Abstract
V gene assembly, class switch recombination, and somatic hypermutation are gene-modifying processes essential to the development of an effective Ab response. If inappropriately applied, however, these processes can mediate genetic changes that lead to disease (e.g., lymphoma). A series of control elements within the Ig H chain (Igh) locus has been implicated in regulating these processes as well as in regulating IgH gene transcription. These include the intronic enhancer (Eµ) and several elements at the 3′ end of the locus (hs1,2, hs3a, hs3b, and hs4) known collectively as the 3′ regulatory region. Although it is clear that the Eµ plays a unique role in V gene assembly, it has not been established whether there are unique functions for each element within the 3′ regulatory region. In earlier studies in mice and in mouse cell lines, pairwise deletion of hs3b and hs4 had a dramatic effect on both class switch recombination and IgH gene transcription; deletion of an element almost identical with hs3b (hs3a), however, yielded no discernible phenotype. To test the resulting hypothesis that hs4 is uniquely required for these processes, we induced the deletion of hs4 within a bacterial artificial chromosome transgene designed to closely approximate the 3′ end of the natural Igh locus. When introduced into an Ig-secreting cell line, an Igα transcription unit within the bacterial artificial chromosome was expressed efficiently and the subsequent deletion of hs4 only moderately affected Igα expression. Thus, hs4 does not play a uniquely essential role in the transcription of a productively rearranged Ig VDJCα transcription unit.
The Ig H chain and L chain (IgH and IgL) loci undergo multiple rounds of DNA recombination and somatic hypermutation over the lifetime of cells in the B lymphocyte lineage. DNA recombination events that result in the deletion of large segments of the genome are required for assembly of the variable region-encoding gene segments (V-D-J and V-J joining) and for the switch from one class of Ab to another (Fig. 1). The latter, class switch recombination (CSR),3 follows Ag stimulation of the B cell and allows for the development of daughter clones that can produce Ag-reactive Ab with the appropriate properties to combat the pathogen in question (e.g., trafficking properties; ability to activate complement, etc.). Somatic hypermutation of the assembled Ig variable region genes is another consequence of Ag stimulation and is the process by which responding B lymphocytes develop a pool of daughter cells producing Abs with higher affinity for the invading pathogen.
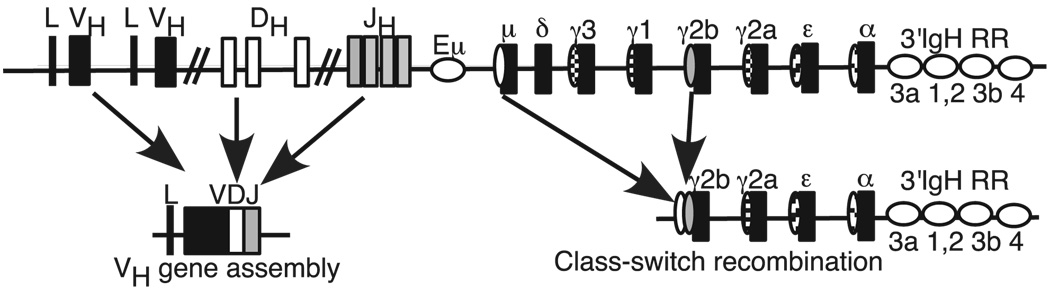
FIGURE 1.
Diagram of the murine IgH locus. Not drawn to scale. Coding sequences (e.g., V, D, J, and C gene segments) are shown as rectangles. C region segments are identified by Greek letters (µ, δ, etc.). Regulatory sequences are shown as horizontal ovals (e.g., Eµ) and the IgH 3′RR shown covers the DNase I HS sites hs3a, hs1,2, hs3b, and hs4. Vertical ovals upstream of each C region gene signify the “switch regions” in which breaks occur during CSR.
The essential and largely lymphoid cell-restricted enzymes involved in these processes have been identified. The recombination-activating genes RAG-1 and RAG-2 produce the proteins that initiate the V region gene assembly process; activation-induced cytidine deaminase initiates both CSR and somatic hypermutation of the DNA surrounding the V region genes (1, 2). Although these processes are fundamental to the development of both a diverse repertoire of B cells and a specific and efficient Ab response to Ag, they also pose a significant threat to the integrity of the genome. It is critical, therefore, that each process is carefully regulated to take place only at the appropriate loci, only within the B cell, and only at the right time relative to the B cell’s stage of maturation or state of activation. It is apparent that this regulatory responsibility falls on a series of cis-acting elements found within and around the IgH and IgL genes.
Five enhancer regions have been identified within the murine IgH locus (Eµ, hs3a, hs1,2, hs3b, and hs4; Fig. 1). The first of these, Eµ, is found in an intron separating the V region and C region coding sequences and, although initially discovered because of its ability to enhance transcription from the promoter of an assembled IgH gene, it has since been shown to be essential for efficient V-D-J recombination (3–9). The other four enhancers lie at the far 3′ end of the IgH locus and have been shown to contribute substantially to IgH gene transcription in Ig-secreting cells as well as play an important role in CSR (10, 11).
Although Eµ is normally present within the JH-Cµ intron of assembled and expressed IgH genes, numerous Ig-secreting cell tumors (myelomas) have been recovered in which Eµ is missing and yet IgH transcription continues unabated (12–15). Reciprocal loss of the 3′ IgH enhancers, however, had a dramatic effect on Ig transcription in an Igα-secreting cell line (LP1.2). Igα transcription fell to 10% of wild-type levels even though Eµ remained within the Igα transcription unit (16, 17). These two findings led to the hypothesis that the role of Eµ in promoting IgH transcription could be supplanted by the 3′ IgH enhancers. In one of the Eµ-deficient cell lines (9921), a technical knockout of the 3′ IgH enhancer region downstream of the active IgH locus (by replacing hs1,2 with a neor gene) abolished IgH transcription, confirming that this region was indeed required for locus expression (10).
In more recent studies we have used an IgH minilocus to study the function of the 3′ IgH enhancers (18). This minilocus, a reporter Igγ2b gene linked to the four 3′ IgH enhancers, was expressed at high levels when stably integrated into the genome of an Ig-secreting cell line (9921). Expression was unaffected by a subsequent in situ deletion of the hs3a/hs1,2 enhancer pair, but an in situ deletion of hs3b/hs4 resulted in a dramatic decrease in reporter gene expression, demonstrating a requirement for one or both of these enhancer elements in sustaining IgH gene transcription (18). In an independent study, the same two enhancers were deleted from the IgH locus of mice with the effect that CSR to all IgH classes except γ1 was impeded to some extent, and sterile transcripts associated with and required for CSR were, concomitantly, reduced (11). The hs3b and hs4 elements have been implicated, therefore, in promoting both CSR and IgH gene transcription.
One of the questions that has directly arisen from both of these studies is whether the effects seen were due to the deletion of hs3b alone, the deletion of hs4 alone, or were achievable only after deletion of both of these regulatory sequences. Several findings have implicated hs4. First, hs4 has several attributes that distinguish it from the other enhancers. In transient transfection assays, hs4 is the most robust of the 3′ IgH enhancers in mature B cell lines and plasmacytoma lines and the only one to show activity in pre-B cell lines (17, 19, 20). It is also the only one of the 3′ IgH enhancers that shows DNase I hypersensitivity in both pro-B and pre-B cells; the other enhancers become DNase I hypersensitive later, at the surface Ig+ cell stage (17, 19, 21, 22). Second, there was reason to doubt that hs3b was required for either CSR or Igh transcription, because the deletion of the highly homologous element hs3a (97% identity) had no effect on either process (18, 23).
In the present studies, we have tested the hypothesis that hs4 is essential for the 3′ regulatory region’s (3′RR) ability to drive IgH gene transcription. For this purpose, a reporter gene system within a bacterial artificial chromosome (BAC) was developed to mimic as closely as possible the endogenous Igh locus (Fig. 2). The BAC was modified to include a fully functional Igα gene with the 3′ IgH enhancers in their natural configuration downstream and with loxP sites surrounding hs4. The Igα-BAC was efficiently expressed when integrated into the genome of Ig-secreting cells (9921 cell line). Subsequent deletion of hs4 from the BAC by cyclization recombination enzyme (CRE)-mediated loxP site recombination, however, gave the surprising result that Igα gene expression was affected only minimally. We conclude that the proposed role of Hs4 in driving IgH gene transcription can be largely supplanted by the activity of the remaining enhancers or of other elements within the locus. As discussed below, this functional redundancy likely serves to safeguard at least the transcriptional role of this regulatory region. It remains to be determined whether CSR, like IgH gene transcription, will similarly show minimal dependence upon hs4.
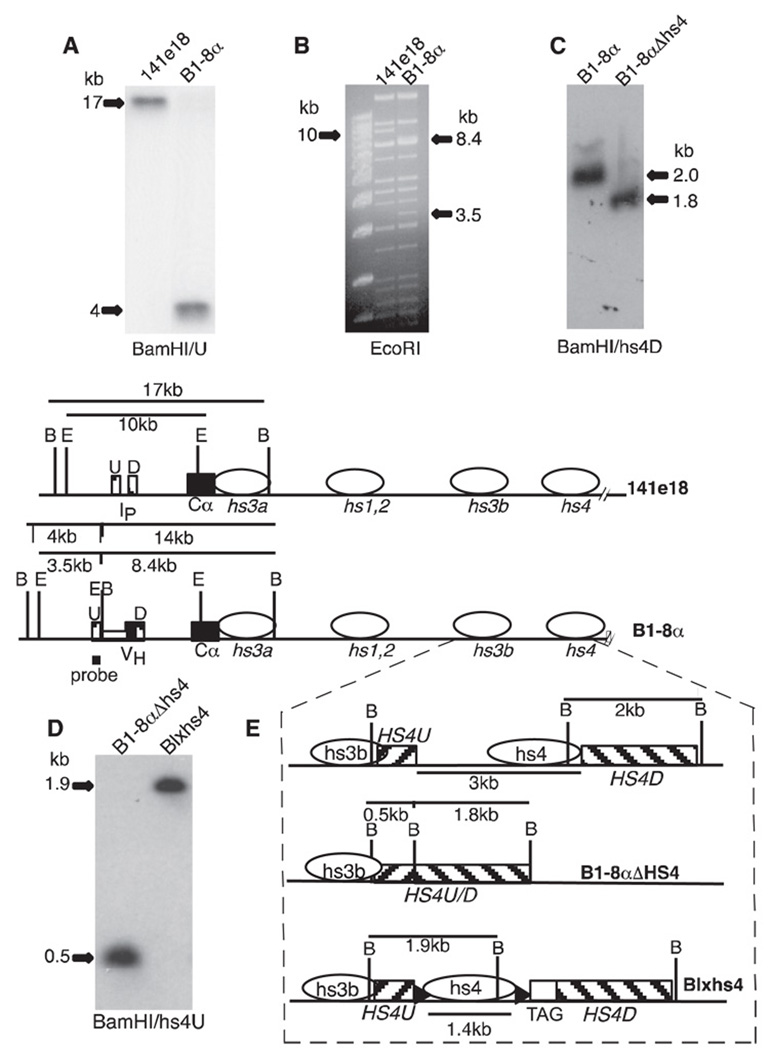
FIGURE 2.
