Fig. 7, fractions 1−9, closed symbols). Adding the second eluting solution containing 150 mM Na + ATP + EDTA resulted in the removal of a large amount of protein from control cells (Fig. 7, fractions 10−18, open symbols) and from cells treated with ANG II (Fig. 7, fractions 10−18, closed symbols). The subsequent addition of 2% SDS also resulted in the elution of a large amount of protein from control and ANG II-treated cells (Fig. 7, fractions 19−22).
Fig. 7.
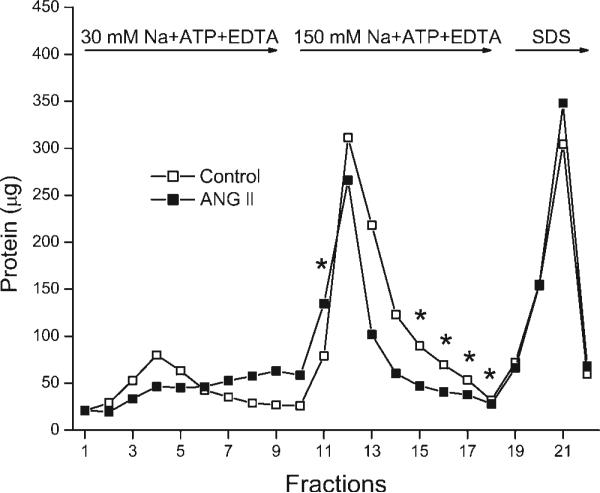
Treating E10A cells with or without 10 nM ANG II significantly altered the subsequent elution of cellular protein from the digoxin-affinity column (P ≤ 0.001). There was also a significant interaction between ANG II and the time of elution (P ≤ 0.001). *P ≤ 0.05 indicates values significantly different from control values as detected by post hoc analysis. These data show that ANG II alters the interaction of the wild-type rat α1-subunit of Na-K-ATPase with the cardiac glycoside digoxin. The experiment was carried out as described in materials and methods and is explained in results and discussion. The data are from 6 separate experiments. Data points shown for each fraction are means of the values from all 6 experiments.
When all of the data in Fig. 7 were analyzed using a two-way ANOVA with repeated measures with two between-subject factors, there was a significant difference in the elution of protein from control and ANG II-treated cells (P ≤ 0.001). A post hoc analysis of the data showed that there were significant differences (P ≤ 0.05) in the amount of protein eluted in fraction 11 and fractions 15−18, as indicated in Fig. 7. There are other fractions, such as fraction 13 in Fig. 7, that appear to be different with and without ANG II but that are not statistically different due to more variation between individual experiments. The ANG II-induced increase in the amount of protein in fraction 11 and the reduction in the amount of protein in fractions 15−18 suggest that there was a leftward shift in the elution of protein in response to the solution containing 150 mM Na + ATP + EDTA (Fig. 7). There were no significant differences in the amount of protein eluted with the reduction in free Mg, but there was a rightward shift in the elution profile (Fig. 7, fractions 1−9). Thus ANG II altered the kinetic response to ligands that affect the amount of E2-P without changing the amount of Na-K-ATPase eluted with SDS. The kinetic response was likely to slow the release of Na-K-ATPase from digoxin when the free Mg was reduced, although this effect was not statistically significant, and to accelerate significantly the release when the concentration of NaCl was increased in the presence of ATP and EDTA.
OK cell membranes containing a truncation mutant of rat α1-subunit missing the first 32 amino acids of the NH2 terminus
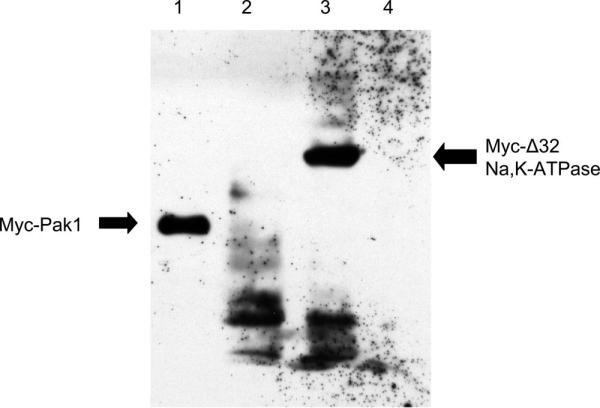
Since the interaction of Na-K-ATPase with other proteins in caveolae are mediated by the NH2 terminus of Na-K-ATPase (36, 38, 41), we wanted to test how ANG II affected the elution of protein in cells expressing a form of the rat α1-subunit missing this part of its NH2 terminus. To carry out these experiments, we transfected T35 cells with the cDNA from a truncation mutant of the rat α1-isoform in which the sequence encoding the first 32 amino acids of the NH2 terminus was replaced by a Myc epitope tag (α-1.Δ32). After selection in 10 μM ouabain, immunoblotting of lysates of these cells showed a band at the expected size with the anti-myc antibody, whereas cells transfected with the full-length rat α1-subunit or the parental OKAT1a cells did not (Fig. 8). The absolute pumping rate for the truncation mutant, measured as the difference in 86Rb influx in the presence of 1 μMand6mM ouabain, was 0.42 ± 0.11 nmol·mg−1·min−1 (n = 4).
