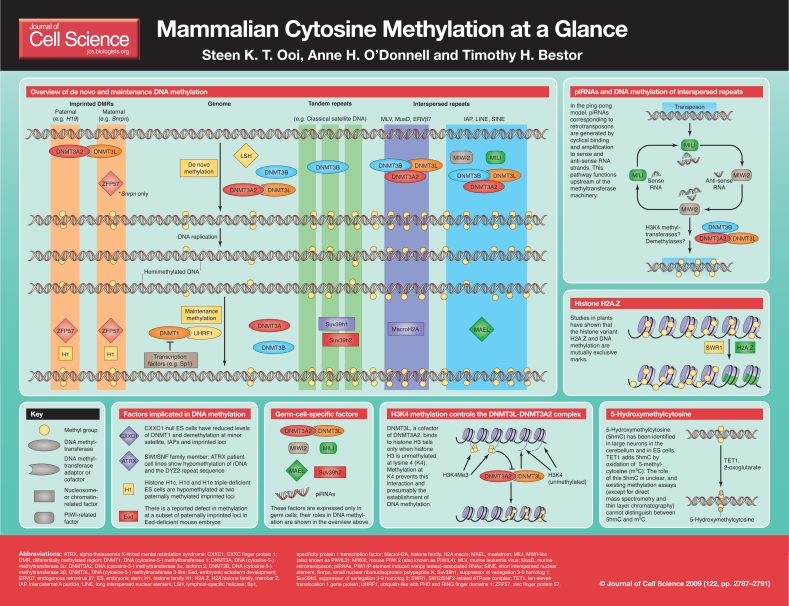
In mammals, methylation at symmetric CpG dinucleotides is necessary for the heritable silencing and regulation of many different classes of DNA sequences. These include highly abundant retrotransposons, such as the long terminal repeat (LTR)-based endogenous retroviruses, [e.g. intercisternal A particles (IAPs)] and non-LTR based elements [e.g. long interspersed nuclear elements (LINEs)]. Tandem repetitive sequences, such as pericentromeric minor and major satellite sequences, are also densely methylated, which is necessary for their stabilization against rearrangements. DNA methylation is also involved in the regulation of genes that are subject to parent-of-origin imprinting and the inactivation of the silent X chromosome in females. These patterns of DNA methylation are established in the germline and are perpetuated with high fidelity throughout the life of the organism. In humans, alterations in genomic methylation patterns are involved in the etiology of imprinting syndromes such as Beckwith-Wiedemann, Prader-Willi and Angelman, and have been implicated in a number of other human conditions including cancer (Feinberg and Tycko, 2004), autoimmune disease (Teitell and Richardson, 2003) and psychiatric disorders (Jiang et al., 2008). Although the DNA methyltransferases that generate 5-methylcytosine are well characterized (for a review, see Goll and Bestor, 2005), the molecular signals responsible for correct targeting during periods of both establishment and maintenance of genomic methylation patterns remain poorly defined. Recent genetic evidence implicates the involvement of adaptors or cofactors that are capable of interacting directly with the methyltransferases themselves, as well as the involvement of nucleosomal components, enzymes that can post-translationally modify histones, and proteins that are involved in the biogenesis of small RNAs.
Figure 1.
In this article and its accompanying poster, we summarize the known factors involved in the establishment and maintenance of methylation profiles in the mammalian genome. It is probable that the chromatin landscape containing these target sequences is dynamic and can be altered by histone-modifying enzymes and by the exchange of histone variants. The recently identified inverse relationship between the genomic distribution of DNA methylation and the methylation of both histone H3 at lysine 4 (H3K4) and presence of the histone variant H2A.Z suggests an interaction between DNA methylation and chromatin (Meissner et al., 2008; Zilberman et al., 2008). Transcription is also known to affect DNA methylation at both promoter and non-promoter sequences (Han et al., 2001), suggesting that, outside the repetitive regions of the genome where most methylation occurs, local changes in gene activity can impact the methylation machinery. An RNA pathway has been discovered that regulates de novo DNA methylation in mammals and has parallels to RNA-directed DNA methylation (RdDM), (Matzke et al., 2007), which is well documented in plants. However, it remains unclear how the mammalian factors that are involved in this process (such as the Argonaute proteins MIWI2 and MILI and nuage-associated Maelstrom protein) mediate their effects. The large number of factors that have been shown over the last few years to be involved in the establishment or maintenance of genomic methylation patterns means that a more complete understanding of the regulation of genomic methylation patterns might soon be a reality.
