Abstract
Background
In recent years, numerous studies have assessed the prevalence of germline mutations in BRCA1 and BRCA2 genes in various cohorts. We here extensively investigated the prevalence and geographical distribution of BRCA1-2 mutations in the entire genetically-homogeneous Sardinian population. The occurrence of phenotypic characteristics which may be predictive for the presence of BRCA1-2 germline mutations was also evaluated.
Methods
Three hundred and forty-eight breast cancer patients presenting a familial recurrence of invasive breast or ovarian carcinoma with at least two affected family members were screened for BRCA1-2 mutations by DHPLC analysis and DNA sequencing. Association of BRCA1 and BRCA2 mutational status with clinical and pathological parameters was evaluated by Pearson's Chi-Squared test.
Results and Conclusion
Overall, 8 BRCA1 and 5 BRCA2 deleterious mutations were detected in 35/348 (10%) families; majority (23/35;66%) of mutations was found in BRCA2 gene. The geographical distribution of BRCA1-2 mutations was related to three specific large areas of Sardinia, reflecting its ancient history: a) the Northern area, linguistically different from the rest of the island (where a BRCA2 c.8764_8765delAG mutation with founder effect was predominant); b) the Middle area, land of the ancient Sardinian population (where BRCA2 mutations are still more common than BRCA1 mutations); and c) the South-Western area, with many Phoenician and Carthaginian locations (where BRCA1 mutations are prevalent). We also found that phenotypic features such as high tumor grading and lack of expression of estrogen/progesterone receptors together with age at diagnosis and presence of ovarian cancer in the family may be predictive for the presence of BRCA1-2 germline mutations.
Background
Breast cancer is the most common malignancy in women in Western countries, currently accounting for one third of all female cancers [1]. In Sardinia, breast cancer represents the principal death-causing malignancy, with an incidence similar to that observed in western countries [2]. Familial aggregation is thought to account for 5–10% of all breast cancer cases, and germline mutations in different genes involved in pathways critical to maintain the genomic integrity have accounted for less than 25% of the inherited breast cancer [3,4]. Part of this familial clustering shows autosomal dominant inheritance with high penetrance due to mutations in the BRCA1 (MIM 113705) and BRCA2 (MIM 600185) breast cancer genes [5,6]. Cumulative breast cancer risks by age 70 are estimated to be 65% for BRCA1 and 45% for BRCA2 mutation carriers. In addition, women with BRCA mutations are at significant risk to develop ovarian cancer and other malignancies [7].
Hundreds of mutations among breast/ovarian cancer families have been found in these two genes. According to the Breast Cancer Information Core (BIC) database [8,9], about two thirds of germline variants identified in BRCA1 and BRCA2 are unique; the remaining ones are recurrent founder mutations which have been described in different ethnic groups and populations. Therefore, the mutation spectrum and prevalence of mutation carriers with breast or ovarian cancer depends on the population studied, and displays considerable variation based on ethnic and geographical diversity. In Italy, 4–27% of the identified mutations recurred among apparently unrelated families, while a regional founder effect has been demonstrated for few mutations [10-13]. In Sardinia, contribution of BRCA1-2 mutations to breast cancer predisposition has been reported for populations from the Northern part of the island [14]. Sardinian population shows strong founder effects for several genetic diseases with some geographical differences within the island [15,16]; indeed, some genetic heterogeneity has been described within the coastal or proximal-to-coast regions, which have suffered repeated invasions over many years, in comparison to the most internal and isolated part of the island [17]. On this regard, the assessment of both spectrum and prevalence of BRCA1 and BRCA2 mutations across the entire island as well as their predictive significance in this population is needed. Nevertheless, assessment of the likelihood of carrying deleterious mutations in BRCA1 or BRCA2 genes among patients who present high-risk cancer evaluation clinics will provide more accurate guidance to women and families about considering genetic testing [7].
Here we report on the prevalence and geographical distribution of BRCA1-2 mutations within the entire Sardinian population. We also investigated the existence of any significant association between mutations in BRCA1-2 genes and phenotypic characteristics of breast cancer patients that could drive specific treatment and influence the process of mutation testing.
Methods
Family selection
A total of 1,085 unselected breast cancer patients was recruited from all clinics at the Universities and Hospitals across the Sardinia island. To avoid any bias, patients were consecutively collected from January 1998 to December 2003 and were enrolled regardless of age at diagnosis, family history status or disease features. Sardinian origin was ascertained in all cases through genealogical studies. Family history of cancer was evaluated through specific questionnaires during the follow-up visits at the participating institutions. Three-hundred-forty-eight (32%) presented a familial recurrence of invasive breast or ovarian carcinoma: 237 originated from North Sardinia, 66 from Middle and 45 from South. Part of the present cohort (112 familial cases with at least three affected members) had been already included in our previous studies [18,19]. The remaining 737 (68%) breast cancer patients were sporadic cases.
Among patients with a family history of breast/ovarian cancer, 131/348 (37.6%) fulfilled the selection criterion for "high risk" families: presence of a breast cancer patient diagnosed before age 50 years and having at least one first- or second-degree relative diagnosed with breast cancer before 50 years of age or ovarian cancer at any age.
Of those, 73 originated from North Sardinia, 21 from the Middle and 37 from the South. The 81% of "high risk" families presented with breast cancer and the 12% with both breast and ovarian cancer. Male breast cancer was present in the 7% of the families (Table 1). The remaining 217 families were categorized as "no high risk" families: presence of a breast cancer patient having at least one first- or second-degree relative who is diagnosed with breast cancer or ovarian cancer at any age.
Table 1.
BRCA1 and BRCA2 mutational analysis
| Characteristics | Cases | BRCA positive Families | ||
|---|---|---|---|---|
| No. | % | No. | % | |
| TOTAL CASES* | 348 | 35 | 10 | |
| "High risk" | 131 | 23 | 17.5 | |
| BC | 106 | 81 | 17 | 16.0 |
| BC/OC | 16 | 12 | 5 | 31.2 |
| MBC | 9 | 7 | 1 | 11.1 |
| Age at diagnosis <40 y | 41 | 31 | 12 | 29.2 |
| "No High risk" | 217 | 12 | 5.5 | |
| BC | 204 | 94 | 11 | 5.4 |
| BC/OC | 8 | 4 | 1 | 12.5 |
| MBC | 5 | 2 | 0 | 0.0 |
| 2 BC/OC in the family | 114 | 4 | 3.5 | |
| ≥ 3 BC/OC in the family | 145 | 17 | 11.7 | |
High risk: presence of a breast cancer patient diagnosed before age 50 years and having at least one first- or second-degree relative who is diagnosed with breast cancer before 50 years of age or ovarian cancer at any age). No high risk: presence of a breast cancer patient having at least one first- or second-degree relative who is diagnosed with breast cancer or ovarian cancer at any age. BC, breast cancer; OC, ovarian cancer; MBC, male breast cancer.