Fig. 8.
An immunoblot prepared using an anti-Myc Tag antibody showing that a truncation mutant of the rat α1-subunit missing the first 32 amino acids of the NH2 terminus (α-1.Δ32), which contains a Myc tag, is expressed in T35 cells (lane 3, Myc-Δ32). Lane 1 is a positive control of COS7 cells transfected with Myc-Pak1 plasmid. Lane 2 is a negative control from E10A cells. Lanes 2 and 3 contain ∼30 μg protein of whole cell lysates.
In experiments with cells expressing α-1.Δ32, ANG II significantly affected the elution of protein (Fig. 9), as shown by a two-way ANOVA using repeated measures with two between-subject factors (P ≤ 0.001). The post hoc analysis showed that ANG II significantly (P ≤ 0.05) decreased the amount of protein eluted with SDS (Fig. 9, fractions 21 and 22) but had no effect on the amount of protein eluted in response to the buffer containing 30 or 150 mM Na + ATP + EDTA. Thus, with the truncation mutant, ANG II reduced the amount of tightly bound Na-K-ATPase eluted with SDS and had no effect on the kinetic response to ligands that reduce the amount of E2-P.
Fig. 9.
Treating OK cells expressing α-1.Δ32 with or without 10 nM ANG II significantly altered the subsequent elution of cellular protein (P ≤ 0.001). The same analysis also showed a significant interaction between ANG II and the time of elution (P ≤ 0.001). A post hoc analysis of the data showed that there were significant differences (P ≤ 0.05) in the amount of protein eluted in fractions 21 and 22.*P ≤ 0.05 indicates values significantly different from control values as detected by post hoc analysis. These results show that ANG II can still affect the interaction of the rat kidney Na-K-ATPase with digoxin when Tyr10, Ser16, and Ser23 are missing. Data points shown for each fraction are means of the values from 5 experiments.
Although we did not test for a significant difference, there appeared to be less total protein eluted from columns loaded with the lysates of cells expressing the truncation mutant (Fig. 9) compared with cells expressing rat wild-type α-subunit (Fig. 7). Since equal amounts of cellular protein were added to all the columns, this apparent reduction is likely related to properties associated with the truncation mutant. It is well established that removing the NH2 terminus shifts the Na-K-ATPase toward E1 (17) and that removing the first 32 amino acids reduces the catalytic turnover by ∼50% (31). We did indeed measure a fourfold lower ouabain-sensitive 86Rb uptake in cells expressing α-1.Δ32 compared with E10A cells, and one would certainly expect an E1-shifted Na-K-ATPase to have reduced binding to digoxin. On the other hand, we did not directly measure the amount of Na-K-ATPase expressed in these two cell lines. Therefore, it is possible that the reduced recovery and lower activity is in part due to less expression of Na-K-ATPase.
Interpreting ANG II-induced changes in elution in terms of Na-K-ATPase properties:elution via change in conformation versus denaturation
The observation that ANG II significantly increased the rate at which the wild-type rat kidney Na-K-ATPase was eluted from the digoxin-affinity column in response to the solution containing 150 mM NaCl + ATP + EDTA without reducing the amount eluted with SDS (Fig. 7) suggests that these two effects can be independent of each other. There was, however, an ANG II-induced reduction in the amount of Na-K-ATPase that was eluted from the ouabainaffinity column with SDS (39). Thus it is possible that in these earlier experiments, ANG II both increased the rate of elution in response to the solution containing 150 mM Na + ATP + EDTA and/or reduced the amount of Na-K-ATPase eluted with SDS (39). The fact that ANG II did not decrease the amount of Na-K-ATPase eluted from the digoxin-affinity column with SDS may be explained by the ∼10-fold higher affinity with which the rat α1-subunit binds digoxin compared with ouabain (24).