DNA methyltransferases
Over 100 modified nucleosides are found in nucleic acids (most of them in RNA), but mammalian DNA contains only 5-methylcytosine (m5C) and a small amount of 5-hydroxymethylcytosine (5hmC) in certain cell types, which is derived from m5C (Kriaucionis and Heintz, 2009; Tahiliani et al., 2009). Compared with plant genomes, which can encode more than ten DNA methyltransferase genes, mammals have only three enzymatically active DNA methyltransferases.
DNMT1
Studies of mice deficient for the enzyme that would eventually become known as DNA methyltransferase 1 (DNMT1) provided insight into the biological function of DNA methylation (Li et al., 1992). Disruption of DNMT1 function results in embryonic lethality and a reduction in global DNA methylation levels to approximately 5% of that found in wild-type mice (Lei et al., 1996). An oocyte-specific isoform, DNMT1o, is required specifically at the eight-cell stage of embryogenesis to maintain methylation patterns at imprinted loci (Howell et al., 2001). DNMT1 has been shown to be active on hemimethylated DNA, which presumably assists the maintenance and methylation activity of DNMT1 (Yoder et al., 1997). However, this preference for hemimethylated DNA alone cannot account for how DNMT1 is correctly targeted. An N-terminal replication-focus-targeting domain is necessary for coordinating DNA methylation and replication (Leonhardt et al., 1992), and this domain has recently been shown to be necessary for the recruitment of DNMT1 to hemimethylated DNA through its interaction with UHRF1 (see below) (Bostick et al., 2007) (reviewed by Ooi and Bestor, 2008).
The DNMT3 family
Screening of expressed-sequence tag (EST) libraries for the characteristic methyltransferase domain sequence motifs led to the identification of the enzymes DNMT3A and DNMT3B (Okano et al., 1998), and subsequent gene disruption studies showed that both of these enzymes are required for de novo methylation. De novo methylation is mediated by a germ-cell-specific isoform of DNMT3, DNMT3A2 (Okano et al., 1999; Okano et al., 1998; Kaneda et al., 2004; Kato et al., 2007). Mutation of the Dnmt3B gene specifically affects methylation at minor satellite repeats (Xu et al., 1999), and this is responsible for the rare human disorder immunodeficiency, centromere instability and facial anomalies (ICF) syndrome. Genetic data obtained using embryonic stem (ES) cells also revealed a role for both DNMT3A and DNMT3B in maintenance methylation (Chen et al., 2003). Together, DNMT1 and DNMT3-family proteins are responsible for establishing and maintaining global patterns of DNA methylation.
TET1
Recent studies have confirmed the existence of 5hmC within mammalian genomes (Kriaucionis and Heintz, 2009), the levels of which are regulated by the protein ten-eleven translocation 1 (TET1), a methylcytosine dioxygenase (Tahiliani et al., 2009). TET1 catalyzes 2-oxoglutarate (2OG)- and Fe (II)-dependent conversion of m5C to 5hmC. The extent and compartments of m5C that undergo conversion to 5hmC remain unknown but, based on its capacity to catalyze this reaction, TET1 could contribute to patterns of DNA methylation.
DNA methyltransferase adaptors and cofactors
Whereas DNA methylation in prokaryotes is a function of the innate sequence specificity of the DNA methyltransferases found in these organisms, mammalian DNA methyltransferases have little or no innate sequence specificity beyond the CpG dinucleotide. Rather, specific genomic methylation patterns in mammals are the result of interaction of DNMTs with diverse regulatory factors.
DNMT3L
Sequence similarity to both DNMT3A and DNMT3B led to the identification of DNMT3L (Aapola et al., 2000), which plays a crucial, sexually dimorphic role in establishing DNA methylation in the germline. In females, DNMT3L is necessary for the establishment of methylation imprints at maternally imprinted loci in growing oocytes (Bourc'his et al., 2001). In males, DNMT3L functions mainly in establishing methylation at retrotransposons in non-dividing prospermatogonia (Bourc'his and Bestor, 2004). DNMT3L lacks active methyltransferase activity, and functions instead as a regulator of both DNMT3A and DNMT3B (Suetake et al., 2004; Suetake et al., 2006; Kareta et al., 2006). The finding that DNMT3L associates in a complex with both DNMT3A and DNMT3B, and is capable of interacting with the N-terminal tail of histone H3 when it is not modified by methylation at lysine 4 (Ooi et al., 2007), indicates that DNMT3L directs de novo methylation to nucleosomal DNA that is unmethylated at H3K4.