Igα gene within a BAC. Diagrams (shown in E) of BACs used in this study include the initial BAC (141e18), a BAC with an Igα transcription unit (B1-8α), a BAC intermediate lacking hs4 (B1-8αAHS4), and BAC with loxP-flanked hs4 (Blxhs4). The 3′ IgH enhancers (hs3a, etc.) are represented by horizontal ovals. Striped boxes (U, D, HS4U, and HS4D) identify regions of homology used in homologous recombination. Ip is the intronic promoter for sterile, Cα germline transcripts that precede CSR. The BamHI (B) and EcoRI (E) recognition sites are shown only in the relevant regions (note: the B site at the junction of HS4U/HS4D in B1-8αAHS4 (enlarged portion of panel E set off by dashed lines, middle diagram) was created by cloning). VH and Cα coding sequences are filled rectangles, loxP sites are filled arrowheads, and human β-globin sequence is marked as TAG. A, BAC DNA cut with BamHI and hybridized with U probe. B, Ethidium bromide stain of BAC DNA cut with EcoRI. Leftmost lane is a 1-kb DNA ladder (m.w. marker). EcoRI fragments modified by the insertion of VH in B1–8α are indicated (compare with BAC maps). C, BAC DNA cut with BamHI and hybridized with hs4D. D, BAC DNA cut with BamHI and hybridized with hs4U.
Materials and Methods
Cell lines
9921 is an IgG2a-producing class switch variant that was derived, through an intermediate, from the IgG2b-producing plasmacytoma MPC11 (24). In the course of the H chain class switch, Eµ was deleted from the γ2a H chain transcription unit in 9921. The Igh allele carrying the γ2a H chain transcription unit is present as a single copy per cell. The nonproductive Igh allele in this cell line (chromosome 12) has undergone reciprocal translocation with c-myc (chromosome 15), and the product carrying the IgH 3′RR has been replicated to three copies per cell (data not shown, and 25). J558 is an IgA-producing myeloma obtained from American Type Culture Collection (ATCC item no. TIB-6). 9921 and J558 were maintained in DMEM (Invitrogen Life Technologies; catalog no. 12100-061) with 10% bovine calf serum (Gemini, catalog no 100–506). The medium contained 50 U/ml penicillin/streptomycin (Atlanta Biologicals, catalog no. B21110), 2 mM L-glutamine (Atlanta Biologicals, catalog no. B90310), and MEM nonessential amino acids (Invitrogen Life Technologies, catalog no. 11140050). Both cell lines were maintained at 37°C in an atmosphere of 8% CO2.
Modifications of BACs
BACs were modified by the described methods (26). The starting BAC (141el8) is ~92 kb in size, contains part of Cε (starting from position 1430 in GenBank sequence X01857.1), and extends 80 kb downstream of Cα (ending at position 91450 in GenBank sequence AF450245) (see Fig. 2). This BAC was produced by Genome Systems (BAC embryonic stem cell mouse library, 129/SVJ mouse strain) and provided by Dr. S. Janz (National Cancer Institute, National Institutes of Health, Bethesda, MD).
B1-8α (~95 kb)
This BAC carries an assembled VH gene upstream of Cα. The VH gene was isolated from plasmid pIVHB1-8L2neor as a 2.2-kb ClaI fragment (27). The latter plasmid was a gift of Dr. K. Rajewsky (Harvard Medical School, Boston, MA). The VH was inserted upstream of Cα, using two regions of homology (U and D in Fig. 2) surrounding the intronic or cryptic promoter for sterile Cα transcripts (induced just before CSR to this CH gene). The primers used to generate “U” by PCR were 5′-ACGCGTCGACCAGTAGGATGTGTAGAGGAT-3′ and 5′-CCGCTCGAGCCAGGACTCCACATGCAT-3′. The underlined sequences are restriction endonuclease recognition sites (SalI and XhoI, respectively) added for sequence cloning. Sequences downstream of these added recognition sites derive from GenBank accession no. U08933. The primers used to generate “D” were 5′-CCGCTCGAGCTCAGTCTGACCCATCCACA-3′ and 5′-ACGCGTCGACAGCCACAACAGCCTGAGT-3′, with XhoI and SalI sites added, respectively (underlined sequences), and the remaining sequences were derived from GenBank accession no. U08933.
B1–8αΔhs4
To construct a BAC with loxP-flanked hs4, hs4 was first removed from B1-8α to generate BAC B1-8αΔhs4 (Fig. 2). The two arms of homology used for this deletion were HS4U and HS4D. HS4U is a 768-bp fragment that lies directly downstream of hs3b (bp 25041–25809 in GenBank accession no.AF450245). This sequence was chosen instead of one closer to hs4 because it has not been possible to clone or amplify the entire ~3-kb sequence lying between hs3b and hs4, probably due to the highly repetitive sequences within this region. Primers generating HS4U included sequences (underlined) derived from GenBank accession no. AF450245: 5′-ATGCAAGCTTAATGGATGTGAGATGAGG-3′ (HS3bF) and 5′-GATCCTAGTCTAGAACTCATTGTGTAGACC-3′ (HS4UR2). A 3-kb fragment downstream of hs4 (starting at bp 28,364 in GenBank accession no.AF450245) was amplified with the following primer pairs: 5′-CCTACCCACCTAACTCCAAGC-3′ (HS4DF) and 5′-GGTAGGAAGGGCGAATTCGC-3′ (HS4DR). A smaller ~1.8-kb fragment mapping from the 5′ end of this 3-kb fragment, was used as HS4D (nt 28382–30156 from GenBank accession no. AF450245). Homologous recombination, using the HS4U and HS4D arms resulted in a deletion extending from nt 25810 to nt 28381, GenBank accession no. AF450245 (actual deletion is ~500 bp greater than this 2571 bp because of missing nucleotides in the GenBank sequence).
Blxhs4
The BAC Blxhs4 was generated by the insertion of loxP-flanked hs4 and a DNA “tag” into the site of the hs4 deletion in B1–8αΔhs4 (Fig. 2). The insert included a 1381-bp SmaI/HindIII fragment encompassing HS4 (isolated from HS3.4; Ref. 19) surrounded by loxP sites derived from ploxP2neor (18) and a downstream sequence of 420 bp (nt 1–418; Gene Bank Accession no.V00497) derived from the cDNA of a sickle cell anemia form of human β-globin (plasmid pcr2βS; a gift from Dr. K. Drlica, Public Health Research Institute, International Center for Public Health, Newark, NJ). At high stringency, the latter β-globin sequence does not hybridize to murine genomic DNA. The final size of the insert was ~1.8 kb. It was inserted into B1-8αΔhs4 with HS4U and HS4D arms of homology similar to those used to generate B1-8αΔhs4 (in Blxhs4 the HS4U ends at nt 25766 and the HS4D begins at nt 28406; GenBank accession no.AF450245).
BEµ
A 1-kb XbaI fragment containing Eµ (15) was added to B1-8α (Fig. 6) using VH-derived sequences and D as homology “arms” for homologous recombination in bacteria.
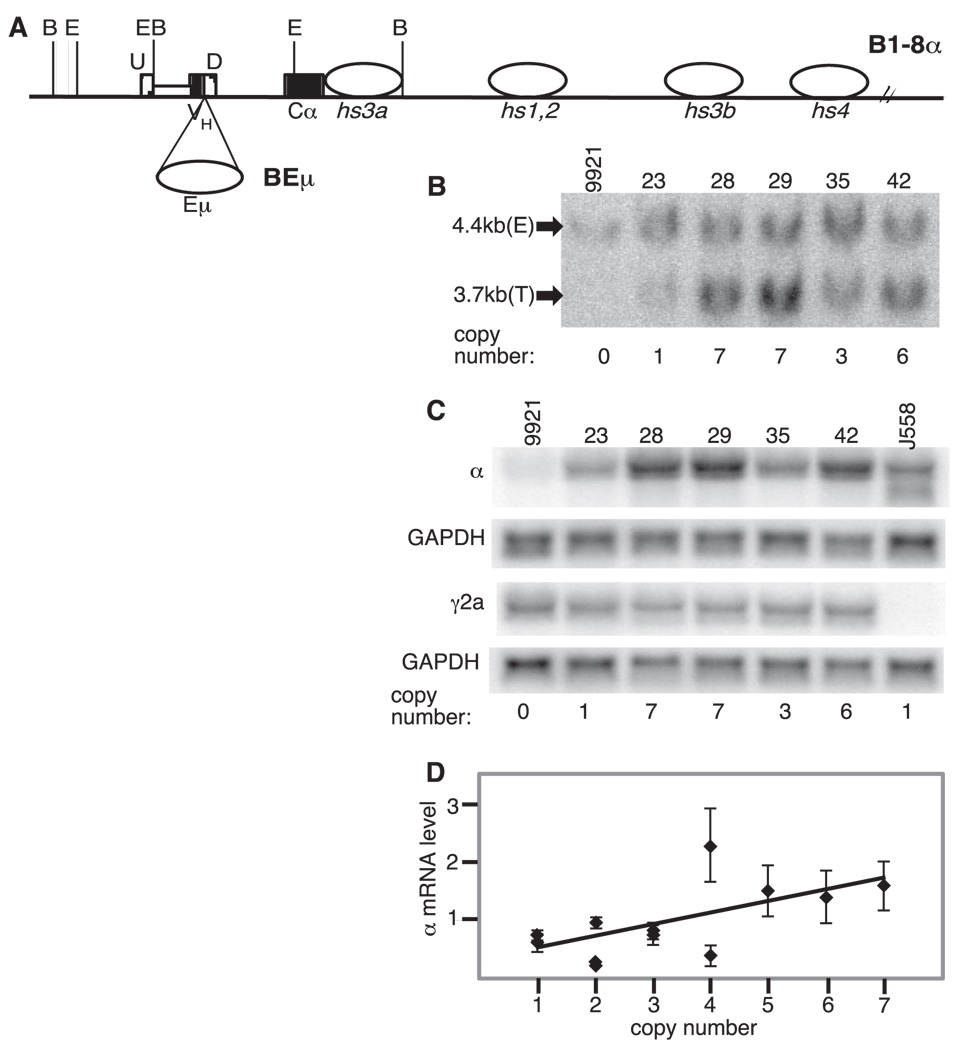
FIGURE 6.