The observation that ANG II significantly reduced the amount of the truncated α1-subunit eluted from the digoxin-affinity column with SDS confirms that ANG II can affect the amount of tightly bound Na-K-ATPase. The fact that we only observed an effect of ANG II on the SDS-eluted Na-K-ATPase in cells expressing the truncation mutant suggests that the NH2 terminus plays a critical role in mediating how Na-K-ATPase remains tightly bound to digoxin. Others have shown that the NH2 terminus of Na-K-ATPase likely plays a central role in the interaction of Na-K-ATPase with proteins in caveolae (36, 38, 41). Perhaps these other proteins restrain the ability of Na-K-ATPase to change conformation and release digoxin. In any event, we suggest that the absence of part of the NH2 terminus weakened the interaction of Na-K-ATPase with other proteins so that ANG II induced a more complete removal of the bound protein before the SDS was added.
Analysis of the kinetic response in terms of expected Na-K-ATPase activity
Since the effect of ANG II on the elution of Na-K-ATPase in response to ligands could in part be due to changes in the kinetic properties of Na-K-ATPase, we developed the following analysis of our results. Based on our analysis of how reducing the free Mg eluted rat kidney microsomes (Fig. 4A), the rightward shift in the elution curve that occurred when the buffer containing 30 mM Na + ATP + EDTA was applied to the column (Fig. 7, fractions 1−9) suggests that ANG II could be slowing the decay of E2-P → E2 → E1 → E1·ATP → Na·E1·ATP. We would expect that this effect would inhibit Na-K-ATPase activity. The observation that reducing the free Mg eluted no protein from columns loaded with protein from cells expressing α-1.Δ32 (Fig. 9) is consistent with a shift toward E1, which would be expected to slow the spontaneous decay of E2-P. Likewise, the leftward shift observed in Fig. 7 in response to increasing the NaCl concentration from 30 to 150 mM can be explained by ANG II increasing the amount of E1-P, based on our conclusion that the solution containing 150 mM Na + ATP + EDTA was eluting Na-K-ATPase via the Hofmeister effect (27). The normal reaction mechanism of Na-K-ATPase proceeds from E1-P to E2-P. Therefore, increasing the amount of E1-P would be consistent with an inhibition of Na-K-ATPase activity. The conclusion that 10 nM ANG II was producing changes in the elution of the rat kidney α1-subunit from the digoxin-affinity column, which is consistent with an inhibition of Na-K-ATPase activity was unexpected, because others have reported that 10 nM ANG II stimulates transcellular sodium transport in OK cells that stably express the AT1a receptor and the endogenous OK Na-K-ATPase (35). On the other hand, concentrations of ANG II in the 10 nM range are often associated with the inhibition of sodium reabsorption in the rat proximal tubule (9) and an inhibition of rat kidney Na-K-ATPase activity (2). Furthermore, when the rat kidney α1-subunit is expressed in OK cells, it is also directly stimulated by picomolar concentrations of ANG II (5).
Since the ratio of E2-P to E1 is one of the factors that determines the affinity of Na-K-ATPase for intracellular sodium and extracellular potassium (16, 31), our results suggest that ANG II could be altering the apparent affinity for sodium and or potassium. This conclusion is consistent with the observation that phosphorylation of Ser23 by PKC alters the E1-E2 poise (19) and evidence that ANG II increases the affinity of Na-K-ATPase for intracellular Na (1).
Conclusions
On the basis of the above-described analysis, we conclude that ANG II stimulates the rate of elution from the digoxin-affinity column by increasing the kinetic response to ligands that trigger a decay in E2-P by a mechanism that involves the first 32 amino acids of the NH2 terminus of Na-K-ATPase. The increased kinetic response could be due to either a change in a rate-limiting step in the reaction mechanism of Na-K-ATPase or a change in how the Na-K-ATPase interacts with proteins that affect the ability of Na-K-ATPase to change conformation, or some combination of these two mechanisms. Elucidating how ANG II regulates the interaction of Na-K-ATPase with cardiac glycosides is relevant to developing a more complete understanding of the molecular mechanisms by which ANG II directly regulates the activity of Na-K-ATPase in the proximal tubule. The concept that ANG II can affect the interaction of Na-K-ATPase with cardiac glycosides should also be explored to determine whether ANG II can regulate the sensitivity of Na-K-ATPase to endogenous ouabain and to understand the clinical implications of treating patients with both ACE inhibitors and cardiac glycosides (3, 12).
