Abstract
We have previously observed that replication and nuclear location of the murine Igh locus are developmentally regulated during B cell differentiation. In non-B, B, and plasma cells, sequences near the 3′ end of the Igh locus replicate early in S while upstream Vh sequences replicate late in S, and the Igh locus is located near the nuclear periphery. In fact, in MEL non-B cells, replication of a 500-kb segment containing Igh-C and flanking sequences occurs progressively later throughout S by 3′ to 5′ unidirectional fork movement. In contrast, in pro- and pre-B cells, the entire 3-Mb Igh locus is located away from the nuclear periphery and replicates early in S by forks progressing in both directions. In this study, using an 18–81 (pre-B) × BW5147 (T) cell fusion system in which Igh expression is extinguished, we found that in all Igh alleles, Vh sequences replicated later in S than 3′ Igh sequences (similar to that detected in BW5147), but the Igh locus was situated away from the nuclear periphery (similar to that observed in 18–81). Thus, pre-B cell-derived Igh genes had changes in replication timing, but not in nuclear location, whereas T cell-derived Igh genes changed their nuclear location but not their replication timing. These data are consistent with the silencing of a pre-B cell-specific replication program in the fusion hybrid cells and independent regulation of the nuclear location of Igh loci.
Replication of the Igh locus is associated with its transcriptional and rearrangement status, subnuclear localization, and stage of B cell development (1, 2). In non-B cells and in B cells at later stages of B cell development, the Igh locus is situated near the nuclear periphery (2, 3). In these cells, an early replicating 3′ domain is connected to a late-replicating Vh domain by a temporal transition region (1, 2). The sequence composition of the temporal transition region is unique for each cell line due to varying deletions of intervening sequences during VDJ joining and class switch recombination. In MEL non-B cells, replication forks progress in a single direction through the temporal transition region, suggesting that any potential bidirectional origin is silent within this 500-kb region (1, 2). The origin of replication of the Igh replicon in MEL cells has been localized ~76–79 kb downstream of the Igh-Cα gene (4).
A very different nuclear location and replication pattern for Igh genes has been observed in pro- and pre-B cells (2, 3). In this study, the entire Igh locus is situated away from the nuclear periphery, and it replicates early in S phase. Replication forks progress in both directions through the examined constant region genes, consistent with activation of at least one additional developmentally regulated origin. In fact, several origins have been identified using the novel single molecule analysis of replicated DNA technique (5) (P. Norio et al., manuscript in preparation). Collectively, these data suggest that a position away from the nuclear periphery is associated with replication of the Igh locus early in S, while a position at the nuclear periphery is associated with a temporal transition region generated by replication of 3′ Igh sequences early in S and Vh sequences later in S. Sequences 3′ of the Igh locus always replicate early in S, regardless of their nuclear position (1, 2, 4).
Previous studies (6) have shown that Ig expression is extinguished in an Ig-producing myeloma cell upon fusion with a T lymphoma cell. Moreover, other myeloma-specific genes are coordinately silenced (7). These changes in gene expression are likely associated with regulated expression of T and B cell-specific transcription factors. In this study, we take advantage of this well-characterized B and T cell fusion hybrid system to investigate the possible mechanism that regulates DNA replication and subnuclear localization of the Igh locus in B cells at early stages of B cell development. By analysis of a fusion hybrid of 18–81 pre-B cells with BW5147 T cells, we found that the location of Igh alleles in the nucleus and their relative replication timing in S are independently regulated.
Materials and Methods
Generation of 18–81 × BW5147 fusion hybrids
Puromycin resistant 18–81 cells (8) were electrofused with ouabain-resistant BW5147 T cells (ATCC CRL 1588) as previously described (8). Selection of fusion hybrids with puromycin and ouabain was begun 48 h postfusion, resulting in >30 subclones after ~3 wk. Subclones were analyzed by PCR for the presence of the 18–81 Vh region and the BW5147 TCR rearranged V region. None of 12 fusion subclones that contained the 18–81 Vh gene expressed IgM, as analyzed by ELISA. Three clones that contained 18–81-derived Igh alleles were maintained, and genomic Southern blot analysis confirmed the presence of Jh-containing alleles from both 18–81 and BW5147. Fluorescence in situ hybridization (FISH)3 studies, as described below (Fig. 1), confirmed the presence of two alleles from 18–81 and two from BW5174. It is highly likely that the 18–81-derived Igh allele is transcriptionally silent in the 18–81 × BW5147 fusion hybrids based on previous studies of myeloma × T cell fusion hybrids, which showed that Igh protein and mRNA expression were always coordinated (9) and that the extinguished allele was methylated de novo, consistent with its being completely silent (6). One clone was selected for detailed analysis of replication and nuclear location, as described in this article. This clone was grown in RPMI 1640 supplemented with 10% FCS, 1% penicillin-streptomycin and 50 mM 2-ME.