UHRF1
Ubiquitin-like, containing PHD and RING finger domains 1 (UHRF1; also known as NP95) is the mammalian homolog of variant in methylation 1 (VIM1), a protein that was identified in a genetic screen for DNA methylation defects in the flowering plant Arabidopsis (Woo et al., 2007). Mice that are null for UHRF1 phenocopy those that are DNMT1 deficient (Bostick et al., 2007; Sharif et al., 2007). Further studies showed that UHRF1 interacts with hemimethylated DNA through a SET- and RING-associated (SRA) domain and that UHRF1 is necessary to target DNMT1 to hemimethylated DNA (Arita et al., 2008; Avvakumov et al., 2008; Hashimoto et al., 2008). UHRF1 greatly increases the preference of DNMT1 for hemimethylated DNA and reconciles the faithful transmission of genomic methylation patterns with a small (5- to 30-fold) inherent preference of DNMT1 for hemimethylated DNA.
Nucleosome- or chromatin-related factors
Histones are subject to numerous post-translational modifications that can modulate gene activity. Although these are usually referred to as epigenetic modifications, there is no compelling evidence that histone modifications are subject to mitotic inheritance. By contrast, genomic methylation patterns are faithfully inherited. Intersections between DNA methylation and histone modifications have recently been uncovered. In the sections below, we describe some of the nucleosome- and chromosome-related factors that have been shown to have a role in DNA methylation.
Suv39h1 and Suv39h2
ES cells that are deficient for both of the histone H3 lysine 9 methyltransferases, suppressor of variegation 3-9 homologs 1 and 2 (Suv39h1 and Suv39h2), display slight demethylation of mouse major satellite tandem repeat sequences (Lehnertz et al., 2003), suggesting that these enzymes or the histone H3K9 methylation mark have a role in the maintenance of DNA methylation in one class of satellite DNA.
Eed
There is a report of biallelic expression and distinct changes in the DNA methylation profiles of the Cdkn1c, Ascl2, Grb10 and Meg3 paternally imprinted genes in mouse embryos that are deficient for the polycomb repressive complex 2 (PRC2) member, embryonic ectoderm development (Eed) (Mager et al., 2003). Components of the PRC2 complex mediate methylation at lysine 27 of histone H3. It is unclear whether the DNA methylation defects are in the maintenance or establishment of paternal imprints.
MacroH2A
MacroH2A is a variant of histone H2A and is defined by the presence of a C-terminal macro domain (Bernstein and Hake, 2006). It is enriched on the inactive X chromosome. There are two macroH2A genes in the mammalian genome, and mice deficient for the more abundant form, MacroH2A1, are viable, fertile and have normal X chromosome inactivation (Changolkar et al., 2007). However, analysis of liver DNA from mutant animals revealed a slight demethylation and reactivation of an LTR retrotransposon (Changolkar et al., 2008).
Histone H1
The linker histone H1 is involved in chromatin structure and organization, and the study of its biological function in mammals has been a challenge due the presence of at least eight genes encoding H1 subtypes. H1c, H1d and H1e triple-deficient ES cells display hypomethylation within the imprinting control regions (ICRs) of at least two paternally methylated imprinted loci (H19-Igf2 and Gtl2-Dlk1) (Fan et al., 2005b). The mutant cells also show altered nucleosome repeat length.
H2A.Z
The histone H2A variant H2A.Z plays a key role in chromatin function and has an essential role during mammalian development, although its mechanisms of action are unknown. Recent work in plants has shown that DNA methylation and H2A.Z are mutually exclusive chromatin marks (Zilberman et al., 2008). Mutation of the plant maintenance CpG methyltransferase MET1 (a DNMT1 homolog) results in gains and losses of DNA methylation and reciprocal changes in H2A.Z localization. Mutation of SWR1, which encodes an ATPase that deposits H2A.Z into nucleosomes, results in mild hypermethylation of the genome. It is not yet known whether H2A.Z is involved in DNA methylation in mammals but, given the strong similarity of the methylation machinery in plants and mammals, DNA methylation in mammals is probably affected by the loss of H2A.Z.