ACKNOWLEDGMENTS
We thank Haiping Chen for the preparation of rat kidney microsomes, Abe Dakhlallah for doing protein assays, Teodora C. Palcu for data analysis and preparation of figure, Jonathan W. Wojtkowiak for culturing cells, Bulent Ozkan for consulting on statistical analysis, and Linda McCraw for help in preparing the manuscript.
GRANTS
This work was supported by National Institutes of Health Grant R01 DK-60752 (to D. R. Yingst) and 1R01 HL-079102 (to N. F. Rossi).
REFERENCES
- 1.Aperia A, Holtbaäck U, Syrén ML, Svensson LB, Fryckstedt J, Greengard P. Activation/deactivation of renal Na+,K+-ATPase: a final common pathway for regulation of natriuresis. FASEB J. 1994;8:436–439. doi: 10.1096/fasebj.8.6.8168694. [DOI] [PubMed] [Google Scholar]
- 2.Bharatula M, Hussain T, Lokhandwala MF. Angiotensin II AT1 receptor/signaling mechanisms in the biphasic effect of the peptide on proximal tubular Na+,K+-ATPase. Clin Exp Hypertens. 1998;20:465–480. doi: 10.3109/10641969809053225. [DOI] [PubMed] [Google Scholar]
- 3.Dagenais DR, Pogue J, Fox K, Simoons ML, Yusuf S. Angiotensin-converting-enzyme inhibitors in stable vascular disease without left ventricular systolic dysfunction or heart failure: a combined analysis of three trials. Lancet. 2006;368:581–588. doi: 10.1016/S0140-6736(06)69201-5. [DOI] [PubMed] [Google Scholar]
- 4.Efendiev R, Pedemonte CH. Contrary to rat-type, human-type Na,K-ATPase is phosphorylated at the same amino acid by hormones that produce opposite effects on enzyme activity. J Am Soc Nephrol. 2006;17:31–38. doi: 10.1681/ASN.2005070681. [DOI] [PubMed] [Google Scholar]
- 5.Efendiev R, Budu CE, Cinelli AR, Bertorello AM, Pedemonte CH. Intracellular Na+ regulates dopamine and angiotensin II receptors availability at the plasma membrane and their cellular responses in renal epithelia. J Biol Chem. 2003;278:28719–28726. doi: 10.1074/jbc.M303741200. [DOI] [PubMed] [Google Scholar]
- 6.Forbush B. Assay of Na,K-ATPase in plasma membrane preparations: increasing the permeability of membrane vesicles using sodium dodecyl sulfate buffered with bovine serum albumin. Anal Biochem. 1983;128:159–163. doi: 10.1016/0003-2697(83)90356-1. [DOI] [PubMed] [Google Scholar]
- 7.Garvin JL. Angiotensin stimulates bicarbonate transport and Na+/K+ ATPase in at proximal straight tubules. J Am Soc Nephrol. 1991;1:1146–1152. doi: 10.1681/ASN.V1101146. [DOI] [PubMed] [Google Scholar]
- 8.Hansen O. Interaction of cardiac glycosides with (Na+ + K+)-activated ATPase. A biochemical link to digitalis-induced inotrophy. Pharmacol Rev. 1984;36:143–163. [PubMed] [Google Scholar]
- 9.Harris PJ, Young JA. Dose-dependent stimulation and inhibition of proximal tubular sodium reabsorption by angiotensin II in the rat kidney. Pflügers Arch. 1977;367:295–297. doi: 10.1007/BF00581370. [DOI] [PubMed] [Google Scholar]
- 10.Hasenauer J, Huang WH, Askari A. Allosteric regulation of the access channels to the Rb+ occlusion sites of (Na+ + K+)-ATPase. J Biol Chem. 1993;268:3289–3297. [PubMed] [Google Scholar]
- 11.Hoffman JF, Potapova O, Wickreme A, Yingst DR. Na pump isoforms in human red cells. Proc Natl Acad Sci USA. 2002;99:14572–14577. doi: 10.1073/pnas.222539999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hoppe UC, Erdmann E. Digitalis in heart failure! Still applicable? Z Kardiol. 2005;94:307–311. doi: 10.1007/s00392-005-0219-0. [DOI] [PubMed] [Google Scholar]
