Summary
Klarsicht, ANC-1 and Syne homology (KASH) proteins localize to the outer nuclear membrane where they connect the nucleus to the cytoskeleton. KASH proteins interact with Sad1-UNC-84 (SUN) proteins to transfer forces across the nuclear envelope to position nuclei or move chromosomes. A new KASH protein, KDP-1, was identified in a membrane yeast two-hybrid screen of a Caenorhabditis elegans library using the SUN protein UNC-84 as bait. KDP-1 also interacted with SUN-1. KDP-1 was enriched at the nuclear envelope in a variety of tissues and required SUN-1 for nuclear envelope localization in the germline. Genetic analyses showed that kdp-1 was essential for embryonic viability, larval growth and germline development. kdp-1(RNAi) delayed the entry into mitosis in embryos, led to a small mitotic zone in the germline, and caused an endomitotic phenotype. Aspects of these phenotypes were similar to those seen in sun-1(RNAi), suggesting that KDP-1 functions with SUN-1 in the germline and early embryo. The data suggest that KDP-1 is a novel KASH protein that functions to ensure the timely progression of the cell cycle between the end of S phase and the entry into mitosis.
Keywords: KASH protein, Outer nuclear membrane, Nuclear envelope, Cell cycle, SUN protein, C. elegans
Introduction
The nuclear envelope (NE) is a double-membrane structure that provides an essential physical barrier between the cytoplasm and nucleoplasm (Gruenbaum et al., 2005; Stewart et al., 2007). In addition, the NE plays many other important, if less well known, functions in the cell. For example, the inner nuclear membrane (INM) provides a docking site for specific loci in interphase to control transcription and for telomeres in meiosis (Bupp et al., 2007; Conrad et al., 2008; Ding et al., 2007; Heessen and Fornerod, 2007). The outer nuclear membrane (ONM) communicates and interacts with components of the cytoskeleton to control nuclear positioning and structure. The ONM, INM and endoplasmic reticulum (ER) all form a single continuous membranous structure, but each is specialized and enriched with specific proteins. The ER lumen and perinuclear space between the ONM and INM is continuous as well, allowing soluble proteins to diffuse between the ER lumen and perinuclear space, and transmembrane proteins to diffuse between the ONM and ER. The INM connects to the ONM at nuclear pore complexes, allowing transmembrane proteins to be targeted to the INM. Transport between the ONM and INM is thought to be active in at least some cases and is restricted by the size of the protein (King et al., 2006; Ohba et al., 2004). Proteins can be sequestered at the INM by interacting with proteins that underlie the nucleus, such as lamins (reviewed by Lusk et al., 2007).
Some transmembrane proteins are specifically localized to the ONM by interacting in the perinuclear space with integral INM proteins, which prevents diffusion to the ER (Crisp et al., 2006; McGee et al., 2006; Padmakumar et al., 2005). Many ONM proteins are involved in connecting forces generated by the cytoskeleton to the nucleus to position nuclei within the cell and to move chromosomes inside the nucleus (reviewed by Starr, 2009). Two problems must be overcome for the nucleus to interact with the cytoskeleton: first, the ONM must be specified; and second, structural connections between the INM and ONM must be formed to transfer forces between the cytoskeleton and the nuclear lamina.
To date, the only proteins known to localize specifically to the ONM contain a domain similar to the C-termini of klarsicht, ANC-1 and Syne (KASH). KASH proteins are C-tail-anchored proteins with a luminal KASH domain following the single transmembrane domain (Starr and Fischer, 2005). KASH proteins, like all C-tail-anchored proteins, are post-translationally inserted into membranes; the C-terminal tail (with less than 30 amino acids) is inserted into the lumen of the target organelle (Borgese et al., 2007). The KASH and transmembrane domains are necessary and sufficient for localization to the ONM (McGee et al., 2006; Padmakumar et al., 2005; Starr and Han, 2002; Zhang et al., 2001). KASH proteins are then restricted to the ONM by directly interacting with a Sad1-UNC-84 (SUN) protein in the perinuclear space. KASH-less isoforms of KASH proteins have been reported at the INM, cytoplasm, centrosomes, Golgi and lipid droplets (Gough et al., 2003; Guo et al., 2005; Libotte et al., 2005; Malone et al., 2003; Starr and Han, 2002).
SUN proteins are type II transmembrane proteins that localize to the INM (reviewed by Starr, 2009). They have a conserved C-terminal luminal SUN domain, which interacts with KASH domains. SUN proteins also have a second, less-conserved KASH-interaction domain (McGee et al., 2006; Padmakumar et al., 2005). Besides these domains, SUN proteins share little or no sequence similarity with each other.
The KASH-SUN interaction in the perinuclear space forms the central link of a KASH-SUN nuclear envelope bridge that connects structural components of the nucleus (such as lamins or chromatin) to components of the cytoplasm (such as actin or the centrosome). This is especially important during nuclear positioning, because it allows the cytoskeleton to connect to a structural component of the nucleus rather than a fluid membrane. KASH-SUN NE bridging proteins are conserved across eukaryotes from Giardia to yeast to humans (reviewed by Starr, 2009). The best-understood system for these important proteins is the model roundworm Caenorhabditis elegans. C. elegans has three known KASH proteins, ANC-1, UNC-83 and ZYG-12, and two SUN proteins, UNC-84 and SUN-1 (also called matefin). UNC-83 directly interacts with UNC-84 to facilitate nuclear migration in the embryonic hypodermal hyp7 precursors, intestinal primordial cells and larval P-cells (McGee et al., 2006; Starr et al., 2001). UNC-83 recruits the plus-end directed microtubule motor kinesin-1 to the outer nuclear envelope, which provides the forces for nuclear migration (Meyerzon et al., 2009a). UNC-84 also interacts with the KASH protein ANC-1 to anchor nuclei and organize mitochondria. ANC-1 is a large, filamentous protein that physically tethers the ONM to the actin cytoskeleton (Starr and Han, 2002). The KASH protein ZYG-12 interacts with the SUN protein SUN-1 to connect centrosomes to the paternal pronucleus during pronuclear migration (Malone et al., 2003). The ZYG-12–SUN-1 NE bridge also functions during apoptosis, nuclear positioning in the germline, and meiotic homolog pairing (Fridkin et al., 2004; Penkner et al., 2007; Tzur et al., 2006).
KASH domains are short and often have divergent sequences. Thus, bio-informatic approaches are of limited use in identifying KASH proteins. For example, ZYG-12 was identified as a KASH protein due to its function in pronuclear migration and SUN-1-dependent NE localization, despite having a highly divergent KASH domain (Malone et al., 2003; Starr and Fischer, 2005). Schizosaccharomyces pombe Kms1 also has a divergent KASH domain (Miki et al., 2004; Shimanuki et al., 1997). Therefore, it is highly likely that unidentified KASH proteins exist, even in the well-studied C. elegans system. Unidentified KASH proteins probably play roles in processes that take place on the ONM. Thus, the identification of KASH proteins will provide mechanistic insight for cellular events occurring on the cytoplasmic surface of the nucleus.
In this study, we identified a novel C. elegans KASH protein, KDP-1, with essential functions throughout development. The kdp-1(RNAi) phenotypes described here suggest that KDP-1 functions to promote the timely transition of the mitotic and meiotic cell cycles from S to M phase. KDP-1 meets all of the criteria of a KASH protein: it contains a C-terminal KASH domain, it interacts with one or more SUN proteins, and it is localized to the NE in a SUN-protein dependent manner. Furthermore, kdp-1(RNAi) partially phenocopies sun-1 germline disruptions. Our data suggest that KDP-1 and SUN-1 partner to form a bridge across the nuclear envelope that functions to promote cell-cycle progression.
Results
Identification of a novel SUN-domain-interacting protein
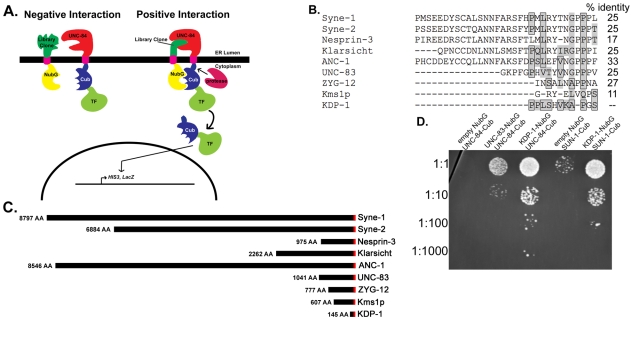
To identify novel C. elegans KASH proteins, we performed a split-ubiquitin membrane yeast two-hybrid (MYTH) screen (Gisler et al., 2008; Paumi et al., 2007; Stagljar et al., 1998; Thaminy et al., 2003) with the SUN protein UNC-84 as bait (Fig. 1A). In this system, the C-terminal moiety of ubiquitin (Cub) along with an artificial transcription factor (TF), which consists of the bacterial LexA-DNA binding domain and the Herpes simplex VP16 transactivator protein, is fused to the integral membrane protein of interest (the bait). The prey protein is fused to the N-terminal moiety of ubiquitin (Nub). Wild-type Nub has an isoleucine at position 13 (NubI). NubI and Cub have high affinity for each other and reassemble spontaneously in the cell to be recognized by the cytosolic ubiquitin-specific proteases (UBPs). By replacing Ile-13 of wild-type NubI with glycine (NubG), the affinity between NubG and Cub is decreased and the two halves only reconstitute as a `pseudo-ubiquitin' protein if they are brought into proximity through an interaction between the bait and prey proteins. Pseudo-ubiquitin is recognized by UBPs that cleave the covalent bond between ubiquitin and the protein of interest. This releases the transcription factor, which enters the nucleus and activates reporter genes (Fig. 1A). The MYTH system was previously used to show a direct interaction between the SUN protein UNC-84 and the KASH protein UNC-83 (McGee et al., 2006). We screened 2×106 colonies transformed with the UNC-84 bait and a C. elegans cDNA prey library, isolating 114 positives, representing 32 genes.
