Abstract
Dupuytren’s disease (DD) is a benign fibroproliferative tumor with an unknown etiology and high recurrence postsurgery. Several observations suggest the possible involvement of skin overlying nodule (SON) and the subcutaneous fat in the pathogenesis of DD. This study aims to (1) compare the gene expression levels of SON and subcutaneous fat in DD and normal subjects and (2) to compare transverse palmar fascia (Skoog’s fibers) from DD patients as internal control tissue, with palmar fascia (transverse carpal ligament) from patients undergoing carpal tunnel release as external control. Skin, fat, and fascia were obtained from five DD patients of Caucasian origin (age = 66 ± 14) and from five control subjects (age = 57 ± 19) undergoing carpal tunnel release. Total ribonucleic acids was extracted from each sample and used for complementary deoxyribonucleic acid synthesis. Real-time quantitative polymerase chain reaction was used to assess the gene expression levels of six candidate genes: A disintegrin and metalloproteinase domain (ADAM12), aldehyde dehydrogenase 1 family member A1 (ALDH1A1), iroquois homeoboxprotein 6 (IRX6), periostin, osteoblast specific factor, proteoglycan 4, and tenascin C. Using independent t test, ADAM12, ALDH1A1, and IRX6 expression levels in DD fats were significantly (p < 0.05) higher than those in the controls. There is no significant difference in the gene expression levels of all six genes when comparing disease and control fascia and skin. Interestingly, ADAM12 up-regulation has also been observed in several other fibrotic and proliferative disorders. In conclusion, this study demonstrates potential roles for subcutaneous fat in DD pathogenesis as well as supports the use of transverse palmar fascia as appropriate control tissues in DD research.
Keywords: Dupuytren’s disease, Dupuytren’s contracture, Subcutaneous fat, Skin overlying nodule, Transverse palmar fascia, Recurrence, Gene expression, Transcriptomics
Introduction
Dupuytren’s disease (DD) is a common fibroproliferative disorder that affects palmar and digital fascia, leading to disability in affected individuals due to progressive and permanent flexion contracture of the digits [14].
Typically, a nodule appears first, followed by the development of cord, which progressively and irreversibly shortens and leads to the contracture of the digits [29]. It is believed that the contracture in DD is caused by the myofibroblast [10], a cell type believed to play an important role in contraction of connective tissues post injury and fibrosis [15]. However, the origin of the causative cells, the original fibroblast that myofibroblasts differentiate from, for DD formation and contracture remains unknown.
While surgery remains the mainstay of treatment for DD, the recurrence rate is high [36]. Several groups have reported lower postsurgery DD recurrence with dermofasciectomy, a procedure that involves replacing the overlying affected skin as well as subcutaneous fat adjacent to nodule with a skin graft [13, 40, 41]. Several other observations also suggest possible involvements of skin overlying nodule (SON) or subcutaneous fat in DD pathogenesis [2, 13, 27, 28, 40, 41, 46]. Despite this, there are no studies that have investigated the possible role of skin and fat in DD by transcriptomic analysis.
Molecular analysis has become an essential component in many areas of modern medical research [34]; however, there have been few molecular studies to date carried out in molecular dissection of DD. Limited number of previous microarray and linkage studies have demonstrated lists of genes that may potentially be involved in DD pathologenesis; however, these potential findings require further validation with more sensitive experimental approaches [18, 35]. Quantitative polymerase chain reaction (qPCR) is a sensitive, rapid, and accurate method of quantifying the transcript levels of genes of interest in a study differentiating between disease and healthy tissue [43].
Therefore, in order to further investigate the possible involvement of fat and skin in DD pathology, transcriptomic or differential gene expression analysis of various candidate genes was performed in skin, fat, and fascia from DD cases and control subjects.
Materials and Methods
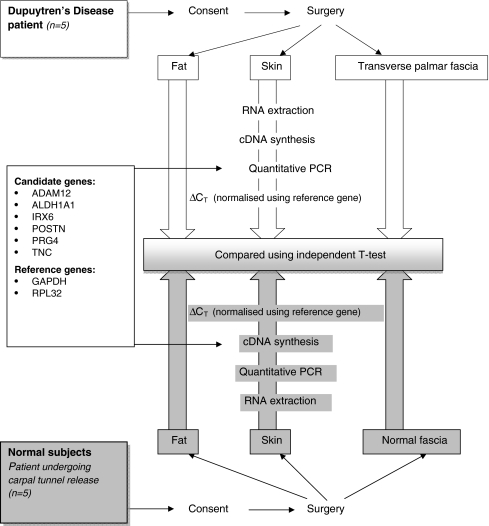
The steps taken in the study were summarized in Fig. 1. In brief, ribonucleic acids (RNA) were extracted from biopsies of skin, fat, and fascia (Fig. 2) from five DD and five control subjects. Using the extracted RNA, complementary DNA (cDNA) was synthesized and used in quantitative polymerase chain reactions. The results from qPCR were analyzed using independent t test.