- 13.Huang WH, Askari A. Red cell Na+,K+-ATPase: a method for estimating the extent of inhibition of an enzyme sample containing an unknown amount of bound cardiac glycoside. Life Sci. 1975;16:1253–1261. doi: 10.1016/0024-3205(75)90310-0. [DOI] [PubMed] [Google Scholar]
- 14.Humphrey PA, Lupfert C, Apell HJ, Cornelius F, Clarke RJ. Mechanism of the rate-determining step of the Na+,K+-ATPase pump cycle. Biochemistry. 2002;41:9496–9507. doi: 10.1021/bi025836o. [DOI] [PubMed] [Google Scholar]
- 15.Jorgensen PL. Isolation and characterization of components of the sodium pump. Q Rev Biophys. 1975;7:239–274. doi: 10.1017/s0033583500001426. [DOI] [PubMed] [Google Scholar]
- 16.Jorgensen PL. Transmission of E1–E2 structural changes in response to Na+ or K+ binding in Na,K-ATPase. Ann NY Acad Sci. 2003;986:22–30. doi: 10.1111/j.1749-6632.2003.tb07135.x. [DOI] [PubMed] [Google Scholar]
- 17.Jorgensen PL, Andersen JP. Structural basis for E1–E2 conformational transitions in Na,K-pump and Ca-pump proteins. J Membr Biol. 1988;103:95–120. doi: 10.1007/BF01870942. [DOI] [PubMed] [Google Scholar]
- 18.Liang M, Tian J, Liu L, Pierre S, Liu J, Shapiro J, Xie ZJ. Identification of a pool of non-pumping Na/K-ATPase. J Biol Chem. 2007;282:10585–10593. doi: 10.1074/jbc.M609181200. [DOI] [PubMed] [Google Scholar]
- 19.Logvinenko NS, Dulubova I, Fedosova N, Larsson SH, Nairn AC, Esmann M, Greengard P, Aperia A. Phosphorylation by protein kinase C of serine-23 of the α-1 subunit of rat Na+,K+-ATPase affects its conformational equilibrium. Proc Natl Acad Sci USA. 1996;93:9132–9137. doi: 10.1073/pnas.93.17.9132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lücking K, Nielsen JM, Pedersen PA, Jørgensen PL. Na-K-ATPase isoform (α3, α2, α1) abundance in rat kidney estimated by competitive RT-PCR and ouabain binding. Am J Physiol Renal Fluid Electrolyte Physiol. 1996;271:F253–F260. doi: 10.1152/ajprenal.1996.271.2.F253. [DOI] [PubMed] [Google Scholar]
- 21.Mansier P, Charlemagne D, Rossi B, Preteseille M, Swynghedauw B, Lelievre L. Isolation of impermeable inside-out vesicles from an enriched sarcolemma fraction of rat heart. J Biol Chem. 1983;258:6628–6635. [PubMed] [Google Scholar]
- 22.Mattingly RR. Mitogen-activated protein kinase signaling in drug-resistant neuroblastoma cells. Methods Mol Biol. 2003;218:71–83. doi: 10.1385/1-59259-356-9:71. [DOI] [PubMed] [Google Scholar]
- 23.Morth JP, Pedersen BP, Toustrup-Jensen MS, Sorensen TLM, Petersen J, Andersen JP, Vilsen B, Nissen P. Crystal structure of the sodium-potassium pump. Nature. 2007;450:1043–1049. doi: 10.1038/nature06419. [DOI] [PubMed] [Google Scholar]
- 24.Noel F, Fagoo M, Godfraind T. A comparison of the affinities of rat (Na+ + K+)-ATPase isozymes for cardioactive steroids, role of lactone ring, sugar moiety and KCl concentration. Biochem Pharmacol. 1990;40:2611–2616. doi: 10.1016/0006-2952(90)90578-9. [DOI] [PubMed] [Google Scholar]
- 25.Pedemonte CH, Pressley TA, Cinelli AR, Lokhandwala MF. Stimulation of protein kinase C rapidly reduces intracellular Na+ concentration via activation of the Na+ pump in OK cells. Mol Pharmacol. 1997;52:88–97. doi: 10.1124/mol.52.1.88. [DOI] [PubMed] [Google Scholar]
- 26.Periyasamy SM, Huang WH, Askari A. Origins of the different sensitivities of (Na+ + K+)-dependent adenosinetriphosphatase preparations to ouabain. Comp Biochem Physiol B. 1983;76:449–454. doi: 10.1016/0305-0491(83)90274-2. [DOI] [PubMed] [Google Scholar]
