Abstract
Neural prostheses aim to provide treatment options for individuals with nervous-system disease or injury. It is necessary, however, to increase the performance of such systems before they can be clinically viable for patients with motor dysfunction. One performance limitation is the presence of correlated trial-to-trial variability that can cause neural responses to wax and wane in concert as the subject is, for example, more attentive or more fatigued. If a system does not properly account for this variability, it may mistakenly interpret such variability as an entirely different intention by the subject. We report here the design and characterization of factor-analysis (FA)–based decoding algorithms that can contend with this confound. We characterize the decoders (classifiers) on experimental data where monkeys performed both a real reach task and a prosthetic cursor task while we recorded from 96 electrodes implanted in dorsal premotor cortex. The decoder attempts to infer the underlying factors that comodulate the neurons' responses and can use this information to substantially lower error rates (one of eight reach endpoint predictions) by ≲75% (e.g., ∼20% total prediction error using traditional independent Poisson models reduced to ∼5%). We also examine additional key aspects of these new algorithms: the effect of neural integration window length on performance, an extension of the algorithms to use Poisson statistics, and the effect of training set size on the decoding accuracy of test data. We found that FA-based methods are most effective for integration windows >150 ms, although still advantageous at shorter timescales, that Gaussian-based algorithms performed better than the analogous Poisson-based algorithms and that the FA algorithm is robust even with a limited amount of training data. We propose that FA-based methods are effective in modeling correlated trial-to-trial neural variability and can be used to substantially increase overall prosthetic system performance.
INTRODUCTION
Neural prostheses, which are also termed brain–machine and brain–computer interfaces (BCIs), offer the potential to substantially increase the quality of life for people suffering from motor disorders, including paralysis and amputation. Such devices translate electrical neural activity from the brain into control signals for guiding paralyzed upper limbs, prosthetic arms, and computer cursors. A few research groups have now demonstrated that monkeys (e.g., Carmena et al. 2003; Musallam et al. 2004; Santhanam et al. 2006a; Serruya et al. 2002; Taylor et al. 2002; Velliste et al. 2008) and humans (e.g., Hochberg et al. 2006; Kennedy et al. 2000; Leuthardt et al. 2004; Schalk et al. 2008; Wolpaw and McFarland 2004) can learn to move computer cursors and robotic arms to various target locations simply by activating neural populations that participate in natural arm movements. Although encouraging, even these compelling proof-of-concept, laboratory-based systems fall short of exhibiting the level of performance needed for many everyday behaviors and for achieving clinical viability.
We previously demonstrated the design and implementation of a neural prosthetic system based on neural activity from an electrode array implanted in dorsal premotor cortex (PMd) (Santhanam et al. 2006a). Activity in this region is known to correlate with an upcoming reach endpoint, including both direction and extent (Messier and Kalaska 2000). Using this information, we demonstrated the ability to predict the subject's intended reach (e.g., one of eight potential targets) at a performance much higher than previously reported. The improvements were achieved by systematically designing the neural analysis epochs and implementing standard maximum likelihood estimators (Zhang et al. 1998) based on simple Gaussian and Poisson statistical models of neural firing rate. These models offer ease of computation, have been used in similar studies, and are fairly standard in the field (e.g., Brockwell et al. 2004; Hatsopoulos et al. 2004; Maynard et al. 1999; Shenoy et al. 2003). We now revisit the choice of these models to further increase prosthetic performance and, perhaps, to gain insight into the modeling of neural activity.
Reach endpoint is the “signal” that we seek to extract from the neural recordings. Other systems attempt to predict continuous arm kinematics (see Velliste et al. 2008, among others) and, although the techniques presented here can apply to those systems, we currently restrict ourselves to discrete reach-endpoint classifiers. Reach endpoint is a primary influence on PMd activity during the planning of upcoming movements, although there is also a variety of other factors that modulate neural observations from experimental trial to trial. For example, there is evidence that PMd activity can depend on other behavioral aspects of movement control, including the type of grasp (Godschalk et al. 1985), the required accuracy (Gomez et al. 2000), reach curvature (Hocherman and Wise 1991), reach speed (Churchland et al. 2006a), and (to some degree) force (Riehle et al. 1994). Additionally, there can be other types of unobserved influences (e.g., attentional states and biophysical “spiking noise”) (Chestek et al. 2007; Musallam et al. 2004) that further modulate cortical activity, even when observable behavior is held fixed. These various influences may inadvertently mask the signal of interest (reach endpoint) and thereby reduce prediction (decoding) accuracy.
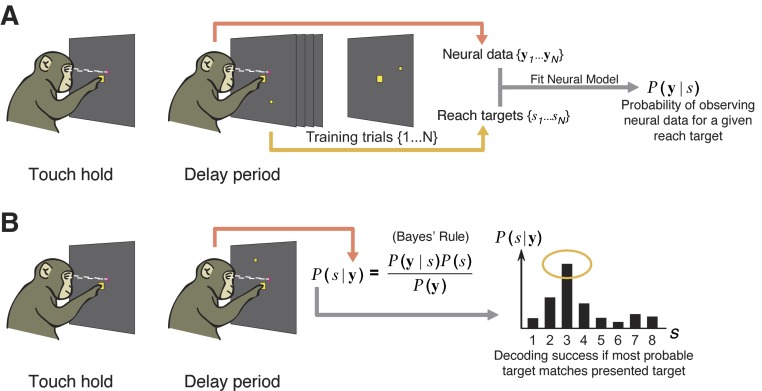
Figure 1 A provides an example of the trial-to-trial variability present in the activity of two recorded neurons. On each trial, a subject is randomly presented one of eight reach targets and is instructed to make a reach to that target after some delay.1 Trials were first grouped by upcoming reach endpoint and, for each neuron, the emitted action potentials (spikes) were counted in a fixed time interval after the endpoint was cued to the subject. This overall neural response is the well-known “tuning curve” (e.g., Georgopoulos et al. 1982). Note the large variability in spike counts within each endpoint's set of trials, which results in overlap between the ranges of spike counts for reach endpoint. The challenge for any prediction algorithm is to avoid mistaking these variations as being the signature for an entirely different reach endpoint.
FIG. 1.
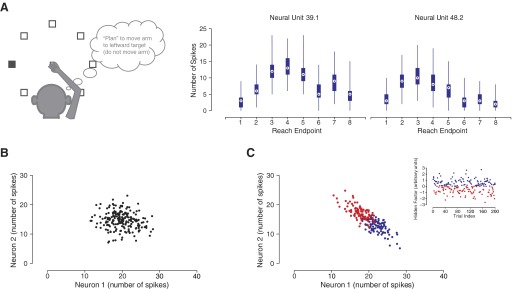
A: schematic of experimental task where subject reaches to one of 8 reach endpoints. Data from 2 real neurons are shown where action potentials were counted over a 200-ms window after target presentation for each experimental trial. Trials were segregated by reach endpoint and a box plot was generated denoting the median spike count (white circles with black dots), the 25th to 75th percentiles of the spike counts (blue rectangles), and the ranges of the outliers (thin “whiskers”) for each endpoint. B: simulation of 2 neurons whose trial-to-trial spike counts were perturbed independent of one another. C: simulation of 2 neurons whose spike counts were primarily perturbed by a shared hidden factor (see inset). The diagonal orientation relative to the main axes is a telltale sign of a correlated relationship between these 2 neurons.
Consider now the neural response when reaching to a single reach endpoint. It is useful to frame this variability as falling into two classes: “independent” and “shared” variability, across the neural population. Independent variability can come from many sources. Electrophysiologists are perhaps most familiar with channel noise in the spiking process (i.e., spiking noise). Other sources include variability in active dendritic integration, synaptic failure, general quantal (in the vesicle sense) release variation, and axonal failure in incoming spikes (Faisal et al. 2008). Figure 1B shows simulated data for two neurons that exhibit independent variability and no shared variability for a particular reach endpoint. The spike counts for each neuron are different on each trial, but there is no inherent correlation between the observations with regard to the magnitude or polarity of these perturbations. Critically, when modeling neural data for prosthetic systems, we conventionally make the simplifying assumption that all trial-to-trial variability unrelated to the decoding task falls into the class of independent variability. For classic Gaussian models, this assumption is made to avoid a problem of too little training data for fitting a full covariance matrix (Maynard et al. 1999). For Poisson models, there is no standard multivariate generalization.
In contrast, Fig. 1C shows simulated data from two neurons that exhibit shared variability and independent variability. The inset shows a hypothetical factor, such as intended reach speed, undergoing modulation across trials for the same reach endpoint (blue points correspond to fast reaches and red dots to slow reaches). In this construction, when the speed is greater than average, neuron 1 emits slightly more spikes than average and neuron 2 emits slightly fewer spikes than average. When the factor is below average, the reverse is true. This neuron-by-neuron difference in polarity and magnitude is commonplace among response properties (e.g., Churchland et al. 2006a). Linking the hypothetical factor in this example to a behavioral feature like reach speed helps build intuition on how correlations can arise in the neural observations. However, it is important to realize that shared variability can also arise from internal cognitive states, such as attention or motivation, or even by-products of the neural circuitry, such as shared inputs (Kulkarni and Paninski 2007). Regardless of the source, decoding algorithms can potentially benefit from modeling this shared variability. The algorithm could systematically ignore shared variability, ideally resulting in better decoding performance.
In reality, we do not have direct access to the inset in Fig. 1C: many different uncontrollable factors can be involved and many of them are simply unobservable (e.g., cognitive attentiveness to the task). We cannot explicitly model the factors that influence the neural observations. We instead attempt to infer a set of shared underlying factors for each trial, along with the mapping between these factors and the observed data. By “mapping,” we mean the precise polarity and magnitude by which a factor perturbs the response of a particular neuron. Such a mapping defines the regular relationships between the observed neurons and some common factors, providing a model of how the spike counts of a population of neurons are expected to covary.
We are motivated to pursue such a solution given the observation that the mean neural response associated with planning to a given reach location modulates during a high-speed sequence of target presentations (Kalmar et al. 2005). Initial studies showed that performing a “trial-by-mean” normalization approach (that simply divides the response rate of each neuron by the mean response rate across all measured neurons on that trial) appreciably improved performance (Gilja et al. 2005). Additionally, any attempts to better model neural activity can potentially lead to insights into the neural code, although we do not directly explore this topic here.
We first briefly survey maximum-likelihood techniques for developing reach-endpoint classifiers and we then substitute conventional neural models with a Factor Analysis (FA)–based model. Discovery of the underlying factors and their corresponding mappings to the observed data is the fundamental objective of FA (Everitt 1984). The FA-based model provides the foundation for a better endpoint classifier for our reach-task data set. Next, we extend the relevant models to handle multitarget data to build an even higher-performance classifier. The performance achieved by these new methods is compared with the classic Gaussian and Poisson maximum-likelihood approaches used in our previous work (Santhanam et al. 2006a).
We also explore three central issues associated with such models. First, we look at the influence of the neural data window size (i.e., the time over which neural data are considered for the classification training and prediction) on the efficacy of our new FA methods. As the data window size is reduced, our ability to identify shared underlying factors may be reduced. As such, it is important to quantify whether the FA methods provide as much improvement for short data windows (25–75 ms) as they do for moderate window lengths (150–250 ms). Second, a natural question is whether it is possible to gain even more performance by extending the FA model to use Poisson statistics (instead of the Gaussian statistics in standard FA), especially given that our neural recordings are comprised of low-mean count data. We examine this question by technically modifying our FA models to support Poisson statistics in a manner similar to the work of Yu et al. (2006), which in turn drew inspiration from Smith and Brown (2003) and Brown et al. (1998). Finally, because it may not be possible to retrieve large amounts of training data from disabled clinical patients, it is fruitful to understand how well these new algorithms perform as the training size is varied across real-world ranges. We investigate this question to better qualify the robustness of the multitarget FA algorithm.