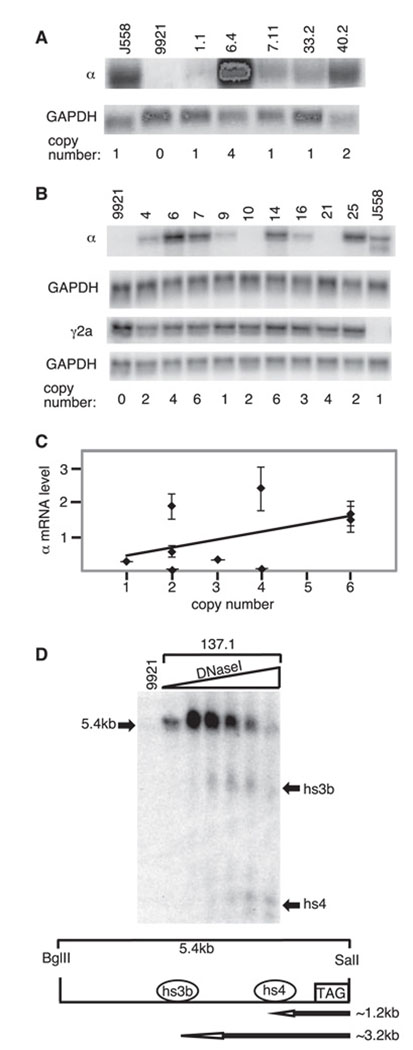
Igα BAC with both Eµ and the 3′RR. A, Diagram of B1-8α and the site of insertion of the intronic enhancer Eµ to generate BEµ. Symbols are as described for Fig. 2. B, Representative genomic Southern blotting was used to determine BAC copy numbers. DNA from the parental line (9921) is included as well as that from five BEµ transformants (23, 28, etc.). Estimates of BAC copy numbers are provided below the lanes. C, Northern blots measuring Igα mRNA levels in BEµ transformants. Representative blots are shown for α, γ2a, and GAPDH mRNA in five BEµ transformants. D, Plot of Igα mRNA levels (with SD) vs transgene copy number for 12 BEµ transformants. Igα mRNA levels were normalized to GAPDH and the level for each clone was divided by that for J558 on the same blot (to normalize among experiments). Igα mRNA levels were determined in at least three independent experiments. The best-fit line of linear regression analysis is shown (p = 0.0002, R2 = 0.34).
Introducing BACs into cell lines
BACs were digested with NotI to purify the insert from the 6.7-kb pBeloBAC11 vector using gel filtration (CL-4B Sepharose beads; Amersham Biosciences, catalog no. 17-0150-01). Ten micrograms of BAC and 400 ng of psk-2loxPneor (18) were mixed with a 1-ml suspension of 107 9921 cells. The neor plasmid was included as a means for selecting cells that had incorporated DNA. The neor gene in this plasmid is linked to the phosphoglycerate kinase-1 promoter and upstream activating element, making it active in eukaryotic cells. Transfection was by electroporation (in a 0.4-cm cuvette at 960 microfarad and 250 V in a Bio-Rad Gene Pulser electroporator with capacitance extender). The cells were then diluted in nonselective medium and plated in 96-well tissue culture plates. Fortyeight hours following transfection the medium was supplemented with G418 (final concentration 1.5 mg/ml; Invitrogen Life Technologies, catalog no. 11811-031). Colonies were visible ~2 wk after transfection and arose in ~30 to ~40% of the wells on each culture plate, indicating that most growing wells contain the progeny of a single transformant. All lines analyzed were further subcloned to assure purity. In prior studies we have shown that presence of psk-2loxPneor neither positively nor negatively influences the expression of a cotransfected IgH reporter gene (18).
After the clones were established, genomic DNA was subjected to both PCR and Southern blot analyses to determine whether the integrated BAC remained intact. To confirm that the 5′ end of the BACs was present, genomic DNA was amplified with a T7 primer (remaining with the BAC insert after NotI digestion) and a primer within Cε: 5′-GTAATACGACT CACTATAGGG-3′ (T7) and 5′-GGACGACATGACTTAACCAG-3′ (CεR; GenBank accession no. X01857.1). The 3′ end of the BAC was detected with primers for Sp6 (again, a vector sequence that remains with the insert after NotI digestion) and a primer mapping 80 kb downstream of Cα: 5′-GCTATTTAGGTGACACTATAG-3′ (Sp6) and 5′-GCATGTGC ATTCAACAGTGG-3′ (Cα80F; GenBank accession no. A450245).
Inducing deletion of hs4 with CRE recombinase
pEGFP-CRE (8.8 kb) was created by ligating a CRE-containing fragment from pBS185 (Invitrogen Life Technologies, catalog no. 10347-011) into the HindIII site of pEGFP-C1 (Clontech Laboratories, catalog no. 6084-1). The resulting plasmid produces both a red-shifted variant of the green fluorescence protein that can be detected by flow cytometry and the CRE recombinase that mediates loxP site-specific recombination.
Blxhs4-containing cell lines were individually transfected with pEGFP-CRE (10 µg plasmid per 107 cells; electroporation was performed as described above) and fluorescent cells were detected and cloned by flow cytometry 48 h later. Cells from individual wells were screened by PCR for hs4 deletion. Primers flanked the site of deletion, lying within HS4U and β-globin, respectively (see Fig. 2, HS4U and TAG): 5′-CTAGCCAGGC AGTGATAGC-3′ (HS4UF) and 5′-TGAGGTTGCTAGTGAACACAG-3′ (β-globinR).
After loxP recombination, the PCR product of these primers is 700 bp. In several cases the 700-bp product was gel purified and sequenced to confirm that the deletion resulted from CRE-mediated loxP recombination. Because the cotransfected neor gene in psk-21oxPneor was also flanked by loxP sites, it was possible for the neor gene to also be deleted in enhanced GFP (EGFP)-CRE-transfected cells. Clones were retested for G418r after transfection, but all of those examined (including those that had undergone hs4 deletion) remained G418r, indicating that at least one copy of neor remained in the cells. As noted above, we have previously demonstrated that this neor gene and its control sequences cannot supplant the 3′RR function in a cotransfected IgH reporter gene (18).
Southern blot analyses
Restriction enzyme-digested DNAs were loaded into individual lanes of 0.7% agarose gels (0.5~1 µg of BAC DNA; ~25 µg of cell line genomic DNA). DNA was then transferred to nylon membranes (Micron Separations, catalog no. N00HY00010) or (Schleicher & Schuell Bioscience, catalog no. 10415296). Blots were hybridized by traditional methods (as previously described; for example, see Ref. 18) or with Quickhyb (Stratagene, catalog no. 201220). Probes were labeled by the random primer method (Megaprime; Amersham Biosciences, catalog no. RPN 1605).
To estimate BAC copy number in cell lines, DNA was digested with HindIII and hybridized with the U probe described above (also see Fig. 3). There are approximately four copies of this region of the IgH locus in 9921 cells (one from the functional IgH locus expressing γ2a and three replicates of the homologous but c-myc translocated Igh allele). The U probe detects a 4.4-kb fragment from these endogenous loci (four copies) and a 3.7-kb fragment from the BAC. Hybridization intensity was quantified by a PhosphorImager and ImageQuant Software (GE Healthcare). The BAC copy number was estimated from two to three Southern analyses per clone. The SD of data obtained for individual clones did not exceed 0.5; the copy number was rounded to the nearest whole number.