PIWI-related factors
Small RNAs have been known to be involved in de novo DNA methylation in plants for many years (Wassenegger et al., 1994) but only recently has RNA-directed DNA methylation been observed in mammals, in which it appears to be restricted to male germ cells.
MILI and MIWI2
Members of the P-element induced wimpy testes (PIWI) class of the highly conserved Argonaute protein family are involved in the repression of retrotransposons and are necessary for gametogenesis in mammals (Aravin and Bourc'his, 2008). Mutations in either MILI (also known as PIWIL2) or MIWI2 (also known as PIWIL4) result in a failure to establish methylation at LINE1 and IAP retrotransposons during male gametogenesis (Aravin et al., 2007; Carmell et al., 2007; Kuramochi-Miyagawa et al., 2008). MILI and MIWI2 are necessary for the biogenesis of a special class of small RNAs known as PIWI-interacting RNAs (piRNAs) and are involved in cyclical binding to, and amplification of, sense and anti-sense species corresponding to retrotransposons. Genetic data suggest that this pathway functions upstream of the methyltransferase machinery (Aravin et al., 2008). The exact mechanism by which MILI and MIWI2 trigger de novo methylation is unclear, but is likely to involve demethylation of H3K4, which would attract the DNMT3L-DNMT3A2-DNMT3B complex.
Other factors involved in DNA methylation
Loss-of-function genetic analyses have identified other factors that are required for the establishment or maintenance of genomic methylation patterns.
ATRX
The human disorder alpha-thalassemia X-linked mental retardation (ATRX) syndrome is characterized by severe mental retardation, facial dysmorphia, urogenital abnormalities and α-thalassaemia. Mutations in patients have been mapped to the ATRX gene, which encodes a switch/sucrose non-fermentable (SWI/SNF) nucleosome remodelling protein. The DNA of cultured cells from ATRX patients shows hypomethylation of rDNA and the Y chromosome satellite DYZ2 repeat sequences (Gibbons et al., 2000). The observed methylation defects could reflect a role of ATRX protein in either de novo or maintenance methylation.
LSH
The SNF2/helicase family member lymphoid-specific helicase (LSH) is necessary for some de novo DNA methylation (Zhu et al., 2006). Loss of this factor leads to demethylation at repeat regions where most DNA methylation occurs, and at some gene promoters (Dennis et al., 2001). In the absence of LSH, imprinting is mostly intact, except that the paternal imprint at the Cdkn1c gene is lost (Fan et al., 2005a). Biochemical studies confirmed an indirect interaction between LSH and the DNMT3A and DNMT3B isoforms; in addition, there is also evidence of a role for LSH, DNMT3A and DNMT3B in the maintenance of DNA methylation, specifically in ES cells (Zhu et al., 2006).
ZFP57
Zinc finger protein 57 (ZFP57) is required for the post-fertilization maintenance of DNA methylation at a subset of imprinted loci within multiple imprinted domains (Li et al., 2008). Imprints cannot be maintained in embryos lacking both maternal and zygotic ZFP57, whereas maternal stores of ZFP57 in ZFP57-null oocytes are sufficient to maintain some methylation imprints. Additionally, ZFP57 is required for the establishment of maternal imprints at the Snrpn locus in the oocyte, although the maternal imprint can be rescued post-fertilization in about half of the heterozygous mice from a ZFP57-null mother. Defects in ZFP57 are reportedly involved in human transient neonatal diabetes, with mosaic hypomethylation at imprinted loci (Mackay et al., 2008).
CXXC1
CXXC1 (also known as CXXC finger protein 1, CFP1) was identified as a protein that specifically binds to unmethylated CpGs (Voo et al., 2000). Disruption of the Cxxc1 gene results in embryonic lethality (Carlone and Skalnik, 2001), although null embryos derived from Cxxc1-deficient mouse ES cells were found to be demethylated at minor satellite DNA, IAP elements and imprinted loci (Carlone et al., 2005). It is unknown how CXXC1 impacts DNA methylation, although Cxxc1-deficient cells have been reported to express lower levels of DNMT1. Hence, the demethylation in Cxxc1-deficient cells could partly be accounted for by reduced stability of DNMT1 protein (Carlone et al., 2005). CXXC1 is a component of the mammalian SETD1A (Lee and Skalnik, 2005) and SETD1B (Lee et al., 2007) histone H3K4 methyltransferase complexes.