METHODS
Delayed-reach task and neural recordings
We trained two rhesus monkeys (G and H) to perform a standard instructed-delay center-out reaching task (e.g., Cisek and Kalaska 2004), while recording neural activity in the arm representation area of monkey PMd. Animal protocols were approved by the Stanford University Institutional Animal Care and Use Committee. Hand and eye positions were tracked optically (Polaris, Northern Digital, Waterloo, Ontario, Canada; Iscan, Burlington, MA). Stimuli were back-projected onto a frontoparallel screen 30 cm from the monkey. As shown in Fig. 2 A, real-reach trials began when the monkey touched a central yellow square and fixated his eyes on a magenta cross. Following a touch hold time (200–400 ms), a visual reach target appeared on the screen. After a randomized (200–1,000 ms) delay period, a “go” cue (central touch and fixation cues were extinguished and the reach target was slightly enlarged) indicated that a reach should be made to the target. As previously mentioned, neural activity during the delay period (time from target appearance until the “go” cue) reflects, among other variables, the endpoint of the upcoming reach (Churchland et al. 2006a; Messier and Kalaska 2000). This task is also known as a “delayed-reach” task.
FIG. 2.
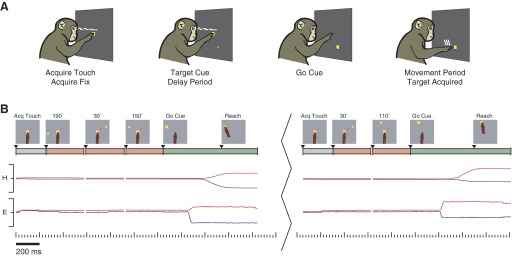
Behavioral task. A: depiction of the standard instructed-delay reach trial. B: example of 5 consecutive trials in our prosthetic cursor data set. Illustration was adapted from Santhanam et al. (2006a) (with prosthetic cursor omitted for clarity). The break between the 3rd and 4th trials constitutes an intertrial interval (500–1,200 ms) that occurs after real reaches, providing time for the monkey to receive his juice reward and return his hand to the center of the screen to start a subsequent trial. The illustrations show the monkey's arm state relative to the visual stimuli. The colored timeline below depicts the various stages of each trial: gray corresponding to the initial period of a trial before the target is shown, orange corresponding to the delay period, and green corresponding to the time after the go cue. The hand (H) and eye (E) positions are graphed throughout all 5 trials, with blue and red lines showing the horizontal and vertical coordinates, respectively. Full range of scale for these data is ±15 cm from the center touch cue. Trials were from experiment H20041106.4.
Eye fixation was enforced throughout the delay period to control for eye-position-modulated activity in PMd (Batista et al. 2007; Cisek and Kalaska 2002). This fixation requirement is appropriate in a clinical setting if targets are near-foveal or imagined as in a virtual keyboard setup. The hand was also not allowed to move until the go cue was presented, providing a proxy for the cortical function of a paralyzed subject. Subsequent to a brief reaction time, the reach was executed, the target was held (∼200 ms), and a juice reward was delivered along with an auditory tone. An intertrial interval (∼250 ms) was inserted before starting the next trial. We presented various target configurations (2, 4, 8, or 16 targets), but for simplicity we restrict ourselves here to the 8-target configurations. We collected between 75 and 150 usable trials per reach endpoint (behavior condition) in a given data set. We also collected a small number of data sets with >200 usable trials per condition.
Our monkeys were well trained before we collected the data from this conventional delayed-reach task. Therefore their behavior was highly stereotyped and the trial-to-trial variability present in the neural responses may not be representative of real-life applications. Fortunately we had access to data sets that were not as stereotyped by virtue of having the following characteristic: interspersed within the delayed-reach trials, we introduced several high-speed reach-target presentations (dubbed prosthetic cursor trials). For these trials, an initial target was presented and displayed for a predetermined time, within about 200–500 ms depending on the particular data set. The usual go cue was then omitted after this time period and another target was almost immediately presented thereafter (∼15 ms). Several rapid trials were sequenced until the go cue was eventually shown, instructing the animal to follow through with a real reach. Since the monkey did not know whether a particular trial will end in a reach, he naturally planned a reach to each of the presented targets.
This experimental setup was originally designed for the purposes of our previous study (Santhanam et al. 2006a). We later observed that the neural responses modulate depending on how many high-speed target presentations are presented in a row (Kalmar et al. 2005) and this variability degraded our ability to decode the subject's intended reach endpoint. These data sets are ideal candidates for applying our FA-based decoding algorithms since they show a reduction in decoding performance due to unaccounted factors. We also shuffle the trials within the day to ensure that the training data set is chosen from the entire data set, as opposed to only some initial set of trials, because we presume the subject's attention and motivation tend to steadily decline throughout the experimental session (Chestek et al. 2007). The shuffling is especially appropriate since the attentional and motivational states of a human subject during clinical usage will surely be more unpredictable than our own highly controlled laboratory setup.
A 96-channel silicon electrode array (Cyberkinetics, Foxborough, MA) was implanted in PMd in the right hemisphere for monkey G and left hemisphere for monkey H, contralateral to the reaching arm. Photographs depicting the anatomical placement of the array and details of the methods by which we discriminated and screened spike waveforms have been published previously (Santhanam et al. 2006a; cf. Supplemental methods). As in the past, we applied the ANOVA tuning test, meaning that a single- or multineuron unit was included in our analyses only if its activity was significantly modulated by reach endpoint during the delay period. This was done for data sets from both monkey G and monkey H for consistency. Similar unit screening procedures have been performed by others (e.g., Musallam et al. 2004) and we use our scheme across all algorithms tested.2 We also removed all units that showed no modulation for any one reach endpoint in the set of training trials. This was to prevent certain degeneracies in the fitting of the FA models. There were only a few units (5–15 units and ≲10% of tuned units) removed for this reason.
For all of the data analyses, the window for counting neural spikes, known commonly as the “integration window” (Tint), started 150 ms after target presentation (i.e., Tskip = 150 ms). We varied Tint within a range of 25 to 250 ms depending on the particular analysis being performed. For each trial, all spiking activity from a particular neural unit that fell within this interval was aggregated to produce a single spike count per neuron per trial. Table 1 provides a perspective as to the number of units that were available after the unit screening procedures. There are naturally far fewer units available when considering very brief Tint periods—some units are tuned for the reach endpoint only later into the delay period and, consequently, do not pass the ANOVA tuning test. Since our recordings were performed with low-impedance (∼100–250 kΩ) electrode arrays, there were significantly more multineuron units than single-neuron units available overall. The relative numbers of these two types of units were consistent with our previous studies (25–35 single-neuron and 60–80 multineuron units for the 100-ms integration window). Furthermore, although the multineuron units were not always of the most pristine quality as they may have been if recorded from high-impedance (>1 MΩ) single electrodes, these units were included as long as they passed the tuning test.
TABLE 1.
Typical number of units tuned for reach endpoint for four example data sets
| Data Set | Number of Units for Given Plan Period, Tint |
|||
|---|---|---|---|---|
| 25 ms | 50 ms | 100 ms | 200 ms | |
| G20040428 | 74 | 99 | 117 | 138 |
| G20040429 | 99 | 126 | 136 | 172 |
| H20040916 | 56 | 87 | 125 | 158 |
| H20040928 | 60 | 93 | 127 | 156 |
The first letter in each data set designates the monkey (monkey G or monkey H) and the subsequent digits denote the calendar date of the particular data set.
Overview of the decoding process
The goal was to develop a system that is able to predict the reach target given only the neural data recorded from the implanted electrode array. The general approach is as follows. First, we exploit the relationship between the neural activity and the reach endpoint; as a simple example, a neuron may be active for reaches to leftward endpoints and inactive for reaches to rightward endpoints. Additionally, the precise activity can be variable on any given trial due to a number of ancillary factors, such as behavioral correlates, attention to the task, and spiking noise. We model this relationship between neural spiking and reach endpoint, encompassing the non-endpoint-related variability, using a probabilistic framework.
An initial set of “training” trials is collected from which both the neural activity and reach endpoint are used to identify the parameters of the probabilistic model. The model defines how likely it is to observe a specific set of neural activity given a selected reach endpoint. Traditionally, this model has been Gaussian (usually with no covariance) or Poisson; here we introduce a new type of constrained correlated Gaussian based on FA. Finally, we decode the reach target for each “test” trial by choosing the most probable target given the observed neural data. During this prosthetic decoding phase, the system is blind to the final reach target and must predict it from neural activity (and the fitted neural model) alone. Decoding error is computed as the percentage of test trials for which the predicted reach endpoint did not match the actual reach endpoint. In our experiments, we reserved approximately half the trials recorded for reaches to each target to serve as test data.
Figure 3 provides a high-level overview of the decoding process. The top panel shows the training procedure and the bottom panel shows the decoding procedure. In the following text, we start by describing the decoding procedure—this process is agnostic to the neural models chosen for the decoding algorithm. Then, we cover the probabilistic neural models that can be plugged into the decoding procedure.
FIG. 3.
Pictorial overview of the decoding process with details provided in methods. A: training stage where the neural spiking data are used in conjunction with the known reach endpoints to fit a probabilistic model of neural data. B: classification stage where the most probable reach endpoint is selected using the recorded neural data.
Reach endpoint classification
We used a standard maximum a posteriori (MAP) decoding (Zhang et al. 1998) to infer the subject's intended reach endpoint given the neural data. Take S to be the total number of reach targets (S = 8 for our eight-target data sets) and s to be an index corresponding to reach endpoint; s can take the values {1, …, S}. For each experimental trial, we counted the number of spikes observed from each neural unit within the neural integration window. These counts were assembled into a q-dimensional vector y, where q is the number of neural units.
The intended reach was decoded from this vector by finding the most probable target ŝ using a probabilistic model linking neural activity and reach targets. Symbolically
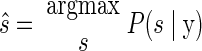 |
(1) |
where P(s|y) represents the probability of the reach target being s given the observed neural data y. Bayes' rule can be used to express Eq. 1 in terms of the neural model P(y|s), which is the probability of observing the current set of spike counts y from the electrode array if the upcoming reach endpoint is indeed s, together with the distribution of reach targets used in the experiment P(s) and the total distribution of spike count vectors P(y)
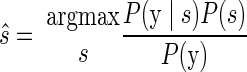 |
(2) |
If each reach target is used with equal probability in the experiment, P(s) is constant. Under these conditions, the most probable target in a trial is that for which the associated distribution of spike-count vectors assigns highest probability to the counts observed
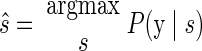 |
(3) |
The success of the decoder clearly depends on the selection of a good model P(y|s) for the spike counts associated with each reach target.
Standard Gaussian and Poisson models
The most commonly used probabilistic models for spike counts are Gaussian and Poisson. For the Gaussian distribution, similar to Maynard et al. (1999), we have the following mathematical expression
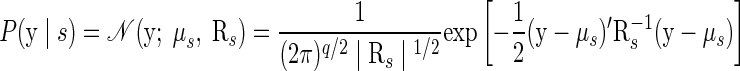 |
(4) |
where, again, y is the vector of spike counts for a single trial, μs is a q-dimensional mean vector, and Rs is a q × q covariance matrix. Here, and throughout, we use the notation A′ for A transpose. The parameters μs and Rs are fitted to the mean and variance of the training data for reach endpoint s. Here, μs is the mean of the distribution associated with target s. If there were no influence of other behavioral correlates, internal cognitive correlates, spiking noise, or other perturbations, this would give the number of spikes that each neuron would reliably produce when the subject reached to that target. The Rs matrix allows for the observed spike counts to be perturbed from μs on a trial-to-trial basis, where large perturbations from the mean are considered less probable.