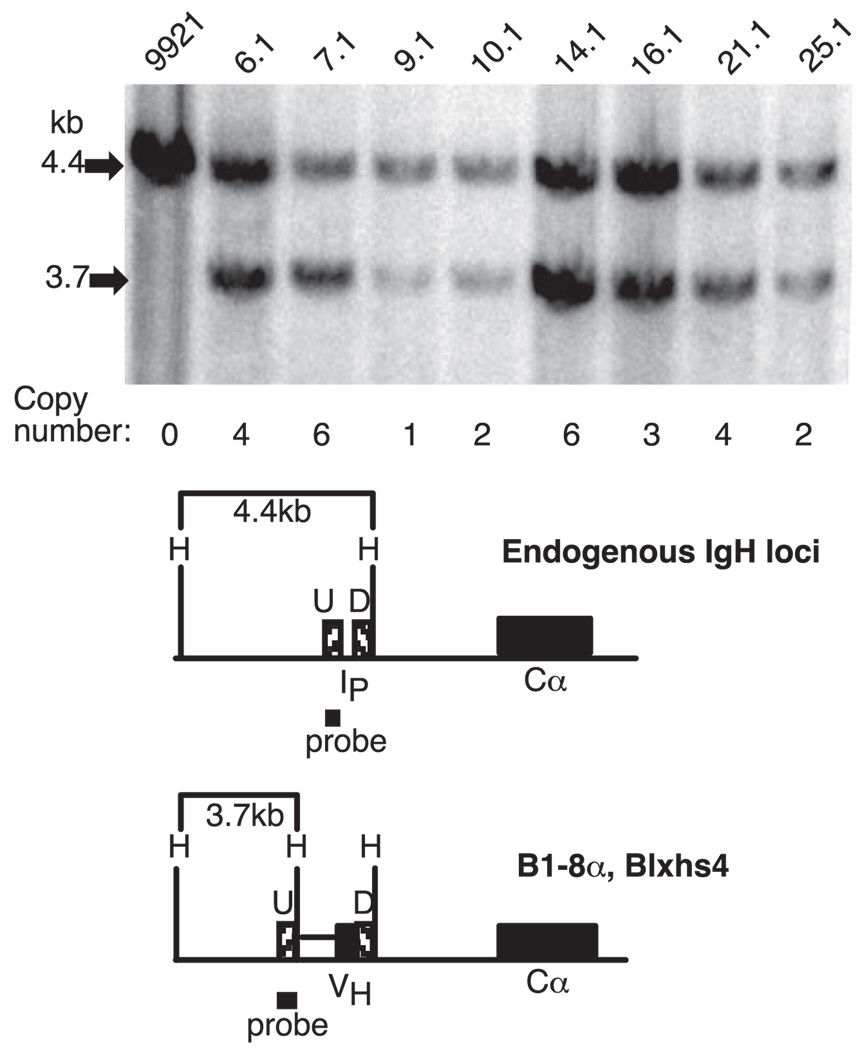
FIGURE 3.
BAC-containing transformants of the IgG2a-secreting cell line 9921. Top, Autoradiograph of DNA from the parental line (9921) and individual transformants (e.g., 6.1, 7.1, etc.) cut with HindIII and hybridized with the probe shown in the diagram below. The estimated number of copies of the BAC in each transformant is shown below the lanes (with the 4.4-kb fragment assigned a value of four copies; see Materials and Methods). Bottom, Diagram of the region upstream of Cα in the endogenous loci of 9921 and in the BACs introduced into this cell line. H (HindIII) recognition sites are shown in the relevant regions.
Northern blot analyses
Total cellular RNA was isolated by a TRIzol reagent (Invitrogen Life Technologies, catalog no. 15596-026) according to the manufacturer’s instructions. Total cellular RNA (~20 µg) was denatured with formamide and size fractionated by electrophoresis through 1% formaldehyde-agarose gels. RNA was then transferred to a GeneScreen nylon membrane (PerkinElmer, catalog no. 881623-1200). Blots were hybridized as previously described (for example, see Ref. 18). The probe used to detect Igα mRNA was a 310-bp Cα fragment (cloned in pBSCα) produced with the following primer pair: 5′-GAATGAGCTCTTGTCCCTGAC-3′ (CaCh3F) and 5′-CATGATCACAGACACGCTGAC-3′ (CαCh3R; GenBank accession no.D11468). The probe for the detection of Igγ2a mRNA was a 312-bp SacI fragment covering the CH3 region of γ2a (from pCH3γ2a) (28). GAPDH was detected with murine GAPDH cDNA (Ambion, catalog no. 7330).
To quantify Igα transgene expression in the transformants, at least three separate experiments were done (three separate sets of Northern blots) for each transformant. In each experiment, two blots were prepared. One was hybridized sequentially with the γ2a and GAPDH probes; the other was hybridized sequentially with the Cα and GAPDH probes. GAPDH was used to normalize the data in each case. In some cases, γ2α and α mRNA levels were compared in individual clones, taking advantage of the fact that the probes for these mRNAs were of approximately the same size and GC content (estimated melting temperatures 77.5 and 80.5°C, respectively) and taking into consideration any differences in specific activity of the labeled probes in each experiment.
Statistical analyses of Northern blot data
Data for nine Blxhs4 transformants and 12 BEµ transformants were subjected to linear regression analyses. BAC copy number data for each transformant were obtained from two or three genomic Southern blots, and Igα, Igγ2a, and GAPDH mRNA levels were measured for each transformant in three independent experiments (Northern blots). For the scatter plots provided in Fig. 4 and Fig. 6, the expression of Igα in each clone is given relative to that of J558 on the same blot (Igα normalized to GAPDH in every case). Because the raw data exhibited a degree of nonnormality that would have brought decreased accuracy in the significance tests, a square root transformation was used to bring the data close to normality before calculating the p values. The p values are a standard statistical test of the null hypothesis that the slope of the regression line is equal to 0. At values below 0.05, there is a >95% probability that the slope is >0 and, therefore, that copy number is a good linear predictor of the expression level. The coefficient of determination (R2) for each data set was also calculated; the closer this value comes to 1, the better the correlation (e.g., in these experiments R2 is the fraction of the variability in the expression level in Igα that is accounted for by its least squares linear regression on the transgene copy number).
FIGURE 4.
Igα expression in BAC transformants. A and B, Northern blots of RNA from an Igα producing plasmacytoma (J558), the γ2a-producing parental line (9921), and the 9921 BAC transformants (1.1, 6.4, etc) (A). Duplicate gels were transferred to nylon and sequentially hybridized with probes for Cα and GAPDH or for Cγ2a and GAPDH, respectively. Representative blots are shown (experiment repeated three times). Estimated BAC copy number shown below each lane (the single-copy J558 endogenous gene is represented as 1). A, 9921 transformants carrying B1-8α. B, 9921 transformants carrying Blxhs4. (Note that specific activities of the GAPDH and Igα probes were not held constant from experiment to experiment, so the relative signals vary among experiments; e.g., compare A and B. This has no adverse effect on normalizing samples to GAPDH within an individual experiment.) C, Plot of Igα mRNA levels vs transgene copy number for nine Blxhs4 transformants. Igα mRNA levels were normalized to GAPDH and the level for each clone was divided by that for J558 in each experiment (to normalize among experiments). The SD for each clone (from three determinations) is shown. The best-fit line of linear regression is shown (linear regression by least squares criterion (Data Desk); p = 0.009, R2 = 0.18). D, DNase I hypersensitivity sites 3b and 4 in the Blxhs4 transgene. Southern blot of BglII/SalI-digested DNA hybridized with the β-globin “tag” as the probe and treated with increasing levels of DNase I. Representative data from one of the Blxhs4 transformants (137.1) is shown. Below the autoradiograph is a diagram of the region analyzed. The approximate sizes of the DNase I fragments expected for hs3b and hs4 are indicated (~3.2 and ~1.2 kb, respectively).
DNase I hypersensitivity assays
The DNase I hypersensitivity assay was performed as described previously (22). Briefly, cell nuclei were isolated from cell lines (9921,137.1, 7.1, and 6.1) by Dounce homogenization and treated with increasing amounts of DNase I (2 × 107 cells per sample; Worthington Biochemical) for 5 min at 37°C. DNA was then isolated and ~20 µg/sample was digested overnight with BglII and SalI (New England Biolabs). Southern blots of the digested DNA were as described above (genomic Southern blots). The probe used was from plasmid pcr2βS (cDNA for human β-globin; TAG in Fig. 2).
Polymerase chain reactions
PCRs were conducted with either HotStart Taq polymerase (Qiagen, catalog no. 203203) (for detection) or PfuTurbo (Stratagene, catalog no. 201220) (for cloning and sequencing) and usually continued for 30 cycles in GeneAmp PCR system 9600 (PerkinElmer).
Results
Development of a BAC with an IgH gene and a 3′ regulatory region
In previous studies, we showed that an IgH reporter gene with linked 3′ IgH enhancers (hs3a, hs1,2, hs3b, and hs4) was efficiently expressed in both a surface Ig+ cell line and in an Ig-secreting cell line but that its expression was not strictly correlated with the copy number and was generally lower than that of an endogenous Igh locus (18). A similar construct tested by others in transgenic mice gave comparable results (29). In an effort to more precisely mimic the endogenous locus, we constructed a functional Igα gene within BAC 141e18, a BAC that extends from 13 kb upstream of Cα (within the second exon of Cε) to a site >40 kb downstream of hs4 (Fig. 2). The reasoning was that this large expanse of DNA might both supply all of the regulatory sequences required for normal IgH gene expression and insulate the reporter gene from the effects of the neighboring chromatin. Although previous reporter constructs contained all of the known 3′ IgH regulatory sequences, the BAC carried these sequences in their natural context. For example, inverted repeats surround hs1,2, and the secondary structure possibly formed by these repeats could play a role in regulating the chromatin structure and gene activity (30, 31). Furthermore, additional control elements not yet identified might lie among or downstream of the known ones. Interestingly, in collaboration with others we have recently identified a region with insulator activity downstream of hs4, and this region was not included in previous minilocus constructs but is present in BAC 141e18 (22).
We generated a BAC in which an assembled V region gene with an associated IgH promoter was inserted upstream of Cα (Bl-8α; see Fig. 2). Modification of this and all other derivative BACs was achieved through homologous recombination in bacteria (Materials and Methods and Ref. 26). Briefly, we used the PCR to amplify regions upstream (U) and downstream (D) of the site of integration. These regions were ligated to the assembled VH gene and introduced into a shuttle vector that was, in turn, transfected into BAC-containing bacteria where recombination took place. The shuttle vector was designed so that integration of the VH gene also led, simultaneously, to deletion of the intronic promoter responsible for Cα sterile transcripts so that transcription of the resulting Igα gene would be entirely dependent upon the natural VH promoter.