* including 112 families already reported in our previous studies
According to number of breast/ovarian cancer in the families, we had 75 families with 3 or more affected members and 47 families with two affected members among the "high risk" cases; 67 families had 3 or more affected members and 70 families had two affected members among the "no high risk" cases. Information on total number of affected family members were uncertain or not available for 9 and 80 patients in the "high risk" and "no high risk" groups, respectively.
Clinical and pathological characteristics of the breast cancer probands including age at diagnosis, mono- or bilateral tumour location, stage of disease, tumour-node-metastasis system status, tumour grading, estrogen/progesterone receptor status, were confirmed by medical records and pathology reports.
Patients were informed about the aims of the study; blood samples for genetic testing were drawn after obtaining their written consent. The study was reviewed and approved by the ethical review boards of the participating Institutions accounting for samples' collection (University of Sassari; Azienda Sanitaria Locale of Sassari, Nuoro and Cagliari).
Mutation detection
Mutation analysis of coding sequences and intron-exon boundaries of the BRCA1 and BRCA2 genes was carried out in the series of 348 probands from families with hereditary breast cancer. Genomic DNA was obtained from isolated white blood cells by salting out procedure [20]. Mutation detection for all coding regions and splice boundaries of BRCA1 and BRCA2 genes was performed by single strand conformation polymorphism (SSCP) for a small fraction of cases (27 family probands); all remaining cases underwent mutational screening by denaturing high performance liquid chromatography (DHPLC), using the WAVE Nucleic Acid Fragment Analysis System (Transgenomic, Omaha, NE), followed by automated sequencing as previously reported [18,19]. To confirm that each germline variant detected by sequencing was a real mutation and not a polymorphism, 103 unrelated normal individuals (corresponding to 206 chromosomes), originating from the same geographical area and with no family history for breast cancer, were used as controls and screened for each gene variant identified. For sequence homology searches, National Center for Biotechnology Information Blast database http://www.ncbi.nlm.nih.gov/BLAST/ has been used [21]. All identified variants were verified for their existence and nomenclature into the BIC database http://research.nhgri.nih.gov/bic/ and the Human Gene Mutation Database http://www.hgmd.cf.ac.uk/ac/[22-24].
Statistical Analysis
The Pearson's Chi-Squared test was used for assessing the existence of any correlation between occurrence of deleterious germline mutations in BRCA1, BRCA2, or either gene and probands' characteristics. The following variables and categories were defined and included in our analyses: presence of ovarian or male breast cancer in family, synchronous or asynchronous bilateral breast carcinoma, age at diagnosis, histological tumor grade as standardized by Elston and Ellis [25], disease stage (pathological primary tumor size, pathological nodal status, presence of metastases) according to TNM classification which describes the anatomical extended of the disease by Sobin et al [26], estrogen and progesterone receptor status. The exact coefficient for sample proportion analysis was calculated to determine all of the significant parameters (below the 0.05 level). Odds ratios of carrying BRCA1-2 mutations were estimated by the logistic regression model and reported with 95% confidence interval (95% CI). All analyses were performed with the statistical package SPSS/7.5 for Windows.
Results
Family screening for BRCA1 and BRCA2 mutations
Genomic DNA from 348 probands of breast-ovarian cancer families was screened for germline mutations in BRCA1 and BRCA2 genes. Overall, deleterious germline mutations were detected in 35/348 (10%) familial cases: 23/131 (17.5%) among "high-risk" and 12/217 (5.5%) among "no high-risk" families (Table 1). Families with 3 or more BC cases, including the 112 reported in our previous studies, showed a prevalence rate of 11.7% (17/145). Presence of ovarian cancer in family increased the occurrence of BRCA1-2 germline mutations in either the subset of "high-risk" families (5/16; 31%) or the group of "no high-risk" families (1/8; 12.5%). No similar variation of mutation rates was observed in families presenting a male member with breast cancer. Stratification of families according to number of breast-ovarian cancer cases showed that BRCA1/2 positivity was significantly higher in families with 3 or more affected members than those with only two cases, with frequencies of 11.7% and 3.5%, respectively. Among 41/131 "high-risk" probands diagnosed at age ≤40 years, 12 (29%) presented a BRCA1-2 germline mutation.
The overall mutation prevalence was 7% in North Sardinia and 17% and 18% in families from Middle and South Sardinia, respectively. BRCA positivity accounts for 11% (8/73), 38% (8/21) and 19% (7/37) in "high risk" families from these geographical areas (data not shown).
Spectrum of BRCA1-2 mutations in Sardinia
Overall, 8 distinct BRCA1 and 5 distinct BRCA2 pathogenic mutations were found in 35 (10%) out of 348 breast-ovarian cancer families; 11 "high-risk" families were positive for BRCA1 and 12 for BRCA2, whereas 1 BRCA1 and 11 BRCA2 mutations were identified in "no high-risk" families. Therefore, majority (23/35; 66%) of such mutations was found in BRCA2, and 12/35 (34%) were in BRCA1. All mutations were absent in normal genomic DNA from 103 unrelated healthy individuals (corresponding to 206 control chromosomes) and were classified as disease-causing variants due to their predicted effects on proteins. In Table 2, all germline mutations detected in our series have been reported. The BRCA1 c.916_917delTT mutation was the most represented BRCA1 variant (three positive families out of 348 cases; 1%), while the variants c.8764_8765delAG and c.3950_3952delTAGinsAT were the most recurrent mutations in BRCA2 among patients with a positive family history [13/348 (4%) and 7/347 (2%), respectively]. These mutations showed similar frequencies in both the "high risk" and no "high risk" groups (6.8% vs 5%).