FIGURE 1.
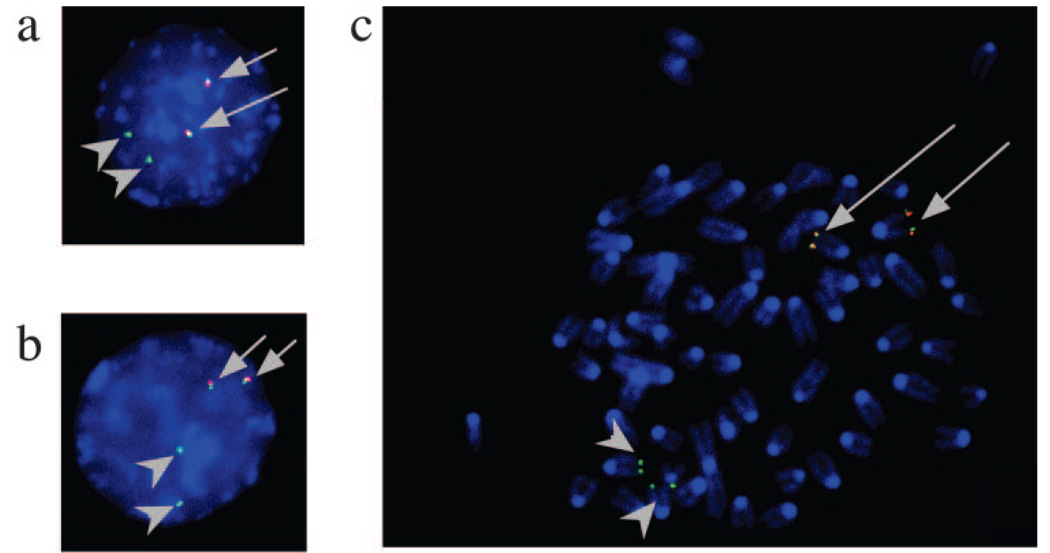
2D FISH images distinguish pre-B cell- and T cell-derived Igh loci in an 18–81 × BW5147 fusion cell line. Interphase nuclei (a and c) and a mitotic chromosome spread were hybridized to BAC71H20 (DJ) (red signal), along with BAC526A21 (images a and c, green signal) or BAC 199M11 (image b, green signal). Nuclei were counterstained with 4′,6′-diamidino-2-phenylindole (blue). Gray arrows indicate the Igh alleles from T cells (both red and green signals are present), and gray arrowheads indicate the Igh alleles from pre-B cells (only green signals can be detected).
Two-dimensional (2D) FISH
2D FISH was performed as previously described (2). For nuclear localization and replication timing studies, bacterial artificial chromosomes (BACs) containing various segments of the Igh cluster were used. These were: BAC 526A21, which contains Ch-proximal VhJ558 genes and flanking sequence; BAC 71H20, which contains ~90 kb of the Dh and Jh segments, and BAC 199M11, which contains sequences downstream of Cα (2). BAC colonies were cultured in Luria-Bertani (LB) broth supplemented with chloramphenicol (12.5 mg/ml). DNA was purified using Qiagen-tip 100 according to the instructions from the company, except that the elution buffer was prewarmed at 65°C before use. BACs were labeled with biotin-16-dUTP or digoxigenin-11-dUTP using a nick-translation kit (Roche) as described previously (2). Slides were visualized by conventional fluorescence microscopy and digital images were taken by a CCD4 camera and combined in Adobe Photoshop.