Figure 1.
Summary flowchart of the steps taken in the study. Summary of the steps taken in the study to determine whether there is differential gene expression levels for genes in DD and control skin, fat, and fascia [39]. Following the consent of all subjects selected for the study, relevant surgical procedures (dermofasciectomy for cases and carpal tunnel release for controls) were carried out, and biopsies of skin, fat and palmar fascia were obtained. The biopsy samples were subjected to RNA extraction, following which cDNA synthesis and quantitative PCR were carried out. Relative gene expression levels were determined using the 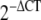 . ΔCT was determined by subtracting the threshold cycle of reference gene from those of target genes. Independent t tests were then carried out on relative gene expression levels to determine genes that are significantly (p < 0.05) differentially expressed. ADAM12 A disintegrin and metalloproteinase domain; ALDH1A1 aldehyde dehydrogenase 1 family member; IRX6 iroquois homeoboxprotein 6; PRG4 proteoglycan 4; TNC tenascin C; POSTN periostin, osteoblast specific factor; RPL32 ribosomal protein L32; GAPDH glyceraldehyde-3-phosphate dehydrogenase; ΔCT delta threshold cycle; PCR polymerase chain reactions; RNA ribonucleic acid; cDNA complementary deoxyribonucleic acid.
. ΔCT was determined by subtracting the threshold cycle of reference gene from those of target genes. Independent t tests were then carried out on relative gene expression levels to determine genes that are significantly (p < 0.05) differentially expressed. ADAM12 A disintegrin and metalloproteinase domain; ALDH1A1 aldehyde dehydrogenase 1 family member; IRX6 iroquois homeoboxprotein 6; PRG4 proteoglycan 4; TNC tenascin C; POSTN periostin, osteoblast specific factor; RPL32 ribosomal protein L32; GAPDH glyceraldehyde-3-phosphate dehydrogenase; ΔCT delta threshold cycle; PCR polymerase chain reactions; RNA ribonucleic acid; cDNA complementary deoxyribonucleic acid.
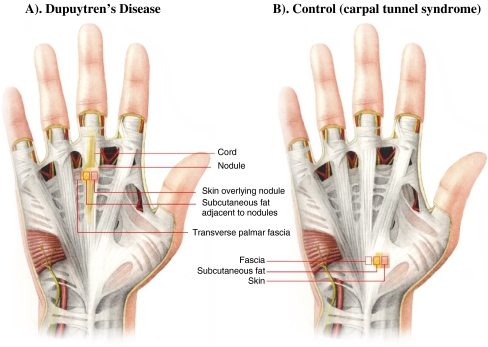
Figure 2.
Tissues subjected to analysis in this study. a This figure demonstrates the palm of the hand of an individual affected with Dupuytren’s disease, where the overlying skin has been removed to demonstrate the position of the palmar fascia in relation to the disease and harvested samples as indicated. Skin overlying palmar nodule, subcutaneous fat adjacent to palmar nodule, and transverse palmar fascia (Skoog’s fibers) were obtained from Dupuytren’s disease patients. b This figure demonstrates the palm of the hand of a control subject, where the overlying skin has been removed to demonstrate the position of the palmar fascia harvested. Skin, palmar fascia (transverse carpal ligament), and fat were obtained from control subjects, individuals undergoing carpal tunnel release.
Patients
Five DD cases and five control subjects were included in the study. All recruited DD cases were diagnosed with advanced stage of DD, which was determined clinically by an experienced hand surgeon. All patients presented flexion contracture of the metacarpophalangeal joint and proximal interphalangeal joint as well as presence of nodules. All participating DD patients were men, of Caucasian origin, with a mean age of 66 ± 14 years who had not previously received treatments for DD. Four of the five control subjects included in the study were Caucasians, with the remaining one being Asian Indian. Three of the control subjects were male, and two were female. The average age of the control subjects was 57 ± 19 years. The study was approved by the institutional review board for human subject research.