In practice it is difficult to fit Rs as a full covariance matrix; there are too many elements in the matrix and too few training trials. This leads to the risk of overfitting the training data and thereby reducing decoding performance for the test data. It is convenient to constrain the covariance matrix (Rs) to be diagonal to avoid this problem. A diagonal covariance matrix can model only independent variation in the spike counts of different cells. This is a clear limitation of such a Gaussian model. Note that Maynard et al. (1999) fit a full covariance matrix for one of their analyses and they were able to gain performance improvements in a classification task similar to ours. However, their data set contained many fewer neurons (10 s vs. 100 s) and also used a substantially larger integration window (600 vs. as little as 25 ms). The approach of fitting a full covariance matrix would not be effective for our data sets. One approach to avoiding overfitting might be “regularization” (e.g., Hastie et al. 2003), in which a penalty term is introduced (sometimes interpreted as a prior distribution) to prevent extreme excursions in the parameter value. An alternative in this case would be to parameterize the covariance matrix, reducing the effective number of degrees of freedom that need to be fit. It is this second approach that we explore later in this section.
Another common approach for modeling the spike counts of neurons is to fit the data to a Poisson distribution (Dayan and Abbott 2001). The probability function can be expressed as
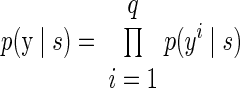 |
(5) |
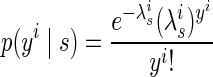 |
(6) |
where yi is the spike count for neural unit i in a single trial and λi is the mean spike count fitted to the training data for reach direction s. By construction, all trial-to-trial variability is independent, and the model cannot account for correlations between neural observations.
Simple verifications with our data sets confirmed that both Gaussian and Poisson models are reasonable fits. Historically, Poisson-based algorithms have been found to perform better than Gaussian-based ones for decoding applications (Brockwell et al. 2004; Hatsopoulos et al. 2004; Santhanam et al. 2006a). Our output variables are the spike counts from the recorded neurons and these counts are by construction nonnegative integers. The spike counts can be relatively low in some cases (e.g., fewer than five spikes) and are conventionally considered to be better fit to a Poisson distribution. One might suspect that the data are not well suited for a Gaussian distribution, especially since a Gaussian distribution has nonzero probability density for noninteger numbers, as well as negative numbers. However, a simple solution of preprocessing the neural spike counts via a square-root transform can make the data a better fit for a Gaussian distribution (Thacker and Bromiley 2001). We use this square-root transform whenever using Gaussian-based decoding algorithms in this study.
Factor-analysis modeling
MODELING SHARED VARIABILITY.
The diagonal Gaussian and Poisson models are constrained to treat all trial-to-trial variation as independent between cells. A model that could successfully capture spike-count correlations within the population might well yield a more accurate decoding algorithm. We argued earlier that to fit a Gaussian model with a full covariance matrix to a large population of cells would require an impractically large number of trials. Instead, we propose to use a parameterized form of covariance matrix, which models correlations in a limited number of principal directions. The form we use derives from a standard statistical technique called Factor Analysis, or FA, where shared variance between observed quantities are modeled as a linear function of unobserved factors.
For our prosthetic system, the observed variables are the neural spiking data that we record from the electrode array. The underlying factors represent the physical, cognitive, or cortical-network states of the subject that are uncontrolled or effectively uncontrollable. We can use the larger number of observed variables to help triangulate the smaller number of unobserved factors in the system. FA is a well-established technique of multivariate statistics (e.g., Everitt 1984; Roweis and Ghahramani 1999). It is a popular tool in the social sciences (Brown 2006) and epidemiology research (e.g., Shmulewitz et al. 2001). We recapitulate the method here in the context of our prosthetic decoding application.
For each experimental trial, as before, we collected the (square root of the) number of spikes observed from each neural unit within the neural integration window into the vector y. We also consider an abstract set of underlying factors (also known as “latent dimensions”), which are assembled into a vector x. This vector lies within a lower-dimensional p-dimensional space; in other words, there are a total of p underlying factors. We take these factors to be Gaussian distributed, independent, and of unit variance around 0 (in fact, assuming any other nondegenerate Gaussian distribution would yield the same probabilistic model). The observation y is also taken to be Gaussian distributed, but with a mean that depends on the hidden factors x. The complete model is as follows
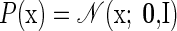 |
(7) |
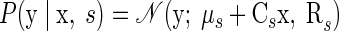 |
(8) |
As in Eq. 4, μs contains the number of spikes that each neuron would produce for endpoint s, if there was only endpoint-related variability in the system, and Rs is a diagonal matrix that captures the independent variability present in y from trial to trial. However, there is now an additional mechanism by which trial-to-trial variability can emerge in the neural observations: specifically, the neural data vector y is perturbed by a linear function of the underlying factors. The matrix Cs is a q × p-dimensional matrix that provides the “mapping” between the factors and the observations. Given that the underlying factors are shared between the observed neural units, there is a predefined correlation structure between the neural units built into the model; integrating over all possible x, we can obtain the likelihood of the data y for a reach endpoint s
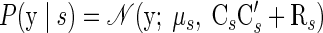 |
(9) |
Equations 7 and 8 are in fact the same form used for a probabilistic principal component analysis (PCA) model (or rather, sPCA3 ). Standard PCA will be familiar to readers as a simple tool used to find the “dimensions that matter” (or rather, the dimensions of greatest variance) within a multidimensional data set. Similarly, the neural model described by Eqs. 7 and 8 attempts to capture the underlying factors that most heavily influence the trial-to-trial shared variability in the neural observations. Both sPCA and FA can be viewed as effective ways to parameterize a full covariance matrix for the high-dimensional observations y. Indeed, Eq. 9 implies that the covariance of y is CsCs + Rs for a reach endpoint s. The first term in the covariance, CsCs, captures the shared (or “common-mode”) variability across the neural population. The second term, Rs, captures the remaining unexplained variability, which is assumed to be independent across neurons. It includes biophysical spiking noise and other nonshared sources of variability. (We briefly discuss the reason for choosing FA over sPCA in the appendix.)
MODEL FITTING.
The model's parameters (μs, Cs, Rs for each endpoint s) are fit by maximum likelihood using the collection of observed neural data (y) and the final reach endpoint (s) across all trials in the training data set.
Although P(y|s) is Gaussian (Eq. 9), the parametric form of the covariance makes direct calculation of the maximum-likelihood parameters difficult. However, they can be found using a standard algorithm for models that include unobserved variables (here, the factors x), called expectation–maximization (EM) (Dempster et al. 1977). The algorithm is iterative and is run until convergence is reached. The exact fitting procedures for FA are fully described elsewhere (Ghahramani and Hinton 1997; Roweis and Ghahramani 1999) and are omitted here for the sake of brevity.
CLASSIFICATION.
When decoding test trials, we can use the maximum-likelihood estimator previously described (cf. Eq. 3)
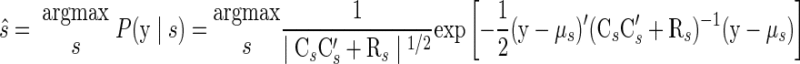 |
(10) |
where Eq. 10 follows directly from Eq. 9. We refer to this approach as FAsep since there is a separate Cs and Rs per reach target s.4 To reiterate, the FA model provides a method by which we can approximate the covariance structure of our observed variables without fitting a full covariance matrix. Once we have fit the model, the decoding procedure does not rely on finding the underlying factor vector (x), but rather operates solely with the distribution P(y|s).
MODEL SELECTION.
One open question is how to select p, the number of underlying factors. With too many factors in the model, the model parameters will be fit to any idiosyncratic correlation structure in the training data and these features may not appear in the test data. If p is too small (i.e., too few factors), the model will be unable to capture all underlying correlation present in the training data. One must ensure that the model is not “overfit” to the training data and that it instead generalizes well for new (test) data. The choosing of the optimal p, or p*, is part of the process of “model selection.” It is important to keep in mind that the underlying factors identified for each model are abstract. The factors constitute a vehicle by which we are able to describe shared variability—the exact number of factors selected does not necessarily have physical significance. The model selection procedure is a simple, necessary step to ensure that a model fit with the training trials will perform well when decoding the test data.
We used the standard approach of partitioning data into training and validation sets, to find the value of p beyond which overfitting became a problem (Hastie et al. 2003). As mentioned earlier, each data set was split approximately into two equal halves, one to serve as a training set and the other as a test set. The trials in the training set were further divided to perform a fivefold or tenfold cross-validation. For example, in fivefold cross-validation, roughly four fifths of the training trials would be used to train the FA model and one fifth to validate the prediction error of the model. This would be repeated five times until all of the original training set was used for validation. A series of models, each with a different number of underlying factors p, were tested using this cross-validation method. The p* was selected by identifying the model with the lowest decoding error. The performance of this one model was then assessed on the test data, to compare the classification performance of the optimal FA model against the traditional Gaussian and Poisson models.
Multitarget extension
The FAsep approach to modeling the multiunit data described by Eqs. 7 and 8 is a straightforward extension to the standard Gaussian model of Eq. 4. However, one might intuit that reach endpoint could simply be another factor (or set of factors) included in the vector x, rather than being treated separately in the output space as in Eq. 8. Also, the FAsep model has the disadvantage of requiring a large number of parameters. There is a separate Cs and Rs matrix per reach endpoint—if there are 100 neural units, 8 targets, and 2 factors, the approach will require the fitting of 100 × 8 × (2 + 1 + 1) parameters, where the terms in the sum correspond to the contributions from Cs, Rs, and μs. The number of training data scales with the number of neural units and targets, so in effect there are four parameters to learn for each unit and reach endpoint; that is, if there were 60 training trials per reach endpoint, we would have about 15 individual data measurements per parameter that needed to be estimated.
As such, we developed a second, novel approach to FA that shares the same output mapping between target locations and incorporates the separation of reach endpoint through the shared latent space. We formalize this model as follows
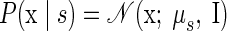 |
(11) |
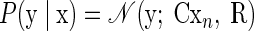 |
(12) |
where μs is now a p-dimensional vector that describes the reach-endpoint influence on the neural data within the lower-dimensional space. The C matrix is shared across reach endpoints, thereby unifying the effect of endpoint-related and non-endpoint-related factors on neural data. For each trial, the endpoint-related effect on the neural observations is Cμs, whereas the non-endpoint-related factors perturb the observations by C(x − μs). As before, R is diagonal and describes the independent variability. We refer to this model as FAcmb, since the output mapping is “combined” across reach targets. We modified the standard FA model-fitting procedure to account for the new form of FAcmb. An outline of the derivation is provided in the appendix.