- 27.Post RL, Klodos I. Interpretation of extraordinary kinetics of Na+-K+-ATPase by a phase change. Am J Physiol Cell Physiol. 1996;271:C1415–C1423. doi: 10.1152/ajpcell.1996.271.5.C1415. [DOI] [PubMed] [Google Scholar]
- 28.Post RL, Kume S, Tobin T, Orcutt B, Sen AK. Flexibility of an active center in sodium-plus-potassium adenosine triphosphate. J Gen Physiol. 1969;54:306s–326s. doi: 10.1085/jgp.54.1.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ribeiro AB. Angiotensin II antagonists—therapeutic benefits spanning the cardiovascular disease continuum from hypertension to heart failure and diabetic nephropathy. Curr Med Res Opin. 2006;22:1–16. doi: 10.1185/030079905X75041. [DOI] [PubMed] [Google Scholar]
- 30.Schön R, Schönfeld W, Menke KH, Repke KRH. Mechanism and role of Na+/Ca2+ competition in (Na,K)-ATPase. Acta Biol Med Ger. 1972;29:643–659. [PubMed] [Google Scholar]
- 31.Segall L, Lane LK, Blostein R. New insights into the role of the N terminus in conformational transitions of the Na,K-ATPase. J Biol Chem. 2002;277:35202–35209. doi: 10.1074/jbc.M206115200. [DOI] [PubMed] [Google Scholar]
- 32.Sen AK, Tobin T, Post RL. A cycle for ouabain inhibition of sodium- and potassium-dependent adenosine triphosphatase. J Biol Chem. 1969;244:6596–6604. [PubMed] [Google Scholar]
- 33.Skou JC. The Na,K-pump. Methods Enzymol. 1988;156:1–25. doi: 10.1016/0076-6879(88)56004-4. [DOI] [PubMed] [Google Scholar]
- 34.Suzuki K, Post RL. The Sodium Pump: Recent Developments. Rockefeller Univ. Press; New York: 1991. Slow and rapid components of dephosphorylation kinetics of Na,K-ATPase. pp. 375–378. [Google Scholar]
- 35.Thekkumkara TJ, Cookson R, Linas SL. Angiotensin (AT1A) receptor-mediated increases in transcellular Na transport in proximal tubule cells. Am J Physiol Renal Physiol. 1998;274:F897–F905. doi: 10.1152/ajprenal.1998.274.5.F897. [DOI] [PubMed] [Google Scholar]
- 36.Tian J, Cai T, Yuan Z, Wang H, Liu L, Haas M, Maksimova E, Huang XY. Binding of Src to Na+/K+-ATPase forms a functional signaling complex. Mol Biol Cell. 2006;17:317–326. doi: 10.1091/mbc.E05-08-0735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Van Hussye JW, Jewell EA, Lingrel JB. Site-directed mutagenesis of a predicted cation binding site of Na,K-ATPase. Biochemistry. 1993;32:819–826. doi: 10.1021/bi00054a012. [DOI] [PubMed] [Google Scholar]
- 38.Wang H, Haas M, Liang M, Cai T, Tian J, Li S, Xie Z. Ouabain assembles signaling cascades through the caveolar Na+/K+-ATPase. J Biol Chem. 2004;279:17250–17259. doi: 10.1074/jbc.M313239200. [DOI] [PubMed] [Google Scholar]
- 39.Yingst DR, Massey KJ, Rossi NF, Mohanty MJ, Mattingly RR. Angiotensin II directly stimulates activity and alters the phosphorylation of Na-K-ATPase in rat proximal tubule with a rapid time course. Am J Physiol Renal Physiol. 2004;287:F713–F721. doi: 10.1152/ajprenal.00065.2004. [DOI] [PubMed] [Google Scholar]
- 40.Yingst DR, Yang SH, Schiebinger R. Purification of active Na+-K+-ATPase using a new ouabain-affinity column. Am J Physiol Cell Physiol. 1998;275:C1167–C1177. doi: 10.1152/ajpcell.1998.275.4.C1167. [DOI] [PubMed] [Google Scholar]
- 41.Zhang S, Malmersjo S, Li J, Ando H, Aizman O, Uhlen P, Mikoshiba K, Aperia A. Distinct role of the N-terminal tail of the Na,K-ATPase catalytic subunit as a signal transducer. J Biol Chem. 2006;281:21954–21962. doi: 10.1074/jbc.M601578200. [DOI] [PubMed] [Google Scholar]