Sample Collection
Three tissue biopsies, including the SON, subcutaneous fat superficial to nodule, and unaffected transverse palmar fascia (Skoog’s fibers), were carefully dissected from each DD patient at the time of surgery (Fig. 2a). Three tissue biopsies, including the skin, subcutaneous fat, and palmar fascia (transverse carpal ligament), were obtained from individuals undergoing carpal tunnel release (Fig. 2b). The harvested biopsy samples were kept in RNAlater (Ambion, UK) at 4°C overnight and stored at −80 until use.
RNA Extraction
For each biopsy sample, approximately 8 mm3 of tissue was finely diced and placed in four separate 2-mL round-bottom Eppendorf tubes, each containing a flame-sterilized steel ball bearing and 1 mL Trizol (Invitrogen, UK). Qiagen Tissue Lyser (Qiagen, UK) was used to homogenize the tissues, at 30 oscillations per second for 12 min. The homogenized tissue suspension in each tube was transferred to a 1.5-mL Eppendorf tube and centrifuged at 13,000 rpm for 10 min. The resulting supernatant was transferred to a new Eppendorf tube, mixed well with 0.2 mL chlorophorm, and left at room temperature for 2 min. The mixture was then spun at 13,000 rpm for 15 min. The upper aqueous layer was collected and mixed with an equal volume of 70% ethanol, which was then further processed with RNeasy kit (Qiagen) according the manufacturer’s instructions. DNase treatment was then carried out using DNAfree kit (Ambion) according to the manufacturer’s protocol. NanoDrop ND-1000 UV-visible spectrophotometer (Labtech International, UK) was used to estimate the total RNA concentration.
Complementary DNA Synthesis
SuperScript II ™ Reverse Transcriptase kit (Invitrogen) was used for complementary DNA synthesis. One microliter of nucleotide mix (10 mM for each nucleotide; Invitrogen), 375 ng oligo-dT (Invitrogen), 62.5 ng random primers (Invitrogen), total RNA, and nuclease-free water (Ambion) were used to make up 12 μL reaction volume. Total RNA (0.5–1 μg) was used in each reaction, and the gene expression levels were normalized with internal reference gene using relative gene expression level method at the stage of quantitative polymerase chain reaction.
The mixture was incubated at 65°C for 5 min. Following rapid cooling on ice, 2 μL of 0.1 M dithiothreitol, 1 μL of RNaseOut (Invitrogen), and 4 μL of First-Strand Buffer (250 mM Tris–hydrochloride, pH 8.3 at room temperature; 375 mM potassium chloride; 15 mM magnesium chloride) were then added to each reaction tube. The mixtures were then incubated 42°C. After 2 min, 1 μL SuperScript ™ II Reverse Transcriptase (Invitrogen) was added to each reaction tube, which was incubated for 10 min at 25°C and then 50 min at 42°C. The reaction was inactivated by incubating at 70°C for 15 min.
Gene Selection
Six candidate genes and two reference genes were taken from Shih et al. [39]. In short, using bioinformatic approaches, including functional clustering with DAVID Bioinformatic Resources, existing microarray data, and linkage analysis, six candidate genes were short-listed on the basis of value of fold changes observed in microarray and functions [18, 35, 39]. The candidate genes were: A disintegrin and metalloproteinase domain (ADAM12), aldehyde dehydrogenase 1 family member (ALDH1) A1, iroquois homeoboxprotein 6 (IRX6), proteoglycan 4 (PRG4), tenascin C (TNC), and periostin, osteoblast specific factor (POSTN). The reference genes were ribosomal protein L32 (RPL32) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) [39].
Quantitative Polymerase Chain Reaction
Quantitative polymerase chain reactions were done in real-time using the LightCycler®480 platform (Roche Diagnostics GmBh, Germany) and corresponding LightCycler® (Roche Diagnoistics, UK) 480 software release 1.5.0 (version 1.5.0.39, Roche Diagnostics).
Each qPCR reaction was carried out in a final volume of 10 μL, consisting of 4 μL diluted template cDNA (approximately 5 ng cDNA), 5 μL LightCycler 480 Probes Master (Roche Diagnostics GmBh, Germany), 0.2 μM of forward primer (Metabion International AG, Martinsried, Germany), 0.2 μM of reverse primer (Metabion International AG, Martinsried, Germany), 0.1 μL probe from Universal Probe Library (Roche Diagnostics GmBh, Germany), and 0.7 μL nuclease-free water (Ambion; see Table 1 for the primer and probe details). In no template controls, water was used instead of cDNA. Each reaction was done in triplicates. Three hundred eighty-four multi-well plates (Roche Diagnostics GmBh, Germany) were used.
Table 1.