Fig. 1.
KDP-1 is a KASH protein that interacts with SUN-1 and UNC-84. (A) Schematic describing the MYTH screen. Left: The bait, a fusion of a transcription factor (TF; LexA-VP16), the C-terminal half of ubiquitin (Cub) and the transmembrane and C-terminal portion of UNC-84, is unable to bind to a random clone from the C. elegans cDNA library fused to a mutated form of the N-terminus of ubiquitin (NubG) that is not able to bind Cub on its own. Right: An interaction occurs between UNC-84 and the library clone, bringing NubG and Cub together in the cytoplasm. This results in the proteolytic cleavage of a transcription factor and subsequent activation of the reporter gene system in yeast. (B) Alignment of the KDP-1 KASH domain with other KASH domains. Boxed amino acids are identical to, and shaded amino acids are similar to, aligned residues in KDP-1. The percent identity to the KDP-1 KASH domain is noted to the right. (C) Schematic showing the size differences between some known KASH proteins: human Syne-1, Syne-2 and Nesprin-3, D. melanogaster Klarsicht, S. pombe Kms1p and C. elegans ANC-1, UNC-83, ZYG-12, and KDP-1. The KASH and transmembrane domains are in red. (D) L40 yeast transformed with the empty prey vector NubG, NubG::unc-83, or NubG::kdp-1 and the bait TF::Cub::unc-84 or TF::Cub::sun-1. Yeast were serial diluted on -Leu-Trp-His media supplemented with 75 mM 3-AT.
Three C-tail-anchored proteins were identified among the positives: a SNARE, a predicted mitochondrial protein, and the undescribed protein Y67H2A.5. Of these, only Y67H2A.5 had a C-terminus with sequence similarity to known KASH domains (Fig. 1B). We therefore call Y67H2A.5 kdp-1 for KASH domain protein. The 11-residue long KDP-1 KASH domain is similar to other KASH domains. For example, it is 27% identical and 55% similar to the KASH domain of UNC-83 (Fig. 1B). This level of similarity is about the same as that between the known KASH domains of ZYG-12 and UNC-83 (Malone et al., 2003; McGee et al., 2006; Starr and Fischer, 2005). Besides its KASH and transmembrane domain, KDP-1 shares no homology with other known KASH proteins and has no conserved domains. This is not unexpected, as most KASH proteins share no primary sequence homology outside of their KASH domain. However, like other KASH proteins (Starr and Fischer, 2005), the secondary structure of KDP-1 is predicted to be mostly helical with extended loops. There are no predicted coiled-coil regions. Although there are kdp-1 homologs in the closely related nematodes Caenorhabditis briggsae (CBG01794), Caenorhabditis remanei (CRE10281), and Pristionchus pacificus (PPA13939) and the more distant nematode Brugia malayi (BM1_55030), there are no obvious kdp-1 homologs outside of nematodes. KDP-1 is only 145 amino acids, much smaller than other known KASH proteins, some of which (ANC-1, Syne-1 and MSP-300) can be over 8000 amino acids long (Fig. 1C) (Starr and Fischer, 2005).
KASH proteins bind to SUN proteins in the intermembrane space of the NE. KASH proteins are known to promiscuously interact with different SUN proteins (Crisp et al., 2006; Ketema et al., 2007; McGee et al., 2006). Therefore, we tested whether the other known SUN protein in C. elegans, SUN-1, could interact with KDP-1 in the MYTH system. KDP-1 interacted with SUN-1 to activate the HIS3 reporter, as evidenced by growth on medium lacking histidine (Fig. 1D). Thus, in the MYTH system, KDP-1 interacted with both SUN-1 and UNC-84.
KDP-1 is an NE protein
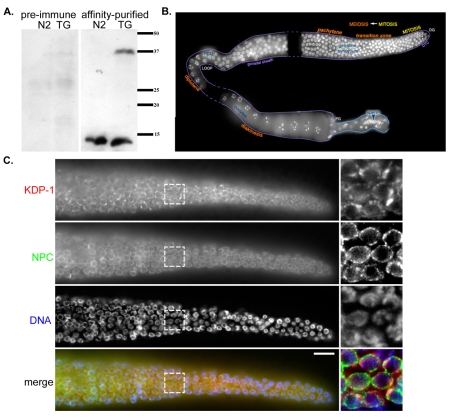
A key characteristic of KASH proteins is that they localize to the ONM (reviewed by Starr, 2007; Starr and Fischer, 2005; Wilhelmsen et al., 2006). To follow KDP-1 expression and localization, we created transgenic lines expressing a GFP-tagged KDP-1 fusion protein and raised polyclonal antibodies against the predicted cytoplasmic domain of KDP-1. A western blot of whole-worm extracts from wild-type and transgenic animals expressing GFP::KDP-1 was probed with the anti-KDP-1 antibody (Fig. 2A). The wild-type extract lane had a distinct band at the approximate expected size of KDP-1 (∼15 kDa). The transgenic extract lane had this band as well as one at the approximate expected size of GFP::KDP-1 (∼42 kDa), which was also recognized by anti-GFP (data not shown). No bands were visible when probed with pre-immune serum (Fig. 2A). These data suggest that our antibody is specific and that the kdp-1 gene is expressed. Expression of kdp-1 was also confirmed by genome-wide studies that identified over 100 independent expressed sequence tags (Bieri et al., 2007) and demonstrated that kdp-1 was expressed in embryonic, late larval and adult worms by high-throughout in situ hybridization [Nematode Expression Pattern Database, http://nematode.lab.nig.ac.jp/db2/index.php (Shin-i and Kohara, 2005)].
Fig. 2.
Antibodies against KDP-1 localize to the NE of germline nuclei. (A) Western blot of worm lysates from wild-type (N2) or transgenic (TG) worms expressing GFP::KDP-1. Blot was probed with pre-immune serum (left) or the affinity-purified KDP-1 antibody (right). Size in kDa is shown on the right. (B) Dissected adult hermaphrodite gonad stained with DAPI to show nuclei. The mitotic zone is the most distal part of the gonad (DG). Stages of meiotic prophase I are marked in orange. Individual bivalents can be seen in diakinesis oocytes at the proximal end of the gonad (PG). DTC is the region covered by the distal tip cell and PCD is the region of the gonad where most programmed cell death occurs. Image is a composite of three gonads; dashed lines correspond to intervening areas not covered by individual gonad images. Adapted from (Lints and Hall, 2005). (C) Single focal plane of fixed wild-type germline nuclei from the mitotic zone through early pachytene probed with anti-KDP-1 (row 1 and red in merge) and anti-nuclear pore complex (row 2 and green in merge) antibodies and stained with DAPI (row 3 and blue in merge). Scale bar: 10 μm. Box represents zoomed region to the right.
To test whether KDP-1 localized to the NE, wild-type hermaphrodite gonads were immunostained with the KDP-1 antibody. The C. elegans gonad is a highly ordered structure that contains mitotic nuclei at the distal end followed by nuclei progressing through the stages of meiotic prophase (Fig. 2B) (reviewed by Hubbard and Greenstein, 2005) [see also Worm Atlas, http://www.wormatlas.org/handbook/reproductivesystem/reproductivesystem.htm (Lints and Hall, 2005)]. Punctate NE enrichment of KDP-1 was observed in nuclei in the distal gonad in the mitotic zone, through the transition zone, and in pachytene meiotic prophase nuclei (Fig. 2C). Similar NE staining patterns in these same nuclei has been shown by SUN-1 immunolocalization (Penkner et al., 2007). KDP-1 puncta alternated with puncta from a nuclear pore marker (Fig. 2C). Unfortunately, the anti-KDP-1 antibody was not robust and immunolocalization experiments in other tissues were not sufficient to determine subcellular localization (data not shown).