Table 2.
BRCA1 and BRCA2 pathogenetic mutationsa
| N° of positive families | GENE | EXON | DNA LEVEL | PROTEIN LEVEL | Mutation type | BIC |
|---|---|---|---|---|---|---|
| 1 | BRCA1 | 5 | c.300 T>G | p.Cys61Gly | missense | YES |
| 3 | BRCA1 | 11 | c.916_917delTT | p.Val266SerfsX19 | frameshift | YES |
| 1 | BRCA1 | 11 | c.1099_1100delCA | p.Thr327fsX1 | frameshift | YES |
| 1 | BRCA1 | 11 | c.1499insA | p.Phe461fsX18 | frameshift | YES |
| 2 | BRCA1 | 11 | c.1632 A>T | p.Lys505X | non sense | NO |
| 1 | BRCA1 | 11 | c.1638 A > T | p.Arg507X | non sense | NO |
| 2 | BRCA1 | 11 | c.3823_3826delACAA | p.Asn1235fsX27 | frameshift | NO |
| 1 | BRCA1 | 14 | c.4575delA | p.Ser1486fsX18 | frameshift | NO |
| 7 | BRCA2 | 11 | c.3950_3952delTAGinsAT | p.Phe1241fsX17 | frameshift | NO |
| 1 | BRCA2 | 11 | c.6586 C>G | p.Ser2120X | non sense | NO |
| 1 | BRCA2 | 11 | c.6023_6024delTA | p.His1932X12 | frameshift | YES |
| 13 | BRCA2 | 20 | c.8764_8765delAG | p.2846X22 | frameshift | YES |
| 1 | BRCA2 | 18 | c.8559+1G>T | - | splicing | YES |
As indicated in Table 2, 4 out of 8 BRCA1 mutations and 2 out of 5 BRCA2 mutations have not been previously reported in either the BIC database. http://research.nhgri.nih.gov/bic/ or in the Human Gene Mutation Database http://www.hgmd.cf.ac.uk/ac/[23,24]. Besides deleterious mutations, complete BRCA1-2 screening identified 32 missense mutations and 16 intronic variations: 21 already described polymorphisms, 14 variants of unknown clinical significance and 13 novel variants not reported in the BIC database (Table 3).
Table 4.
Logistic regression analysis to identify predictors of BRCA mutations
| Characteristics | No. of cases | BRCA+ carriers | OR (95% CI) | P |
|---|---|---|---|---|
| Age of Onset | ||||
| < 45 years | 139 | 21 (15%) | 1.8 (1.05–4.28) | 0.046 |
| > 45 years | 209 | 14 (7%) | ||
| Risk criteria | ||||
| high-risk family | 131 | 23 (18%) | 3.2 (1.51–6.67) | 0.002 |
| no high risk family | 217 | 12 (6%) | ||
| Ovarian cancer in family | ||||
| presence | 23 | 6 (26%) | 3.7 (1.44–9.40) | 0.006 |
| absence | 244 | 27 (11%) | ||
| Bilaterality | ||||
| presence | 36 | 8 (22%) | 2.4 (1.05–5.39) | 0.038 |
| absence | 228 | 25 (11%) | ||
| T | ||||
| T1 | 151 | 12 (8%) | 2.23 (0.57–8.82) | 0.251 |
| T2 | 70 | 10 (14%) | ||
| T3-4 | 26 | 4 (15%) | ||
| N | ||||
| N- | 126 | 15 (12%) | 1.02 (0.39–2.67) | 0.963 |
| N+ | 112 | 13 (12%) | ||
| M | ||||
| M0 | 201 | 20 (10%) | 1.33 (0.15–11.7) | 0.797 |
| M1 | 24 | 3 (12%) | ||
| Tumor Grading | ||||
| G1-2 | 171 | 9 (5%) | 11.3 (3.38–37.6) | < 0.001 |
| G3-4 | 33 | 12 (36%) | ||
| Estrogen Receptor (ER) | ||||
| ER- | 55 | 16 (29%) | 0.18 (0.06–0.53) | 0.002 |
| ER+ | 102 | 10 (10%) | ||
| Progesteron Receptor (PR) | ||||
| PR- | 81 | 21 (26%) | 0.22 (0.07–0.66) | 0.007 |
| PR+ | 67 | 6 (9%) |
Table 3.
Other genetic variants in BRCA1 and BRCA2
| Gene | Exon | Nucleotide | Codon | Intron | Base change | Mutation type | Nomenclature | BIC citation/Clinical importance |
|---|---|---|---|---|---|---|---|---|
| BRCA1 | 561–34 | 7 | T > C | IVS7-34 T>C | YES/no | |||
| 667–57 | 8 | delT | IVS8-57 delT | YES/no | ||||
| 11 | 2196 | 693 | G > A | missense | D693N | YES/no | ||
| 11 | 2731 | 871 | C > T | missense | P871L | YES/no | ||
| 11 | 3232 | 1038 | A > G | missense | E1038G | YES/no | ||
| 11 | 3667 | 1183 | A > G | missense | K1183R | YES/no | ||
| 15 | 4654 | 1512 | G > T | missense | S1512I | YES/no | ||
| 16 | 4956 | 1613 | A > G | missense | S1613G | YES/no | ||
| 16 | A > G | IVS16-68 A>G | YES/no | |||||
| 16 | A > G | IVS16-92 A>G | YES/no | |||||
| 11 | 1186 | 356 | A > G | missense | Q356R | YES/unknown | ||