The replication timing of Igh genes was estimated by FISH, as used previously (10–12). Data obtained by FISH for the Igh locus have been shown to be consistent with data obtained by the BrdU S-phase fractionation technique (12). The number of doublets (indicating replication) for each allele in the nuclei of cells in log phase was calculated. To determine the subnuclear localization of Igh sequences, the distance from the center of the nucleus to the individual signal and the radius of each nucleus were measured using the NIH Image program (Scion Image 1.62). The relative position of the Igh locus was specified by the ratio of the distance from the signal to the center of the nucleus divided by the radius of the nucleus. A relative distance of >0.8 was considered to be located near the nuclear periphery. Data were processed in an Excel program. At least 50 nuclei were analyzed in each experiment.
Three-dimensional (3D) FISH
FISH on 3D-preserved nuclei was done as described elsewhere (13). In brief, cells were attached to glass coverslips using poly-l-lysine, fixed for 30 min with fixation permeabilization buffer (20 mM KH2PO4, 130 mM NaCl, 20 mM KCl, 10 mM EGTA, 20 mM MgCl2, 0.1% (v/v) Triton X-100, and 0.5% (v/v) glutaraldehyde (grade 1, 70% aqueous; Sigma-Aldrich)). The coverslips were then washed twice (15 min/wash) with sodium borohydride solution (1 mg/ml prepared freshly in water). The samples were sequentially incubated with 5% rabbit serum/5% FCS, goat anti-lamin B (M20) antisera (Santa Cruz Biotechnology), and Cy5-coupled anti-goat Ig (Jackson ImmunoResearch). Postfixation cross-linking of proteins was done by incubating the coverslips with a 50 mM solution of ethylene glycol bis(succinimidyl succinate) in PBS for 30 min 37°C. Cells were incubated for 1 h with RNase A (100 µg/ml in 2× SSC), washed in PBS, and the chromosomal DNA was denatured in 0.1 M NaOH (pH 12.7) for 2 min, followed immediately by rinsing in ice-cold PBS before applying DNA probes. Hybridization was performed overnight in humid chambers at 37°C. Biotinylated probes were detected with FITC-avidin (primary Ab; Roche) and FITC anti-avidin D (secondary Ab; Vector Laboratories). Digoxigenin-labeled probes were detected with Cy3-conjugated monoclonal mouse anti-digoxin (primary Ab; Jackson ImmunoResearch) and Cy3-conjugated goat anti-mouse IgG (secondary Ab; Jackson ImmunoResearch). Images were captured using a Provis AX70 microscope equipped with a Cool-SNAP digital camera, and the data were analyzed in Slidebook 3.0 (Intelligent Imaging Innovations).
Results
Replication timing of the Ig H chain variable region sequences in the 18–81 pre-B cell line is delayed after fusion with the BW5147 T cell line
We used a 2D FISH technique to compare the replication timing of the Igh locus in the 18–81 pre-B cell line and the BW5147 T cell line with that in the 18–81 × BW5147 fusion hybrid. Replication was determined by measuring the proportion of alleles displaying doublets as a result of replication and sister chromatid resolution (10–12). A gene replicated at the beginning of S will show doublets of each of its alleles throughout the S phase, whereas a gene replicated late in the S phase will show doublets only briefly before cell division. This will be detected as a greater (replication early in S) vs lesser (replication later in S) frequency of unsynchronized cells displaying doublets for the contrasting genes. A delay in sister-chromatid resolution has been observed for a number of leukocyte-specific genes in lymphocytes (14), although not reported for Ig genes. Such a delay could falsely indicate that a gene sequence was replicated later in S than actually occurred. However, it is unlikely that individual Igh alleles would be subject to differential sister-chromatid resolution in the same nuclear milieu. We, therefore, propose that the singlet/doublet approach is satisfactory to assess relative replication timing of Igh gene sequences. Two BAC probes were used. The first, BAC 199M11, contains a 125-kb region beginning immediately downstream of the Cα gene and extending through Crip, Crp2, and Mta1 gene sequences. This segment replicates early in S in each of the cell lines we previously examined. The second, BAC 526A21, contains the Ch-proximal VhJ588 genes: these Vh genes replicate early in the S phase in pre-B cell lines but late in the S phase in other types of cell lines examined (2).
In accord with our previous studies of pro- and pre-B cells (2), we find that the entire Igh locus appears to replicate early in the S phase in 18–81. The percentages of total alleles showing doublets for the J558- and the 3′ Igh-hybridizing regions are similar (27.1 and 29.2%, respectively; Table I). In addition, we found that in BW5147, as in other non-B cell lines and in mature B and plasma cells, Igh-C sequences are replicated earlier in S than Igh-V sequences. The percentage of total alleles showing doublets for the Vh region in BW5147 (11.9%) is less than for the downstream 3′ Igh region (28.2%).