Primer and probe details.
| Gene | Transcript ID | Forward primer | Reverse primer | Probe |
|---|---|---|---|---|
| ADAM12 | NM_003474.3 NM_021641.3 | tggaagaaggagaggagtgtg | cattgcagcagcgattcata | ttcctctg |
| ALDH1A1 | NM_000689.3 | ccaaagacattgataaagccataa | cacgccatagcaattcacc | ctcctctg |
| IRX6 | NM_024335.2 | ctcactgtatggggcactga | gccaggctggatgtaaaact | ggaggctg |
| POSTN | NM_006475.1 | atgggagacaaagtggcttc | ctgctcctcccataatagactca | tccagtgt |
| PRG4 | NM_005807.2 | tcgtgattcagcaagtttcatc | cagttgcaggtggcatctc | tggggaag |
| TNC | NM_002160.1 | ccttgctgtagaggtcgtca | ccaacctcagacacggcta | ctgggaga |
ADAM12 A disintegrin and metalloproteinase domain, ALDH1A1 aldehyde dehydrogenase 1 family member, IRX6 iroquois homeoboxprotein 6, PRG4 proteoglycan 4, TNC tenascin C, POSTN periostin, osteoblast specific factor
The qPCR reactions were initiated at 95°C for 5 min to activate the Hot Start Taq polymerase. Each of the 45 amplification cycles consisted of a 10-s denaturation step at 95°C and a 30-s annealing and elongation step at 60°C. The fluorescence intensity was recorded at the end of the 30-s annealing and elongation step in each cycle. After the 45 cycles of amplification, a cooling step at 40°C was carried out.
Data Analysis
In order to determine the significant difference between the gene expression levels of DD and control fat, skin, and fascia, the relative threshold cycle (CT) method was used [25]. CT values were obtained from qPCR. The CT values for DD fascia (n = 4) were previously published in Shih et al. [39]. ΔCT was calculated by deducting CT of the internal controls, which was the averaged CT of the reference genes, from the CT of the target genes. Levels of target gene expression normalized using reference genes or relative gene expression levels were represented by 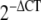 [37].
[37].
Independent t tests on the 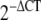 of DD and control fat, skin, and fascia were carried out using SPSS 15.0 (SPSS, USA), as suggested by Schmittgen and Livak [37]. Fold change of the genes between the normal and DD tissues were calculated using the
of DD and control fat, skin, and fascia were carried out using SPSS 15.0 (SPSS, USA), as suggested by Schmittgen and Livak [37]. Fold change of the genes between the normal and DD tissues were calculated using the 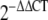 method [25].
method [25].
Results
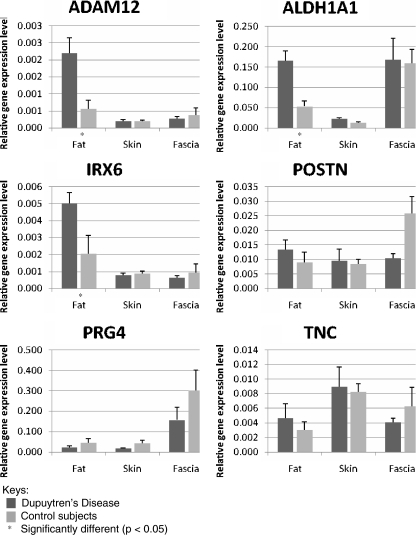
As described in Table 2 and Fig. 3, using independent t test, ADAM12, ALDH1A1, and IRX6 expression were significantly (p < 0.05) higher in DD fat than in control fat (Table 2). The average up-regulation for ADAM12, ALDH1A1, and IRX6 in DD fat was 3.9-, 3.1-, and 2.5-fold, respectively (Table 2; Fig. 3). However, there was no significant difference in the relative gene expression levels of the six investigated genes between DD and control skin or fascia (Table 2; Fig. 3).
Table 2.