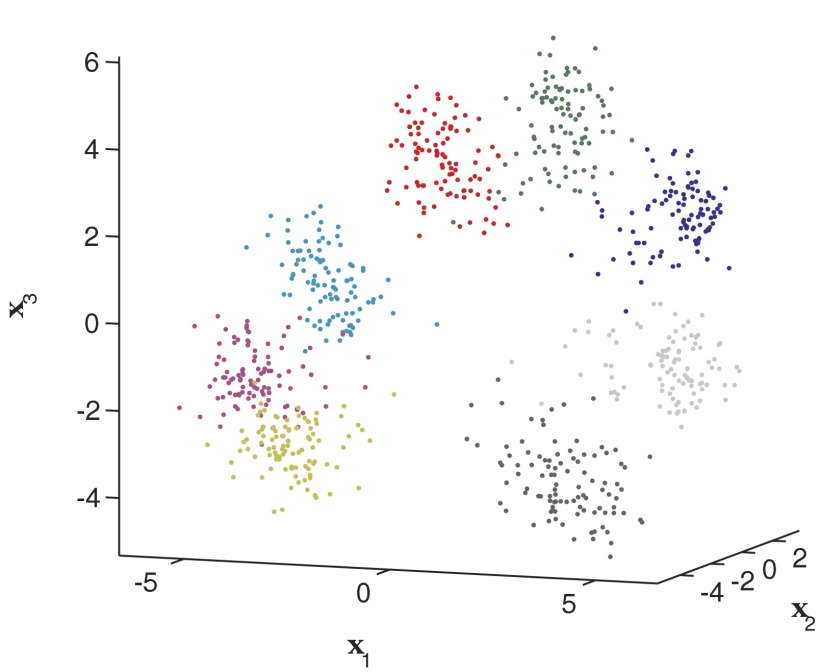
Figure 4 shows an example of how these unified factors might manifest themselves over many trials.5 There are three abstract factors that capture both endpoint and non-endpoint-related influences on the neural data y (which is not shown and is much higher dimensional). Each colored cloud of points corresponds to a different reach endpoint, with trials to a given endpoint clustered next to one another. Note that even in this three-dimensional instantiation of FAcmb, there is visible separation between the clouds, suggesting that it will be possible to discriminate between endpoints.
FIG. 4.
Visualization of a combined FA model (FAcmb) with a 3-dimensional latent space. Each point corresponds to the inferred x for a given trial. Axis scaling is determined by the fitted model. The coloring of the data points denotes the upcoming reach target. Clusters correspond to different reach endpoints. Analysis performed on data set G20040428.
Although such a visualization is helpful in building intuition, to decode the reach endpoint for test trials, we express Eqs. 11 and 12 as P(y|s) and use the same maximum-likelihood techniques from Eq. 3
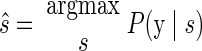 |
(13) |
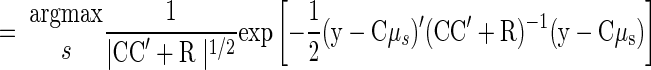 |
(14) |
The number of parameters in this model for 100 neural units, 8 targets, and 16 latent dimensions6 is 100 × (16 + 1) + 8 × 16, where the second term corresponds to the parameters arising from the latent-space means μs. This yields about two parameters per unit per target. That is, if there are 60 training trials per reach endpoint, there are now on average roughly 30 individual data measurements available to estimate each parameter. For interested readers, we also provide a brief discussion of signal-dependent variability in the context of FAsep and FAcmb in the appendix.
Poisson likelihood model
Earlier, we discussed how traditional Poisson-based models can be more effective than Gaussian-based models of cortical neural data (i.e., Eq. 6 vs. Eq. 4). Might this also be the case for the above-cited FA approaches? In the following text, we briefly describe how we adapted the FA models to accommodate Poisson statistics. We call this family of models “Factor Analysis with Poisson Output” (FAPO). Deviating from Gaussian statistics can be mathematically challenging. The short summary is that Eqs. 15 and 16 describe a model analogous to FAsep, which we call FAPOsep. Similarly, Eqs. 18 and 19 constitute the FAPOcmb model.
To provide one potential motivation for Poisson statistics, note how the lower-dimensional x in Eq. 7 is projected to the higher-dimensional observational space in Eq. 8; on a given trial, the mean of the observed data vector y is determined by the particular x on that trial. However, the variance of the neural vector y does not change (R is fixed regardless of x), and therefore the model is unable to capture the Poisson-like relationship between the mean and the variance as the underlying factors exert their influence. Furthermore, because the neural spike counts are small nonnegative integers, the Gaussian distribution may not be the best description of the data.
One strategy is to replace the Gaussian observational model of FA with a point-process or Poisson likelihood model (Smith and Brown 2003; Yu et al. 2006). Taking this approach, the model can be written as follows
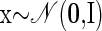 |
(15) |
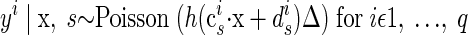 |
(16) |
As before, x represents the abstract underlying factors. The exact mapping from factors to observations follows from a linear transformation on x using the p-dimensional scaling vector ci and the scalar offset di. This provides a proxy for the instantaneous firing rate for the neural observations yi, where i indexes the particular neural unit of interest. The constant Δ is the nonnegative time-bin width. The function h ensures that the mean firing rate argument to the Poisson distribution is nonnegative. This particular instantiation is referred to as FAPOsep, since there is a separate ci and di for each reach target s.
The Poisson distribution—along with the nonlinear mapping function h—makes an analytic solution to the EM algorithm intractable. Thus we must use approximations when performing the EM algorithm; we refer to previous work that uses the same techniques to fit such functional forms (Yu et al. 2006, 2007). To find the most probable reach endpoint, we must compute P(y|s) and apply the maximum-likelihood formula of Eq. 3. In general form, the formulation is
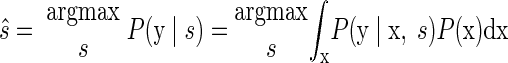 |
(17) |
An analogous model to FAcmb can be formulated and is dubbed FAPOcmb
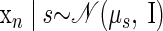 |
(18) |
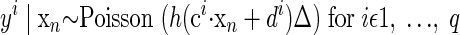 |
(19) |
Since these Poisson models are not the central thrust of this work, we refer the reader to the full derivation of FAPOcmb, available in an on-line Technical Report (Santhanam et al. 2006b).
RESULTS
Model selection
We first choose the number of abstract factors (p) that will result in optimal decoding performance. Conceptually, we must ensure that we have enough factors to sufficiently describe the shared variability in the data but not too many factors that we overfit the training data. This a necessary step before we can compare FA models to the traditional Gaussian and Poisson models in the next subsection. We examined two data sets, G20040401 and H20040916, for which there were 185 and 200 total trials per reach target, respectively. This yielded around 80 training and around 20 validation trials per reach target for each cross-validation fold. A neural integration window of 250 ms was used for this particular analysis, although we saw similar results for smaller integration windows as well.
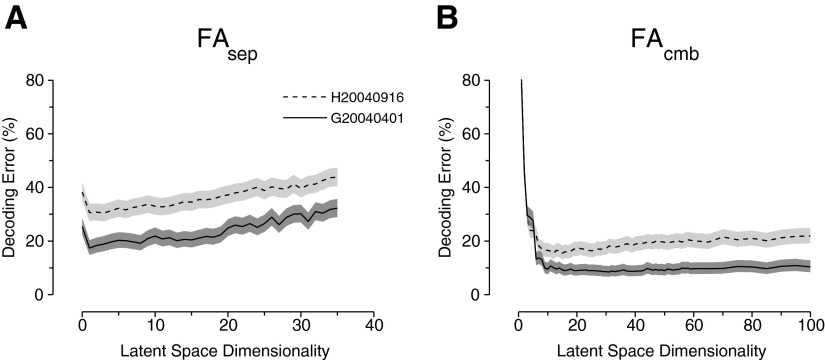
Figure 5 A shows the validation performance for these two data sets as a function of p when fitting the FAsep algorithm. The performance for p = 0 corresponds to a classic Gaussian model where there is no Cs matrix and only a diagonal covariance matrix, Rs, per reach endpoint. The best validation performance for FAsep was obtained for low p: p* = 1 for G20040401 and p* = 3 for H20040916. The results show that as p is increased beyond p*, the performance steadily declines. Overfitting is an issue for even relatively small values of p. Why might this be? There are surely many factors that influence the upcoming reach—for example, reach direction, distance, curvature, speed, and force. However, for our data set, reach trajectories to the same endpoint were highly similar. Second, the shared variability (captured by trial-to-trial modulation of the factors) may be small relative to the independent variability. Given a relatively small number of neurons (∼100) and training trials for each reach target (∼80), it is likely that we are unable to identify more hidden factors without poorly estimating the model parameters and overfitting. The penalty for overfitting is evident; by p = 30, all of the benefits from using FAsep over the classic Gaussian approach (p = 0) are lost.
FIG. 5.
Model selection for separate FA model (FAsep) and FAcmb. The shaded area denotes the 95% confidence interval (Bernoulli process) around the mean performance (embedded line). A: optimal cross-validated performance for FAsep occurs for a low number of factors. B: optimal cross-validated performance for FAcmb occurs between 10 and 20 factors.
We performed the same sweep of p for FAcmb and the validation performance is plotted in Fig. 5B. The best validation performance for the combined FA model was obtained for higher p than for FAsep: p* = 31 for G20040401 and p* = 12 for H20040916. Although at first it seems that the optimal number of factors for G20040401 is significantly higher than that for H20040916, one can see that the validation performance reaches a floor around p* = 15 for the G20040401 data set. There is negligible improvement when increasing the dimensionality past that point that, combined with the fact that performance degradation due to overfitting in the H20040916 data set is relatively minor, demonstrates that the FAcmb model is less sensitive to the exact choice of p than in FAsep. For interested readers, we provide a brief discussion on why p* might be larger for FAcmb than that for FAsep in the appendix. This is a side issue since the end goal is to optimize decoding performance and we chose the best p in that regard for each of FAsep and FAcmb.
Comparison to classic decoding algorithms
Next, we compared the performance of four different decoders grouped into two pairs. One pair consisted of the classic, independent-Gaussian and -Poisson models. The second pair was the FA-based models, FAsep and FAcmb. We computed decoding accuracy based on an integration window length of 250 ms. These are the central results of this study. For the classic Gaussian and Poisson algorithms, half the trials in each data set were used to train the model parameters and the model was used to predict the reach endpoint for each of the remaining trials in the data set. For the FA-based algorithms, the training set was first used to select the optimal dimensionality p* using fivefold cross-validation and the training set was then used in its entirety to fit a model with p* dimensionality. As with the classic algorithms, the fitted model was finally applied to the test data to compute the average decoding performance.
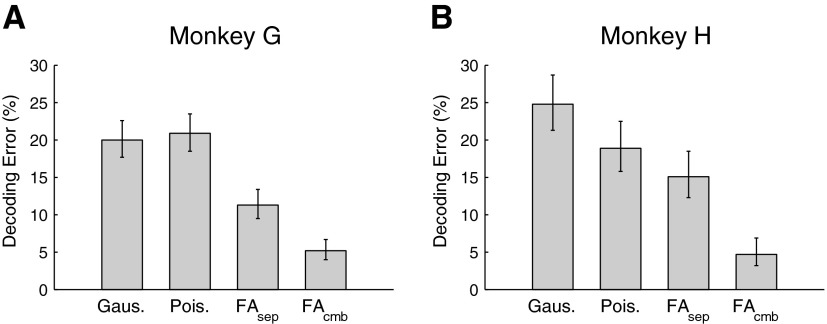
Figure 6 shows the relative performances of these four algorithms. There were grand totals of 1,024 and 528 test trials for the G20040429 and H20040928 data sets, respectively. For data set G20040429, the classic Gaussian7 and Poisson algorithms have roughly the same performance, whereas for data set H20040928 the classic Gaussian algorithm has the worst performance, with roughly 5% worse accuracy than the classic Poisson algorithm. (Most often, especially with data sets from monkey H, the classic Gaussian algorithm showed worse performance than the classic Poisson algorithm, likely due to lower firing rates observed in the monkey H data sets.) The FAsep algorithm reduced the decoding error for monkey G's data set by nearly 10%, but had only a modest improvement for monkey H's data set. The FAcmb algorithm showed by far the best performance. Our novel decoder was able to drastically reduce the decoding error down to only about 5%.