| 11 | 1822 | 568 | C > T | missense | P568L | YES/unknown | ||
| 11 | 2596 | 826 | C > A | missense | T826K | YES/unknown | ||
| 11 | 4158 | 1347 | A > G | missense | R1347G | YES/unknown | ||
| 11 | 4173 | 1352 | G > A | missense | E1352K | YES/unknown | ||
| 16 | 5075 | 1652 | G > A | missense | M1652I | YES/unknown | ||
| 17 | C > T | IVS17-53 C>T | YES/unknown | |||||
| 18 | G > A | IVS18+65 G>A | YES/unknown | |||||
| 5 | 287 | 56 | A > T | missense | K56N | NO/unknown | ||
| 5 | 288 | 57 | G > A | missense | G57R | NO/unknown | ||
| 17 | G > T | IVS17-53 G>T | NO/unknown | |||||
| 20 | T > C | IVS20+34 T>C | NO/unknown | |||||
| 22 | 5469 | 1784 | G > A | missense | W1784I | NO/unknown | ||
| BRCA2 | 5'UTR | 203 | G > A | missense | 203G>A | YES/no | ||
| 10 | 1379 | 384 | C > T | missense | S384F | YES/no | ||
| 10 | 1093 | 289 | A > C | missense | N289H | YES/no | ||
| 11 | 3199 | 991 | A > G | missense | N991D | YES/no | ||
| 11 | 4469 | 1414 | C > T | missense | T1414M | YES/no | ||
| 11 | 4486 | 1420 | G > T | missense | D1420Y | YES/no | ||
| 16 | T > C | IVS16-14 T>C | YES/no | |||||
| 20 | 8731 | 2835 | T > C | missense | S2835P | YES/no | ||
| 22 | 21 | T > C | IVS21-66 T>C | YES/no | ||||
| 27 | 10204 | 3326 | A > T | nonsense | K3326X | YES/no | ||
| 1 | 353 | 42 | C > T | missense | Y42C | YES/unknown | ||
| 10 | 1409 | 394 | A > C | missense | E394A | YES/unknown | ||
| 11 | 3623 | 1132 | A > G | missense | K1132R | YES/unknown | ||
| 11 | 6328 | 2034 | C > T | missense | R2034C | YES/unknown | ||
| 11 | 6359 | 2044 | G > C | missense | G2044A | YES/unknown | ||
| 25 | 24 | T > C | IVS24-16 T>C | YES/unknown | ||||
| 10 | 1343 | 372 | A > C | missense | W372N | NO/unknown | ||
| 2 | G > A | IVS2+65 G>A | NO/unknown | |||||
| 4 | A > C | IVS4+67 A>C | NO/unknown | |||||
| 1042 | 272 | A > T | missense | N272I | NO/unknown | |||
| 11 | 5967 | 1910 | A > T | missense | N1910I | NO/unknown | ||
| 14 | C > T | IVS14+53 C>T | NO/unknown | |||||
| 14 | 7863 | 2546 | T > C | missense | S2546P | NO/unknown | ||
| 16 | 7975 | 2583 | G > A | missense | D2583N | NO/unknown | ||
| 16 | T > A | IVS16-32 T>A | NO/unknown | |||||
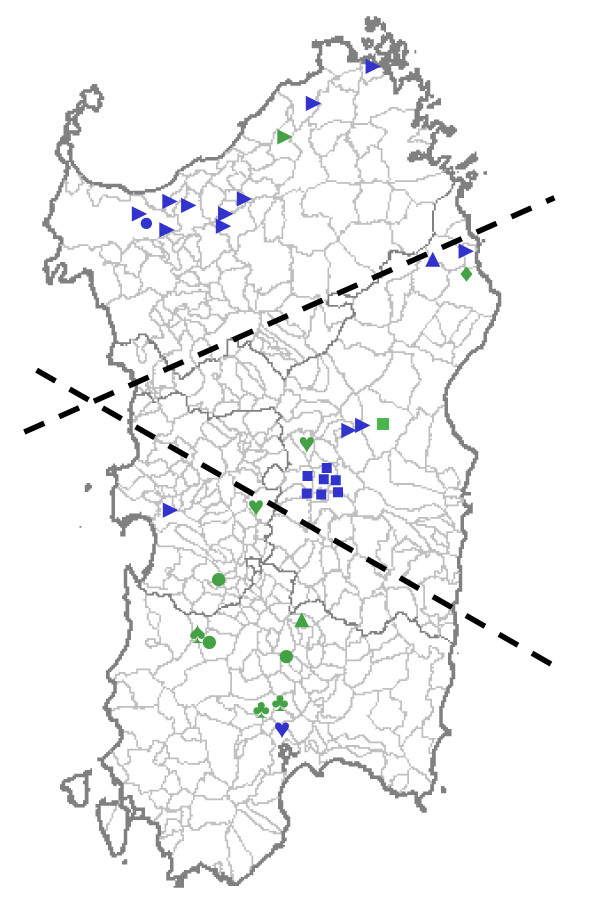
The geographical origin of the families positive for deleterious BRCA1-2 mutations is shown in Figure 1. The BRCA1 variants occurred in families originating from different areas of the island: 7 probands from the South, 2 from the Middle (BRCA1 c.3823_3826delACAA, BRCA1 c.300 T>G) and 1 from the North (BRCA1 c.1632 A>T). Carriers of the BRCA1 c.916_917delTT mutation were identified in three unrelated families from the South area. The BRCA2 c.8764_8765delAG and BRCA2 c.3950_3952delTAGinsAT variants were found in probands from the North and Middle Sardinia, whereas the remaining BRCA2 variants were detected in single families from the Central-East coast and South.
Figure 1.
Geographical distribution of BRCA1-2 mutations carriers in Sardinia. Symbols indicate villages of origin for the patients presenting deleterious germline mutations in either BRCA1 or BRCA2 genes: "green square" BRCA1 c.300 T>G; green circle BRCA1 c.916_917delTT; "green triangle" BRCA1 c.1099_1100delCA; "green arrow" BRCA1c.1499insA; "green heart"BRCA1 c.1632 A>T; "green diamond" BRCA1 c.1638 A>T;"green clubs" BRCA1 c.3823_3826delACAA; "green spades" BRCA1 c.4575delA; "blue square" BRCA2 c.3950_3952delTAGinsAT; "blue triangle" BRCA2 c.6586 C>G; "blue circle" BRCA2 c.6023_6024delTA; "blue arrow" BRCA2 c.8764_8765delAG; "blue heart"BRCA2 c.8559+1G>T. Dashed lines delimit the three geographical regions reported in the text.