Table I.
FISH analysis of relative timing of replication of Cα downstream and VhJ558 sequences
| % of Total Igh alleles showing doublets for |
||
|---|---|---|
| 3′ Igha | VhJ558b | |
| Origin of allele | ||
| 18–81 | 29.2 | 27.1 |
| BW5147 | 28.2 | 11.9 |
| Fusion hybridc | ||
| 18–81-derived | 34.0 | 19.3 |
| BW5147-derived | 37.0 | 14.4 |
Identified by CT7–199M11, which contains 125 kb immediately downstream of the Cα gene.
Identified by CT7–526A21, which contains proximal Vh J558 sequences.
The Igh alleles from 18–81 pre-B cells and from BW5147 T cells are distinguished by BAC 71H20 (DJ region), which can hybridize to the T cell-derived Igh locus.
Of the >30 18–81 × BW5147 fusion hybrids examined, none expressed the 18–81 IgM H chain. These data are in accordance with previous observations showing that when T cells are fused with B cells, the B cell transcription program is extinguished (6). One clone was selected for detailed analysis of replication and nuclear location. To distinguish the Igh alleles from BW5147 T cells and pre-B cells, we used BAC 71H20, which contains the Dh and Jh segments. Dh and Jh segments are present in BW5147, but are deleted in 18–81 pre-B cells as a consequence of VDJ joining. Thus, when BAC 71H20 (red) is used along with either BAC199M11 (3′ Igh; green) or BAC526A21 (VhJ558; green), there are two closely associated FISH signals (red and green) that identify the T cell-derived Igh alleles, while only one (green) FISH signal can be detected for each of the two pre-B cell-derived Igh alleles (see FISH images in Fig. 1).
In our examination of the 18–81 × BW5147 fusion hybrid, we found that replication of the Vh region is delayed for the pre-B cell-derived alleles when compared with 18–81 (Table I). Only 19.3% of all pre-B cell-derived alleles displayed a doublet for the Vh region, and 34.0% of these alleles showed a doublet for the downstream 3′ Igh region. The replication timing of the Igh locus did not change for the alleles derived from the BW5147 T cell line (14.4% doublets for VhJ558 segments on T cell-derived alleles, see Table I). Therefore, the replication program of the pre-B cell-derived Igh alleles in the 18–81 × BW5147 fusion hybrid resembled that of non-B, B, and plasma cells. There is no apparent difference in cell growth of fusion hybrids as compared with parental cells that would indicate a difference in cell cycle. Small differences in portions of the cell cycle might accentuate or diminish differences in replication timing between 3′ Ch and Vh sequences, but they would be unlikely to prevent us from detecting a profile like that observed in pro- and pre-B cells, where both sequences replicate at the same time.
The Igh locus is located away from the nuclear periphery in the 18–81 × BW5147 fusion cell line
Since replication timing changed for the pre-B cell-derived Igh alleles in the fusion hybrid, we wanted to know whether the location of the Igh locus also changed. We first confirmed that data collected from 2D FISH are consistent with those collected from 3D-preserved nuclei by examining the subnuclear localization of the Igh locus in 18–81 pre-B cells using both techniques. The same two BACs that were used for the replication timing studies were used here. Both 2D and 3D FISH showed that the Igh-V region was located away from the periphery of the nucleus in the 18–81 pre-B cells (79% and 84.4% of the signals from 2D and 3D FISH studies, respectively). Similar results were also detected for the region located immediately downstream of the Cα gene (data not shown). We, therefore, used 2D FISH to examine the subnuclear localization of the Igh loci in the fusion cell line. Again, the pre-B cell allele and T cell allele were distinguished using the BAC 71H20(DJ) probe.