FIG. 6.
Comparison of decoding algorithms on 2 example data sets. Error bars denote the 95% confidence interval (Bernoulli process) for each measurement. A: data set G20040429. B: data set H20040928.
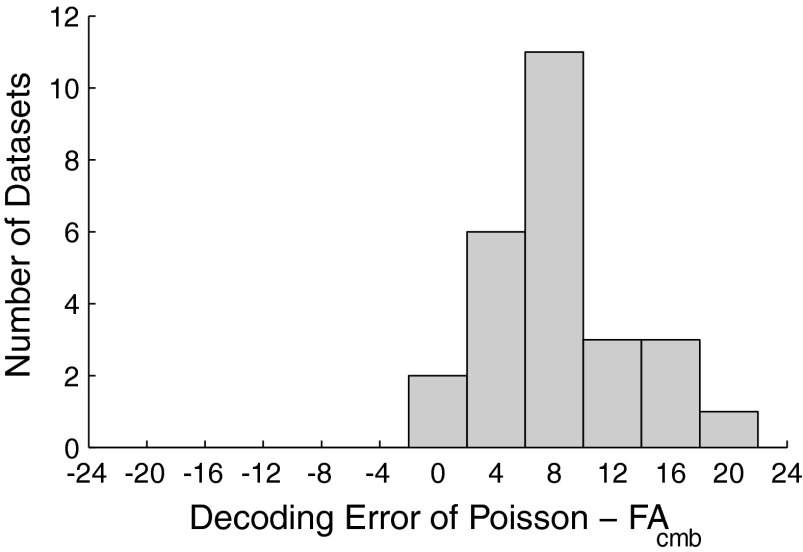
We further explored whether FAcmb was capable of predicting reach endpoint more accurately in several other data sets. For this purpose, we used an integration window of 200 ms, which was a similar window length to the 250 ms previously used. We examined 8 data sets from monkey G and 18 data sets from monkey H. Baseline performance was taken to be the average decoding error from the classic Poisson algorithm. We applied the FAcmb algorithm to the same data sets and computed the reduction in decoding error or improvement in overall decoding accuracy. The results are plotted in Fig. 7. The FAcmb algorithm improved performance by 6–10% in 11 data sets and >10% in 7 data sets. No data sets showed a degradation in performance when using FAcmb over the classic Poisson algorithm. This was not the case for FAsep, where for a few data sets in monkey H, the algorithm performed worse than even the classic Poisson decoding approach, again possibly due to the lower firing rates observed in the monkey H data sets.
FIG. 7.
Performance improvement with FAcmb across many data sets. Plotted is a histogram of the difference in reach endpoint decoding error between a classic Poisson-based algorithm and the FAcmb algorithm.
We also performed a simple control to confirm that the shared variability that we are modeling was not simply an uninteresting artifact of poor spike classification. If the spike separation on each electrode were not perfect, there could be an increased amount of correlated noise between neural units on the same electrode. To better explore this question, we restricted our analyses to include only the best unit (either single neuron or multineuron) from each electrode. For data set H20040928, we found that a classic Poisson algorithm exhibited a decoding error of 26.3% for an integration window of 200 ms. The FAcmb algorithm was able to achieve a decoding error of 16.9% using the same data. We saw significant improvement in performance even in this restricted scenario, reducing the potential concern that the model was simply capturing correlations that are a by-product of poor spike classification.
Finally, we investigated the benefits of FAcmb for conventional delayed-reach data sets (i.e., data sets in which there was no intermixing of high-speed reach endpoint cues). For one data set (G20040508), where we used an integration window of 200 ms, there was a modest performance improvement for using FAcmb (reduction of 5.3% error). This is relatively unsurprising for two reasons. First, there is not as much variability in the presentation of the target cue, thereby leading to a relatively nonvariable experimental environment for the subject. Second, for these data sets, performance is often already high with classic Poisson decoding (91.6% decoding accuracy). We saw similar results for three such data sets from monkey H.
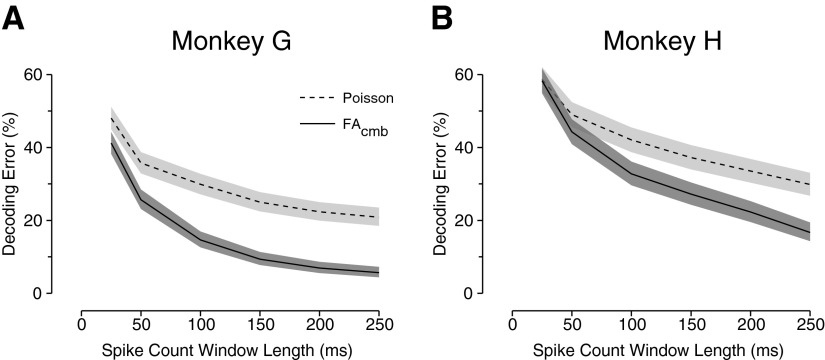
Integration window size
All of the performance results reported so far have been based on neural integration windows of 200 and 250 ms. In our previous work (Santhanam et al. 2006a), however, we showed that even shorter integration windows are preferable when optimizing the information throughput of a prosthetic system. As such, we tested the performance of the FAcmb decoder as a function of the integration time (Tint), beginning 150 ms after target onset. Figure 8 shows a comparison between the simple Poisson-based decoder and the FAcmb decoder. Integration window durations of 25, 50, 100, 150, 200, and 250 ms were tested. A separate FAcmb model was fit for each Tint, first using 10-fold cross-validation to select the appropriate p* for each.
FIG. 8.
Performance of the FAcmb algorithm as a function of the data integration window length. Shading and embedded line are the same as in Fig. 5. The performance with the classic Poisson-based algorithm is plotted as a reference. A: data set G20040429. B: data set H20040916.
The performance differential between simple Poisson-based decoding and FAcmb decoding was noticeable across integration window lengths, but most appreciable with the longest integration windows. For monkey G, the Poisson-based algorithm has performance roughly similar to that of FAcmb for Tint = 25 ms, although for monkey H, this relationship extends up to Tint = 50 ms. For these smaller windows, the spike counts for the neural units can be rather low. For example, in a time interval of [150, 200] ms after target presentation, the spike counts for a unit are most often only 0, 1, or 2. How might this affect the performance of FAcmb? The FA model assumes that the observed neural variability comprises a shared component CC′ (from the uncontrolled or unobserved factors) and an independent component R (from the biophysical process of generating action potentials and other nonshared noise sources). Reducing the window length degrades our ability to separate shared from independent contributions.
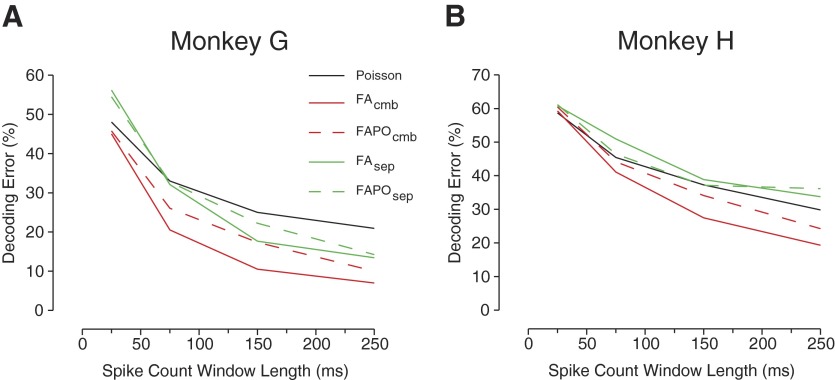
FAPOsep and FAPOcmb
Next, we compared FAPOsep and FAPOcmb to their counterparts, FAsep and FAcmb, to see whether there might be advantages to a Poisson-based FA approach. We again varied the integration window length to explore the regime of low spike counts, in which Poisson models usually perform better than Gaussian models. Figure 9 shows the performance from the four FA decoding algorithms. The performance of the classic Poisson method is also shown as a baseline. The results show that both FAsep and FAPOsep fare worse than the classic Poisson algorithm for small windows in monkey G and for all window lengths in monkey H. Conversely, the combined approaches, FAcmb and FAPOcmb, show substantial performance improvement over the alternatives, especially for larger window lengths.
FIG. 9.
Performance of new Poisson-based FAPOsep and FAPOcmb algorithms. The performances with the classic Poisson-based algorithm, FAsep, and FAcmb are plotted for reference. The data integration window was varied to see whether there was any advantage to FAPOsep or FAPOcmb as lower spike counts were inputted into the models. A: data set G20040429. B: data set H20040916.
Surprisingly, the Poisson-based FA algorithms do not perform as well as the Gaussian-based versions. This is somewhat counterintuitive since a Poisson distribution is thought to better model neural data than a Gaussian, especially for small time windows. The effects we saw could be due to one of two main reasons.
1. The Poisson fitting procedure includes several approximations, which could result in a suboptimal fit, even when the training likelihood increased with every EM iteration. Furthermore, in some cases, for FAPOsep, the training likelihood would in fact start to drop after close to 100 iterations, albeit slowly.
2. Ordinarily when spike counts are modeled using a Poisson distribution, there is an implicit lumping together of shared and independent variability. In both FAPOsep and FAPOcmb models, the Poisson distribution is used to model only the independent variability. After separating out the shared variability, the independent variability may no longer be Poisson distributed. Furthermore, spike counts can sometimes be sub- or super-Poisson (Churchland et al. 2006b; Shadlen and Newsome 1998). Gaussian-based models may better characterize the data by not overconstraining the mean–variance relationship, although it is difficult to directly assess this question since we do not have a ground truth for the underlying data.
Ultimately, we were able to achieve good performance improvements with the simpler FAcmb algorithm. There was no strong motivation to use the more complicated FAPOcmb approach for our data sets. Investigation into the exact breakdown and characteristics of shared and independent variability is left to future work.
Training set size
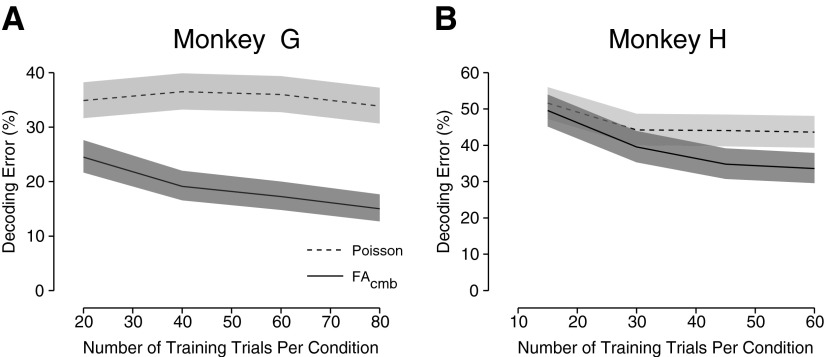
Our last analysis was to examine the effects of training size on system performance. Given that there can be a large number of parameters to learn for the FA-based models, it is possible that these models do not provide such dramatic performance improvements over the classic models when the training set size is restricted. This is especially of interest since one may not be able to collect a voluminous training set from paralyzed patients, since the patients can suffer from fatigue or other medical issues. To investigate this question, we divided each data set into three subsets. The first subset was used to train the FAcmb model with p dimensions. It is the size of this subset that was varied to assess performance across different training set sizes. The second set was used as a validation set to find the optimal p*. The model with p* dimensions was then used to decode the test trials in the third set. As before, the classic Poisson algorithm was also used as a baseline comparison. For the Poisson algorithm, the model was simply trained on the first subset of trials and performance was computed with the third set. The validation set was not needed since there is no model selection phase for the Poisson algorithm. The data integration window size for this analysis was 100 ms.