Correlation between BRCA1-2 mutations and probands' phenotype characteristics
As shown in Table 4, univariate analysis indicated that presence of a BRCA1-2 mutation correlated significantly with earlier diagnosis age (P = 0.046), classification of the family as "high risk" (P = 0.002), recurrence in the family of ovarian cancer (P = 0.006), and development of a bilateral disease (P = 0.038). Considering the pathological parameters of primary tumors, no statistically significant correlation between BRCA1-2 mutations and primary tumor size (T) or axillary nodal status (N) or distant localization of the disease (M) was observed. However, the BRCA1-2 mutation rates were significantly higher in the subsets of patients with a more undifferentiated primary carcinoma (tumor grading 3–4, P < 0.001) and lack of expression of estrogen receptor (ER-, P = 0.002) or progesterone receptor (PR-, P = 0.007).
Discussion
In this study, we have reported the prevalence of BRCA1-2 germline mutations in patients with a positive family history of breast and/or ovarian cancer from Sardinia, whose population shows genetic peculiarity due to geographical isolation and strong genetic drift [27,28]. Prevalence of BRCA1-2 mutations may indeed vary among distinct populations due to concurrence of different environmental factors and genetic backgrounds; in other words, patients origin may strongly account for different mutation rates in candidate genes.
In the present study, a germline pathogenic mutation in either BRCA1 or BRCA2 was identified in 10% of screened breast-ovarian cancer families (BRCA1 mutations were detected in about 3% of cases, while BRCA2 mutations were identified in about 7% of families). BRCA positivity reached 17.5% when considering the "high risk" families, 29% in families with probands diagnosed at age ≤40 years and 31% in the presence of ovarian cancer in the family. Prevalence rate in "high risk" families was higher than that of families with 3 or more affected member regardless of age at onset (11.7%). These results confirm that clinical characteristics such as ovarian cancer in the family, age at diagnosis and number of cases are good predictors for the likelihood to be a BRCA mutation carrier.
The overall prevalence of BRCA1-2 mutations among breast cancer patients from the entire Sardinia was quite similar to that previously reported for breast-ovarian cancer families originating from the Northern part of the island (for both studies, 15% BRCA-positive carriers were observed in breast-ovarian cancer families [14,18]. However prevalence of BRCA deleterious mutations was higher in Middle Sardinia (38% vs 11% in the North. and 19% in the South). When compared to many other Italian and European studies, the frequency is relatively low [10-13,29-32]. Since the sensitivity of mutation detection methods is not complete, some mutations may have remained undetected in the present study. Large genomic deletions, which do escape detection by both DHPLC and direct sequencing may account for a fraction of mutation-negative breast and ovarian cancer families in Sardinia. Overall and even considering a lack of sensitivity of the screening approach used in the present study, the prevalence of BRCA mutations in Sardinian families remains low. The selection criterion of two close relatives with breast cancer before the age of 50 years, used in this study, may somehow explain this low prevalence rate, though it is not possible to exclude that a specific genetic background may play a role on breast cancer susceptibility among Sardinian population. Noteworthy, a similar low prevalence of BRCA mutations was reported in the Finnish population which has genetic features comparable to the Sardinian one, where the historical, cultural and geographical isolation may have selected specific genetic variants as susceptibility genes for breast cancer [33].
The geographical origin of the families positive for deleterious BRCA1-2 mutations is shown in Figure 1. The BRCA2 c.8764_8765delAG and c.3950_3952delTAGinsAT variants were previously described as founder mutations in North and Middle Sardinia, respectively. In particular, cases carrying the BRCA2 c.8764_8765delAG mutation belonged to unrelated families originating from different villages in the northern part of the island [11,14]; most of families genotyped with markers flanking the BRCA2 gene at 13q12-q13 locus were demonstrated to share a large haplotype, not found in control chromosomes from the same geographical area [11]. Conversely, the BRCA2 c.3950_3952delTAGinsAT, which was previously reported as a founder mutation, was instead running in families belonging to a single extended pedigree confined to a small village of the central part of Sardinia [19]. The BRCA1 variants occurred in families originating from different areas of the island without a defined geographical clustering, though about half of the cases carrying the BRCA1 mutations originated from the South Sardinia. Interestingly, in three unrelated families from the South and Middle Sardinia we identified carriers of the BRCA1 c.916_917delTT mutation that has been reported in breast-ovarian cancer families from South Italy [34]. Altogether, the geographical distribution of the genetic variants in the island, suggests that BRCA1-2 mutations are related to the specific three large areas of Sardinia, which reflects its ancient history: a) the North area, delimited by the mountain chain crossing Sardinia and linguistically different from the rest of the island (where the c.8764_8765delAG variant acts as founder mutation); b) the Middle area, land of the ancient Sardinian population and domain of pastoral culture (where BRCA2 mutations are still more common than BRCA1 mutations); and c) the South-West area, with many Phoenician and Carthaginian archeological sites (where BRCA1 mutations are prevalent).
Majority of BRCA1-2 germline mutations identified in Sardinian families is unique: most of the variants were detected in single families and 6 were novel mutations.
The frequencies of BRCA1 or BRCA2 mutations among breast-ovarian cancer families varies widely between populations world-wide: Icelandic breast cancer families present almost exclusively BRCA2 mutations [30], roughly equal numbers of BRCA1 and BRCA2 mutations have been described in French Canadian or British breast cancer families [35,36], while a clear prevalence of BRCA1 mutations has been observed in the United States [37,38]. In this study BRCA2 mutations were more frequent than BRCA1 mutations, although it should be taken into account that a high proportion of families are carrying the BRCA2 c.8764_8765delAG founder mutation. Mutations in BRCA2 gene have been detected in up to 40% of male breast cancers in Iceland [39]; in our isolated population, we detected a BRCA2 mutation in 1/14 (7%) men with breast cancer with a frequency similar to that reported for male breast cancers in USA (4%) [40]. Again, these findings suggest that other environmental and/or genetic factors are contributing to the susceptibility to male breast cancer in Sardinia.
Specific pathological features have been reported in hereditary breast cancer with differences between BRCA1 and BRCA2 associated tumors [41]. In our series, the analysis of clinico-pathological characteristics showed some differences between BRCA1-2 wild type and mutation carriers. Though no correlation with the stage of disease (the main prognostic factor for breast cancer patients) was observed, the presence of BRCA1-2 mutations were significantly associated with some pathological characteristics (higher tumor grading, lack of expression of estrogen/progesterone receptors) which are recognized to have a negative impact on prognosis. Unfortunately, data from mutation screening of BRCA1 and BRCA2 were combined for the statistical analysis due to the low number of positive cases for each single gene in our series, although distinct differences in both molecular pathology and histopathology have been shown according to the specific carrier status.
