Abstract
BACKGROUND:
Both environmental and genetic factors are involved in the etiology of neural tube defects (NTDs). Inadequate folate intake and obesity are important environmental risk factors. Several folate-related genetic variants have been identified as risk factors; however, little is known about how genetic variants relate to the increased risk seen in obese women. Uncoupling Protein 2 (UCP2) is an attractive candidate to screen for NTD risk because of its possible role in obesity as well as energy metabolism, type-2 diabetes, and the regulation of reactive oxygen species. Interestingly, a previous study found that a common UCP2 compound homozygous genotype was associated with a threefold increase in NTD risk.
METHODS:
We evaluated three polymorphisms, −866G>A, A55V, and the 3′UTR 45bp insertion/deletion, as risk factors for NTDs in Irish NTD cases (N=169), their mothers (N=163), their fathers (N=167) and normal control subjects (N=332).
RESULTS:
Allele and genotype frequencies were not significantly different when comparing NTD mothers, NTD fathers, or affected children to controls. Additionally, the previously reported risk genotype (combined homozygosity of 55VV and 3′UTR 45bp deletion/deletion) was not present at a higher frequency in any NTD group when compared to controls.
CONCLUSIONS:
In our Irish study population, UCP2 polymorphisms do not influence NTD risk. Moreover, the prevalence of this allele in other populations was similar to the Irish prevalence but far lower than reported in the previous NTD study, suggesting that this previous finding of an association with NTDs might have been due to an unrepresentative study sample.
Keywords: neural tube defects, spina bifida, UCP2, obesity
INTRODUCTION
Uncoupling proteins are mitochondrial membrane transporters that disrupt the coupling between the mitochondrial proton gradient and ATP synthesis. UCP1, found in the inner mitochondrial membrane of brown adipose tissue, serves as a proton leak channel that allows cells to release stored energy as heat. While the physiological role of UCP1 in thermogenesis is relatively well understood, the function of its homologs, UCP2 and UCP3, remains unclear. Suggested models hypothesize roles for these genes in thermogenesis and body weight regulation, the attenuation of mitochondrial reactive oxygen species (ROS) production, insulin regulation, and fatty acid export (Krauss et al., 2005). Such putative functions have justified the evaluation of common variants in these genes for a role in a variety of human diseases and phenotypes such as body composition and resting energy expenditure (Yanovski et al., 2000), energy metabolism (Kovacs et al., 2005), obesity (Liu et al., 2005; Ochoa et al., 2007), multiple sclerosis (Otaegui et al., 2007; Vogler et al., 2005), diabetic neuropathy (Rudofsky et al., 2006; Yamasaki et al., 2006), coronary artery disease (Humphries et al., 2007), and schizophrenia (Yasuno et al., 2007).
Case-control association studies have provided inconsistent and sometimes contradictory results, but seem to implicate UCP2 polymorphisms in energy metabolism, obesity, and type-2 diabetes. A 45 base pair insertion polymorphism in the 3′UTR of exon 8 has shown association with energy expenditure (Kovacs et al., 2005) and obesity in some studies (Marti et al., 2004; Yanovski et al., 2000), but not all (Dalgaard et al., 2003). The more common G allele of the UCP2 −866G>A promoter polymorphism has been associated with a modest increased risk for obesity in a Caucasian European population (Esterbauer et al., 2001; Gable et al., 2007; Krempler et al., 2002), but was paradoxically associated with a decreased risk for type-2 diabetes (Krempler et al., 2002) and lower fasting plasma levels of insulin (Gable et al., 2007). The −866AA genotype was associated with an increased risk for type-2 diabetes in Italian women (D'Adamo et al., 2004), while another study identified the −866GG genotype as a risk factor in a separate Italian cohort (Bulotta et al., 2005).
Functional studies have been variable. Reports that the −866A allele is associated with enhanced in vivo adipose UCP2 mRNA expression (Esterbauer et al., 2001) and that UCP2 negatively regulates insulin secretion (Zhang et al., 2001) support the finding that individuals with the −866AA genotype have reduced B-cell function and lower insulin secretion in response to glucose stimulation (Sesti et al., 2003). However, Wang et al. (2004) found that −866AA individuals had decreased adipose UCP2 mRNA levels. Cleary the role for UCP2 polymorphisms in the etiology of obesity and type-2 diabetes is still uncertain; nonetheless these studies suggest that common variants in this gene may influence disease susceptibility and gene expression levels.