Statistical comparison of the gene expression levels in Dupuytren’s disease and control fat, skin, and fascia.
| Gene symbol | Gene name | p value | Average fold change | ||||
|---|---|---|---|---|---|---|---|
| Fat | Skin | Fascia | Fat | Skin | Fascia | ||
| ADAM12 | A disintegrin and metalloproteinase domain 12 | 0.012* | 0.937 | 0.648 | 3.9* | 1.0 | 0.7 |
| ALDH1A1 | Aldehyde dehydrogenase family 1 member A1 | 0.004* | 0.067 | 0.892 | 3.1* | 1.7 | 1.1 |
| IRX6 | Iroquois homeobox protein 6 | 0.044* | 0.608 | 0.607 | 2.5* | 0.9 | 0.7 |
| POSTN | Periostin, osteoblast specific factor | 0.377 | 0.820 | 0.056 | 1.5 | 1.1 | 0.4 |
| PRG4 | Proteoglycan 4 | 0.309 | 0.147 | 0.299 | 0.5 | 0.4 | 0.5 |
| TNC | Tenascin C | 0.493 | 0.824 | 0.801 | 1.5 | 1.1 | 0.7 |
The table indicates the fold changes and statistical probabilities (p values) of each gene being differentially expressed in tissue from Dupuytren’s disease when compared to those from control subject. If the fold change value is 1, there is no difference in average gene expression levels between disease and control tissues. If the value is larger than 1, that indicates the gene is up-regulated in the tissues from DD. If the value is lower than 1, that indicates the gene is down-regulated. The fold changes that are statistically significant (p < 0.05) are indicated with an asterisk. The CT values for DD fascia (n = 4) were previously published in Shih et al. [39]
*Statistically significant (p < 0.05)
Figure 3.
Relative gene expression in each sample. The relative gene expression levels, obtained by normalizing candidate gene expression levels to reference genes, were obtained from quantitative polymerase chain reactions. The dark-colored bars represent the data for Dupuytren’s disease samples, and the light colored bars represent the data for control samples. Statistically significant (p < 0.05) differential gene expression for ADAM12, ALDH1A1, and IRX6 were observed in DD fat, which are marked with asterisk in the graphs. ADAM12 A disintegrin and metalloproteinase domain; ALDH1A1 aldehyde dehydrogenase 1 family member; IRX6 iroquois homeoboxprotein 6; PRG4 proteoglycan 4; TNC tenascin C; POSTN periostin, osteoblast specific factor. The CT values for DD fascia (n = 4) were previously published in Shih et al. [39].
Discussion
This study has demonstrated differential gene expression levels in subcutaneous fat, superficial and adjacent to nodules, compared to normal nonaffected tissue. ADAM12, IRX6, and ALDH1A1 were found to be up-regulated in DD fat when compared to normal fat. However, of the six selected genes, there were no statistically significant differences in their expression levels between DD and control skin and fascia.
ADAM12 belongs to the group of disintegrin and metalloproteases (ADAMs). There has been an increasing interest in ADAM12, which has been suggested to be involved in several fibrotic and neoplastic disorders. Up-regulation of ADAM12 has been observed in nodules of DD [39], and in addition, ADAM12 dysregulation or mutation has also been reported in various fibrotic conditions or abnormal cell growth, including keloids [38], liver fibrosis, carcinoma [19] and cancer of the breast [7, 22], liver [23], stomach [4], bladder [9], colon [19], glioblastoma [21], and prostate [31]. ADAM12 is a multifunctional protein that is involved in various cellular process and pathways, including TGF-beta and epidermal growth factor signaling pathway [22].
ALDH1A1 is an aldehyde dehydrogenase, which is responsible for the oxidation of acetaldehyde, the metabolic product of ethanol [20]. Other substrates of ALDH1A1 include acetaldehyde, benzaldehyde, 4-hydroxynonenal, malondialdehyde, and retinaldehyde [5]. Although alcoholism has been reported to be associated to DD, there is discrepancy in this behavioral association [3, 6, 16]. Furthermore, our previous study showed no significant difference in ALDH1A1 expression in nodule or cord [39]. Whether the observed overexpression of ALDH1A1 in DD fat may be associated with DD pathology requires further characterization.
Up-regulation in of IRX6, which is a gene that locates within the linkage identified by Hu et al. [18], has been observed in fat in this study and in nodule by Shih et al. [39]. The function of IRX6 is currently not well-characterized in humans. The gene family of Iroquois homeobox gene (IRX) are involved in embryonic patterning, morphogenesis, growth, and differentiation [17, 30, 44]. By using overexpression and knockdown of IRX6 in prostate cancer cell lines, Myrthue et al. [30] demonstrated IRX5 may be involved in the regulation of cell cycle and apoptosis in prostate cancer cells.
There are three possible mechanisms through which subcutaneous fat may potentially be involved in DD pathogenesis. Firstly, as suggested by Flint [8], subcutaneous fat may provide shock-absorbing effect, and reduced fat in the palm may result in higher risk of repeated trauma damaging the palmar fascia. The following observations support this hypothesis. Subcutaneous fat tissues thickness is negatively correlated to age [32], which coincides with the higher prevalence of DD in older populations [1]. Bergenudd et al. [2] have reported that, within a 55-year-old population, lower skinfold index, a measurement for subcutaneous fat thickness, was observed in DD patients.