Conclusion
In summary, our results on the entire Sardinian population demonstrated that: a) prevalence of BRCA1 and BRCA2 mutations is 10% in familial breast-ovarian cancer; b) the spectrum of mutations among Sardinians is unique; c) some specific phenotypic features may be predictive for the presence of BRCA1-2 germline mutations and should be therefore considered when counselling Sardinian patients about undergoing genetic testing. On this regard, the fulfilment of one or more of the following criteria seems to increase the probability to detect a predisposing BRCA1-2 mutation among Sardinian breast cancer patients: a) high-risk classification of the patient's family; b) presence of at least one family member with ovarian cancer; c) occurrence of synchronous or asynchronous bilateral breast carcinoma.
Our findings further confirm that mutation frequency for BRCA1 and BRCA2 as cancer susceptibility genes for breast-ovarian carcinoma needs to be evaluated in each distinct geographical area. Finally, the absence of any BRCA1 or BRCA2 mutation in majority of "high-risk" families with breast-ovarian cancer support the general hypothesis that additional breast cancer susceptibility genes remain to be identified.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
GP, AL, AU, carried out the molecular genetic studies, participated in the sequence alignment and drafted the manuscript. PF, GP, MP, ML, participated to the DHPLC analysis and resequencing. AG, AC, AC, FT participated in the collection and assembly of clinical data. MB performed the statistical analysis. AF, SO, CF, MCS, GL participated in the provision of patients and revision of the histological records. LC participated in the design and coordination of the study, GP participated in the design and coordination of the study and helped to draft the manuscript, MM contributed to the study conception, obtained financial support and revised the manuscript. All authors read and approved the final manuscript.
Pre-publication history
The pre-publication history for this paper can be accessed here:
Contributor Information
Grazia Palomba, Email: graziap68@yahoo.it.
Angela Loi, Email: loiangela@hotmail.com.
Antonella Uras, Email: labematologia.hsf@aslnuoro.it.
Patrizia Fancello, Email: labematologia.hsf@aslnuoro.it.
Giovanna Piras, Email: labematologia.hsf@aslnuoro.it.
Attilio Gabbas, Email: labematologia.hsf@aslnuoro.it.
Antonio Cossu, Email: cossu@uniss.it.
Mario Budroni, Email: mariobudroni@tiscali.it.
Antonio Contu, Email: oncomedss@tiscali.it.
Francesco Tanda, Email: tandaf@uniss.it.
Antonio Farris, Email: afarris@uniss.it.
Sandra Orrù, Email: sandraorru@asl8cagliari.it.
Carlo Floris, Email: carlofloris2@virgilio.it.
Marina Pisano, Email: marina.pisano@icb.cnr.it.
Mario Lovicu, Email: mlovicu@uniss.it.
Maria Cristina Santona, Email: labematologia.hsf@aslnuoro.it.
Gennaro Landriscina, Email: labematologia.hsf@aslnuoro.it.
Laura Crisponi, Email: laura.crisponi@inn.cnr.it.
Giuseppe Palmieri, Email: lgpalmieri@yahoo.com.
Maria Monne, Email: labematologia.hsf@aslnuoro.it.
Acknowledgements
We are grateful to the patients and their families who partecipated to this study. Work was supported by Italian Ministry of Health "Progetto Ricerca Finalizzata", Sardinia Regional Government (Regione Autonoma Sardegna), Italian Association for Research on Cancer (AIRC) and Fondazione Banco di Sardegna.
References
- Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics 2002. CA Cancer J Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- Budroni M, Cesaraccio R, Pirino D, Sechi O, Oggiano M, Piras D, Sechi A, Cossu A, Palmieri G, Tanda F. In: Cancer Incidence in Five Continents. Curado MP, Edwards B, Shin HR, Storm H, Ferlay J, Heanue M, Boyle P, editor. IX. IARC Scientific Publications, No. 160, Lyon, IARC; 2007. Cancer incidence in Sassari Province (1998–2002) [Google Scholar]
- Easton DF, Pooley KA, Dunning AM, Pharoah PD, Thompson D, Ballinger DG, Struewing JP, Morrison J, Field H, Luben R, Wareham N, Ahmed S, Healey CS, Bowman R. SEARCH collaborators. Meyer KB, Haiman CA, Kolonel LK, Henderson BE, Le Marchand L, Brennan P, Sangrajrang S, Gaborieau V, Odefrey F, Shen CY, Wu PE, Wang HC, Eccles D, Evans DG, Peto J, Fletcher O. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447:1087–1093. doi: 10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stacey SN, Manolescu A, Sulem P, Rafnar T, Gudmundsson J, Gudjonsson SA, Masson G, Jakobsdottir M, Thorlacius S, Helgason A, Aben KK, Strobbe LJ, Albers-Akkers MT, Swinkels DW, Henderson BE, Kolonel LN, Le Marchand L, Millastre E, Andres R, Godino J, Garcia-Prats MD, Polo E, Tres A, Mouy M, Saemundsdottir J, Backman VM, Gudmundsson L, Kristjansson K, Bergthorsson JT, Kostic J, Frigge ML, Geller F, Gudbjartsson D, Sigurdsson H, Jonsdottir T, Hrafnkelsson J, Johannsson J, Sveinsson T, Myrdal G, Grimsson HN, Jonsson T, von Holst S, Werelius B, Margolin S, Lindblom A, Mayordomo JI, Haiman CA, Kiemeney LA, Johannsson OT, Gulcher JR, Thorsteinsdottir U, Kong A, Stefansson K. Common variants on chromosomes 2q35 and 16q12 confer susceptibility to estrogen receptor-positive breast cancer. Nat Genet. 2007;39:865–869. doi: 10.1038/ng2064. [DOI] [PubMed] [Google Scholar]