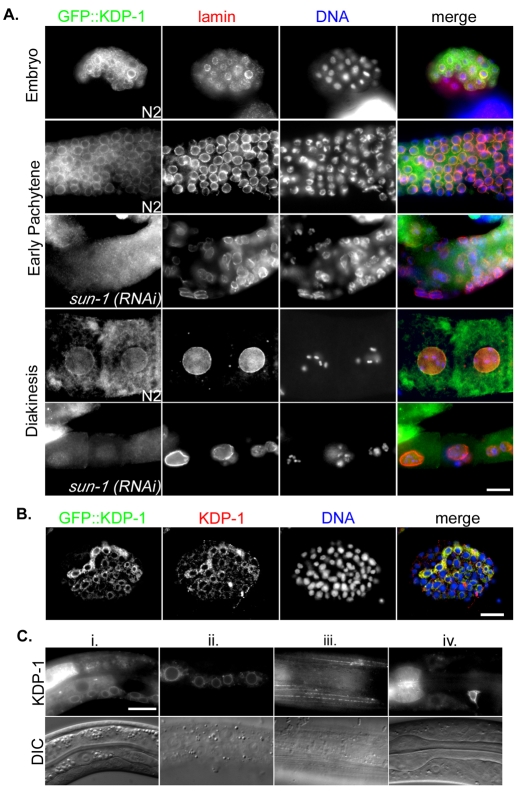
To confirm the immunolocalization pattern of KDP-1 to the NE and the in situ expression pattern of kdp-1, a GFP::KDP-1 fusion protein was expressed in wild-type animals under the control of the endogenous kdp-1 promoter. Several transgenic lines with extra-chromosomal arrays were created by particle bombardment. The brightest transgenic lines were examined by staining with antibodies against lamin to mark the NE and against GFP to observe the fusion protein (Fig. 3A). GFP::KDP-1 was enriched at the NE in both the embryo and germline (Fig. 3A). In the germline, GFP::KDP-1 was NE-enriched in the distal mitotic zone and all stages of meiotic prophase, but most prominent on nuclei in the transition zone between the distal mitotic zone and meiotic prophase, and in early pachytene (Fig. 3A). As nuclei moved into more proximal regions of the germline, the NE-enrichment weakened, although enrichment could still sometimes be seen at the NE and in unidentified cytoplasmic bodies of unfertilized oocytes before entering the spermatheca (Fig. 3A). After the ∼30-cell stage embryo, GFP::KDP-1 NE enrichment was visible. Punctate cytoplasmic expression was also observed in the germline (Fig. 3A). When transgenic embryos were stained with anti-KDP-1 antibodies, similar localization patterns were observed (Fig. 3B). Live imaging of GFP fluorescence in GFP::KDP-1 larvae showed expression in many tissue types, often enriched at the NE (Fig. 3C). Transgenic lines were also made with GFP::KDP-1 driven by a heat-shock promoter. Similar patterns of GFP::KDP-1 at the NE were observed after heat shock (data not shown).
Fig. 3.
GFP::KDP-1 localizes to the NE. (A) Single focal planes of fixed embryos or gonads expressing GFP::KDP-1 were probed with anti-GFP (first column and green in merge) and anti-lamin (second column and red in merge) antibodies and stained with DAPI to observe chromatin (third column and blue in merge). Images of an embryo are in the first row, pachytene regions of gonads in rows 2-3, and maturing oocytes in the proximal gonad in rows 4-5. Samples in rows 3 and 5 were from sun-1(RNAi) animals. Scale bar: 10 μm. (B) Single focal plane of a GFP::KDP-1 embryo probed with the GFP (green in merge) and KDP-1 (red in merge) antibodies and stained with DAPI (blue in merge). Scale bar: 10 μm. (C) Live larvae expressing GFP::KDP-1 in various tissues including (i) the ventral cord, (ii) hypodermal cells, (iii) body wall muscles, (iv) neurons and pharynx. NE enrichment is visible in the ventral cord and hypodermal cells. Scale bar: 10 μm.
The NE localization of KASH proteins is dependent on SUN proteins (Crisp et al., 2006; Ketema et al., 2007; Kracklauer et al., 2007; Malone et al., 2003; Padmakumar et al., 2005; Starr and Han, 2002; Starr et al., 2001). SUN-1 plays an essential role in the C. elegans germline and early embryo, whereas UNC-84 is expressed in the early germline, late embryo, and in most larval and adult tissues (Fridkin et al., 2004; Lee et al., 2002; Malone et al., 2003). To test whether NE localization of KDP-1 is SUN-1 dependent, gonads from sun-1(RNAi) transgenic worms expressing GFP::KDP-1 were stained for GFP and lamin. In sun-1(RNAi) gonads, the NE enrichment of GFP::KDP-1 was significantly reduced compared with untreated worms (Fig. 3A). Low levels of NE-enriched GFP::KDP-1 were sometimes observed, which was probably due to incomplete penetrance of the RNAi treatment. unc-84(n369), a null allele (Malone et al., 1999), and unc-84(RNAi) larvae and embryos did not have noticeably different GFP::KDP-1 localization from wild-type larvae and embryos (data not shown). Therefore, KDP-1 localization to the NE in the germline is dependent on SUN-1. KDP-1 localization to the NE of late embryonic, larval and adult tissues did not require UNC-84. Because SUN-1 is not expressed in these tissues (Fridkin et al., 2004), the data suggest that KDP-1 can localize to the NE independently of a SUN protein, or more likely, that other SUN proteins remain to be identified.
kdp-1 is an essential gene
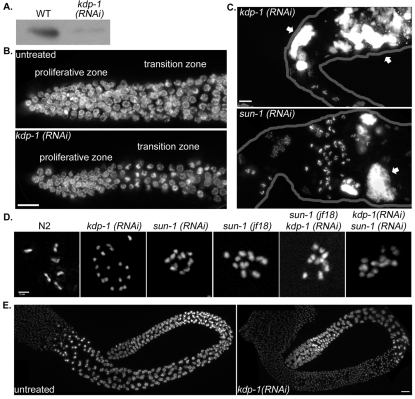
To test the function of kdp-1, animals were fed or injected with dsRNA against kdp-1. A number of phenotypes were observed, depending on the RNAi treatment window, including embryonic and larval lethality, reduced growth rates, and defects in gonadal development. Efficient knockdown of KDP-1 was demonstrated by western analysis of wild-type and kdp-1(RNAi) lysates (Fig. 4A). Some 96% of embryos laid by hermaphrodites fed dsRNA against kdp-1 for 72 hours failed to hatch (n=169 embryos laid from five parents; Table 1). The rare escapers were arrested in early larval stages. Mating kdp-1(RNAi) hermaphrodites to untreated males did not rescue the embryonic lethality. After hermaphrodites were fed dsRNA against kdp-1 for 48 hours, 100% of embryos failed to hatch whether they were mated to untreated males (n=210 embryos laid from four parents) or selfed (n=188 embryos laid from four parents). Therefore, kdp-1 is an essential gene in the germline.
Fig. 4.
kdp-1(RNAi) causes germline defects. (A) Western blot of lysates from wild-type (WT; N2) or kdp-1(RNAi) mixed-stage animals probed with anti-KDP-1. Equal loading of the lysates was confirmed by staining with Coomassie blue. (B) Partial projections of DAPI-stained proliferative and transition zones of wild-type (untreated) and kdp-1(RNAi) gonads. Scale bar: 10 μm. (C) Partial projections of DAPI-stained gonads from kdp-1(RNAi), or sun-1(RNAi) animals. Arrows mark abnormal large chromatin masses. Outlines of the gonads have been drawn in grey. Scale bar: 10 μm. (D) Projections of deconvolved images of DAPI-stained diakinesis nuclei of the indicated genotypes. Scale bar: 2 μm. (E) Projection of a DAPI-stained untreated male gonad and a kdp-1(RNAi) male gonad. Scale bar: 10 μm.
Table 1.
kdp-1(RNAi) embryonic lethality
| kdp-1(RNAi) exposure (hours) | Number of embryos laid (in 24 hours) | Number of embryos unhatched | Lethality (%) |
|---|---|---|---|
| 24 | 369 | 0 | 0 |
| 48 | 284 | 50 | 18 |
| 72 | 169 | 164 | 96 |
kdp-1(RNAi) L1 larvae fed dsRNA after hatching were examined to determine whether kdp-1 functions outside early embryonic development. Indeed, severe growth defects were observed. Only 24% of L1 worms grown on kdp-1 RNAi plates grew to young adults in 3 days at 20°C (n=128). As a comparison, after 3 days 100% of animals fed control bacteria grew to young adults (n=122). After 7 days of eating dsRNA, 99% of the kdp-1(RNAi) animals reached adulthood. Thus, besides its essential role in early embryogenesis, kdp-1 is required for normal larval growth.
kdp-1 is required for meiotic progression of nuclei through the gonad
Severe defects in the organization of the germline were observed in adults fed dsRNA against kdp-1 from the L1 stage (Fig. 4). Because development is slowed in kdp-1(RNAi) larvae and because kdp-1(RNAi) gonads looked small, we hypothesized that the number of actively dividing nuclei in the proliferative zone of kdp-1(RNAi) gonads might be reduced. We counted nuclei in mitotic zone, distal to the transition zone. There were significantly fewer nuclei in the mitotic region of kdp-1(RNAi) gonads (143.29±22.32, n=7 gonads) than in untreated gonads (208.00±10.96, n=8 gonads, P=0.009, unpaired one-tailed t-test) (Fig. 4B). The same number of mitotic nuclei, as identified by a phospho-histone antibody that labels metaphase through telophase chromosomes (Hsu et al., 2000), were seen in kdp-1(RNAi) gonads (5.47±0.82, n=15, gonads) as compared to wild type (4.79±0.80, n=32 gonads, P=0.3, unpaired one-tailed t-test). kdp-1(RNAi) male gonads also had smaller proliferative zones (44.2±7.5 μm, n=5 gonads) compared to untreated male gonads (95.1±4.1 μm, n=7 gonads, P=0.00004). We used an apoptosis marker, CED-1::GFP, to determine whether kdp-1(RNAi) germline nuclei were undergoing ectopic apoptosis. Ectopic apoptosis was not detected in the proliferative zone. The total number of CED-1::GFP positive nuclei was actually less in kdp-1(RNAi) adult hermaphrodite gonads (1.2±0.2, n=22 gonads) than in untreated gonads (3.7±0.5, n=18). This was probably due to fewer total nuclei in the gonad. Given some of the cell cycle delay phenotypes described below, we hypothesize that mitotic nuclei in kdp-1(RNAi) gonads are dividing at a slower rate than in wild type.