As shown in the maps of Fig. 2 before BAC recombination with the shuttle vector, probe U detects a 17-kb BamHI fragment that, after recombination, becomes 4 kb in size. This change was confirmed in B1-8α (Fig. 2A). Because the bacterial RecA enzyme is transiently expressed in this system and the BAC is a very large DNA molecule with several repeated DNA sequences, it was possible that changes other than those we intended would take place during homologous recombination in the bacteria. To look for evidence of this, we digested the original BAC (141e18) and the BAC with inserted VH (Bl-8α) with the restriction enzyme EcoRI and compared the resulting pattern of fragments detected by ethidium bromide staining. There were no changes in restriction pattern apart from those predicted: a 10-kb band in 141e18 was absent in B1-8α, replaced by 8.4- and 3.5-kb bands (Fig. 2B).
BAC Igα expression in an Ig-secreting cell line
Once confirmed as having the desired structure, the murine sequences of B1-8α were released from the bacterial vector and introduced into the plasmacytoma cell line 9921 by electroporation (Materials and Methods). 9921 is a plasmacytoma that secretes IgG2a. The single, functional Igγ2a gene lacks Eµ (as the result of an aberrant CSR event) and so its expression is entirely dependent upon 3′ IgH regulatory sequences (10, 15), making it a particularly appropriate cell line for studying 3′RR function. This cell line also carries three copies of a translocated chromosome that juxtaposes the oncogene c-myc with Cγ2a and all Igh locus sequences downstream (including the 3′RR) (25).
Genomic DNAs from 9921 and from B1-8α transformants were cut with HindIII and hybridized with a probe that corresponded to the region just upstream of the VH gene insertion in B1-8α (probe U in Fig. 3). This probe detects a 4.4-kb HindIII fragment that is generated from both the functional Igh locus and the Igh/c-myc translocated chromosomes in 9921 and detects a 3.7-kb fragment from the BAC transgene. A representative blot is shown for the parental line and BAC-containing transformants (Fig. 3). The intensity of the 4.4- and 3.7-kb bands was used to estimate transgene copy number, with the 4.4-kb band set at four copies as in the 9921 parental line (two or three independent experiments for each transformant; see Materials and Methods). Each transformant was also examined by PCR to confirm that both ends of the BAC were intact (Materials and Methods, data not shown).
Transgene expression in the B1-8α transformant lines was determined by Northern blot (steady-state mRNA analyses). Igα mRNA levels (transgene) were measured by using a probe for gapdh mRNA to normalize total RNA levels in each sample (Fig. 4A). Although a clone with four copies of the BAC transgene produced substantially more Igα mRNA than others with one or two copies (compare clone 6.4 with others), not all single-copy clones produced the same amount of Igα mRNA (e.g., Igα mRNA was barely detectable in clone 1.1; see Fig. 4A). This suggested to us that the presence of all of the known 3′ IgH enhancers in their natural configuration and at a natural distance from an IgH promoter could not fully shield the Igα transgene from integration site effects.
Development of a BAC with loxP sites flanking the 3′ IgH enhancer hs4
Because the per copy expression level differed somewhat among independent B1-8α transformants, we elected to generate several independent BAC transformants and then test the effect of hs4 deletion from the BACs stably integrated in the genome of the recipient cells. In this way, each transformant would provide information about BAC gene activity at a particular site of integration, both before and after hs4 deletion.
To achieve this goal, we first deleted hs4 from B1-8α and then replaced it with a loxP-flanked version of hs4. A 3-kb region encompassing hs4 was deleted by homologous recombination in bacteria to generate B1-8αΔhs4 (Fig. 2). A 1.4-kb region encompassing hs4 was flanked by loxP sites, and a 420-bp portion of human β-globin cDNA was appended downstream to serve as a DNA “tag” for subsequent DNase I hypersensitivity assays (see below). Again, through homologous recombination in bacteria this ~1.8-kb segment of DNA was inserted into B1-8αΔhs4, reconstituting the hs4 element but with loxP sites on either side (Fig. 2). The resulting BAC was designated Blxhs4. Apart from the addition of loxP sites and the DNA tag, Blxhs4 differs from B1-8a by virtue of a deletion (~1.6 kb) between hs3b and hs4 (Materials and Methods). The BACs were cut with appropriate restriction enzymes to confirm that the expected modifications (and no other changes) had taken place (Fig. 2 and data not shown). For example, BamHI digestion was used to distinguish the three BACs. Using a DNA probe that corresponded to the 3′ homology region for BAC modification (hs4D), B1-8α generated a 2.0-kb BamHI fragment while B1-8αΔhs4 generated a fragment of 1.8 kb (Fig. 2C). Similarly, B1-8αΔhs4 and Blxhs4 could be distinguished by 0.5- and 1.9-kb BamHI fragments, respectively, detected by the 5′ homology region (hs4U; see Fig. 2D).
Blxhs4 was transfected into 9921 cells and individual clones were isolated, subcloned, and tested for intact BAC ends. RNAs from transformants carrying the intact BAC were analyzed for Igα, γ2a, and GAPDH mRNA expression. In each of three experiments, duplicate blots were hybridized with probes for α and γ2a mRNA, respectively, and then both blots were “erased” of the initial probe and rehybridized with a probe for GAPDH mRNA. Representative results are shown in Fig. 4B. As was seen with the B1-8α transformants (Fig. 4A), the Blxhs4 transformants expressed Igα mRNA at varying levels with a general trend upward with increasing BAC copy number (Fig. 4, B and C). The modifications surrounding hs4 (e.g., the addition of loxP sites) led to no obvious change in expression of the associated Igα transcription unit. As plotted in Fig. 4C, Igα mRNA levels in nine independent Blxhs4 clones generally correlated with copy number (p = 0.009; R2 = 0.18); but, as in the B1-8α transformants, there were differences in expression level among clones with the same copy number (e.g., compare clones 4, 10, and 25, all of which carried two copies of the transgene).
hs3b and hs4 were discovered (and named) by virtue of the fact that they constituted DNase I hypersensitive regions in the IgH locus. We examined the Blxhs4 transformants to determine whether both regions showed the characteristic DNase I hypersensitivity within the context of this BAC. DNase I hypersensitivity assays were conducted using the “tag” (human β-globin sequences) introduced downstream of hs4 as a DNA probe (see Fig. 2E) because the DNA fragments it detects are limited to those within the BAC transgene. As shown in Fig. 4D, DNase I hypersensitivity sites corresponding to hs3b and hs4 were present in the Blxhs4 transformants. The chromatin structure downstream of the Igα gene in Blxhs4, therefore, mimics that of the endogenous locus, and hs4 in this BAC, as in the endogenous locus, is hypersensitive to DNase I.
hs4 deletion does not eliminate Igα expression
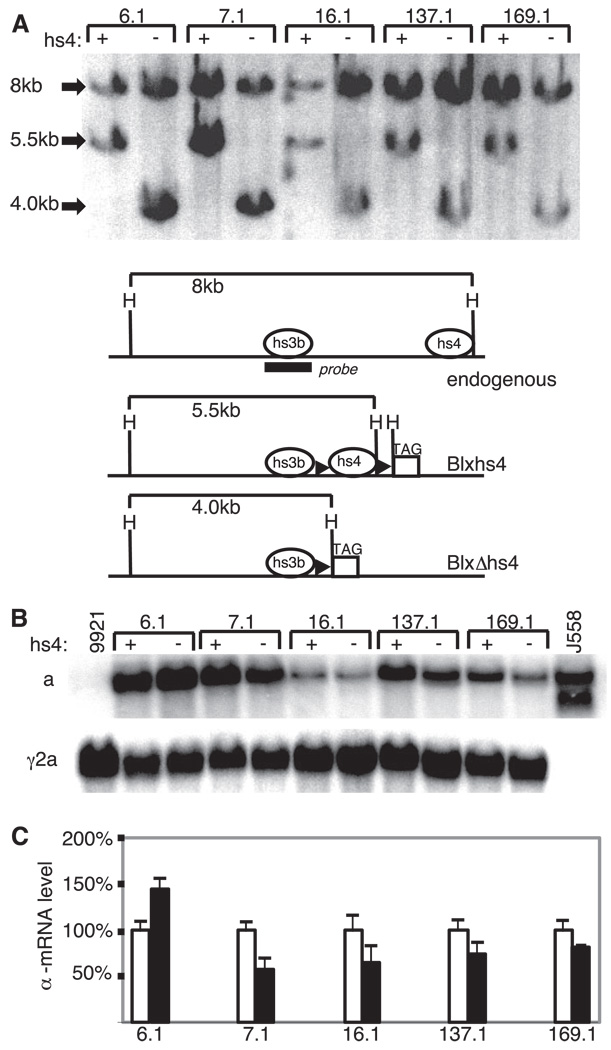
Five independent Blxhs4 transformants (clones 6.1, 7.1, 16.1, 137.1, and 169.1) were then transfected with a plasmid that encodes both EGFP and the bacterial CRE enzyme (required for loxP-mediated DNA deletion). These five clones each carried from one to six copies of Blxhs4 (clone 6.1, three copies; clone 7.1, six copies; clone 16.1, three copies; clone 137.1, one copy; and clone 169.1, one copy). Forty-eight hours after transfection, cells expressing EGFP were identified by flow cytometry and cloned. The selection of EGFP-expressing cells was done to enrich for cells that had taken up the EGFP/CRE plasmid and, therefore, might have undergone loxP-mediated deletion of hs4 in the integrated Blxhs4 BAC. Growing clones were screened for hs4 deletion by PCR (Materials and Methods; data not shown), and the deletion was further confirmed by genomic Southern blot analyses. As shown in Fig. 5A, a probe homologous to hs3b detects a 5.5-kb HindIII fragment in Blxhs4, but this fragment is reduced to 4-kb after hs4 deletion. The same probe detects an 8-kb HindIII fragment from the endogenous IgH loci. The expected change in the HindIII restriction pattern upon hs4 deletion was detected in genomic DNA isolated from subclones of each of the five original Blxhs4 transformants (Fig. 5A). To confirm that these were, indeed, loxP-mediated deletions, the deletion site was isolated by PCR, cloned, and sequenced (data not shown).
FIGURE 5.