Another possible mechanism for the involvement of subcutaneous fat is through abnormal cellular activities in the tissue. Rabinowitz et al. [33] observed differential lipid composition in DD fat, which was richer in free fatty acids and showed a significantly higher content of octanoate and other short-chain fatty acids than control. In this study, ADAM12 and IRX6 have been shown to be significantly up-regulated in subcutaneous fat, which coincide with their up-regulation previously observed in nodules [39]. Also, several reports of DD fat histology described an infiltration of fibrous tissues or displacement by collagen fibers [27, 28, 46]. The use of dermofasciectomy, a procedure that replaces the skin and subcutaneous fat overlying the DD site, results in a lower recurrence rate [13, 40, 41]. In short, subcutaneous fat may be pathogenic because it has a differential lipid composition, fibrotic infiltrate, and similarity in gene expression pattern with nodules. The removal of subcutaneous fat during dermofasciectomy may therefore account for the lower recurrence rate.
A third possible mechanism is the presence of an abnormal population of progenitor cells in the subcutaneous fat, which then in turn may act as a source of cells that cause DD pathology. Mesenchymal stem cells and multipotent progenitor cells have been characterized in subcutaneous fat [26, 47]. It has been speculated that stem cells or progenitor cells may be the origins of cells which cause fibrogenesis in peyronie’s disease [11], liver fibrosis following mesenchymal stem cell transplantation [42], and fibrotic disorder of the skeletal muscles [24]. Stem cells are also involved in wound healing [12]. Involvement of stem-like cell in abnormal cell growth, such as cancers, has also been increasingly well characterized [45].
Of the six candidate genes studied, there were no significantly different gene expressions between the external fascia from samples obtained from patient undergoing carpal tunnel release in subjects suffering from carpal tunnel syndrome (CTS) and the internal fascia from DD cases. Using whole genome microarray analysis, Rehman et al. [35] demonstrated some differential gene expression between external and internal control fascia. However, only seven genes, out of approximately 14,500 genes, were reported to show significantly different gene expression [35]. Further studies may need to be carried out to investigate the use of transverse-palmar fascia as internal control.
While no differential gene expression was observed for the six selected candidate genes between skin or normal fascia from DD and control subjects, differential gene expression for three of the six genes was shown in DD subcutaneous fat. As well as suggesting a possibility of using internal nonaffected fascia from DD patients as internal control, these results also support the hypothesis that fat may be involved in DD. Further work involving DD subcutaneous fat should be encouraged to elucidate and confirm the role of fat in DD pathology or recurrence.
References
- 1.Bayat A, McGrouther DA. Management of Dupuytren’s disease—Clear advice for an elusive condition. Ann R Coll Surg Engl 2006;88:3–8. [DOI] [PMC free article] [PubMed]
- 2.Bergenudd H, Lindgarde F, Nilsson BE. Prevalence of dupuytren’s contracture and its correlation with degenerative changes of the hands and feet and with criteria of general health. J Hand Surg 1993;18 B:254–7. [DOI] [PubMed]
- 3.Burke FD, Proud G, Lawson IJ, et al. An assessment of the effects of exposure to vibration, smoking, alcohol and diabetes on the prevalence of dupuytren’s disease in 97,537 miners. J Hand Surg: European Volume 2007;32:400–6. [DOI] [PubMed]
- 4.Carl-McGrath S, Lendeckel U, Ebert M, et al. The disintegrin-metalloproteinases ADAM9, ADAM12, and ADAM15 are upregulated in gastric cancer. Int J Oncol 2005;26:17–24. [PubMed]
- 5.Collard F, Vertommen D, Fortpied J, et al. Identification of 3-deoxyglucosone dehydrogenase as aldehyde dehydrogenase 1A1 (retinaldehyde dehydrogenase 1). Biochimie 2007;89:369–73. [DOI] [PMC free article] [PubMed]
- 6.Degreef I, Steeno P, De Smet L. A survey of clinical manifestations and risk factors in women with Dupuytren’s disease. Acta Orthop Belg 2008;74:456–60. [PubMed]
- 7.Dyczynska E, Syta E, Sun D, et al. Breast cancer-associated mutations in metalloprotease disintegrin ADAM12 interfere with the intracellular trafficking and processing of the protein. Int J Cancer 2008;122:2634–40. [DOI] [PMC free article] [PubMed]
- 8.Flint M. The genesis of the palmar lesion. In: Mcfarlane RM, Mcgrouther DA, Flint MH, editors. Dupuytren’s disease: biology and treatment. New York: Churchill Livingstone; 1990. p. 140–5.