We observed abnormal, large chromatin masses in the proximal gonad in oocytes both distal and proximal to the spermatheca in 60% of kdp-1(RNAi) gonads (n=48 gonads) (Fig. 4C). These chromatin masses were not observed in male gonads (Fig. 4E). Similar large chromatin masses can result from a failure in meiosis, fertilization (Detwiler et al., 2001) or checkpoint activation (Kitagawa and Rose, 1999), leading to endomitosis. Normally, six bivalents are observed at diakinesis of meiosis I (Hubbard and Greenstein, 2005). However, more than six chromatin bodies of varying size and number were observed in five out of 48 kdp-1(RNAi) gonads, often in nuclei in diakinesis (Fig. 4D; supplementary material Fig. S1). The number of chromatin bodies did not match the amount that would be expected for univalents (12) or sister chromatid separation (24), and individual fragments were often irregularly sized and difficult to distinguish (supplementary material Fig. S1). Similar polyploidy in diakinesis has been observed in the mdf-1/mdf-2 spindle assembly checkpoint mutants prior to endomitosis (Kitagawa and Rose, 1999).
Large chromatin masses were also observed in sun-1(RNAi) gonads in the proximal germline (Fig. 4C). Mild disorganization defects of meiotic nuclei in kdp-1(RNAi) were rarely observed (data not shown), but never as severe as for sun-1 gonads, in which nuclei fall into the rachis (Penkner et al., 2007). Therefore, kdp-1 partially phenocopies sun-1 in the germline.
The 12 chromatin bodies in diakinesis sun-1(RNAi) nuclei (Fig. 4D) have been attributed to pairing defects, leading to univalents (Penkner et al., 2007). Although the extra chromatin bodies in kdp-1(RNAi) diakinesis nuclei could be partially due to a combination of pairing defects and aneuploidy from mitotic defects, sun-1(RNAi) diakinesis nuclei defects are at least partially distinct from the kdp-1(RNAi) defects. Double RNAi of sun-1 and kdp-1 failed to cause more than 12 visible chromatin bodies (Fig. 4D). Additional chromatin bodies could be present in double sun-1(RNAi); kdp-1(RNAi) gonads, but were not detected due to the severe sun-1(RNAi) gonad disorganization.
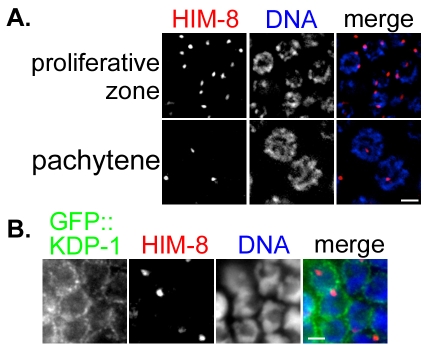
To determine whether KDP-1 has defects in meiotic homolog pairing, we stained wild-type and kdp-1(RNAi) gonads with the HIM-8 X chromosome pairing center antibody. Unpaired proliferative zone nuclei in kdp-1(RNAi) gonads have two visible HIM-8 foci. By early pachytene, a single HIM-8 focus was visible, corresponding to overlapping signals from two paired X chromosomes (Fig. 5A). HIM-8 foci did not colocalize with GFP::KDP-1 (Fig. 5B), as observed with GFP::ZYG-12 (Penkner et al., 2007). This suggests that kdp-1 does not play a role in pairing of the X chromosome.
Fig. 5.
kdp-1 is not required for X chromosome pairing in meiosis. (A) Partial projection of kdp-1(RNAi) germline nuclei probed with a HIM-8 (red) antibody and stained with DAPI (blue). Proliferative zone nuclei are shown on the top row and pachytene nuclei on the bottom row. Scale bar: 2 μm. (B) Focal plane of a GFP::KDP-1 gonad stained with HIM-8 (red) and GFP (green) antibodies and DAPI (blue). No overlap is seen between GFP::KDP-1 and HIM-8 staining. Scale bar: 2 μm.
kdp-1 is required for normal cell-cycle progression in the early embryo
The cell cycle in the early blastomeres of C. elegans switches rapidly between S and M phases with no G1 or G2 gaps. Progression through the cell cycle in early embryos can be followed by observing the nucleus by DIC optics or GFP::histone fusion proteins. After fertilization, the female pronucleus completes meiosis and both pronuclei form a nuclear envelope. Pronuclear migration occurs and the two pronuclei meet in the posterior of the embryo near the end of S phase, as the chromosomes begin to condense. Prior to nuclear envelope breakdown (NEBD), the centrosome-pronuclear complex moves to the center of the embryo and rotates 90°, a process called centration. Normally, chromosomes begin to condense before centration, and prior to NEBD.
The first cell divisions of kdp-1(RNAi) embryos were examined for cell-cycle defects in fixed embryos. Embryos from hermaphrodites injected with dsRNA against kdp-1 displayed the most severe phenotypes after 48 hours. Many of such kdp-1(RNAi) embryos had large, lobulated chromatin masses (Fig. 6A). Embryos from hermaphrodites injected with dsRNA against kdp-1 had a less severe phenotype 24-48 hours after injection. Chromosomes failed to completely align on the metaphase plate and lagging chromatid fragments were seen during anaphase (Fig. 6B).
Fig. 6.
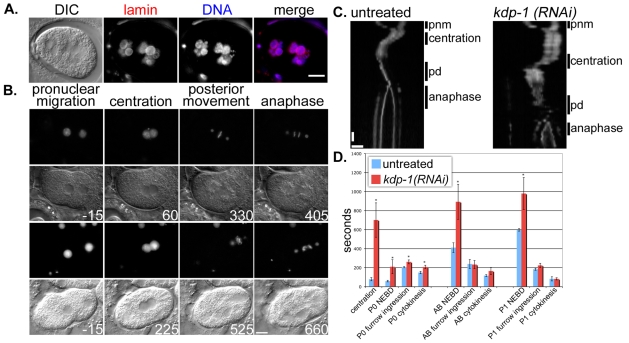
kdp-1(RNAi) causes delays in the embryonic cell cycle. (A) Large chromatin masses are shown in a kdp-1(RNAi) embryo probed with a lamin antibody and stained with DAPI. Scale bar: 10 μm. (B) DIC and GFP fluorescence of TH32 (histone H2B::GFP, gamma tubulin::GFP) single-cell embryos at selected stages of the first mitotic cell cycle. Top two panels are the GFP and DIC frames from an untreated embryo. Bottom two frames are from a kdp-1(RNAi) embryo. Times are recorded in seconds, with time 0 being the moment the pronuclei meet. Scale bar: 10 μm. (C) GFP fluorescence kymographs of the first mitotic division of representative TH32 untreated and kdp-1(RNAi) embryos. The kymographs are vertically aligned at the moment the pronuclei meet. Each slice is 2 μm. Cell-cycle stages are labeled along the side as pnm (pronuclear migration), centration, pd (posterior displacement), or anaphase. Horizontal scale bar: 10 μm. Vertical scale bar: 60 seconds. (D) Comparison of the length of individual cell-cycle events in untreated and kdp-1(RNAi) embryos for the first three mitotic divisions. Centration was defined as the first frame in which the maternal and paternal pronuclei touched until centration was completed. NEBD was the moment the NE began to lose its clear definition by DIC. Furrow ingression was the first frame in which a furrow could be seen. Cytokinesis was the last frame in which the furrow advanced. Error bars denote s.e.m. P-values were calculated by performing a one-tailed unpaired t-test (*P<0.05).
To analyze the kdp-1(RNAi) phenotype during early cell cycles and early embryonic nuclear migrations, we filmed the first two rounds of mitosis, from pronuclear migration through the AB division, in kdp-1(RNAi) embryos from hermaphrodites injected with dsRNA against kdp-1, 24-48 hours prior to analysis. Pronuclear migration occurred normally in kdp-1(RNAi) embryos, but significant delays in cell-cycle events were observed. Most apparent were a delay in centration and a subsequent delay in NEBD after the male and female pronuclei met (Fig. 6B-D; supplementary material Movies 1-5). The time from pronuclear meeting to complete centration was 78.75±17 seconds (average ± s.e.m., n=4) in wild-type embryos, but 701.25±181 seconds (n=5, P=0.007) in kdp-1(RNAi) embryos. Kymographs of representative wild-type and kdp-1(RNAi) embryos show that the delay primarily occurred before centration began, with the centrosome-pronuclear complex remaining at the posterior for an extended time (Fig. 6C). Maximum centration rates were not significantly different in kdp-1(RNAi) embryos once centration did begin. The wild-type centration velocity averaged 0.098±0.034 μm/second and the average kdp-1(RNAi) centration velocity was 0.077±0.011 μm/second (P=0.4, unpaired one-tailed t-test). Consistent with a cell-cycle delay in the entry of M phase, chromosomes remained mostly uncondensed during the delay. In fact, kdp-1(RNAi) chromosomes remain uncondensed until posterior displacement (Fig. 6B; supplementary material Movies 3 and 5). In untreated embryos, chromosomes condensed immediately after pronuclear migration (Fig. 6B; supplementary material Movies 2 and 4). The cell-cycle delay marked by a delay in centration was similar to defects caused by disruption of various components of the DNA replication machinery (Brauchle et al., 2003; Encalada et al., 2000) (Ed Munro, University of Washington, Friday Harbor, WA, personal communication). Following centration, there were significant, but less severe delays at other times during the first two mitotic cell cycles (Fig. 6D). These data show that kdp-1 is required for proper progression through the cell cycle and appears to be very important for the entry into mitosis.