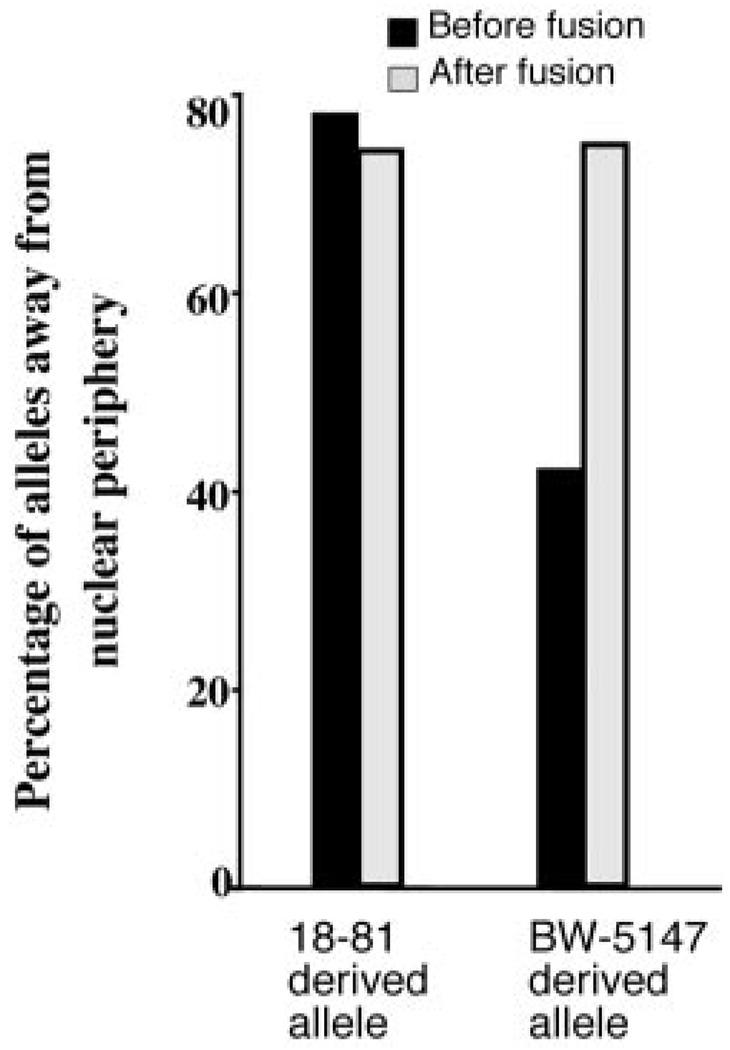
Surprisingly, we found that despite the change in replication timing, the Igh locus from the pre-B cell-derived alleles was maintained away from the perinuclear region: compare 79% of the Vh region BAC signals located away from the periphery of the nucleus in the 18–81 pre-B cell line with 74% of the 18–81-derived Vh region BAC signals after fusion (Fig. 2). Moreover, we found that the T cell alleles were also located away from the periphery of the nucleus in the fusion cell line. Before cell fusion, 57.6% of the variable region signals were found close to the nuclear periphery in BW5147 T cells. In contrast, after cell fusion, only 25.5% of the variable region signals derived from the T cells were found close to the nuclear periphery (Fig. 2). Similar results were found for the downstream region sequences.
FIGURE 2.
Location of the Igh locus within the nucleus in BW5147 T cells and 18–81 pre-B cells before and after cell fusion. In 18–81 pre-B cells, both 2D and 3D FISH data show that the Igh locus is situated away from the periphery of the nucleus (both data sets show 0.8). The location of the Igh locus from pre-B cells was not changed after fusion with T cells. More than 75% of the alleles were located away from the periphery of the nucleus both before and after cell fusion. However, the location of the Igh locus from T cells was changed after fusion with 18–81, from <40% of the alleles away from the periphery of the nucleus before fusion to >75% of the alleles away from the periphery of the nucleus after fusion. The difference between the location of the BW5147-derived allele in BW5147 cells as compared with the 18–81 × BW5147 fusion hybrid is highly significant (p < 0.0001).
Discussion
In the 18–81 pre-B × BW5147 T cell fusion hybrid studied here, the pre-B cell-specific Igh expression program was extinguished, and there were changes in replication timing and nuclear position of the Igh alleles. Pre-B cell-derived alleles in the hybrid line acquired the replication pattern that is evident in BW5147 and other non-B cells, as well as in B and plasma cells. Despite this change in replication, the pre-B cell-derived alleles did not change position within the nucleus: they remained at a distance from the nuclear periphery. There was a reciprocal effect on the T cell-derived alleles. These alleles continued to replicate as in the T cell parent line. The position of these alleles within the nucleus, however, changed to more closely resemble that of Igh alleles in pre- and pro-B cells (away from the nuclear periphery). In summary, all of the Igh alleles in the hybrid were unexpressed, replicated the Ch genes early in S and the Vh genes late in S, and all were located away from the nuclear periphery.