Maelstrom
Maelstrom is a homolog of a gene originally identified in Drosophila that is necessary for the repression of transposons and for gametogenesis in females (Klattenhoff and Theurkauf, 2008). In male mice, Maelstrom is a component of nuage, a germ-cell-specific perinuclear structure that might be involved in transposon repression, among other functions. Maelstom deficiency results in defective spermatogenesis, which is associated with de-repression and demethylation of LINE1 elements in spermatocytes. (Soper et al., 2008). Both the molecular mechanism of Maelstrom function and how nuage specifically regulates the maintenance of DNA methylation remain unknown.
Conclusions
The rate of discovery of the factors required for the establishment and maintenance of genomic methylation patterns is accelerating rapidly, but the nature of the actual cues that trigger de novo methylation remain elusive. The recent discovery of a role for histone H3K4 methylation and small RNAs in retrotransposon methylation is of great importance, and we can anticipate a resolution of the long-standing question as to the nature of the mechanisms that control de novo methylation.
We thank Mathieu Boulard and John Edwards for comments on the manuscript. Supported by grants from the NIH (to T.H.B.) and a DOD BCRP predoctoral fellowship (to A.H.O.). Deposited in PMC for release after 12 months.
References
- Aapola, U., Kawasaki, K., Scott, H. S., Ollila, J., Vihinen, M., Heino, M., Shintani, A., Kawasaki, K., Minoshima, S., Krohn, K. et al. (2000). Isolation and initial characterization of a novel zinc finger gene, DNMT3L, on 21q22.3, related to the cytosine-5-methyltransferase 3 gene family. Genomics 65, 293-298. [DOI] [PubMed] [Google Scholar]
- Aravin, A. A. and Bourc'his, D. (2008). Small RNA guides for de novo DNA methylation in mammalian germ cells. Genes Dev. 22, 970-975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravin, A. A., Sachidanandam, R., Girard, A., Fejes-Toth, K. and Hannon, G. J. (2007). Developmentally regulated piRNA clusters implicate MILI in transposon control. Science 316, 744-747. [DOI] [PubMed] [Google Scholar]
- Aravin, A. A., Sachidanandam, R., Bourc'his, D., Schaefer, C., Pezic, D., Toth, K. F., Bestor, T. and Hannon, G. J. (2008). A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol. Cell 31, 785-799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arita, K., Ariyoshi, M., Tochio, H., Nakamura, Y. and Shirakawa, M. (2008). Recognition of hemi-methylated DNA by the SRA protein UHRF1 by a base-flipping mechanism. Nature 455, 818-821. [DOI] [PubMed] [Google Scholar]
- Avvakumov, G. V., Walker, J. R., Xue, S., Li, Y., Duan, S., Bronner, C., Arrowsmith, C. H. and Dhe-Paganon, S. (2008). Structural basis for recognition of hemi-methylated DNA by the SRA domain of human UHRF1. Nature 455, 822-825. [DOI] [PubMed] [Google Scholar]
- Bernstein, E. and Hake, S. B. (2006). The nucleosome: a little variation goes a long way. Biochem. Cell Biol. 84, 505-517. [DOI] [PubMed] [Google Scholar]
- Bostick, M., Kim, J. K., Esteve, P. O., Clark, A., Pradhan, S. and Jacobsen, S. E. (2007). UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science 317, 1760-1764. [DOI] [PubMed] [Google Scholar]
- Bourc'his, D. and Bestor, T. H. (2004). Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature 431, 96-99. [DOI] [PubMed] [Google Scholar]
- Bourc'his, D., Xu, G. L., Lin, C. S., Bollman, B. and Bestor, T. H. (2001). Dnmt3L and the establishment of maternal genomic imprints. Science 294, 2536-2539. [DOI] [PubMed] [Google Scholar]