Discussion
Identification of a new KASH protein
Here, we have identified a new C. elegans protein, KDP-1, that fits all the criteria of a KASH protein. KDP-1 interacted directly with two known SUN proteins, SUN-1 and UNC-84, in a MYTH system. Like other KASH proteins, KDP-1 localized to the NE in a SUN-dependent manner. sun-1(RNAi) disrupted the localization of KDP-1 to the NE, suggesting that the KDP-1 and SUN-1 interaction is relevant in vivo. Finally, analysis of the KDP-1 primary amino acid sequence suggested that it is a C-tail-anchored protein and that the C-tail of KDP-1 is homologous to other KASH domains. Given the data presented here, we conclude that KDP-1 is a newly identified KASH protein. Based on what is known about other KASH proteins, KDP-1 probably localizes to the ONM and, along with its partner SUN-1, completes a bridge across the NE.
KDP-1 was identified through a MYTH screen that identified interactions between integral membrane proteins that take place within the lumen of the ER or some other topologically equivalent compartment (Fetchko and Stagljar, 2004; Stagljar et al., 1998). Our MYTH screen only identified KDP-1, and failed to find known KASH proteins ANC-1, UNC-83 or ZYG-12. This could be explained if the library was not complex enough to contain the other KASH proteins, the KASH proteins were out of frame with NubG, or their interactions were weaker. However, because this system has been used to show a direct interaction between the KASH domain of UNC-83 and the SUN domain of UNC-84 (Fig. 1D) (McGee et al., 2006), it is unlikely that the interactions were too weak to be detected.
To our knowledge, this is the first successful MYTH screen using a NubG-tagged C. elegans cDNA library and the first MYTH screen using an integral nuclear membrane protein bait. The MYTH system was successful in identifying KDP-1 as a KASH protein, whereas the low level of similarity of the KDP-1 KASH domain to other known KASH domains had prevented bioinformatic approaches from identifying KDP-1 as a KASH protein. Thus, the MYTH system should prove valuable for identifying other new KASH proteins from other libraries. Just how many KASH proteins might exist is an open question. KDP-1 is not the most divergent KASH protein identified. ZYG-12 in C. elegans, Kms1p in S. pombe, and Cms4p in S. cerevisiae are less similar (Conrad et al., 2008; King et al., 2008; Malone et al., 2003; Miki et al., 2004; Starr, 2009), suggesting that other divergent KASH proteins remain to be identified. There are about 325 C-tail-anchored proteins in the human genome (Kalbfleisch et al., 2007). As C-tail-anchored proteins are targeted to a variety of organelles, the number that are targeted to the ONM and retained by SUN proteins will be much smaller. However, there are almost certainly more KASH proteins awaiting discovery than those currently known.
KDP-1 is required for normal cell cycle progression
kdp-1 is required for multiple processes throughout development, including germline fertility, early embryo viability and larval growth. We propose that kdp-1 functions to facilitate meiotic and mitotic cell-cycle progression. Animals treated with dsRNA against kdp-1 displayed a wide variety of phenotypes, most of which can be explained by a defect or delay in cell-cycle progression. At a gross phenotypic level, kdp-1(RNAi) slowed larval growth. This severe delay could be explained by a mitotic cell-cycle delay in post-embryonic lineages, including hypodermal seam cells and somatic gonad cells, which normally divide in each larval stage (Mohler et al., 2002; Tilmann and Kimble, 2005).
Specific delays in the cell cycle were most clearly seen when we filmed the first two zygotic divisions. We observed the strongest defects at more than 48 hours after injecting hermaphrodites with dsRNA against kdp-1 and a less severe defect 24-48 hours after injection. The most telling cell-cycle delays were observed with the mild RNAi treatments. After pronuclear migration and before centration in the one-cell embryo, kdp-1(RNAi) embryos paused for about 10 minutes; similar delays were seen in the P1 and AB cells (Fig. 6D). In addition to this defect in centration of the centrosome-pronuclear complex, the pseudo-cleavage furrows persisted and chromosomes did not condense. Defects in the DNA replication machinery cause similar phenotypes (Brauchle et al., 2003; Encalada et al., 2000) (Ed Munro, personal communication). We often saw failures in cytokinesis in kdp-1(RNAi) embryos, which could be due to cytokinesis proceeding before mitosis was completed. Along these lines, we observed poorly formed metaphase plates and lagging chromosomes at anaphase in early mitotic events (Fig. 6B; supplementary material Movie 5). Embryos from the strong RNAi treatment showed a more severe phenotype in which large lobulated chromatin masses were observed (Fig. 6A). These large masses are probably the result of endomitosis occurring after a delay or arrest in meiosis. Endomitosis, the unregulated division of a nucleus without cytokinesis, occurs when sperm do not fertilize the oocyte or when the cell cycle is inappropriately arrested in meiotic prophase I (Detwiler et al., 2001; Iwasaki et al., 1996; Sadler and Shakes, 2000). We observed no obvious defects in spermatogenesis in males, which suggests that endomitosis is not due to fertilization defects alone. Thus, we propose that kdp-1 functions in the timely progression of the cell cycle.
The phenotypes observed in kdp-1(RNAi) gonads are consistent with the hypothesis that kdp-1 normally promotes progression through the cell cycle. The major phenotype seen in kdp-1(RNAi) gonads was a combination of large chromatin masses and extra chromatin bodies in diakinesis nuclei (Fig. 4; supplementary material Fig. S1). The large chromatin masses seen in kdp-1(RNAi) gonads, like the ones seen in severe kdp-1(RNAi) embryos, are probably due to mitotic defects in the proliferative zone or meiotic defects leading to endomitosis. The chromatin fragments observed in diakinesis are probably a precursor to endomitotis, as they are often associated with large chromatin masses (supplementary material Fig. S1). Similar chromatin fragments have also been observed in endomitotic gonads in mdf-1/mdf-2 checkpoint mutants (Kitagawa and Rose, 1999). Wild-type levels of mitotic cells were observed in kdp-1(RNAi) distal gonads, despite the observation that the size of the mitotic zone in kdp-1(RNAi) gonads was reduced. This could reflect a delayed mitosis similar to that seen in the early embryo in which defects in metaphase and anaphase could lead to aneuploidy. These data suggest that kdp-1 could be required for cell-cycle progression in the germline in the same way that it is required in the early embryo.
sun-1(RNAi) gonads had similar proximal germline endomitotic and diakinesis defects to kdp-1(RNAi). However, the germline disorganization phenotypes sometimes seen in kdp-1(RNAi) gonads are not nearly as severe as those seen in sun-1(RNAi) gonads. The extra chromatin fragments occasionally observed in kdp-1(RNAi) diakinesis nuclei were not observed in sun-1 diakinesis nuclei, but this could be due to a detection problem in the severely disorganized gonads. This, combined with data showing KDP-1 interacting with SUN-1 and the sun-1-dependent NE localization of KDP-1 in the germline, supports a model in which KDP-1 on the ONM interacts with SUN-1 on the INM. In this model, SUN-1 functions though KDP-1 to regulate cell-cycle progression, but also functions through other KASH proteins, such as ZYG-12, to position nuclei in the syncytial gonad and to facilitate meiotic pairing. Because of the inability of sun-1 to completely phenocopy kdp-1, KDP-1 probably also functions independently of SUN-1.
A model for KDP-1 function in cell-cycle progression
How could a KASH protein presumably localized to the ONM regulate the cell cycle? Other KASH proteins function with SUN proteins to transmit forces generated in the cytoplasm across the NE to the nuclear lamina or chromatin (reviewed by Starr, 2009). The severely lobulated nuclei sometimes seen in kdp-1(RNAi) embryos (Fig. 6A) suggest that a lack of force transmitted across the NE could lead to defects in the nuclear lamina. In other systems, defects in the nuclear lamina lead to similarly misshapen nuclei (Sullivan et al., 1999). Although we cannot exclude the possibility that KDP-1 maintains nuclear integrity and that the cell-cycle defect could be due to some factor inappropriately leaking out of a defective NE, disruptions of the nuclear lamina are probably not the cause of the cell cycle phenotypes observed. Defects in the nuclear lamina induce a different early embryonic phenotype in which pronuclei remain abnormally condensed. The small pronuclei fail to keep centrosomes attached, leading to defects in pronuclear migration and eventually to catastrophic chromosomal segregation defects, which lead to embryonic lethality (Meyerzon et al., 2009b). We did not observe small nuclei, detached centrosomes or pronuclear migration defects in kdp-1(RNAi) embryos (Fig. 6). Furthermore, mutations in SUN proteins do not cause similar small-nuclei phenotypes. We therefore propose that the primary role for KDP-1 is in regulating the cell cycle, independently of transferring forces across the NE to maintain NE integrity.
We propose that KDP-1 functions on the cytoplasmic surface of the NE to transmit signals from the nucleus at the end of S phase to the cytoplasm in order to promote M phase. In kdp-1(RNAi) embryos, no delays were seen in decondensing of pronuclei after meiosis or entry into the first mitotic S phase. However, a delay did occur at the end of S phase, before chromosomes condensed and NEBD. Thus, our model predicts that S phase completes normally in kdp-1(RNAi) cells, but that communication between the nucleoplasm and the cytoplasm of this event is disrupted. Agreeing with this model, the cell-cycle delay in kdp-1(RNAi) embryos was not dependent on the chk-1 DNA replication checkpoint (data not shown). This is in contrast to mutations that disrupt the completion of S phase, which lead to similar delays in centration, chromosome condensation and NEBD (Brauchle et al., 2003). kdp-1(RNAi) does not cause a complete block of entry into M phase, because eventually cells attempt to segregate their chromosomes.