These observations reveal that replication timing and the nuclear position of Vh sequences are two, independently regulated phenomena. VhJ558 sequences on the pre-B-derived Igh alleles shift from early to late replication in the hybrid line and yet they remain away from the nuclear periphery. VhJ558 sequences on the T cell-derived Igh alleles continue to be replicated late in the hybrid line, but they move away from the nuclear periphery. As described previously, there is a temporary change in both the replication pattern and the nuclear location of Igh loci in pro-B and pre-B cells. The findings presented here suggest that these are developmentally related events that are independently regulated. It is possible that chromosome loss that occurs upon fusion of these two heteroploid lines has contributed to revealing independent regulation.
Recent studies have provided evidence for a relationship between nuclear location, compaction of the Igh locus, use of Vh genes for VDJ joining, and Pax5 expression. In pro-B cells, before Pax5 expression, the Igh locus is subject to association with histone H3 that is methylated on K9 (di-me K9 H3) (15), is located away from the nuclear periphery and is in an extended conformation. Upon Pax5 expression, the locus remains located away from the nuclear periphery, loses its association with di-me K9 H3 (15), and contracts (16), presumably making it possible for distal as well as proximal Vh gene segments to be accessible to VDJ joining. The expression of ectopic Pax5 in T cells resulted in the movement of the Igh locus away from the periphery, but the locus was maintained in an extended state (16). In these T cells, only the proximal Vh genes were available for VDJ joining. Movement of the Igh locus away from the periphery, therefore, may be necessary for but is not sufficient to ensure locus compaction. It is proposed that both Igh locus movement and compaction are required for the process of VDJ joining of distal Vh genes.
A shift to early replication of the Vh genes is yet another change that takes place in pro- and pre-B cells able to undergo VDJ joining, but its relationship to locus movement within the nucleus, compaction, and the process of VDJ joining itself is unclear. A shift in Igh locus position away from the nuclear periphery may be necessary for but not sufficient to generate a shift to early replication of the Vh genes. In the hybrid line of this study, the BW5147-derived Igh alleles moved away from the nuclear periphery, perhaps in response to transient Pax5 expression in the newly formed hybrid. Neither of these alleles (nor the 18–81-derived Igh alleles), however, underwent early replication of the Vh genes, suggesting that this likely required other factors expressed in the pre-B cell but not in the T cell parent or the hybrid line. Igh locus compaction and VDJ joining of distal Vh genes in pre-B cells have been suggested to require not only Pax5 but an additional B cell-specific factor, X, which presumably is active only in pro- and pre-B cells.
In summary, the experiments presented here suggest that the mechanisms that promote replication of the entire Igh locus early in S in pro- and pre-B cells are either wholly independent of the shift in location of the Igh locus away from the nuclear periphery or require the latter shift, but involve additional factors/events, as well.
Acknowledgments
Microscopy was performed at the Analytical Imaging Facility of the Albert Einstein College of Medicine.
Footnotes
This work was supported by National Institutes of Health R01 Grants AI13509 (to B.K.B.), CA-62363 (to L.A.E.), and GM45751 (to C.L.S.), and Albert Einstein College of Medicine Cancer Center Core Grant P30CA13330.
Abbreviations used in this paper: 2D, two dimensional; 3D, three dimensional; FISH, fluorescence in situ hybridization; BAC, bacterial artificial chromosome.
Disclosures
The authors have no financial conflict of interest.