- 9.Frohlich C, Albrechtsen R, Dyrskjøt L, et al. Molecular profiling of ADAM12 in human bladder cancer. Clin Cancer Res 2006;12:7359–68. [DOI] [PubMed]
- 10.Gabbiani G, Majno G. Dupuytren’s contracture: fibroblast contraction? An ultrastructural study. Am J Pathol 1972;66:131–46. [PMC free article] [PubMed]
- 11.Gonzalez-Cadavid NF, Rajfer J. Mechanisms of disease: new insights into the cellular and molecular pathology of Peyronie’s disease. Nat Clin Pract Urol 2005;2:291–7. [DOI] [PubMed]
- 12.Gurtner GC, Werner S, Barrandon Y, et al. Wound repair and regeneration. Nature 2008;453:314–21. [DOI] [PubMed]
- 13.Hall PN, Fitzgerald A, Sterne GD, et al. Skin replacement in Dupuytren’s disease. J Hand Surg (Edinburgh, Lothian) 1997;22:193–7. [DOI] [PubMed]
- 14.Hindocha S, Stanley JK, Watson JS, et al. Revised tubiana’s staging system for assessment of disease severity in dupuytren’s disease—Preliminary clinical findings. Hand 2008;3:80–6. [DOI] [PMC free article] [PubMed]
- 15.Hinz B. Formation and function of the myofibroblast during tissue repair. J Invest Dermatol 2007;127:526–37. [DOI] [PubMed]
- 16.Hnanicek J, Cimburova M, Putova I, et al. Lack of association of iron metabolism and Dupuytren’s disease. J Eur Acad Dermatol Venereol 2008;22:476–80. [DOI] [PubMed]
- 17.Houweling AC, Dildrop R, Peters T, et al. Gene and cluster-specific expression of the Iroquois family members during mouse development. Mech Dev 2001;107:169–74. [DOI] [PubMed]
- 18.Hu FZ, Nystrom A, Ahmed A, et al. Mapping of an autosomal dominant gene for Dupuytren’s contracture to chromosome 16q in a Swedish family. Clin Genet 2005;68:424–9. [DOI] [PubMed]
- 19.Iba K, Albrechtsen R, Gilpin BJ, et al. Cysteine-rich domain of human ADAM 12 (meltrin α) supports tumor cell adhesion. Am J Pathol 1999;154:1489–501. [DOI] [PMC free article] [PubMed]
- 20.Jelski W, Szmitkowski M. Alcohol dehydrogenase (ADH) and aldehyde dehydrogenase (ALDH) in the cancer diseases. Clin Chim Acta 2008;395:1–5. [DOI] [PubMed]
- 21.Kodama T, Ikeda E, Okada A, et al. ADAM12 is selectively overexpressed in human glioblastomas and is associated with glioblastoma cell proliferation and shedding of heparin-binding epidermal growth factor. Am J Pathol 2004;165:1743–53. [DOI] [PMC free article] [PubMed]
- 22.Kveiborg M, Frohlich C, Albrechtsen R, et al. A role for ADAM12 in breast tumor progression and stromal cell apoptosis. Cancer Res 2005;65:4754–61. [DOI] [PubMed]
- 23.Le Pabic H, Bonnier D, Wewer UM, et al. ADAM12 in human liver cancers: TGF-β-regulated expression in stellate cells is associated with matrix remodeling. Hepatology 2003;37:1056–66. [DOI] [PubMed]
- 24.Li Y, Huard J. Differentiation of muscle-derived cells into myofibroblasts in injured skeletal muscle. Am J Pathol 2002;161:895–907. [DOI] [PMC free article] [PubMed]
- 25.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 2001;25:402–8. [DOI] [PubMed]
- 26.Mackay DL, Tesar PJ, Liang LN, et al. Characterizing medullary and human mesenchymal stem cell-derived adipocytes. J Cell Physiol 2006;207:722–8. [DOI] [PubMed]
- 27.Martini AK. The anatomical relationship between the skin and pathologically altered aponeurosis in Dupuytren’s disease. Handchir Mikrochir Plast Chir 1985;17:134–8. (DIE ANATOMISCHEN BEZIEHUNGEN ZWISCHEN HAUT UND PATHOLOGISCH VERANDERTER APONEUROSE BEIM MORBUS DUPUYTHEN). [PubMed]
- 28.Martini AK, Puhl W. Micromorphological studies in Dupuytren’s disease. Z Orthop Ihre Grenzgeb 1980;118:291–9. [DOI] [PubMed]
- 29.Moyer KE, Banducci DR, Graham Iii WP, et al. Dupuytren’s disease: physiologic changes in nodule and cord fibroblasts through aging in vitro. Plast Reconstr Surg 2002;110:187–93. [DOI] [PubMed]
- 30.Myrthue A, Rademacher BL, Pittsenbarger J, et al. The iroquois homeobox gene 5 is regulated by 1,25-dihydroxyvitamin D3 in human prostate cancer and regulates apoptosis and the cell cycle in LNCaP prostate cancer cells. Clin Cancer Res 2008;14:3562–70. (An official journal of the American Association for Cancer Research). [DOI] [PubMed]
- 31.Peduto L, Reuter VE, Sehara-Fujisawa A, et al. ADAM12 is highly expressed in carcinoma-associated stroma and is required for mouse prostate tumor progression. Oncogene 2006;25:5462–6. [DOI] [PubMed]
- 32.Petrofsky JS, Prowse M, Lohman E. The influence of ageing and diabetes on skin and subcutaneous fat thickness in different regions of the body. J Appl Res 2008;8:55–61.