We predict that KDP-1 functions to recruit components of the cell cycle to the NE or to sequester cell-cycle proteins at the NE. KDP-1 could receive the signal across the NE through interaction with a SUN protein or from a soluble protein as it emerges from a nuclear pore. There is one other published report of a protein trafficked to the ONM in order to respond to a signal exiting the nucleus. In Arapidopsis, RanGAP is targeted to the ONM where it functions to maintain the Ran GTP gradient used for nuclear import. A plant-specific family of integral ONM proteins called WIPs localize RanGAP to the ONM (Xu et al., 2007). Likewise, the identification of additional proteins that interact with KDP-1 will help elucidate the molecular mechanisms by which KDP-1 regulates timely progression through the cell cycle.
Materials and Methods
C. elegans strains and transgenic lines
C. elegans strains were cultured using standard conditions (Brenner, 1974). Some nematode strains used in this work were provided by the Caenorhabditis Genetics Center, which is funded by the NIH National Center for Research Resources (NCRR). The strain UD227 carries an extrachromosomal array ycEx110. The array was introduced by particle bombardment of the unc-119(ed3) strain with pSL307 (gfp::kdp-1) and co-transfection with an unc-119-rescuing plasmid as a transformation marker (Praitis et al., 2001). Because kdp-1 is the second gene in an operon, the genomic region up to the next gene upstream of the operon was included to make pSL307, a vector with GFP fused to the N-terminus of KDP-1 and under the control of the native promoter. First, a 3274 bp genomic fragment containing the promoter region and kdp-1, which lacks introns, was amplified with KpnI and SacI overhangs and cloned into the vector pBS SK+ (Stratagene). Then, a PacI site was engineered just after the start codon of kdp-1 using strand overlap extension (SOEing) (Horton et al., 1990). Finally, GFP was amplified from pPD118.26 (Fire et al., 1990) with PacI overhangs and inserted in frame with kdp-1 at the engineered PacI site. The strain UD259 carries the ycEx124 that was made by co-microinjection of the wild-type N2 strain with pSL201 (hsp-16/2::gfp::kdp-1) and a plasmid containing the dominant rol-6(su1006) allele as a transformation marker (Mello et al., 1991). pSL201 was constructed by amplifying genomic kdp-1 DNA with EcoRI overhangs, which was cloned into the pPD118.26 vector.
RNA interference
dsRNA-treated animals were fed bacteria expressing dsRNA on NGM + IPTG + ampicilin plates (Timmons and Fire, 1998) seeded with the sun-1 feeding vector (V-156A07), or the zyg-12 feeding vector (II-4I13) (Kamath et al., 2003) in HT115 bacteria. pSL265 was constructed by amplifying genomic kdp-1 with EcoRI overhangs and cloning the PCR product into pPD129.36 (Timmons and Fire, 1998). L4 larvae were fed on sun-1 RNAi plates and the offspring were analyzed. L1 larvae were fed on kdp-1 RNAi plates and analyzed after reaching adulthood. For time-lapse microscopy, young adults were injected with dsRNA corresponding to kdp-1 as in (Fire et al., 1998). dsRNA was synthesized in vitro from a kdp-1 PCR product template using T7 RNA polymerase. Parents were dissected 24-48 hours post-injection and the embryos visualized.
Raising antibodies against KDP-1
To raise antibodies against KDP-1, the N-terminal 111 amino acids of KDP-1 were fused to the C-terminus of maltose-binding protein (MBP), expressed in E. coli, and purified on an amylose column according to the manufacturer's protocols (New England Biolabs). The MBP–KDP-1 expression vector (pSL212) was constructed by amplifying the predicted cytoplasmic region of kdp-1 and cloning it into the BamHI and EcoRI sites of pMAL-c2X (New England Biolabs). Purified MBP::KDP-1 was injected into rats four times and the sera tested by western blot against fusion protein (data not shown). The affinity-purified KDP-1 antibody from the rat with the strongest immune response was used for subsequent study. For affinity purification, a GST::KDP-1 plasmid (pSL213) was constructed by amplifying the cytoplasmic region of kdp-1 with BamHI and EcoRI overhangs and cloning the PCR product into the pGEX-2T vector (GE Healthcare). GST::KDP-1 was expressed in bacteria and purified on a glutathione Sepharose column (GE Healthcare); 18 mg was bound to an Affigel-10 column (Bio-Rad); 1 ml of serum was purified on the column using the included protocol. Affinity-purified serum was adsorbed against kdp-1(RNAi) bacteria as described (Miller and Shakes, 1995).
Immunofluorescence microscopy and western blotting
For immunolocalization studies, embryos were freeze-cracked and fixed with methanol according to previously published protocols (McGee et al., 2006). Gonads were dissected and immunostained as published (Martinez-Perez and Villeneuve, 2005), except that blocking was performed in 5% milk in PBS with Tween 20. Affinity-purified KDP-1 antibody was used at a dilution of 1:100. The anti-GFP rabbit polyclonal antibody ab6556 was used at a dilution of 1:500 (Novus Biologicals, Littleton, CO). The anti-HIM-8 polyclonal guinea pig antibody (gift of Abby Dernburg, University of California, Berkeley, CA) was used at a dilution of 1:500 (Phillips et al., 2005). The anti-LMN-1 polyclonal guinea pig antibody was used at a dilution of 1:1000 (gift of Jun Kelly Liu, Cornell University). The mouse monoclonal antibody mAb414 against nuclear pore complexes was used at a dilution of 1:1000 (Covance). The mouse monoclonal antibody against tubulin, DM1A, was used at a dilution of 1:500 (Sigma-Aldrich). The rabbit polyclonal antibody against histone 3 phospho-serine 10 (H3 P-S10) was used at a dilution of 1:100 (Upstate USA, Charlottesville, VA). Donkey anti-rabbit conjugated to Cy2, donkey anti-mouse conjugated to Cy2, donkey anti-rat conjugated to Cy3, and donkey anti-guinea pig conjugated to Cy3 were all used at a dilution of 1:200 for secondary staining (Jackson ImmunoResearch Laboratories).
Images were collected on a Leica DM 6000 with a 63× PLAN APO 1.40 objective, a Leica DC350 FX camera, and Leica FW4000 software. Images were uniformly manipulated using the remove background and levels controls in ImageJ (Abramoff et al., 2004). Deconvolution was performed in ImageJ using the Iterative Deconvolve 3D plugin (Optinav). Mitotic zone nuclei were counted from the distal tip until the first visible crescent-shaped transition zone nucleus. For time-lapse imaging, DIC images were taken at 5-second intervals and fluorescent images taken at 10-second intervals. Movies were 150× real time. Movies were processed in Final Cut Express (Apple Computer) and ImageJ. Kymographs were made using the ImageJ Kymograph or reslice plugin.
Mixed-stage animals were collected by centrifugation and boiled for 10 minutes in equal volume 2× SDS buffer. Lysates were spun down and the supernatants boiled, subjected to SDS-PAGE, transferred to an immobilon-P PVDF membrane (Millipore), and hybridized with KDP-1 antibody at a dilution of 1:1,000. For a loading control, parallel gels with the same lysates were stained with Coomassie blue.
Membrane yeast two-hybrid system
Details on the MYTH system have been published previously (Gisler et al., 2008; Paumi et al., 2007; Stagljar et al., 1998; Thaminy et al., 2003). An oligo(dT)-primed, size-selected (0.5-2.5 kb; average insert size of 1.2 kb) C. elegans whole adult-stage cDNA library with 7×106 independent clones was constructed in the prey vector pPR3-N (Dualsystems Biotech) by directional cloning into SfiI restriction sites. The library was engineered for type II transmembrane proteins so that potential KASH proteins will have a topology with the KASH domain in the ER lumen and the NubG fusion in the cytoplasm. The library was transformed into the S. cerevisiae L40 strain [MATa his3-200 trp-901, leu2-3 112 ade2, LYS2::(4xlexAop)-HIS3, URA3::(8xlexAop)-lacZ GAL4] containing the TF::Cub::UNC-84 bait plasmid pSL36 (Fetchko and Stagljar, 2004; McGee et al., 2006). It was previously shown that pSL36 does not self-activate and interacts with NubI, suggesting that it is targeted to the membrane and has proper topology with SUN domain in the ER lumen (McGee et al., 2006). Transformants were screened on -Leu-Trp-His medium supplemented with 5 mM 3-AT (3-amino-1H-1,2,4 triazole). A total of 2×106 clones were screened. The 198 positives were retested on -Leu-Trp-His + 3AT to confirm growth. A β-galactosidase filter lift assay (Fetchko and Stagljar, 2004) was also performed. Some 114 plasmids were extracted from isolates (Ausubel, 1987) and digested; 65 plasmids with unique digestion patterns were sequenced, representing 32 genes.
Targeted MYTH was performed by transforming the kdp-1 prey plasmid into L40 yeast containing either the UNC-84 bait pSL36 (McGee et al., 2006) or a full-length SUN-1 bait (gift of Il Minn and Chris Malone, Penn State University, PA). The UNC-83 prey plasmid pSL37 was the positive control (McGee et al., 2006). The empty prey NubG was used as negative control. All cultures were grown to an OD600 of 0.8 and 1 ml was spun down, washed and resuspended in 1 ml of water. Tenfold serial dilutions were performed and plated onto a -Leu-Trp-His + 75 mM 3-AT plate. The 75 mM 3-AT was required to suppress self-activation of the SUN-1 bait vector, whereas the pSL36 bait vector required only 5 mM 3-AT. Because low membrane targeting efficiency results in self-activation, it is probable that the SUN-1 bait is not targeting efficiently to the membrane. As a result of this higher self-activation, β-galactosidase filter lifts could not be used for the SUN-1 bait.