- Miki Y, Swensen J, Shattuck-Eidens D, Futreal PA, Harshman K, Tavtigian S, Liu Q, Cochran C, Bennet LM, Ding W. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science. 1994;266:66–71. doi: 10.1126/science.7545954. [DOI] [PubMed] [Google Scholar]
- Wooster R, Bignell G, Lancaster J, Swift S, Mangion J, Collins N, Gregory S, Gumbs C, Micklem G. Identification of the breast cancer susceptibility gene BRCA2. Nature. 1995;378:789–792. doi: 10.1038/378789a0. [DOI] [PubMed] [Google Scholar]
- Easton D. Breast cancer genes – what are the real risks? Nat Genet. 1997;16:210–211. doi: 10.1038/ng0797-210. [DOI] [PubMed] [Google Scholar]
- Szabo C, Masiello A, Ryan JF. The BIC Consortium. Brody LC. The Breast Cancer Information Core: Database Design, Structure, and Scope. Hum Mut. 2000;16:123–131. doi: 10.1002/1098-1004(200008)16:2<123::AID-HUMU4>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- The Breast Cancer Information Core. http://research.nhgri.nih.gov/bic/
- Nedelcu R, Liede A, Aubé J, Finch A, Kwan E, Jack E, NaroD SA, Randall S, Hugel L, Clark K. BRCA mutations in Italian breast/ovarian cancer families. Eur J Hum Genet. 2002;10:150–152. doi: 10.1038/sj.ejhg.5200755. [DOI] [PubMed] [Google Scholar]
- Pisano M, Cossu A, Persico I, Palmieri G, Angius A, Casu G, Palomba G, Sarobba MG, Rocca PC, Dedola MF, Olmeo N, Pasca A, Budroni M, Marras V, Pisano A, Farris A, Massarelli G, Pirastu M, Tanda F. Identification of a founder BRCA2 mutation in Sardinia. Br J Cancer. 2000;82:553–9. doi: 10.1054/bjoc.1999.0963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marroni F, Cipollini G, Peissel B, D'Andrea E, Pensabene M, Radice P, Caligo MA, Presciuttini S, Bevilacqua G. Reconstructing the Genealogy of a BRCA1 Founder Mutation by Phylogenetic Analysis. Ann Hum Genet. 2008;72:310–318. doi: 10.1111/j.1469-1809.2007.00420.x. [DOI] [PubMed] [Google Scholar]
- Papi L, Putignano AL, Congregati C, Zanna I, Sera F, Morrone D, Falchetti M, Turco MR, Ottini L, Palli D, Genuardi M. Founder mutations account for the majority of BRCA1-attributable hereditary breast/ovarian cancer cases in a population from Tuscany, Central Italy. Breast Cancer Res Treat. 2008. in press . [DOI] [PubMed]
- Palmieri G, Palomba G, Cossu A, Pisano M, Dedola MF, Sarobba MG, Farris A, Olmeo N, Contu A, Pasca A, Satta MP, Persico I, Carboni AA, Cossu-Rocca P, Contini M, Mangion J, Stratton MR, Tanda F. BRCA1 and BRCA2 germline mutations in Sardinian breast cancer families and their implications for genetic counseling. Ann Oncol. 2002;13:1899–1907. doi: 10.1093/annonc/mdf326. [DOI] [PubMed] [Google Scholar]
- Rosatelli MC, Dozy A, Faà V, Meloni A, Sardu R, Saba L, Kan YW, Cao A. Molecular characterization of beta-thalassemia in the Sardinian population. Am J Hum Genet. 1992;50:422–446. [PMC free article] [PubMed] [Google Scholar]
- Loudianos G, Dessi V, Lovicu M, Angius A, Altuntas B, Giacchino R, Marazzi M, Marcellini M, Sartorelli MR, Sturniolo GC. Mutation analysis in patients of Mediterranean descent with Wilson disease: identification of 19 novel mutations. J Med Genet. 1999;36:833–836. [PMC free article] [PubMed] [Google Scholar]
- Lampis R, Morelli L, Macis MD, Mulargia A, Loddo M, De Virgiliis S, Marrosu MG, Todd JA, Cucca F. The inter-regional distribution of HLA class II haplotypes indicates the suitability of the Sardinian population for case-control association studies in complex diseases. Hum Mol Genet. 2000;9:2959–2965. doi: 10.1093/hmg/9.20.2959. [DOI] [PubMed] [Google Scholar]
- Palomba G, Pisano M, Cossu A, Budroni M, Dedola MF, Farris A, Contu A, Baldinu P, Tanda F, Palmieri G. Spectrum and prevalence of BRCA1 and BRCA2 germline mutations in Sardinian breast cancer patients through a hospital-based screening. Cancer. 2005;104:1172–1179. doi: 10.1002/cncr.21298. [DOI] [PubMed] [Google Scholar]
- Monne M, Piras G, Fancello P, Santona MC, Uras A, Landriscina G, Mastio G, Gabbas A. Identification of a founder BRCA2 mutation in Sardinian breast cancer families. Fam Cancer. 2007;6:73–79. doi: 10.1007/s10689-006-9107-7. [DOI] [PubMed] [Google Scholar]
- Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16:1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Center for Biotechnology Information Blast. http://www.ncbi.nlm.nih.gov/BLAST/
- den Dunnen JT, Antonarakis SE. Nomenclature for the description of human sequence variations. Hum Genet. 2001;109:121–124. doi: 10.1007/s004390100505. [DOI] [PubMed] [Google Scholar]
- Human Gene Mutation Database. http://www.hgmd.cf.ac.uk/ac/
- National Human Genome Research Institute. http://research.nhgri.nih.gov/bic/
- Elston CW, Ellis IO. Pathological prognostic factors in breast cancer. The value of histological grade in breast cancer: experience from a large study with long-term follow-up. Histopathology. 1991;19:403–410. doi: 10.1111/j.1365-2559.1991.tb00229.x. [DOI] [PubMed] [Google Scholar]