Figure 10 shows the results of varying the amount of training data used to fit the model for two data sets, one from each monkey. For monkey G, the classic Poisson algorithm shows a consistent level of performance regardless of the training size. The variation in decoding error is within the confidence intervals and is therefore not statistically significant. Similarly, for monkey H, the classic Poisson algorithm shows a fairly consistent level of performance, excepting the very initial drop representing a change from 15 to 30 training trials per condition. The FAcmb algorithm, however, shows steady improvement as the number of available training trials increases. Nevertheless, by 60 trials per condition there is a substantial benefit for using FAcmb over the classic Poisson algorithm in both monkeys: reduction of about 20% error for monkey G and a reduction of about 10% error for monkey H.
FIG. 10.
Effect of training size on performance of FAcmb. The number of trials used to fit the initial model was varied, the dimensionality of the model was chosen based on a fixed set of validation trials, and the final performance was computed based on a separate fixed set of test trials. Shading and embedded line are the same as in Fig. 5. The performance with the classic Poisson-based algorithm is shown for reference. A: data set G20040429. B: data set H20040916.
DISCUSSION
We investigated here the use of a more sophisticated decoding algorithm in the hopes of achieving higher neural prosthetic performance. FA techniques were proposed as a method to help better account for shared, correlated variability arising from uncontrolled and unobserved aspects of the prosthetic task and two methods were offered to help improve reach endpoint prediction. Results showed that using an entirely separate FA model for each reach endpoint (FAsep) was not as effective as our novel approach of fitting a combined model to the entire data set (FAcmb). The latter strategy requires fewer model parameters and is able to aggregate data from all reach endpoints to fit these parameters. As such, it may be less prone to estimation error and overfitting. Surprisingly, the more complicated extensions to support Poisson distributions appeared to be unnecessary because the Gaussian-based models performed better (even for Tint as low as 25 ms). This may have been due to one or more reasons: the square-root transform may mitigate the potential pitfalls with using a Gaussian model, the approximations used for fitting the Poisson-based models may have been detrimental, and/or the independent variability may not be well described by a Poisson distribution.
The full utility of the FA methodology was demonstrated with our BCI data sets where the task design had different operating modes (BCI vs. reach trials). This task contained a sufficient amount of shared variability such that conventional algorithms were unable to achieve high levels of performance. We could have fit an explicit model that accounted for the speed of target presentations, but we instead chose to infer abstract factors to demonstrate how effective this technique can be when the nature of the covariates is unknown. FAcmb was able to describe the shared variability in the task and significantly outperform conventional methods. For a clinical prosthetic setup, the situation of mixing BCI and reach trials would not be realistic since the patient would be paralyzed. However, in a real-world setting, there can be many underlying factors that modulate the neural observations, including the subject altering the pace at which he or she uses the BCI, environmentally induced distractions, or subconscious changes in the subject's motivation. FA can be one tool by which the system designer can combat performance degradation. The computational overhead to using FAcmb primarily exists during the training of the model. When decoding test trials, the decoding procedure is nearly identical to decoding with a simple Gaussian algorithm, except that the output covariance is no longer diagonal [i.e., Cov (y) = CC′ + R instead of simply R].
The FAcmb algorithm also appears to be relatively robust. The process of model selection to choose the optimal number of factors did not show any sharp peaks in performance. The performance increased rapidly as the number of factors p was increased, but then did not drop drastically as p moved beyond the optimum p*. In fact, the decline in performance past p* is sufficiently gentle that a system designer could potentially choose to skip the model selection process entirely (if it was deemed too computationally expensive) and simply fix the number of factors to, for example, p = 20. As seen in Fig. 5B, such a choice of p, although not optimal, would still be able to achieve close to optimal performance. In fact, the differences in performance for values of p between p = 12 and p = 20 was within the 95% confidence interval. We also found that it is possible to achieve significant performance improvements with a reasonable number of training trials per condition, demonstrating that a human subject will not be required to perform thousands of training trials to justify the use of FAcmb.
For those already familiar with other methodological studies in the field of cortical neural prosthesis, we place our work in context with these past studies here. Other work has explored the use of state-space models as a foundation for decoding the subject's motor intentions. Primarily this has been applicable to the decoding of arm trajectories (e.g., Brockwell et al. 2004; Truccolo et al. 2005; Wu et al. 2006; Yu et al. 2007). These studies start with a model that consists of a linear Gaussian model of the arm state, followed by an either linear or nonlinear mapping to the space of observed neural spiking data. The models are trained based on trials in which both the underlying arm state and the neural data are known. In the case of our work, we are not using the latent state model to directly capture the relationship between the arm state and the neural firing, but rather we are using it to capture the variability induced by the unobserved and uncontrolled parameters. When training our model, we do not have access to the ground truth of x and must infer it along with the model parameters C and R through the EM procedure. The FA latent variables reflect aggregate network activity. This stands in contrast to other models of correlated activity that posit direct connections between observed neurons or from a small set of unobserved neural units (Kulkarni and Paninski 2007; Okatan et al. 2005).
Recently we published a Hidden Markov Model (HMM) technique (Kemere et al. 2008) and either FAsep or FAcmb can be complementary to this work. The primary objective of the HMM algorithm is to help identify “neural-state transitions” between baseline, plan, and move states, modeling the various states in a discrete framework. The HMM model includes no concept of continuous factor variables that create correlations in the neural observations. In theory, one could potentially use an FA approach to model correlation in the neural data, while simultaneously using the HMM algorithm to identify state transitions; we reserve this exploration for future work.
Our multitarget extension to FAsep is certainly specific to the behavioral task performed by our subject. However, the concept itself is a simple one: there are underlying behavioral and cognitive parameters that can diminish the performance of a prediction algorithm, whether it be in the context of predicting reach endpoint or the entire reach trajectory. The exact details of how to extend an existing algorithm to use FA to “learn” these underlying factors is of course specific to each algorithm. Furthermore, provided that the factors are modeled as Gaussian random variables and affect the observables through a linear mapping, it is likely that one can rather easily incorporate such a technique within other decoding frameworks. Although there is no guarantee that the achievable performance improvements in other contexts will be comparable to those seen in our data sets, our work suggests that the overall strategy espoused here is worth investigating if increasing prosthetic performance is of paramount interest. Indeed, a similar approach has been taken by Wu et al. (2008) to extend the Kalman filter framework so that it can account for unobserved factors. The study shows an improved performance that, like our own, is quite promising.
We chose to use a probabilistic framework and construct a generative model a priori that we felt reasonably describes how our reaching task relates to the neural data. A potential disadvantage, however, is that the training procedure maximizes the likelihood of the observed data without assigning a cost for how well or how poorly the model can classify the data. Despite this drawback, the fact that our FA approach improves the performance indicates that we may be revealing something intrinsic about the system. (For now, we treat the factors as abstract, although FA can potentially be a useful tool to help discover new, or confirm existing, correlates in an experimental setting.) An alternate algorithmic strategy would be to train a classifier that expressly accounts for misclassifications during the fitting process (e.g., Ng and Jordan 2002). This is the approach taken by several standard algorithms in the field of machine learning, including neural network classifiers, support vector machines, and Gaussian process classifiers. Furthermore, there is also the possibility of using a hybrid between discriminative and generative methods (Raina et al. 2004). An eventual comparison between the FA approach and these other approaches would be fruitful and will help better frame the broader influence of what we have shown here.
The performance improvement that we were able to achieve, especially with the FAcmb method, was substantial. For several data sets, the baseline performance was a ≥20% classification error rate when using the classic Poisson algorithm. These error rates were routinely reduced to 5–10% when using our FAcmb algorithm. For prosthetic tasks in which achieving high single-trial accuracy is the primary objective, being able to reduce the error rate to a mere 5% can have a dramatic impact on the clinical utility of these systems for subjects with motor disabilities. Combined with other improvements, including HMMs for facilitating free-paced prosthetic control (Kemere et al. 2008) and strategies for optimally placing the virtual targets on the computer screen (Cunningham et al. 2008), we hope that ongoing research will yield a substantial improvement in the quality of life of patients.
GRANTS
This work was supported by National Defense Science and Engineering Graduate Fellowships and National Science Foundation (NSF) Graduate Research Fellowships to G. Santhanam, B. M. Yu, and V. Gilja; Medical Scientist Training Program grant and Bio-X Fellowship at Stanford to A. Afshar; Christopher and Dana Reeve Foundation grant to S. I. Ryu and K. V. Shenoy; Gatsby Charitable Foundation grant to M. Sahani and B. M. Yu; National Institute of Neurological Disorders and Stroke/Collaborative Research in Computational Neuroscience Grant 5-R01-NS-054283-02 to K. V. Shenoy and M. Sahani; and the following awards and grants to K. V. Shenoy: Burroughs Wellcome Fund Career Award in the Biomedical Sciences, Center for Integrated Systems at Stanford, NSF Center for Neuromorphic Systems Engineering at Caltech, Office of Naval Research, Sloan Foundation, and Whitaker Foundation.
Acknowledgments
We thank J. Cunningham for valuable discussions, S. Eisensee for administrative assistance, M. Howard for surgical assistance and veterinary care, and D. Haven for computing support.
APPENDIX
sPCA versus FA
Although sPCA and FA are very similar in form, sPCA constrains the independent variances (i.e., elements of Rs) to be identical for each neuron, whereas FA allows each neuron to have a different independent variance. This distinction is important. The variance of windowed spike counts is often considered to be proportional to the mean spike count (e.g., Shadlen and Newsome 1998), where the proportionality constant is about 1. Neurons with higher mean firing rates often exhibit higher independent spiking variances. Although applying a square-root transform to the spike counts can help stabilize this variance relationship (Kihlberg et al. 1972), this process is imperfect. Ultimately, sPCA allows for only a single independent variance parameter across all neurons, whereas FA has the benefit of more parameters in Rs to capture the richness in the data; as such, we chose to focus on FA for our work.
Signal-dependent variability
Do the FA-based models contend with the possibility that trial-to-trial variability could be signal-dependent? Recall that the reach endpoint is the signal of interest. For FAsep there is a different FA model per reach endpoint; thus the model of variability is, by construction, already signal dependent. For FAcmb there is a single set of factors for all reach endpoints. The nonreach-endpoint factors perturb the reach endpoint signal (μs) with the same covariance, irrespective of s (cf. I in Eq. 11). Similarly, the independent variability described by R in FAcmb is also the same regardless of s. Thus FAcmb does not directly allow for signal-dependent sources of variability.
Comparison of p* between FAsep and FAcmb
For FAsep we found usually p* ≤3. For FAcmb p* most often lay within the range of 10–35. Why might this be? We provide one technically oriented explanation here. Intuition suggests that a well-fit model of the form described by Eq. 12 should have the property that the observation mean for a given target [i.e., E(y|s)] closely matches the empirical mean of the data for that same target. In the case of FAcmb the observation mean is Cμs for target location s. The observations lie in a q-dimensional space, whereas the vector μs is in a lower p-dimensional space and C is rank p. Thus the vector Cμs lies within a p-dimensional subspace spanned by the columns of C. Across S targets, the S observation means can at most lie in an S-dimensional subspace that includes the origin. Thus p may need to be at least equal to S for the model to capture the appropriate target-specific observation means. For our data sets, where S = 8, we found that the measured means were in fact linearly independent—thus ≥8 latent dimensions (p ≥ 8) were needed to describe them accurately. The even higher values of p* that we find for FAcmb may possibly arise from the greater statistical power afforded by combining data across all the reach targets.