In situ deletion of hs4 in Blxhs4 transformants. A, Genomic Southern blotting of transformants carrying the Blxhs4 transgene with (+) and without (−) hs4. The BAC copy numbers for these clones are three (clone 6.1), six (clone 7.1), three (16.1), one (137.1), and one (169.1). The probe (hs3b) is indicated in the maps in A. This probe also detects a smaller HindIII fragment surrounding hs3a, but the blot was cut before hybridization to remove this fragment. Diagrams of the endogenous IgH locus and the Blxhs4 transgene before and after loxP-mediated deletion of hs4 are shown. Relevant HindIII (H) sites are shown; other symbols are as described for Fig. 2. Quantification of the blots suggests that hs4 deletion was accompanied by loss of one BAC copy in clones 7.1 and 16.1. B, Northern blot of RNA from five independent Blxhs4 transformants (6.1, 7.1, etc.) before and after hs4 deletion. The probes used were specific for α and γ2a mRNA. The parental cell line 9921 (expressing Igγ2a only) and the α-expressing J558 cell line are included as controls. C, Bar graph of data obtained from three independent experiments (Northern blots) measuring α-mRNA expression before (□) and after (■) hs4 deletion. The data are plotted for each transformant relative to initial expression level (before hs4 deletion) of 100%.
To determine the effect of the hs4 deletion on Igα expression, mRNA levels were compared before and after the hs4 deletion. As shown in Fig. 5C, Igα mRNA levels decreased in four of the transformants and increased in one. Three independent experiments for each transformant pair (before and after the hs4 deletion) were performed (see representative blot in Fig. 5B) and the results were quantified (Fig. 5C). Although there is some variation in effect at independent integration sites (most notably the increase seen with hs4 deletion in clone 6.1), the common result is only a small change in Igα mRNA levels. Clones 7.1 and 16.1 each lost a single copy of the transgene as well as all copies of hs4 (copy number was reduced from six to five in clone 7.1 and from three to two in clone 16.1), perhaps explaining some of the reduction in Igα levels in these two lines. We conclude that although hs4 likely contributes to the expression of the Igα gene present in the BAC, its loss is largely compensated by other elements within this locus.
Addition of Eµ does not dramatically improve the correlation between BAC copy number and expression level
In reporter genes transiently or stably introduced into Ig-secreting cells, the 3′RR can drive IgH promoter activity in the absence of Eµ and vice versa (for example, Refs. 15, 18, and 20). To eliminate this functional redundancy and to therefore be able to examine the functional roles of subelements of the 3′RR more directly, we have used transgenes that lack Eµ (present study and Ref. (18). In the present study, we found that an IgH gene in BACs containing >90 kb of DNA from the IgH locus (including all known 3′ regulatory sequences) was efficiently expressed but not entirely shielded from integration site effects (Fig. 4). We also found in our analyses of the Blxhs4 transformants carrying one or two copies of the Igα transgene that the transgene was generally expressed at a fraction of the level of the endogenous Igγ2a gene in the same cells and at a fraction of the level of Igα produced by J558 (representative data in Fig. 4, B and C). Although we circumvented the problem of integration site effects by examining gene expression before and after hs4 deletion at each of several different integration sites, we were intrigued by the implication that this large expanse of DNA directly derived from the Igh locus could not reconstitute, in full, all of the attributes of the endogenous locus.
To study this issue further, we considered the possibility that Eµ serves a unique locus-activating function that is no longer required by the already active Igh loci in Ig-secreting cells but that might be required by an Igh locus newly introduced into them (e.g., the BAC transgene). To explore this possibility, a BAC was constructed in which Eµ was placed between the VH and Cα coding sequences in B1-8α (Fig. 6). This new BAC, BEµ, was confirmed in structure by genomic Southern and ethidium bromide staining of BamHI- and EcoRI-digested BAC DNA (data not shown).
9921 cells were transfected with BEµ (isolated insert of this BAC). The transgene copy number in the resulting clones was determined as described earlier (representative data in Fig. 6B), Igα mRNA levels were assessed by Northern blotting and quantitative data were obtained from the blots (triplicate experiments on 12 independent transformants; representative data in Fig. 6C). As shown in Fig. 6D, there is a linear relationship between the transgene copy number and the transgene expression level (p = 0.0002). The slope of the line correlating the copy number and the expression level in these BEµ transformants was not significantly different from that obtained for the Blxhs4 transformants (BAC lacking Eµ) (p = 0.001; compare plots, Fig. 4 and Fig. 6), but the regression correlation coefficient (coefficient of determination) is better in the BEµ transformants (R2 = 0.34 vs 0.18). This could mean that Eµ3 or an interaction between Eµ and the 3′RR, somehow further shields the BAC from integration site effects, but this is not certain because the correlation in the two types of transformants is very similar when one of the “outliers” among the Blxhs4 transformants (four-copy transformant with unusually low expression) is eliminated from the analyses (R2 = 0.31).
Discussion
The discovery of an enhancer (hs1,2) downstream of the rodent IgH constant region genes was the result of a search for regulatory sequences that could explain IgH gene expression in Eµ-deficient, Ig-secreting cells (32–34). Chromosome translocations that juxtaposed the oncogene c-myc with the 3′ end of the Igh locus, resulting in a promoter shift and deregulated c-myc expression, also compelled this search (35, 36). Indeed, since the discovery of this first 3′ IgH enhancer, there has accrued ample evidence that it, along with its neighbors (3′RR), play a role in both situations. In a cell line that already lacks Eµ, enhancer-replacement within the 3′RR by a selectable marker gene completely arrested IgH gene transcription, supporting the hypothesis that IgH transcription in an Eµ-deficient locus is 3′RR dependent (10). In other studies, c-myc was juxtaposed with the 3′RR both in transgenes and by gene-replacement within the natural IgH and c-myc loci (19, 36, 37). In all of these cases, the result was deregulated expression of c-myc, supporting the notion that the 3′RR regulates not only the IgH promoters of assembled IgH genes but can also regulate the c-myc promoter(s) when c-myc insinuates itself into the Igh locus by chromosome translocation.
Studies of mice in which either individual or pairs of 3′ IgH enhancers were removed from the endogenous Igh locus by homologous recombination have revealed a role for the 3′RR in regulating the activity of the cryptic promoters upstream of each of the IgH constant region genes. These promoters are responsible for the so-called “sterile transcripts” that immediately precede CSR. In one study, the removal of both hs3b and hs4 resulted in a moderate (2-fold) decrease in surface IgM expression by resting B cells but in an even more pronounced decrease in the expression of many of the other Ig H chain isotypes (11). The most dramatic effects were seen for IgG2b and IgG3, and the decrease in cells switching to these isotypes in appropriately stimulated splenic cell cultures was correlated with a decrease in sterile transcripts from the respective constant region genes. Class switching to γ1 was unaffected by the hs3b/hs4 double deletion. A more recent study, however, suggests that switching even to this class is 3′RR dependent (38).
The goal of the present study has been to more precisely define the role of hs4 in regulating the promoter of an assembled IgH gene. In previous work we showed that the deletion of both hs3b and hs4 arrested IgH reporter gene expression in an Ig-secreting cell line (9921), but the deletion of both hs3a and hs1,2 did not (18). In the present study we have developed an IgH reporter gene within a BAC so that it more closely resembles the natural Igh locus and have tested its activity in the same Ig-secreting cell line of our previous studies (9921). We found that the DNA regions encompassing hs3b and hs4 do indeed become DNase I hypersensitive in the transfected cell line but that hs4 deletion has only a moderate effect on reporter gene expression. Although four of five cell lines examined showed some reduction in Igα expression after hs4 deletion (Fig. 5), the decreases seen (20–43% reduction) did not approach those seen previously upon hs3b/hs4 deletion (92–100% reduction, ref. 18). In clones 7.1 and 16.1, deletion of hs4 was accompanied by the deletion of one copy of the BAC. This loss might explain some of the reduction in Igα expression seen in these particular Δhs4 clones, but because expression is not strictly copy number dependent it is not possible to ascribe a precise value to each BAC copy in these transformants. In any case, the effect of an hs4 deletion on IgH transcription is small, suggesting that other elements within the BAC are sufficient to sustain IgH transcription in the absence of hs4. This finding does not support the generally held notion that hs4 serves a unique and critical role within the Igh locus, at least with regard to IgH promoter activity.
Candidates for driving IgH gene transcription in the absence of both Eµ and hs4 are the other 3′RR enhancers (hs1,2, hs3a, and hs3b), the newly discovered DNase I hypersensitive sites downstream of hs4, and, perhaps, other (as yet unidentified) control elements that may lie farther downstream. As discussed earlier, numerous studies, both in vivo (endogenous Igh locus) and in vitro (Igh transgenes) implicate the 3′RR as critical to both IgH gene transcription and to CSR (10, 11, 16–18, 38, 39). Deletion of any one of the individual 3′RR elements, however, has yet to yield a phenotype (present study and Refs. 11 and 23). We suggest that this finding reflects extensive functional redundancy among these elements and/or among these and other elements in the locus. The BAC of the present studies extends ~80 kb downstream of Cα (~45 kb downstream of the recently described hs5–7). Additional deletion studies with this and related BACs will aid in more precisely identifying the elements contributing to IgH transcription and unraveling their functional interactions.
It should be noted that in the present experiments, as in those undertaken earlier with a minilocus (18), a loxP-flanked neor gene was cotransfected with the IgH transgenes (Materials and Methods). Importantly, we have shown that this neor gene and its associated phosphoglycerate kinase promoter is not able to drive the expression of a cotransfected IgH gene; none of 25 independent transformants carrying both this neor gene and an IgH gene lacking the 3′RR were able to express the IgH transgene (18). This argues against there being any compensatory effect of the neor gene upon hs4 deletion in the BACs of the present studies. Certainly, when a neor gene is inserted directly into the 3′RR it can affect locus activity (10), but the present studies do not involve insertion of the neor gene into the BAC. Rather, in this study the neor gene remains on a plasmid that has no homology to the BAC and, at best, will integrate at the 5′ or 3′ end of the BAC. This means that it never intervenes between the IgH promoter and the 3′RR and, should it lie adjacent to the BAC, it would lie >40 kb upstream of the 3′RR (if it integrates 5′ of the BAC) or >40 kb 3′ of the 3′RR (if it integrates 3′ of the BAC). In studies by others, the insertion of a pgk-neor cassette 2 kb downstream of hs4 in vivo (in mice) had no effect on either IgH transcription or CSR (40).