- Carlone, D. L. and Skalnik, D. G. (2001). CpG binding protein is crucial for early embryonic development. Mol. Cell. Biol. 21, 7601-7606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlone, D. L., Lee, J. H., Young, S. R., Dobrota, E., Butler, J. S., Ruiz, J. and Skalnik, D. G. (2005). Reduced genomic cytosine methylation and defective cellular differentiation in embryonic stem cells lacking CpG binding protein. Mol. Cell. Biol. 25, 4881-4891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmell, M. A., Girard, A., van de Kant, H. J., Bourc'his, D., Bestor, T. H., de Rooij, D. G. and Hannon, G. J. (2007). MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev. Cell 12, 503-514. [DOI] [PubMed] [Google Scholar]
- Changolkar, L. N., Costanzi, C., Leu, N. A., Chen, D., McLaughlin, K. J. and Pehrson, J. R. (2007). Developmental changes in histone macroH2A1-mediated gene regulation. Mol. Cell. Biol. 27, 2758-2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Changolkar, L. N., Singh, G. and Pehrson, J. R. (2008). macroH2A1-dependent silencing of endogenous murine leukemia viruses. Mol. Cell. Biol. 28, 2059-2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, T., Ueda, Y., Dodge, J. E., Wang, Z. and Li, E. (2003). Establishment and maintenance of genomic methylation patterns in mouse embryonic stem cells by Dnmt3a and Dnmt3b. Mol. Cell. Biol. 23, 5594-5605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis, K., Fan, T., Geiman, T., Yan, Q. and Muegge, K. (2001). Lsh, a member of the SNF2 family, is required for genome-wide methylation. Genes Dev. 15, 2940-2944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan, T., Hagan, J. P., Kozlov, S. V., Stewart, C. L. and Muegge, K. (2005a). Lsh controls silencing of the imprinted Cdkn1c gene. Development 132, 635-644. [DOI] [PubMed] [Google Scholar]
- Fan, Y., Nikitina, T., Zhao, J., Fleury, T. J., Bhattacharyya, R., Bouhassira, E. E., Stein, A., Woodcock, C. L. and Skoultchi, A. I. (2005b). Histone H1 depletion in mammals alters global chromatin structure but causes specific changes in gene regulation. Cell 123, 1199-1212. [DOI] [PubMed] [Google Scholar]
- Feinberg, A. P. and Tycko, B. (2004). The history of cancer epigenetics. Nat. Rev. Cancer 4, 143-153. [DOI] [PubMed] [Google Scholar]
- Gibbons, R. J., McDowell, T. L., Raman, S., O'Rourke, D. M., Garrick, D., Ayyub, H. and Higgs, D. R. (2000). Mutations in ATRX, encoding a SWI/SNF-like protein, cause diverse changes in the pattern of DNA methylation. Nat. Genet. 24, 368-371. [DOI] [PubMed] [Google Scholar]
- Goll, M. G. and Bestor, T. H. (2005) Eukaryotic cytosine methyltransferases. Annu. Rev. Biochem. 74, 481-514. [DOI] [PubMed] [Google Scholar]
- Han, L., Lin, I. G. and Hsieh, C. L. (2001). Protein binding protects sites on stable episomes and in the chromosome from de novo methylation. Mol. Cell. Biol. 21, 3416-3424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto, H., Horton, J. R., Zhang, X., Bostick, M., Jacobsen, S. E. and Cheng, X. (2008). The SRA domain of UHRF1 flips 5-methylcytosine out of the DNA helix. Nature 455, 826-829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell, C. Y., Bestor, T. H., Ding, F., Latham, K. E., Mertineit, C., Trasler, J. M. and Chaillet, J. R. (2001). Genomic imprinting disrupted by a maternal effect mutation in the Dnmt1 gene. Cell 104, 829-838. [DOI] [PubMed] [Google Scholar]
- Jiang, Y., Langley, B., Lubin, F. D., Renthal, W., Wood, M. A., Yasui, D. H., Kumar, A., Nestler, E. J., Akbarian, S. and Beckel-Mitchener, A. C. (2008). Epigenetics in the nervous system. J. Neurosci. 28, 11753-11759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneda, M., Okano, M., Hata, K., Sado, T., Tsujimoto, N., Li, E. and Sasaki, H. (2004). Essential role for de novo DNA methyltransferase Dnmt3a in paternal and maternal imprinting. Nature 429, 900-903. [DOI] [PubMed] [Google Scholar]