Mathematical derivations for FAcmb
This section provides the derivation of FAcmb. It is recommended that the reader be first familiar with the process of using expectation–maximization (EM) techniques for standard FA. Excellent overviews are provided by Roweis and Ghahramani (1999) and Ghahramani and Hinton (1997). Again, the model for FAcmb is
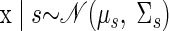 |
(A1) |
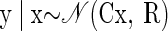 |
(A2) |
The random variable s is the reach target and has a discrete probability distribution over {1, …, S} [i.e., P(s) = πs]. Given s, the latent state vector x ∈ ℝp×1 is Gaussian distributed with mean μs and covariance Σs.8 The outputs y ∈ ℝq×1 are generated from a Gaussian distribution, where C ∈ ℝq×p provides the mapping between latent state and observations and R ∈ ℝq×q is a diagonal covariance matrix. The variables xn and yn denote independent draws from this generative model over N trials. The set of all trials are denoted as {x} and {y}, respectively. The random variable s is assumed to be known during the training of the model.
E step. The E step of EM requires computing the expected log joint likelihood, E [log P({x}, {y}, {s}|θ)], over the posterior distribution of the hidden state vector, P({x}|{y}, {s}, θk), where θk are the parameter estimates at the kth EM iteration. Since the observations are i.i.d., this is the sum of the individual expected log joint likelihoods, E [log P(xn, yn, sn|θ)].
The latent state and output observations are jointly Gaussian given s
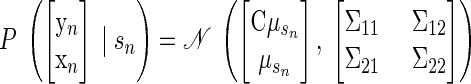 |
(A3) |
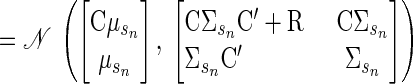 |
(A4) |
Thus the posterior distribution of the hidden state can be written as
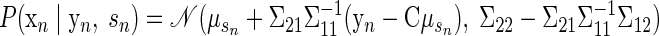 |
(A5) |
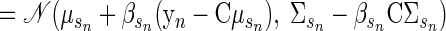 |
(A6) |
where βsn = ΣsnC′(R + CΣsnC′)−1. This conditional is a standard property of the Gaussian distribution. The inverse in βsn can be computed efficiently using the matrix inversion lemma (Sherman–Woodbury–Morrison formula)
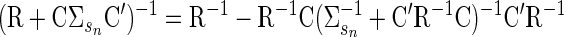 |
(A7) |
For observation n, let Qn be the Gaussian posterior latent state distribution P(x|y, s, θk), which has mean ξn and second moment Ξn
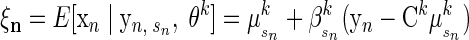 |
(A8) |
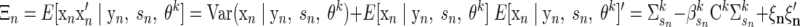 |
(A9) |
The expectation of the log joint likelihood for a given observation can be expressed as follows
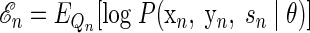 |
(A10) |
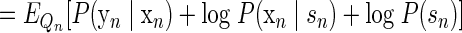 |
(A11) |
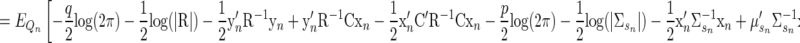 |
(A12) |
The terms that do not depend on xn or any component of θ can be grouped as a constant C, outside the expectation. Doing so, and simplifying further, we have
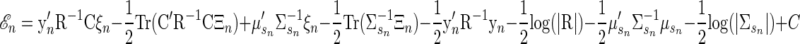 |
(A13) |
The expectation of the log joint likelihood over all of the N observations is simply the sum of the individual ℰn terms
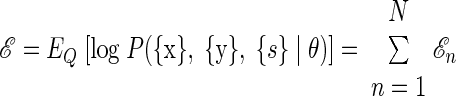 |
M step. The M step requires finding (learning) the θ̂k+1 that satisfies
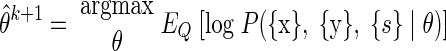 |
(A14) |
This can be achieved by differentiating ℰ with respect to the parameters θ, as shown in the following text. The indicator function, I(sn = s) will prove useful. Also, let Ns = ∑n=1N I(sn = s).
State vector mean, for target s
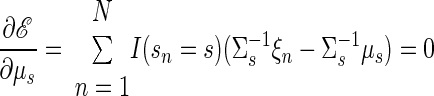 |
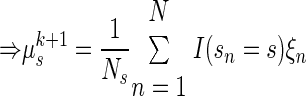 |
(A15) |
• State vector covariance, for target s
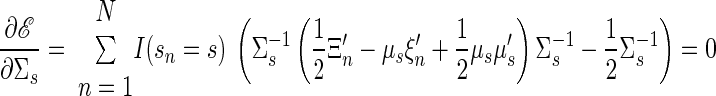 |
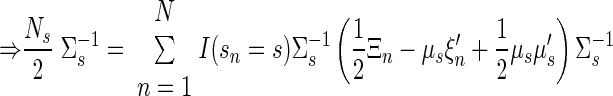 |
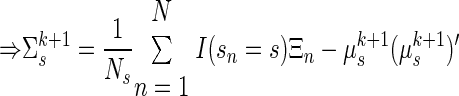 |
(A16) |
• Mapping matrix
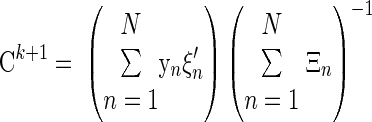 |
(A17) |
Independent variance matrix
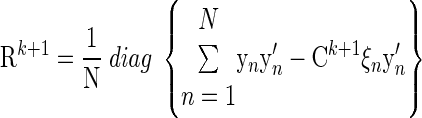 |
(A18) |
where the diag operator sets all of the off-diagonal elements of a matrix to zero.
Inference. Once the model parameters have been chosen, the generative model can be used to make inferences on the training data or new observations. For the training data, the hidden state vector x is the only variable that must be inferred. The posterior distribution of x is a Gaussian, exactly as described previously. This yields a distribution Q with mean μsn + βsn(yn − Cμsn) and covariance Σsn − βsCΣsn. Therefore the maximum a posteriori of x is simply μsn + βsn(yn − Cμsn). The vector x is not required for decoding reach endpoint but was useful in generating Fig. 4.
When performing inference for a new observation, the reach target s is now unknown. The posterior distributions of both s and x, given the data y, are of interest for decoding. The first of these distributions can be expressed as follows
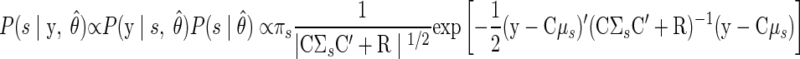 |
(A19) |
To infer x given the data, the following derivation applies
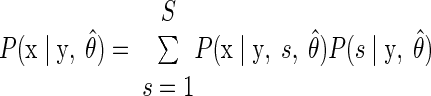 |
(A20) |
where the first factor in the summation is the conditional Gaussian (see Eq. A6) and the second is a weighting as shown earlier. Thus the distribution of x given y (but not conditioned on s) is a mixture of Gaussians.
Footnotes
Exact specifics of our experimental setup are provided in methods and are not essential for this overview.
We use the same screening procedure when comparing performance of classic Gaussian, classic Poisson, and FA-based algorithms, even though units that do not pass the ANOVA test may potentially capture information about other behavioral or cognitive factors. These units could provide further benefit for the FA-based algorithms, but we forgo that possibility so that all algorithms use exactly the same data.
The “sensible” principal component analysis (sPCA) model is a probabilistic approach to PCA and yields the same mapping between latent states and observations as that of conventional PCA. Roweis (1998) provides one mathematical demonstration of this. This algorithm is also known as probabilistic PCA (pPCA) (Tipping and Bishop 1999).
Note that the overall model for y is very similar to the “mixture of factor analyzers” proposed by Ghahramani and Hinton (1997), except that FAsep allows for a separate covariance Rs per mixture component.
We fix the number of factors in this example to be three to allow for convenient plotting of the data; ordinarily the number of factors (p) would be optimized using the model selection procedure described earlier.
As will be shown in results, more latent dimensions are required for FAcmb than for FAsep.
Recall that the data are first square-rooted before being used to fit the Gaussian model.
The use of Σs in the latent space distribution affords a more general model than the choice of a fixed covariance I in the main text, but the simpler model performed as well, and sometimes better, with fewer parameters to be fit.
REFERENCES
- Batista et al. 2007.Batista AP, Santhanam G, Yu BM, Ryu SI, Afshar A, Shenoy KV. Reference frames for reach planning in macaque dorsal premotor cortex. J Neurophysiol 98: 966–983, 2007. [DOI] [PubMed] [Google Scholar]
- Brockwell et al. 2004.Brockwell AE, Rojas AL, Kass RE. Recursive Bayesian decoding of motor cortical signals by particle filtering. J Neurophysiol 91: 1899–1907, 2004. [DOI] [PubMed] [Google Scholar]
- Brown et al. 1998.Brown EN, Frank LM, Tang D, Quirk MC, Wilson MA. A statistical paradigm for neural spike train decoding applied to position prediction from the ensemble firing patterns of rat hippocampal place cells. J Neurosci 18: 7411–7425, 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown 2006.Brown TA Confirmatory Factor Analysis for Applied Research (Methodology in the Social Sciences Series). New York: Guilford, 2006.
- Carmena et al. 2003.Carmena JM, Lebedev MA, Crist RE, O'Doherty JE, Santucci DM, Dimitrov DF, Patil PG, Henriquez CS, Nicolelis MAL. Learning to control a brain–machine interface for reaching and grasping by primates. PLoS Biol 1: 193–208, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chestek et al. 2007.Chestek CA, Batista AP, Santhanam G, Yu BM, Afshar A, Cunningham JP, Gilja V, Ryu SI, Churchland MM, Shenoy KV. Single-neuron stability during repeated reaching in macaque premotor cortex. J Neurosci 27: 10742–10750, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchland et al. 2006a.Churchland MM, Santhanam G, Shenoy KV. Preparatory activity in premotor and motor cortex reflects the speed of the upcoming reach. J Neurophysiol 96: 3130–3146, 2006a. [DOI] [PubMed] [Google Scholar]
- Churchland et al. 2006b.Churchland MM, Yu BM, Ryu SI, Santhanam G, Shenoy KV. Neural variability in premotor cortex provides a signature of motor preparation. J Neurosci 26: 3697–3712, 2006b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cisek and Kalaska 2002.Cisek P, Kalaska JF. Modest gaze-related discharge modulation in monkey dorsal premotor cortex during a reaching task performed with free fixation. J Neurophysiol 88: 1064–1072, 2002. [DOI] [PubMed] [Google Scholar]
- Cisek and Kalaska 2004.Cisek P, Kalaska JF. Neural correlates of mental rehearsal in dorsal premotor cortex. Nature 431: 993–996, 2004. [DOI] [PubMed] [Google Scholar]
- Cunningham et al. 2008.Cunningham JP, Yu BM, Gilja V, Ryu SI, Shenoy KV. Toward optimal target placement for neural prosthetic devices. J Neurophysiol 100: 3445–3457, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayan and Abbott 2001.Dayan P, Abbott LF. Theoretical Neuroscience: Computational and Mathematical Modeling of Neural Systems. Cambridge, MA: MIT Press, 2001.