- Sobin LH, Fleming ID. TNM classification of malignant tumours, 5th edition(1997). Union Internationale Contre le Cancer and the American Joint Committee on Cancer. Cancer. 1997;80:1803–1804. doi: 10.1002/(SICI)1097-0142(19971101)80:9<1803::AID-CNCR16>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- Caramelli D, Vernesi C, Sanna S, Sampietro L, Lari M, Castrì L, Vona G, Floris R, Francalacci P, Tykot R, Casoli A, Bertranpetit J, Lalueza-Fox C, Bertorelle G, Barbujani G. Genetic variation in prehistoric Sardinia. Hum Genet. 2007;122(3–4):327–36. doi: 10.1007/s00439-007-0403-6. [DOI] [PubMed] [Google Scholar]
- Service S, Deyoung J, Karayiorgou M, Roos JL, Pretorious H, Bedoya G, Ospina J, Ruiz-Linares A, Macedo A, Palha JA, Heutink P, Aulchenko Y, Oostra B, van Duijn C, Jarvelin MR, Varilo T, Peddle L, Rahman P, Piras G, Monne M, Murray S, Galver L, Peltonen L, Sabatti C, Collins A, Freimer N. Magnitude and distribution of linkage disequilibrium in population isolates and implications for genome-wide association studies. Nature Genetics. 2006;38:556–560. doi: 10.1038/ng1770. [DOI] [PubMed] [Google Scholar]
- Cipollini G, Tommasi S, Paradiso A, Aretini P, Bonatti F, Brunetti I, Bruno M, Lombardi G, Schittulli F, Sensi E, Tancredi M, Bevilacqua G, Caligo MA. Genetic alterations in hereditary breast cancer. Ann Oncol. 2004;15:I7–I13. doi: 10.1093/annonc/mdh651. [DOI] [PubMed] [Google Scholar]
- Thorlacius S, Olafsdottir G, Tryggvadottir L, Neuhausen S, Jonasson JG, Tavtigian SV, Tulinius H, Ogmundsdottir HM, Eyfjörd JE. A single BRCA2 mutation in male and female breast cancer families from Iceland with varied cancer phenotypes. Nat Genet. 1996;13:117–119. doi: 10.1038/ng0596-117. [DOI] [PubMed] [Google Scholar]
- Górski B, Byrski T, Huzarski T, Jakubowska A, Menkiszak J, Gronwald J, Pluzañska A, Bebenek M, Fischer-Maliszewska L, Grzybowska E, Narod SA, Lubiñski J. Founder mutations in the BRCA1 gene in Polish families with breast-ovarian cancer. Am J Hum Genet. 2000;66:1963–1968. doi: 10.1086/302922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhoog LC, Ouweland AM van den, Berns E, van Veghel-Plandsoen MM, van Staveren IL, Wagner A, Bartels CC, Tilanus-Linthorst MM, Devilee P, Seynaeve C, Halley DJ, Niermeijer MF, Klijn JG, Meijers-Heijboer H. Large regional differences in the frequency of distinct BRCA1/BRCA2 mutations in 517 Dutch breast and/or ovarian cancer families. Eur J Cancer. 2001;37:2082–2090. doi: 10.1016/S0959-8049(01)00244-1. [DOI] [PubMed] [Google Scholar]
- Vehmanen P, Friedman LS, Eerola H, McClure M, Ward B, Sarantaus L, Kainu T, Syrjäkoski K, Pyrhönen S, Kallioniemi OP, Muhonen T, Luce M, Frank TS, Nevanlinna H. Low proportion of BRCA1 and BRCA2 mutations in Finnish breast cancer families: evidence for additional susceptibility genes. Hum Mol Genet. 1997;6:2309–2315. doi: 10.1093/hmg/6.13.2309. [DOI] [PubMed] [Google Scholar]
- Russo A, Calò V, Valentina Agnese V, Bruno Corsa S. BRCA1 genetic testing in 106 breast and ovarian cancer families from southern Italy (Sicily): a mutation analyses. Breast. 2007;105:267–276. doi: 10.1007/s10549-006-9456-9. [DOI] [PubMed] [Google Scholar]
- Tonin PN, Mes-Masson AM, Futreal PA, Morgan K, Mahon M, Foulkes WD, Cole DE, Provencher D, Ghadirian P, Narod SA. Founder BRCA1 and BRCA2 mutations in French Canadian breast and ovarian cancer families. Am J Hum Genet. 1998;63:1341–1351. doi: 10.1086/302099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peto J, Collins N, Barfoot R, Seal S, Warren W, Rahman N, Easton DF, Evans C, Deacon J, Stratton MR. Prevalence of BRCA1 and BRCA2 gene mutations in patients with early-onset breast cancer. J Natl Cancer Inst. 1999;91:904–905. doi: 10.1093/jnci/91.11.943. [DOI] [PubMed] [Google Scholar]
- Ford D, Easton DF, Stratton M, Narod S, Goldgar D, Devilee P, Bishop DT, Weber B, Lenoir G, Chang-Claude J, Sobol H, Teare MD, Struewing J, Arason A, Scherneck S, Peto J, Rebbeck TR, Tonin P, Neuhausen S, Barkardottir R, Eyfjord J, Lynch H, Ponder BA, Gayther SA, Zelada-Hedman M. the Breast Cancer Linkage Consortium. Genetic heterogeneity and penetrance analysis of the BRCA1 and BRCA2 genes in breast cancer families. The Breast Cancer Linkage Consortium. Am J Hum Genet. 1998;62:676–689. doi: 10.1086/301749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone KE, Daling JR, Neal C, Suter NM, O'Brien C, Cushing-Haugen K, Jonasdottir TJ, Thompson JD, Ostrander EA. Frequency of BRCA1/BRCA2 mutations in a population-based sample of young breast carcinoma cases. Cancer. 2000;88:1393–1402. doi: 10.1002/(SICI)1097-0142(20000315)88:6<1393::AID-CNCR17>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- Mikaelsdottir EK, Valgeirsdottir S, Eyfjord JE, Rafnar T. The Icelandic founder mutation BRCA2 999del5: analysis of expression. Breast Cancer Res. 2004;6:R284–290. doi: 10.1186/bcr785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liede A, Karlan BY, Narod SA. Cancer Risks for Male Carriers of Germline Mutations in BRCA1 or BRCA2: A Review of the Literature. Journal of Clinical Oncology. 2004;22:735–742. doi: 10.1200/JCO.2004.05.055. [DOI] [PubMed] [Google Scholar]
- Rennert G, Bisland-Naggan S, Barnett-Griness O, Naomi Bar-Joseph N, Zhang S, Hedy S, Rennert HS, Narod SA. Clinical Outcomes of Breast Cancer in Carriers of BRCA1 and BRCA2 Mutations. N Engl J Med. 2007;357:115–122. doi: 10.1056/NEJMoa070608. [DOI] [PubMed] [Google Scholar]