It remains to be seen whether hs4 serves a unique and critical role in CSR. It is already apparent that the cryptic or intronic promoters upstream of each of the CH genes are differentially dependent on subregions of the 3′RR (i.e., Iγ1 is not dependent on hs3b/hs4 whereas the other intronic promoters, to different degrees, are). But most of these intronic promoters responded to hs3b/hs4 deletion within the endogenous Igh locus in the same manner that a natural IgH promoter responded to hs3b/hs4 deletion in a transgene (significant reduction in activity) (11, 18). It is possible, therefore, that, as with the natural IgH promoter, these intronic promoters will remain active in the absence of hs4.
The present study involved the manipulation of a >90-kb BAC spanning part of Cε and extending downstream >40 kb beyond hs4. It was surprising to us that this large expanse of DNA was not sufficient to achieve wild-type levels of IgH gene expression. Rather, cells carrying a single copy of the BAC often expressed significantly less IgH mRNA than was produced at the single endogenous IgH locus. Adding Eµ to the BAC led to a slightly higher average level of expression in single-copy clones (Fig. 6D); but, again, the overall effect was minimal.
In a recent transgenic mouse study involving a BAC that, like this one, includes all of the 3′RR but extends much farther upstream (11 kb 5′ of the JH region), the authors describe copy number-dependent expression in the independent mouse lines, but the data do not significantly differ from those presented here (41). There was generally more expression when more copies of the BAC were present, but there was considerable variation in expression among lines with the same copy number, and mice with only one copy of the BAC expressed it at considerably lower levels than the endogenous locus (41). In these mouse studies Eµ, as well as the 3′RR, was present.
There are at least two alternate explanations for these findings. Either we have yet to identify and isolate all of the elements required to establish a wild-type Igh locus, or transgenes (integrations into random sites within the genome) face special problems not ordinarily encountered by the endogenous locus (where neighboring genes/DNA is unchanging) and problems for which the locus itself can provide no solution. If the first of these explanations is correct and the BAC is missing essential elements, perhaps these elements lie within the VH genes. Interestingly, an ~1-Mb region covering the distal VH genes is required for Igh locus association with the nuclear periphery (42). Transient loss of this association appears essential to V to DJ rearrangements involving the distal VH genes, but the Igh locus reforms its attachment to the nuclear lamina in B cells and Ig-secreting cells (43–46). The effect of this association on Ig transcription is not yet clear. Although an attractive possibility is that this association is required for proper programming of the Igh locus for transcription (an association not afforded by any of the BACs or transgenes used to date), there are examples of Ig-secreting cells whose Igh loci lack the 1-Mb attachment region and thus are found predominantly away from the nuclear periphery but yet continue to express the Igh locus at high levels (42).
If the second of these possibilities is correct, cloned Igh locus DNA, no matter how extensive, will never ensure integration site-independent expression. Consistent with this hypothesis is the finding that even the prototype for locus control regions, the β-like globin locus control region, is affected by the site of integration and does not ensure expression levels equal to those of the endogenous locus (47–49). This has been made even more apparent by studies in which the same gene is repeatedly integrated into several, distinct chromosomal sites by CRE recombinase-mediated cassette exchange (50, 51).
Acknowledgments
We thank Dr. Siegfried Janz (National Cancer Institute, National Institutes of Health, Bethesda, MD) for providing the BAC 141e18, Dr. Klaus Rajewsky (Harvard Medical School, Boston, MA) for providing the B1-8 VH gene (in plasmid pIVHB1-8L2neor), and Dr. Karl Drlica (Public Health Research Institute, International Center for Public Health, Newark, NJ) for providing the human β-globinS cDNA plasmid pcr2βS. We also thank the laboratory of Dr. Nathaniel Heinz (Rockefeller University, New York, NY) both for protocols and for guidance in carrying out BAC modifications by homologous recombination. Dr. William Williams (Hunter College of the City University of New York, New York, NY) was invaluable as a consultant for the statistical analyses of transgene expression and copy number. Steven Williams provided expert technical assistance. We thank Dr. Barbara K. Birshtein (Albert Einstein College of Medicine, Bronx, NY), Dr. Michael W. Young (Rockefeller University), and Dr. Benjamin Ortiz (Hunter College of the City University of New York) for critical review of the manuscript.
Footnotes
This work was supported by National Institutes of Health Grant AI30653 (to L.A.E.). The infrastructure and instrumentation at Hunter College are supported in part by a Research Centers in Minority Institutions award from the National Institutes of Health (RR-0307).
Abbreviations used in this paper: CSR, class switch recombination; BAC, bacterial artificial chromosome; CRE, cyclization recombination enzyme; EGFP, enhanced GFP; 3′RR, 3′ regulatory region of the IgH locus.
Disclosures
The authors have no financial conflict of interest.
References
- 1.Oettinger MA, Schatz DG, Gorka C, Baltimore D. RAG-1 and RAG-2, adjacent genes that synergistically activate V(D)J recombination. Science. 1990;248:1517–1523. doi: 10.1126/science.2360047. [DOI] [PubMed] [Google Scholar]
- 2.Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y, Honjo T. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–563. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- 3.Banerji J, Olson L, Schaffner W. A lymphocyte-specific cellular enhancer is located downstream of the joining region in immunoglobulin heavychain genes. Cell. 1983;33:729–740. doi: 10.1016/0092-8674(83)90015-6. [DOI] [PubMed] [Google Scholar]
- 4.Gillies SD, Morrison SL, Oi VT, Tonegawa S. A tissue-specific transcriptional enhancer element is located in the major intron of a rearranged immunoglobulin heavy-chain gene. Cell. 1983;33:717–728. doi: 10.1016/0092-8674(83)90014-4. [DOI] [PubMed] [Google Scholar]
- 5.Neuberger MS. Expression and regulation of immunoglobulin heavy chain genes transfected into lymphoid cells. EMBO J. 1983;2:1373–1378. doi: 10.1002/j.1460-2075.1983.tb01594.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen J, Young F, Bottaro A, Stewart V, Smith RK, Alt FW. Mutations of the intronic IgH enhancer and its flanking sequences differentially affect accessibility of the JH locus. EMBO J. 1993;12:4635–4645. doi: 10.1002/j.1460-2075.1993.tb06152.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Serwe M, Sablitzky F. V(D)J recombination in B cells is impaired but not blocked by targeted deletion of the immunoglobulin heavy chain intron enhancer. EMBO J. 1993;12:2321–2327. doi: 10.1002/j.1460-2075.1993.tb05886.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Perlot T, Alt FW, Bassing CH, Suh H, Pinaud E. Elucidation of IgH intronic enhancer functions via germ-line deletion. Proc. Natl. Acad. Sci. USA. 2005;102:14362–14367. doi: 10.1073/pnas.0507090102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Afshar R, Pierce S, Bolland D, Corcoran A, Oltz EM. Regulation of IgH gene assembly: role of the intronic enhancer and 5′DQ52 region in targeting DHJH recombination. J. Immunol. 2006;176:2439–2447. doi: 10.4049/jimmunol.176.4.2439. [DOI] [PubMed] [Google Scholar]
- 10.Lieberson R, Ong J, Shi X, Eckhardt LA. Transcription of an immunoglobulin gene ceases upon deletion of a distant enhancer. EMBO J. 1995;14:6229–6238. doi: 10.1002/j.1460-2075.1995.tb00313.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pinaud E, Khamlichi A, Le Morvan C, Drouet M, Nalesso V, Le Bert M, Cogne M. Localization of the 3′ IgH locus elements that effect longdistance regulation of class switch recombination. Immunity. 2001;15:187–199. doi: 10.1016/s1074-7613(01)00181-9. [DOI] [PubMed] [Google Scholar]
- 12.Klein S, Sablitsky F, Radbruch A. Deletion of the IgH enhancer does not reduce immunoglobulin heavy chain production of a hybridoma IgD class switch variant. EMBO J. 1984;3:2473–2476. doi: 10.1002/j.1460-2075.1984.tb02158.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wabl MR, Burrows PD. Expression of immunoglobulin heavy chain at a high level in the absence of a proposed immunoglobulin enhancer element in cis. Proc. Natl. Acad. Sci. USA. 1984;81:2452–2455. doi: 10.1073/pnas.81.8.2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aguilera RJ, Hope TJ, Sakano H. Characterization of immunoglobulin enhancer deletions in murine plasmacytomas. EMBO J. 1985;4:3689–3693. doi: 10.1002/j.1460-2075.1985.tb04136.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zaller DM, Eckhardt LA. Deletion of a B-cell-specific enhancer affects transfected, but not endogenous, immunoglobulin heavy-chain gene expression. Proc. Nat. Acad. Sci. USA. 1985;82:5088–5092. doi: 10.1073/pnas.82.15.5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gregor PD, Morrison SL. Myeloma mutant with a novel 3′ flanking region: loss of normal sequence and insertion of repetitive elements leads to decreased transcription but normal processing of the α heavy-chain gene products. Mol. Cell. Biol. 1986;6:1903–1916. doi: 10.1128/mcb.6.6.1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Michaelson JS, Giannini SL, Birshtein BK. Identification of 3′ α-HS4, a novel Ig heavy chain enhancer element regulated at multiple stages of B cell differentiation. Nucleic Acids Res. 1995;23:975–981. doi: 10.1093/nar/23.6.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shi X, Eckhardt LA. Deletional analyses reveal an essential role for the hs3b/hs4 IgH 3′enhancer pair in an Ig-secreting but not an earlier-stage B cell line. Int. Immunol. 2001;13:1003–1012. doi: 10.1093/intimm/13.8.1003. [DOI] [PubMed] [Google Scholar]