References
- 1.Ermakova OV, Nguyen LH, Little RD, Chevillard C, Riblet R, Ashouian N, Birshtein BK, Schildkraut CL. Evidence that a single replication fork proceeds from early to late replicating domains in the IgH locus in a non-B cell line. Mol. Cell. 1999;3:321–330. doi: 10.1016/s1097-2765(00)80459-1. [DOI] [PubMed] [Google Scholar]
- 2.Zhou J, Ermakova OV, Riblet R, Birshtein BK, Schildkraut CL. Replication and subnuclear location dynamics of the immunoglobulin heavy-chain locus in B-lineage cells. Mol. Cell. Biol. 2002;22:4876–4889. doi: 10.1128/MCB.22.13.4876-4889.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kosak ST, Skok JA, Medina KL, Riblet R, Le Beau MM, Fisher AG, Singh H. Subnuclear compartmentalization of immunoglobulin loci during lymphocyte development. Science. 2002;296:158–162. doi: 10.1126/science.1068768. [DOI] [PubMed] [Google Scholar]
- 4.Zhou J, Ashouian N, Delepine M, Matsuda F, Chevillard C, Riblet R, Schildkraut CL, Birshtein BK. The origin of a developmentally regulated Igh replicon is located near the border of regulatory domains for Igh replication and expression. Proc. Natl. Acad. Sci. USA. 2002;99:13693–13698. doi: 10.1073/pnas.212392399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Norio P, Schildkraut CL. Visualization of DNA replication on individual Epstein-Barr virus episomes. Science. 2001;294:2361–2364. doi: 10.1126/science.1064603. [DOI] [PubMed] [Google Scholar]
- 6.Zaller DM, Yu H, Eckhardt LA. Genes activated in the presence of an immunoglobulin enhancer or promoter are negatively regulated by a T lymphoma cell line. Mol. Cell. Biol. 1988;8:1932–1939. doi: 10.1128/mcb.8.5.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lieberman SA, Hines MD, Bergsagel PL, Kuehl WM, Eckhardt LA. Coordinate silencing of myeloma-specific genes in myeloma X T lymphoma hybrids. J. Immunol. 1993;151:2588–2600. [PubMed] [Google Scholar]
- 8.Saleque S, Singh M, Birshtein BK. IgH expression and class switching in vitro from an allele lacking the 3′ enhancers DNase I-hypersensitive hs3A and hs1,2. J. Immunol. 1999;162:2791–2803. [PubMed] [Google Scholar]
- 9.Radomska HS, Shen C-P, Kadesch T, Eckhardt LA. Constitutively expressed Oct-2 prevents immunoglobulin gene silencing in myeloma T cell hybrids. Immunity. 1994;1:1. doi: 10.1016/1074-7613(94)90034-5. [DOI] [PubMed] [Google Scholar]
- 10.Selig S, Okumura K, Ward DC, Cedar H. Delineation of DNA replication time zones by fluorescence in situ hybridization. EMBO J. 1992;11:1217–1225. doi: 10.1002/j.1460-2075.1992.tb05162.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boggs BA, Chinault AC. Analysis of DNA replication by fluorescence in situ hybridization. Methods. 1997;13:259–270. doi: 10.1006/meth.1997.0525. [DOI] [PubMed] [Google Scholar]
- 12.Simon I, Tenzen T, Reubinoff BE, Hillman D, McCarrey JR, Cedar H. Asynchronous replication of imprinted genes is established in the gametes and maintained during development. Nature. 1999;401:929–939. doi: 10.1038/44866. [DOI] [PubMed] [Google Scholar]
- 13.Brown KE, Guest SS, Smale ST, Hahm K, Merkenschlager M, Fisher AG. Association of transcriptionally silent genes with Ikaros complexes at centromeric heterochromatin. Cell. 1997;91:845–854. doi: 10.1016/s0092-8674(00)80472-9. [DOI] [PubMed] [Google Scholar]
- 14.Azuara V, Brown KE, Williams RR, Webb N, Dillon N, Festenstein R, Buckle V, Merkenschlager M, Fisher AG. Heritable gene silencing in lymphocytes delays chromatid resolution without affecting the timing of DNA replication. Nat. Cell Biol. 2003;5:668–674. doi: 10.1038/ncb1006. [DOI] [PubMed] [Google Scholar]
- 15.Johnson K, Pflugh DL, Yu D, Hesslein DG, Lin KI, Bothwell AL, Thomas-Tikhonenko A, Schatz DG, Calame K. B cell-specific loss of histone 3 lysine 9 methylation in the VH locus depends on Pax5. Nat. Immunol. 2004;5:853–861. doi: 10.1038/ni1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fuxa M, Skok J, Souabni A, Salvagiotto G, Roldan E, Busslinger M. Pax5 induces V-to-DJ rearrangements and locus contraction of the immunoglobulin heavy-chain gene. Genes Dev. 2004;18:411–422. doi: 10.1101/gad.291504. [DOI] [PMC free article] [PubMed] [Google Scholar]