- 33.Rabinowitz JL, Ostermann Jr L, Bora FW, et al. Lipid composition and de novo lipid biosynthesis of human palmar fat in Dupuytren’s disease. Lipids 1983;18:371–4. [DOI] [PubMed]
- 34.Rea S, O’Sullivan ST. The polymerase chain reaction and its application to clinical plastic surgery. J Plas Rec Aesthet Surg 2006;59:113–21. [DOI] [PubMed]
- 35.Rehman S, Salway F, Stanley JK, et al. Molecular phenotypic descriptors of Dupuytren’s disease defined using informatics analysis of the transcriptome. J Hand Surg 2008;33:359–72. [DOI] [PubMed]
- 36.Ross DC. Epidemiology of Dupuytren’s disease. Hand Clin 1999;15:53–62. [PubMed]
- 37.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative CT method. Nat Protocols 2008;3:1101–8. [DOI] [PubMed]
- 38.Seifert O, Bayat A, Geffers R, et al. Identification of unique gene expression patterns within different lesional sites of keloids. Wound Repair Regen 2008;16:254–65. [DOI] [PubMed]
- 39.Shih B, Wijeratne D, Armstrong DJ, et al. Identification of biomarkers in Dupuytren’s disease by comparative analysis of fibroblasts versus tissue biopsies in disease-specific phenotypes. J Hand Surg 2009;34:124–36. [DOI] [PubMed]
- 40.Tonkin MA, Lennon WP. Dermofasciectomy and proximal interphalangeal joint replacement in Dupuytren’s disease. J Hand Surg (Edinburgh, Lothian) 1985;10:351–2. [DOI] [PubMed]
- 41.Tonkin MA, Burke FD, Varian JPW. Dupuytren’s contracture: a comparative study of fasciectomy and dermofasciectomy in one hundred patients. Br J Oral Maxillofac Surg 1984;9:156–62. [DOI] [PubMed]
- 42.Valfrè Di Bonzo L, Ferrero I, Cravanzola C, et al. Human mesenchymal stem cells as a two-edged sword in hepatic regenerative medicine: engraftment and hepatocyte differentiation versus profibrogenic potential. Gut 2008;57:223–31. [DOI] [PubMed]
- 43.VanGuilder HD, Vrana KE, Freeman WM. Twenty-five years of quantitative PCR for gene expression analysis. BioTechniques 2008;44:619–26. [DOI] [PubMed]
- 44.Van Tuyl M, Liu J, Groenman F, et al. Iroquois genes influence proximo-distal morphogenesis during rat lung development. Am J Physiol, Lung Cell Mol Physiol 2006;290(4):L777–89. [DOI] [PubMed]
- 45.Visvader JE, Lindeman GJ. Cancer stem cells in solid tumours: accumulating evidence and unresolved questions. Nat Rev, Cancer 2008;8:755–68. [DOI] [PubMed]
- 46.Warren RF. The pathology of Dupuytren’s contracture. Br J Plast Surg 1953;6:224–30. [DOI] [PubMed]
- 47.Wu X, Zvonic S, Floyd ZE, et al. Induction of circadian gene expression in human subcutaneous adipose-derived stem cells. Obesity 2007;15:2560–70. [DOI] [PubMed]