- Kareta, M. S., Botello, Z. M., Ennis, J. J., Chou, C. and Chedin, F. (2006). Reconstitution and mechanism of the stimulation of de novo methylation by human DNMT3L. J. Biol. Chem. 281, 25893-25902. [DOI] [PubMed] [Google Scholar]
- Kato, Y., Kaneda, M., Hata, K., Kumaki, K., Hisano, M., Kohara, Y., Okano, M., Li, E., Nozaki, M. and Sasaki, H. (2007). Role of the Dnmt3 family in de novo methylation of imprinted and repetitive sequences during male germ cell development in the mouse. Hum. Mol. Genet. 16, 2272-2280. [DOI] [PubMed] [Google Scholar]
- Klattenhoff, C. and Theurkauf, W. (2008). Biogenesis and germline functions of piRNAs. Development 135, 3-9. [DOI] [PubMed] [Google Scholar]
- Kriaucionis, S. and Heintz, N. (2009). The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science 324, 929-930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa, S., Watanabe, T., Gotoh, K., Totoki, Y., Toyoda, A., Ikawa, M., Asada, N., Kojima, K., Yamaguchi, Y., Ijiri, T. W. et al. (2008). DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Genes Dev. 22, 908-917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J. H. and Skalnik, D. G. (2005). CpG-binding protein (CXXC finger protein 1) is a component of the mammalian Set1 histone H3-Lys4 methyltransferase complex, the analogue of the yeast Set1/COMPASS complex. J. Biol. Chem. 280, 41725-41731. [DOI] [PubMed] [Google Scholar]
- Lee, J. H., Tate, C. M., You, J. S. and Skalnik, D. G. (2007). Identification and characterization of the human Set1B histone H3-Lys4 methyltransferase complex. J. Biol. Chem. 282, 13419-13428. [DOI] [PubMed] [Google Scholar]
- Lehnertz, B., Ueda, Y., Derijck, A. A., Braunschweig, U., Perez-Burgos, L., Kubicek, S., Chen, T., Li, E., Jenuwein, T. and Peters, A. H. (2003). Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr. Biol. 13, 1192-1200. [DOI] [PubMed] [Google Scholar]
- Lei, H., Oh, S. P., Okano, M., Juttermann, R., Goss, K. A., Jaenisch, R. and Li, E. (1996). De novo DNA cytosine methyltransferase activities in mouse embryonic stem cells. Development 122, 3195-3205. [DOI] [PubMed] [Google Scholar]
- Leonhardt, H., Page, A. W., Weier, H. U. and Bestor, T. H. (1992). A targeting sequence directs DNA methyltransferase to sites of DNA replication in mammalian nuclei. Cell 71, 865-873. [DOI] [PubMed] [Google Scholar]
- Li, E., Bestor, T. H. and Jaenisch, R. (1992). Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell 69, 915-926. [DOI] [PubMed] [Google Scholar]
- Li, X., Ito, M., Zhou, F., Youngson, N., Zuo, X., Leder, P. and Ferguson-Smith, A. C. (2008). A maternal-zygotic effect gene, Zfp57, maintains both maternal and paternal imprints. Dev. Cell 15, 547-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay, D. J., Callaway, J. L., Marks, S. M., White, H. E., Acerini, C. L., Boonen, S. E., Dayanikli, P., Firth, H. V., Goodship, J. A., Haemers, A. P. et al. (2008). Hypomethylation of multiple imprinted loci in individuals with transient neonatal diabetes is associated with mutations in ZFP57. Nat. Genet. 40, 949-951. [DOI] [PubMed] [Google Scholar]
- Mager, J., Montgomery, N. D., de Villena, F. P. and Magnuson, T. (2003). Genome imprinting regulated by the mouse Polycomb group protein Eed. Nat. Genet. 33, 502-507. [DOI] [PubMed] [Google Scholar]
- Matzke, M., Kanno, T., Huettel, B., Daxinger, L. and Matzke, A. J. (2007). Targets of RNA-directed DNA methylation. Curr. Opin. Plant Biol. 10, 512-519. [DOI] [PubMed] [Google Scholar]