Supplementary Material
Supplementary material available online at http://jcs.biologists.org/cgi/content/full/122/16/2895/DC1
We thank Jun Kelly Liu, Il Minn and Chris Malone for sharing unpublished reagents; Ed Munro, Abby Dernburg, and Adam Hayashi for helpful advice; Lesilee Rose, JoAnne Engebrecht, and members of the Starr laboratory for comments on the manuscript and helpful discussions. The Stagljar laboratory is supported by grants from the Canadian Foundation for Innovation (CFI), the Canadian Institute for Health Research (CIHR), the Canadian Cancer Society, the Heart and Stroke Foundation of Canada, and Novartis. This research was supported by grant R01GM073874 to D.A.S. from the National Institutes of Health. Deposited in PMC for release after 12 months.
References
- Abramoff, M. D., Magelhaes, P. J. and Ram, S. J. (2004). Image processing with ImageJ. Biophotonics Int. 11, 36-42. [Google Scholar]
- Ausubel, F. M. (1987). Current Protocols in Molecular Biology. NY: John Wiley and Sons.
- Bieri, T., Blasiar, D., Ozersky, P., Antoshechkin, I., Bastiani, C., Canaran, P., Chan, J., Chen, N., Chen, W. J., Davis, P. et al. (2007). WormBase: new content and better access. Nucleic Acids Res. 35, D506-D510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borgese, N., Brambillasca, S. and Colombo, S. (2007). How tails guide tail-anchored proteins to their destinations. Curr. Opin. Cell Biol. 19, 368-375. [DOI] [PubMed] [Google Scholar]
- Brauchle, M., Baumer, K. and Gonczy, P. (2003). Differential activation of the DNA replication checkpoint contributes to asynchrony of cell division in C. elegans embryos. Curr. Biol. 13, 819-827. [DOI] [PubMed] [Google Scholar]
- Brenner, S. (1974). The genetics of Caenorhabditis elegans. Genetics 77, 71-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bupp, J. M., Martin, A. E., Stensrud, E. S. and Jaspersen, S. L. (2007). Telomere anchoring at the nuclear periphery requires the budding yeast Sad1-UNC-84 domain protein Mps3. J. Cell Biol. 179, 845-854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad, M. N., Lee, C. Y., Chao, G., Shinohara, M., Kosaka, H., Shinohara, A., Conchello, J. A. and Dresser, M. E. (2008). Rapid telomere movement in meiotic prophase is promoted by NDJ1, MPS3, and CSM4 and is modulated by recombination. Cell 133, 1175-1187. [DOI] [PubMed] [Google Scholar]
- Crisp, M., Liu, Q., Roux, K., Rattner, J. B., Shanahan, C., Burke, B., Stahl, P. D. and Hodzic, D. (2006). Coupling of the nucleus and cytoplasm: role of the LINC complex. J. Cell Biol. 172, 41-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Detwiler, M. R., Reuben, M., Li, X., Rogers, E. and Lin, R. (2001). Two zinc finger proteins, OMA-1 and OMA-2, are redundantly required for oocyte maturation in C. elegans. Dev. Cell 1, 187-199. [DOI] [PubMed] [Google Scholar]
- Ding, X., Xu, R., Yu, J., Xu, T., Zhuang, Y. and Han, M. (2007). SUN1 is required for telomere attachment to nuclear envelope and gametogenesis in mice. Dev. Cell 12, 863-872. [DOI] [PubMed] [Google Scholar]
- Encalada, S. E., Martin, P. R., Phillips, J. B., Lyczak, R., Hamill, D. R., Swan, K. A. and Bowerman, B. (2000). DNA replication defects delay cell division and disrupt cell polarity in early Caenorhabditis elegans embryos. Dev. Biol. 228, 225-238. [DOI] [PubMed] [Google Scholar]
- Fetchko, M. and Stagljar, I. (2004). Application of the split-ubiquitin membrane yeast two-hybrid system to investigate membrane protein interactions. Methods 32, 349-362. [DOI] [PubMed] [Google Scholar]
- Fire, A., Harrison, S. W. and Dixon, D. (1990). A modular set of lacZ fusion vectors for studying gene expression in Caenorhabditis elegans. Gene 93, 189-198. [DOI] [PubMed] [Google Scholar]
- Fire, A., Xu, S., Montgomery, M. K., Kostas, S. A., Driver, S. E. and Mello, C. C. (1998). Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391, 806-811. [DOI] [PubMed] [Google Scholar]
- Fridkin, A., Mills, E., Margalit, A., Neufeld, E., Lee, K. K., Feinstein, N., Cohen, M., Wilson, K. L. and Gruenbaum, Y. (2004). Matefin, a Caenorhabditis elegans germ line-specific SUN-domain nuclear membrane protein, is essential for early embryonic and germ cell development. Proc. Natl. Acad. Sci. USA 101, 6987-6992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gisler, S. M., Kittanakom, S., Fuster, D., Wong, V., Bertic, M., Radanovic, T., Hall, R. A., Murer, H., Biber, J., Markovich, D. et al. (2008). Monitoring protein-protein interactions between the mammalian integral membrane transporters and PDZ-interacting partners using a modified split-ubiquitin membrane yeast two-hybrid system. Mol. Cell Proteomics 7, 1362-1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gough, L. L., Fan, J., Chu, S., Winnick, S. and Beck, K. A. (2003). Golgi localization of Syne-1. Mol. Biol. Cell 14, 2410-2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruenbaum, Y., Margalit, A., Goldman, R. D., Shumaker, D. K. and Wilson, K. L. (2005). The nuclear lamina comes of age. Nat. Rev. Mol. Cell Biol. 6, 21-31. [DOI] [PubMed] [Google Scholar]
- Guo, Y., Jangi, S. and Welte, M. A. (2005). Organelle-specific control of intracellular transport: distinctly targeted isoforms of the regulator klar. Mol. Biol. Cell 16, 1406-1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heessen, S. and Fornerod, M. (2007). The inner nuclear envelope as a transcription factor resting place. EMBO Rep. 8, 914-919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton, R. M., Cai, Z. L., Ho, S. N. and Pease, L. R. (1990). Gene splicing by overlap extension: tailor-made genes using the polymerase chain reaction. Biotechniques 8, 528-535. [PubMed] [Google Scholar]
- Hsu, J. Y., Sun, Z. W., Li, X., Reuben, M., Tatchell, K., Bishop, D. K., Grushcow, J. M., Brame, C. J., Caldwell, J. A., Hunt, D. F. et al. (2000). Mitotic phosphorylation of histone H3 is governed by Ipl1/aurora kinase and Glc7/PP1 phosphatase in budding yeast and nematodes. Cell 102, 279-291. [DOI] [PubMed] [Google Scholar]
- Hubbard, E. J. and Greenstein, D. (2005). Introduction to the germ line. WormBook, 1-4. [DOI] [PMC free article] [PubMed]
- Iwasaki, K., McCarter, J., Francis, R. and Schedl, T. (1996). emo-1, a Caenorhabditis elegans Sec61p gamma homologue, is required for oocyte development and ovulation. J. Cell Biol. 134, 699-714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalbfleisch, T., Cambon, A. and Wattenberg, B. W. (2007). A bioinformatics approach to identifying tail-anchored proteins in the human genome. Traffic 8, 1687-1694. [DOI] [PubMed] [Google Scholar]
- Kamath, R. S., Fraser, A. G., Dong, Y., Poulin, G., Durbin, R., Gotta, M., Kanapin, A., Le Bot, N., Moreno, S., Sohrmann, M. et al. (2003). Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421, 231-237. [DOI] [PubMed] [Google Scholar]
- Ketema, M., Wilhelmsen, K., Kuikman, I., Janssen, H., Hodzic, D. and Sonnenberg, A. (2007). Requirements for the localization of nesprin-3 at the nuclear envelope and its interaction with plectin. J. Cell Sci. 120, 3384-3394. [DOI] [PubMed] [Google Scholar]
- King, M. C., Lusk, C. P. and Blobel, G. (2006). Karyopherin-mediated import of integral inner nuclear membrane proteins. Nature 442, 1003-1007. [DOI] [PubMed] [Google Scholar]
- King, M. C., Drivas, T. G. and Blobel, G. (2008). A network of nuclear envelope membrane proteins linking centromeres to microtubules. Cell 134, 427-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa, R. and Rose, A. M. (1999). Components of the spindle-assembly checkpoint are essential in Caenorhabditis elegans. Nat. Cell Biol. 1, 514-521. [DOI] [PubMed] [Google Scholar]
- Kracklauer, M. P., Banks, S. M. L., Xie, X., Wu, Y. and Fischer, J. A. (2007). Drosophila klaroid encodes a SUN domain protein required for klarsicht localization to the nuclear envelope and nuclear migration in the eye. Fly 1, 75-85. [DOI] [PubMed] [Google Scholar]
- Lee, K. K., Starr, D. A., Cohen, M., Liu, J., Han, M., Wilson, K. L. and Gruenbaum, Y. (2002). Lamin-dependent localization of UNC-84, a protein required for nuclear migration in C. elegans. Mol. Biol. Cell 13, 892-901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libotte, T., Zaim, H., Abraham, S., Padmakumar, V. C., Schneider, M., Lu, W., Munck, M., Hutchison, C., Wehnert, M., Fahrenkrog, B. et al. (2005). lamin A/C dependent localization of nesprin-2, a giant scaffolder at the nuclear envelope. Mol. Biol. Cell 16, 3411-3424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lints, R. and Hall, D. H. (2005). Reproductive system, parts I-IV. In WormAtlas.