- 19.Madisen L, Groudine M. Identification of a locus control region in the immunoglobulin heavy-chain locus that deregulates c-myc expression in plasmacytoma and Burkitt’s lymphoma cells. Genes Dev. 1994;8:2212–2226. doi: 10.1101/gad.8.18.2212. [DOI] [PubMed] [Google Scholar]
- 20.Ong J, Stevens S, Roeder RG, Eckhardt LA. 3′ IgH enhancer elements shift synergistic interactions during B cell development. J. Immunol. 1998;160:4896–4903. [PubMed] [Google Scholar]
- 21.Giannini SL, Singh M, Calvo CF, Ding G, Birshtein BK. DNA regions flanking the mouse Ig 3′α enhancer are differentially methylated and DNase I hypersensitive during B cell differentiation. J. Immunol. 1993;150:1772–1780. [PubMed] [Google Scholar]
- 22.Garrett FE, Emelyanov AV, Sepulveda MA, Flanagan P, Volpi S, Li F, Loukinov D, Eckhardt LA, Lobanenkov VV, Birshtein BK. Chromatin architecture near a potential 3′ end of the Igh locus involves modular regulation of histone modifications during B-cell development and in vivo occupancy at CTCF sites. Mol. Cell. Biol. 2005;25:1511–1525. doi: 10.1128/MCB.25.4.1511-1525.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Manis JP, van der Stoep N, Tian M, Ferrini R, Davidson L, Bottaro A, Alt FW. Class switching in B cells lacking 3′ immunoglobulin heavy chain enhancers. J. Exp. Med. 1998;188:1421–1431. doi: 10.1084/jem.188.8.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eckhardt LA, Birshtein BK. Independent immunoglobulin class-switch events occurring in a single myeloma cell line. Mol. Cell. Biol. 1985;5:856–868. doi: 10.1128/mcb.5.4.856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stanton LW, Yang JQ, Eckhardt LA, Harris LJ, Birshtein BK, Marcu KB. Products of a reciprocal chromosome translocation involving the c-myc gene in a murine plasmacytoma. Proc. Natl. Acad. Sci. USA. 1984;81:829–833. doi: 10.1073/pnas.81.3.829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang X, Model P, Heintz N. Homologous recombination based modification in Esherichia coli and germline transmission in transgenic mice of a bacterial artificial chromosome. Nat. Biotechnol. 1997;15:859–865. doi: 10.1038/nbt0997-859. [DOI] [PubMed] [Google Scholar]
- 27.Sonoda E, Pewzner-Jung Y, Schwers S, Taki S, Jung S, Eilat D, Rajewsky K. B cell development under the condition of allelic inclusion. Immunity. 1997;6:225–233. doi: 10.1016/s1074-7613(00)80325-8. [DOI] [PubMed] [Google Scholar]
- 28.Tilley SA, Birshtein BK. Unequal sister chromatid exchange: a mechanism affecting Ig gene arrangement and expression. J. Exp. Med. 1985;162:675–694. doi: 10.1084/jem.162.2.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chauveau C, Jansson EA, Muller S, Cogne M, Pettersson S. Cutting edge: Ig heavy chain 3′ HS1–4 directs correct spatial position-independent expression of a linked transgene to B lineage cells. J. Immunol. 1999;163:4637–4641. [PubMed] [Google Scholar]
- 30.Chauveau C, Cogne M. Palindromic structure of the IgH 3′ locus control region. Nat. Genet. 1996;14:15–16. doi: 10.1038/ng0996-15. [DOI] [PubMed] [Google Scholar]
- 31.Saleque S, Singh M, Little RD, Giannini SL, Michaelson JS, Birshtein BK. Dyad symmetry within the mouse 3′ IgH regulatory region includes two virtually identical enhancers (Cα3′E and hs3) J. Immunol. 1997;158:4780–4787. [PubMed] [Google Scholar]
- 32.Pettersson S, Cook GP, Bruggemann M, Williams GT, Neuberger MS. A second B cell-specific enhancer 3′ of the immunoglobulin heavy-chain locus. Nature. 1990;344:165–168. doi: 10.1038/344165a0. [DOI] [PubMed] [Google Scholar]
- 33.Dariavach P, Williams GT, Campbell K, Pettersson S, Neuberger MS. The mouse IgH 3′-enhancer. Eur. J. Immunol. 1991;21:1499–1504. doi: 10.1002/eji.1830210625. [DOI] [PubMed] [Google Scholar]
- 34.Lieberson R, Giannini SL, Birshtein BK, Eckhardt LA. An enhancer at the 3′end of the mouse immunoglobulin heavy chain locus. Nucleic Acids Res. 1991;19:933–937. doi: 10.1093/nar/19.4.933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cory S. Activation of cellular oncogenes in hemopoietic cells by chromosome translocation. Adv. Cancer Res. 1986;47:189–234. doi: 10.1016/s0065-230x(08)60200-6. [DOI] [PubMed] [Google Scholar]
- 36.Park SS, Kim JS, Tessarollo L, Owens JD, Peng L, Han SS, Tae Chung S, Torrey TA, Cheung WC, Polakiewicz RD, et al. Insertion of c-Myc into Igh induces B-cell and plasma-cell neoplasms in mice. Cancer Res. 2005;65:1306–1315. doi: 10.1158/0008-5472.CAN-04-0268. [DOI] [PubMed] [Google Scholar]
- 37.Wang J, Boxer L. Regulatory elements in the immunoglobulin heavy chain gene 3′-enhancers induce c-myc deregulation and lymphomagenesis in murine B cells. J. Biol. Chem. 2005;280:12766–12773. doi: 10.1074/jbc.M412446200. [DOI] [PubMed] [Google Scholar]
- 38.Dunnick WA, Shi J, Graves KA, Collins JT. The 3′ end of the heavy chain constant region locus enhances germline transcription and switch recombination of the four γ genes. J. Exp. Med. 2005;201:1459–1466. doi: 10.1084/jem.20041988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cogne M, Lansford R, Bottaro A, Zheng J, Gorman J, Young KF, Cheng HL, Alt FW. A class switch control region at the 3′ end of the immunoglobulin heavy chain locus. Cell. 1994;77:737–747. doi: 10.1016/0092-8674(94)90057-4. [DOI] [PubMed] [Google Scholar]
- 40.Manis JP, Michaelson JS, Birshtein BK, Alt FW. Elucidation of a downstream boundary of the 3′ IgH regulatory region. Mol. Immunol. 2003;39:753–760. doi: 10.1016/s0161-5890(02)00256-0. [DOI] [PubMed] [Google Scholar]
- 41.Dunnick WA, Shi J, Graves KA, Collins JT. Germline transcription and switch recombination of a transgene containing the entire H chain constant region locus: effect of a mutation in a STAT6 binding site in the γ 1 promoter. J. Immunol. 2004;173:5531–5539. doi: 10.4049/jimmunol.173.9.5531. [DOI] [PubMed] [Google Scholar]
- 42.Yang Q, Riblet R, Schildkraut CL. Sites that direct nuclear compartmentalization are near the 5′ end of the mouse immunoglobulin heavy-chain locus. Mol. Cell. Biol. 2005;25:6021–6030. doi: 10.1128/MCB.25.14.6021-6030.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chowdhury D, Sen R. Stepwise activation of the immunoglobulin µ heavy chain gene locus. EMBO J. 2001;20:6394–6403. doi: 10.1093/emboj/20.22.6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kosak ST, Skok JA, Medina KL, Riblet R, Le Beau MM, Fisher AG, Singh H. Subnuclear compartmentalization of immunoglobulin loci during lymphocyte development. Science. 2002;296:158–162. doi: 10.1126/science.1068768. [DOI] [PubMed] [Google Scholar]
- 45.Maes J, O’Neill LP, Cavelier P, Turner BM, Rougeon F, Goodhardt M. Chromatin remodeling at the Ig loci prior to VDJ recombination. J. Immunol. 2001;167:866–874. doi: 10.4049/jimmunol.167.2.866. [DOI] [PubMed] [Google Scholar]
- 46.Zhou J, Ermakova OV, Riblet R, Birshtein BK, Schildkraut CL. Replication and subnuclear location dynamics of the immunoglobulin heavy-chain locus in B-lineage cells. Mol. Cell. Biol. 2002;22:4876–4889. doi: 10.1128/MCB.22.13.4876-4889.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Porcu S, Kitamura M, Witkowska E, Zhang Z, Mutero A, Lin C, Chang J, Gaensler KM. The human β globin locus introduced by YAC transfer exhibits a specific and reproducible pattern of developmental regulation in transgenic mice. Blood. 1997;90:4602–4609. [PubMed] [Google Scholar]
- 48.Kaufman RM, Pham CT, Ley TJ. Transgenic analysis of a 100-kb human β-globin cluster-containing DNA fragment propagated as a bacterial artificial chromosome. Blood. 1999;94:3178–3184. [PubMed] [Google Scholar]
- 49.Alami R, Greally JM, Tanimoto K, Hwang S, Feng YQ, Engel JD, Fiering S, Bouhassira EE. Beta-globin YAC transgenes exhibit uniform expression levels but position effect variegation in mice. Hum. Mol. Genet. 2000;9:631–636. doi: 10.1093/hmg/9.4.631. [DOI] [PubMed] [Google Scholar]
- 50.Feng YQ, Seibler J, Alami R, Eisen A, Westerman KA, Leboulch P, Fiering S, Bouhassira EE. Site-specific chromosomal integration in mammalian cells: highly efficient CRE recombinase-mediated cassette exchange. J. Mol. Biol. 1999;292:779–785. doi: 10.1006/jmbi.1999.3113. [DOI] [PubMed] [Google Scholar]
- 51.Feng YQ, Warin R, Li T, Olivier E, Besse A, Lobell A, Fu H, Lin CM, Aladjem MI, Bouhassira EE. The human β-globin locus control region can silence as well as activate gene expression. Mol. Cell. Biol. 2005;25:3864–3874. doi: 10.1128/MCB.25.10.3864-3874.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]