- Meissner, A., Mikkelsen, T. S., Gu, H., Wernig, M., Hanna, J., Sivachenko, A., Zhang, X., Bernstein, B. E., Nusbaum, C., Jaffe, D. B. et al. (2008). Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature 454, 766-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okano, M., Xie, S. and Li, E. (1998). Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nat. Genet. 19, 219-220. [DOI] [PubMed] [Google Scholar]
- Okano, M., Bell, D. W., Haber, D. A. and Li, E. (1999). DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99, 247-257. [DOI] [PubMed] [Google Scholar]
- Ooi, S. K. and Bestor, T. H. (2008). Cytosine methylation: remaining faithful. Curr. Biol. 18, R174-R176. [DOI] [PubMed] [Google Scholar]
- Ooi, S. K., Qiu, C., Bernstein, E., Li, K., Jia, D., Yang, Z., Erdjument-Bromage, H., Tempst, P., Lin, S. P., Allis, C. D. et al. (2007). DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature 448, 714-717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharif, J., Muto, M., Takebayashi, S. I., Suetake, I., Iwamatsu, A., Endo, T. A., Shinga, J., Mizutani-Koseki, Y., Toyoda, T., Okamura, K. et al. (2007). The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature 450, 908-912. [DOI] [PubMed] [Google Scholar]
- Soper, S. F., van der Heijden, G. W., Hardiman, T. C., Goodheart, M., Martin, S. L., de Boer, P. and Bortvin, A. (2008). Mouse maelstrom, a component of nuage, is essential for spermatogenesis and transposon repression in meiosis. Dev. Cell 15, 285-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suetake, I., Shinozaki, F., Miyagawa, J., Takeshima, H. and Tajima, S. (2004). DNMT3L stimulates the DNA methylation activity of Dnmt3a and Dnmt3b through a direct interaction. J. Biol. Chem. 279, 27816-27823. [DOI] [PubMed] [Google Scholar]
- Tahiliani, M., Koh, K. P., Shen, Y., Pastor, W. A., Bandukwala, H., Brudno, Y., Agarwal, S., Iyer, L. M., Liu, D. R., Aravind, L. et al. (2009). Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324, 930-935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teitell, M. and Richardson, B. (2003). DNA methylation in the immune system. Clin. Immunol. 109, 2-5. [DOI] [PubMed] [Google Scholar]
- Voo, K. S., Carlone, D. L., Jacobsen, B. M., Flodin, A. and Skalnik, D. G. (2000). Cloning of a mammalian transcriptional activator that binds unmethylated CpG motifs and shares a CXXC domain with DNA methyltransferase, human trithorax, and methyl-CpG binding domain protein 1. Mol. Cell. Biol. 20, 2108-2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassenegger, M., Heimes, S., Riedel, L. and Sanger, H. L. (1994). RNA-directed de novo methylation of genomic sequences in plants. Cell 76, 567-576. [DOI] [PubMed] [Google Scholar]
- Woo, H. R., Pontes, O., Pikaard, C. S. and Richards, E. J. (2007). VIM1, a methylcytosine-binding protein required for centromeric heterochromatinization. Genes Dev. 21, 267-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, G. L., Bestor, T. H., Bourc'his, D., Hsieh, C. L., Tommerup, N., Bugge, M., Hulten, M., Qu, X., Russo, J. J. and Viegas-Pequignot, E. (1999). Chromosome instability and immunodeficiency syndrome caused by mutations in a DNA methyltransferase gene. Nature 402, 187-191. [DOI] [PubMed] [Google Scholar]
- Yoder, J. A., Soman, N. S., Verdine, G. L. and Bestor, T. H. (1997). DNA (cytosine-5)-methyltransferases in mouse cells and tissues. Studies with a mechanism-based probe. J. Mol. Biol. 270, 385-395. [DOI] [PubMed] [Google Scholar]
- Zhu, H., Geiman, T. M., Xi, S., Jiang, Q., Schmidtmann, A., Chen, T., Li, E. and Muegge, K. (2006). Lsh is involved in de novo methylation of DNA. EMBO J. 25, 335-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilberman, D., Coleman-Derr, D., Ballinger, T. and Henikoff, S. (2008). Histone H2A.Z and DNA methylation are mutually antagonistic chromatin marks. Nature 456, 125-129. [DOI] [PMC free article] [PubMed] [Google Scholar]