- Lusk, C. P., Blobel, G. and King, M. C. (2007). Highway to the inner nuclear membrane: rules for the road. Nat. Rev. Mol. Cell Biol. 8, 414-420. [DOI] [PubMed] [Google Scholar]
- Malone, C. J., Fixsen, W. D., Horvitz, H. R. and Han, M. (1999). UNC-84 localizes to the nuclear envelope and is required for nuclear migration and anchoring during C. elegans development. Development 126, 3171-3181. [DOI] [PubMed] [Google Scholar]
- Malone, C. J., Misner, L., Le Bot, N., Tsai, M. C., Campbell, J. M., Ahringer, J. and White, J. G. (2003). The C. elegans hook protein, ZYG-12, mediates the essential attachment between the centrosome and nucleus. Cell 115, 825-836. [DOI] [PubMed] [Google Scholar]
- Martinez-Perez, E. and Villeneuve, A. M. (2005). HTP-1-dependent constraints coordinate homolog pairing and synapsis and promote chiasma formation during C. elegans meiosis. Genes Dev. 19, 2727-2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGee, M. D., Rillo, R., Anderson, A. S. and Starr, D. A. (2006). UNC-83 is a KASH protein required for nuclear migration and is recruited to the outer nuclear membrane by a physical interaction with the SUN protein UNC-84. Mol. Biol. Cell 17, 1790-1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello, C. C., Kramer, J. M., Stinchcomb, D. and Ambros, V. (1991). Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 10, 3959-3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyerzon, M., Fridolfsson, H. N., Ly, N., McNally, F. J., and Starr, D. A. (2009a). UNC-83 is a nuclear-specific cargo adaptor for kinesin-1-mediated nuclear migration. Development 136, 2725-2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyerzon, M., Gao, Z., Liu, J., Wu, J. C., Malone, C. J. and Starr, D. A. (2009b). Centrosome attachment to the C. elegans male pronucleus is dependent on the surface area of the nuclear envelope. Dev. Biol. 327, 433-446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki, F., Kurabayashi, A., Tange, Y., Okazaki, K., Shimanuki, M. and Niwa, O. (2004). Two-hybrid search for proteins that interact with Sad1 and Kms1, two membrane-bound components of the spindle pole body in fission yeast. Mol. Genet. Genomics 270, 449-461. [DOI] [PubMed] [Google Scholar]
- Miller, D. M. and Shakes, D. C. (1995). Caenorhabditis Elegans: Modern Biological Analysis Of An Organism, vol. 48 (ed. H. F. Epstein and D. C. Shakes), p. 659. San Diego, CA: Academic Press. [Google Scholar]
- Mohler, W. A., Shemer, G., del Campo, J. J., Valansi, C., Opoku-Serebuoh, E., Scranton, V., Assaf, N., White, J. G. and Podbilewicz, B. (2002). The type I membrane protein EFF-1 is essential for developmental cell fusion. Dev. Cell 2, 355-362. [DOI] [PubMed] [Google Scholar]
- Ohba, T., Schirmer, E. C., Nishimoto, T. and Gerace, L. (2004). Energy- and temperature-dependent transport of integral proteins to the inner nuclear membrane via the nuclear pore. J. Cell Biol. 167, 1051-1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmakumar, V. C., Libotte, T., Lu, W., Zaim, H., Abraham, S., Noegel, A. A., Gotzmann, J., Foisner, R. and Karakesisoglou, I. (2005). The inner nuclear membrane protein Sun1 mediates the anchorage of Nesprin-2 to the nuclear envelope. J. Cell Sci. 118, 3419-3430. [DOI] [PubMed] [Google Scholar]
- Paumi, C. M., Menendez, J., Arnoldo, A., Engels, K., Iyer, K. R., Thaminy, S., Georgiev, O., Barral, Y., Michaelis, S. and Stagljar, I. (2007). Mapping protein-protein interactions for the yeast ABC transporter Ycf1p by integrated split-ubiquitin membrane yeast two-hybrid analysis. Mol. Cell 26, 15-25. [DOI] [PubMed] [Google Scholar]
- Penkner, A., Tang, L., Novatchkova, M., Ladurner, M., Fridkin, A., Gruenbaum, Y., Schweizer, D., Loidl, J. and Jantsch, V. (2007). The nuclear envelope protein Matefin/SUN-1 is required for homologous pairing in C. elegans meiosis. Dev. Cell 12, 873-885. [DOI] [PubMed] [Google Scholar]
- Phillips, C. M., Wong, C., Bhalla, N., Carlton, P. M., Weiser, P., Meneely, P. M. and Dernburg, A. F. (2005). HIM-8 binds to the X chromosome pairing center and mediates chromosome-specific meiotic synapsis. Cell 123, 1051-1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praitis, V., Casey, E., Collar, D. and Austin, J. (2001). Creation of low-copy integrated transgenic lines in Caenorhabditis elegans. Genetics 157, 1217-1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadler, P. L. and Shakes, D. C. (2000). Anucleate Caenorhabditis elegans sperm can crawl, fertilize oocytes and direct anterior-posterior polarization of the 1-cell embryo. Development 127, 355-366. [DOI] [PubMed] [Google Scholar]
- Shimanuki, M., Miki, F., Ding, D. Q., Chikashige, Y., Hiraoka, Y., Horio, T. and Niwa, O. (1997). A novel fission yeast gene, kms1+, is required for the formation of meiotic prophase-specific nuclear architecture. Mol. Gen. Genet. 254, 238-249. [DOI] [PubMed] [Google Scholar]
- Shin-i, T. and Kohara, Y. (2005). NEXTDB (The nematode expression pattern database). Mishima, Japan: National Institute of Genetics.
- Stagljar, I., Korostensky, C., Johnsson, N. and te Heesen, S. (1998). A genetic system based on split-ubiquitin for the analysis of interactions between membrane proteins in vivo. Proc. Natl. Acad. Sci. USA 95, 5187-5192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr, D. A. (2007). Communication between the cytoskeleton and the nuclear envelope to position the nucleus. Mol. Biosyst. 3, 583-589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr, D. A. (2009). A nuclear-envelope bridge positions nuclei and moves chromosomes. J. Cell Sci. 122, 577-586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr, D. A. and Fischer, J. A. (2005). KASH `n Karry: the KASH domain family of cargo-specific cytoskeletal adaptor proteins. BioEssays 27, 1136-1146. [DOI] [PubMed] [Google Scholar]
- Starr, D. A. and Han, M. (2002). Role of ANC-1 in tethering nuclei to the actin cytoskeleton. Science 298, 406-409. [DOI] [PubMed] [Google Scholar]
- Starr, D. A., Hermann, G. J., Malone, C. J., Fixsen, W., Priess, J. R., Horvitz, H. R. and Han, M. (2001). unc-83 encodes a novel component of the nuclear envelope and is essential for proper nuclear migration. Development 128, 5039-5050. [DOI] [PubMed] [Google Scholar]
- Stewart, C. L., Roux, K. J. and Burke, B. (2007). Blurring the boundary: the nuclear envelope extends its reach. Science 318, 1408-1412. [DOI] [PubMed] [Google Scholar]
- Sullivan, T., Escalante-Alcalde, D., Bhatt, H., Anver, M., Bhat, N., Nagashima, K., Stewart, C. L. and Burke, B. (1999). Loss of A-type lamin expression compromises nuclear envelope integrity leading to muscular dystrophy. J. Cell Biol. 147, 913-920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaminy, S., Auerbach, D., Arnoldo, A. and Stagljar, I. (2003) Identification of novel ErbB3-interacting proteins using the split-ubiquitin membrane yeast two-hybrid technology, Genome Res. 13, 1744-1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilmann, C. and Kimble, J. (2005). Cyclin D regulation of a sexually dimorphic asymmetric cell division. Dev. Cell 9, 489-499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmons, L. and Fire, A. (1998). Specific interference by ingested dsRNA. Nature 395, 854. [DOI] [PubMed] [Google Scholar]
- Tzur, Y. B., Margalit, A., Melamed-Book, N. and Gruenbaum, Y. (2006). Matefin/SUN-1 is a nuclear envelope receptor for CED-4 during Caenorhabditis elegans apoptosis. Proc. Natl. Acad. Sci. USA 103, 13397-13402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelmsen, K., Ketema, M., Truong, H. and Sonnenberg, A. (2006). KASH-domain proteins in nuclear migration, anchorage and other processes. J. Cell Sci. 119, 5021-5029. [DOI] [PubMed] [Google Scholar]
- Xu, X. M., Meulia, T. and Meier, I. (2007). Anchorage of plant RanGAP to the nuclear envelope involves novel nuclear-pore-associated proteins. Curr. Biol. 17, 1157-1163. [DOI] [PubMed] [Google Scholar]
- Zhang, Q., Skepper, J. N., Yang, F., Davies, J. D., Hegyi, L., Roberts, R. G., Weissberg, P. L., Ellis, J. A. and Shanahan, C. M. (2001). Nesprins: a novel family of spectrin-repeat-containing proteins that localize to the nuclear membrane in multiple tissues. J. Cell Sci. 114, 4485-4498. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.