- Dempster et al. 1977.Dempster AP, Laird NM, Rubin DB. Maximum likelihood from incomplete data via the EM algorithm (with discussion). J R Stat Soc Ser B 39: 1–38, 1977. [Google Scholar]
- Everitt 1984.Everitt BS An Introduction to Latent Variable Models. London: Chapman & Hall, 1984.
- Faisal et al. 2008.Faisal AA, Selen LPJ, Wolpert DM. Noise in the nervous system. Nat Rev Neurosci 9: 292–303, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgopoulos et al. 1982.Georgopoulos AP, Kalaska JF, Caminiti R, Massey JT. On the relations between the direction of two-dimensional arm movements and cell discharge in primate motor cortex. J Neurosci 2: 1527–1537, 1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghahramani and Hinton 1997.Ghahramani Z, Hinton G. The EM algorithm for mixtures of factor analyzers. Technical Report CRG-TR-96-1. Toronto: Department of Computer Science, Univ. of Toronto, 1997.
- Gilja et al. 2005.Gilja V, Kalmar RS, Santhanam G, Ryu SI, Yu BM, Afshar A, Shenoy KV. Trial-by-trial mean normalization improves plan period reach target decoding. Soc Neurosci Abstr 519.18, 2005.
- Godschalk et al. 1985.Godschalk M, Lemon RN, Kuypers HG, van der Steen J. The involvement of monkey premotor cortex neurones in preparation of visually cued arm movements. Behav Brain Res 18: 143–157, 1985. [DOI] [PubMed] [Google Scholar]
- Gomez et al. 2000.Gomez JE, Fu Q, Flament D, Ebner TJ. Representation of accuracy in the dorsal premotor cortex. Eur J Neurosci 12: 3748–3760, 2000. [DOI] [PubMed] [Google Scholar]
- Hastie et al. 2003.Hastie T, Tibshirani R, Friedman JH. The Elements of Statistical Leaning (2nd ed.). New York: Springer, 2003.
- Hatsopoulos et al. 2004.Hatsopoulos N, Joshi J, O'Leary JG. Decoding continuous and discrete motor behaviors using motor and premotor cortical ensembles. J Neurophysiol 92: 1165–1174, 2004. [DOI] [PubMed] [Google Scholar]
- Hochberg et al. 2006.Hochberg LR, Serruya MD, Friehs GM, Mukand JA, Saleh M, Caplan AH, Branner A, Chen D, Penn RD, Donoghue JP. Neuronal ensemble control of prosthetic devices by a human with tetraplegia. Nature 442: 164–171, 2006. [DOI] [PubMed] [Google Scholar]
- Hocherman and Wise 1991.Hocherman S, Wise SP. Effects of hand movement path on motor cortical activity in awake, behaving rhesus monkeys. Exp Brain Res 83: 285–302, 1991. [DOI] [PubMed] [Google Scholar]
- Kalmar et al. 2005.Kalmar RS, Gilja V, Santhanam G, Ryu SI, Yu BM, Afshar A, Shenoy KV. PMd delay activity during rapid sequential movement plans. Soc Neurosci Abstr 519.17, 2005.
- Kemere et al. 2008.Kemere C, Santhanam G, Yu BM, Afshar A, Ryu SI, Meng TH, Shenoy KV. Detecting neural state transitions using hidden Markov models for motor cortical prostheses. J Neurophysiol 100: 2441–2452, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy et al. 2000.Kennedy PR, Bakay RAE, Moore MM, Adams K, Goldwaithe J. Direct control of a computer from the human central nervous system. IEEE Trans Rehabil Eng 8: 198–202, 2000. [DOI] [PubMed] [Google Scholar]
- Kihlberg et al. 1972.Kihlberg JK, Herson JH, Schotz WE. Square root transformation revisited. Appl Statist 21: 76–81, 1972. [Google Scholar]
- Kulkarni and Paninski 2007.Kulkarni J, Paninski L. Common-input models for multiple neural spike-train data. Network Comput Neural Syst 18: 375–407, 2007. [DOI] [PubMed] [Google Scholar]
- Leuthardt et al. 2004.Leuthardt EC, Schalk G, Wolpaw JR, Ojemann JG, Morann DW. A brain–computer interface using electrocorticographic signals in humans. J Neural Eng 1: 63–71, 2004. [DOI] [PubMed] [Google Scholar]
- Maynard et al. 1999.Maynard EM, Hatsopoulos NG, Ojakangas CL, Acuna BD, Sanes JN, Normann RA, Donoghue JP. Neuronal interactions improve cortical population coding of movement direction. J Neurosci 19: 8083–8093, 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messier and Kalaska 2000.Messier J, Kalaska JF. Covariation of primate dorsal premotor cell activity with direction and amplitude during a memorized-delay reaching task. J Neurophysiol 84: 152–165, 2000. [DOI] [PubMed] [Google Scholar]
- Musallam et al. 2004.Musallam S, Corneil BD, Greger B, Scherberger H, Andersen RA. Cognitive control signals for neural prosthetics. Science 305: 258–262, 2004. [DOI] [PubMed] [Google Scholar]
- Ng and Jordan 2002.Ng AY, Jordan MI. On discriminative vs. generative classifiers: a comparison of logistic regression and naive Bayes. In: Advances in Neural Information Processing Systems 14, edited by Dietterich TG, Becker S, Ghahramani Z. Cambridge, MA: MIT Press, 2002, vol. 2, p. 841–848.
- Okatan et al. 2005.Okatan M, Wilson MA, Brown EN. Analyzing functional connectivity using a network likelihood model of ensemble neural spiking activity. Neural Comput 17: 1927–1961, 2005. [DOI] [PubMed] [Google Scholar]
- Raina et al. 2004.Raina R, Shen Y, Ng AY, McCallum A. Classification with hybrid generative/discriminative models. In: Advances in Neural Information Processing Systems 16, edited by Thrun S, Saul LK, Schölkopf B. Cambridge, MA: MIT Press, 2004, p. 545–552.
- Riehle et al. 1994.Riehle A, MacKay WA, Requin J. Are extent and force independent movement parameters? Preparation- and movement-related neuronal activity in the monkey cortex. Exp Brain Res 99: 56–74, 1994. [DOI] [PubMed] [Google Scholar]
- Roweis and Ghahramani 1999.Roweis S, Ghahramani Z. A unifying review of linear Gaussian models. Neural Comput 11: 305–345, 1999. [DOI] [PubMed] [Google Scholar]
- Roweis 1998.Roweis ST EM algorithms for PCA and SPCA. In: Advances in Neural Information Processing Systems 10, edited by Jordan MI, Kearns MJ, Solla SA. Cambridge, MA: MIT Press, 1998, p. 626–632.
- Santhanam et al. 2006a.Santhanam G, Ryu SI, Yu BM, Afshar A, Shenoy KV. A high-performance brain-computer interface. Nature 442: 195–198, 2006a. [DOI] [PubMed] [Google Scholar]
- Santhanam et al. 2006b.Santhanam G, Yu BM, Shenoy KV, Sahani M. Factor analysis with Poisson output. Technical Report NPSL-TR-06-1. Stanford, CA: Stanford Univ., 2006b.
- Schalk et al. 2008.Schalk G, Miller KJ, Anderson NR, Wilson JA, Smyth MD, Ojemann JG, Moran DW, Wolpaw JR, Leuthardt EC. Two-dimensional movement control using electrocorticographic signals in humans. J Neural Eng 5: 75–84, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serruya et al. 2002.Serruya MD, Hatsopoulos NG, Paninski L, Fellows MR, Donoghue JP. Instant neural control of a movement signal. Nature 416: 141–142, 2002. [DOI] [PubMed] [Google Scholar]
- Shadlen and Newsome 1998.Shadlen MN, Newsome WT. The variable discharge of cortical neurons: implications for connectivity, computation, and information coding. J Neurosci 18: 3870–3896, 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenoy et al. 2003.Shenoy KV, Meeker D, Cao S, Kureshi SA, Pesaran B, Mitra P, Buneo CA, Batista AP, Burdick JW, Andersen RA. Neural prosthetic control signals from plan activity. Neuroreport 14: 591–596, 2003. [DOI] [PubMed] [Google Scholar]
- Shmulewitz et al. 2001.Shmulewitz D, Auerbach SB, Lehner T, Blundell ML, Winick JD, Youngman LD, Skilling V, Heath SC, Ott J, Stoffel M, Breslow JL, Friedman JM. Epidemiology and factor analysis of obesity, type II diabetes, hypertension, and dyslipidemia (syndrome X) on the Island of Kosrae, Federated States of Micronesia. Hum Hered 51: 8–19, 2001. [DOI] [PubMed] [Google Scholar]
- Smith and Brown 2003.Smith AC, Brown EN. Estimating a state-space model from point process observations. Neural Comput 15: 965–991, 2003. [DOI] [PubMed] [Google Scholar]
- Taylor et al. 2002.Taylor DM, Helms Tillery SI, Schwartz AB. Direct cortical control of 3D neuroprosthetic devices. Science 296: 1829–1832, 2002. [DOI] [PubMed] [Google Scholar]
- Thacker and Bromiley 2001.Thacker NA, Bromiley PA. The effects of a square root transform on a Poisson distributed quantity. Technical Report 2001–010. Manchester, UK: Univ. of Manchester, 2001.
- Tipping and Bishop 1999.Tipping ME, Bishop CM. Probabilistic principal component analysis. J R Stat Soc B 61: 611–622, 1999. [Google Scholar]
- Truccolo et al. 2005.Truccolo W, Eden UT, Fellows MR, Donoghue JP, Brown EN. A point process framework for relating neural spiking activity to spiking history, neural ensemble and extrinsic covariate effects. J Neurophysiol 93: 1074–1089, 2005. [DOI] [PubMed] [Google Scholar]
- Velliste et al. 2008.Velliste M, Perel S, Spalding MC, Whitford AS, Schwartz AB. Cortical control of a prosthetic arm for self-feeding. Nature 453: 1098–1101, 2008. [DOI] [PubMed] [Google Scholar]
- Wolpaw and McFarland 2004.Wolpaw JR, McFarland DJ. Control of a two-dimensional movement signal by a noninvasive brain-computer interface in humans. Proc Natl Acad Sci USA 101: 17849–17854, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu et al. 2006.Wu W, Gao Y, Bienenstock E, Donoghue JP, Black MJ. Bayesian population decoding of motor cortical activity using a Kalman filter. Neural Comput 18: 80–118, 2006. [DOI] [PubMed] [Google Scholar]
- Wu et al. 2008.Wu W, Kulkarni JE, Hatsopoulos NG, Paninski L. Neural decoding of goal-directed movements using a linear statespace. In: Computational and Systems Neuroscience (COSYNE) 2008 Workshops, Salt Lake City, UT. New York: Nova Science, 2008, p. 155.
- Yu et al. 2006.Yu BM, Afshar A, Santhanam G, Ryu SI, Shenoy KV, Sahani M. Extracting dynamical structure embedded in neural activity. In: Advances in Neural Information Processing Systems 18, edited by Weiss Y, Schölkopf B, Platt J. Cambridge, MA: MIT Press, 2006, p. 1545–1552.
- Yu et al. 2007.Yu BM, Kemere C, Santhanam G, Afshar A, Ryu SI, Meng TH, Sahani M, Shenoy KV. Mixture of trajectory models for neural decoding of goal-directed movements. J Neurophysiol 97: 3763–3780, 2007. [DOI] [PubMed] [Google Scholar]
- Zhang et al. 1998.Zhang K, Ginzburg I, McNaughton B, Sejnowski TJ. Interpreting neuronal population activity by reconstruction: unified framework with application to hippocampal place cells. J Neurophysiol 79: 1017–1044, 1998. [DOI] [PubMed] [Google Scholar]