Numerous studies have found obese mothers have a two-fold or greater risk for having a child with a neural tube defect (NTDs) (Scialli, 2006; Waller et al., 1994). It is well documented that congenital malformations occur at a higher rate when the mother has diabetes (Becerra et al., 1990; Janssen et al., 1996), and that diabetic and obese women have numerous metabolic abnormalities, some of which, like hyperglycemia, are associated with a high risk for birth defects (Shaw et al., 2003). Common variants in UCP2 are, therefore, attractive candidates to screen as potential risk factors for NTDs since they may increase susceptibility to obesity or diabetes.
Volcik et al. (2003) reported an association between a UCP2 dual genotype and spina bifida in a case-control analysis. Infants homozygous for both the A55V (val/val) and the 3′UTR 45bp insertion/deletion polymorphism (del/del) in exon 8 had more than a three-fold higher risk for spina bifida. Given these and other results that implicate UCP2 variants in obesity, type-2 diabetes and gene expression, we evaluated three polymorphisms, −866G>A, A55V, and the 3′UTR 45bp insertion/deletion, as risk factors for NTDs using case-control and family-based analyses in an Irish population.
MATERIALS AND METHODS
Study Population
Study subjects were recruited and samples were obtained as previously described (Kirke et al., 1993). We elected to study roughly one-third of our cohort and replicate any positive associations in the remainder of the cohort. The primary sample set consisted of 146 NTD case families and 332 control individuals. Except for three NTD family pairs consisting of parents only, NTD case and mother, and NTD case and father, the NTD families all had a mother, father, and affected child triad. One family group had two NTD cases. An additional unrelated 22 NTD cases, 19 NTD mothers and 22 NTD fathers were also included in the analyses.
DNA Isolation
Genomic DNA was extracted from blood samples or buccal swabs using Qiagen Blood Mini Kits (Qiagen, Valencia, CA) or purified using Purgene (Gentra Systems. Minneapolis, MN).
Genotyping
The insertion polymorphism was genotyped using PCR as previously described (Lentes et al., 1999). The A55V (rs660339) and −866G>A (rs659366) variants were genotyped using matrix assisted laser desorption/ionization-time-of-flight (MALDI-TOF) mass spectrometry (Sequenom, San Diego, CA) of PCR products containing the polymorphism. Genotyping success rates ranged from 95-99% and averaged 97.0% across all samples. To ensure genotyping consistency 10% of samples were re-genotyped. Concordance rates were 100%, 94.6%, and 95.9% for the 3′UTR 45bp insertion/deletion polymorphism, −866G>A, and A55V, respectively.
For comparisons of allele, genotype, and haplotype frequencies, we also genotyped panels of Caucasian and African-American samples obtained from the Coriell Cell Repositories (Camden, NJ) at each of the three markers. We observed a 96.5% genotyping call rate and 96.2% concordance rate after regenotyping >5% of samples. Data from additional populations e.g., Yoruba from Ibadan, Nigeria, Japanese from Tokyo, Han Chinese from Beijing, Northern and Western Europeans from Utah (CEPH) were downloaded from HapMap (http://www.hapmap.org/).
Statistical Analysis
Samples with discordant genotypes or that displayed non-Mendelian inheritance were excluded from statistical analysis for that marker, while samples discordant or non-Mendelian for more than one marker were excluded from all analyses (<2.0% of genotypes excluded overall). Odds ratios with a 95% confidence interval were used to test single marker homozygous genotypes and the previously reported compound homozygous risk genotype (V/V, del/del) for an association with NTDs by comparing individual NTD groups with controls.
Additionally, a family-based test was performed for each marker. A log-linear model allowing for direct maternal genetic effects along with case genetic effects was used to test for association. For both cases and mothers, the model allows for a separate relative multiplicative risk due to one or two copies of the index allele, compared to no copies of the index allele. The model was fit by maximizing the likelihood of observed triads given the parental genes using SAS PROC GENMOD (Weinberg et al., 1998).
Three marker haplotypes (−866G>A, A55V, 3′UTR 45bp insertion/deletion) were estimated for all groups and subjected to χ2 analysis with the aid of Haploview (Barrett et al., 2005).
RESULTS
Genotype distributions and allele frequencies are presented in Table 1. Hardy-Weinberg equilibrium was observed for each variant across all groups except for the insertion polymorphism among controls (p= 0.04). Case-control comparisons of genotype frequencies between NTD cases or NTD mothers and controls did not reveal any association with NTDs (Table 2). Family-based tests of each UCP2 variant were also negative as log-linear modeling of case and maternal effects did not show significant association (data not shown). Haplotype frequencies also did not significantly differ between NTD cases and controls or between NTD mothers and controls (data not shown).
Table 1.
Genotype Distributions and Allele Frequencies in Controls and NTD Triads*
| Marker | Genotype/allele | Controls | Affected Children | NTD mothers | NTD fathers |
|---|---|---|---|---|---|
| −866G/A | n = 317 | n = 158 | n = 151 | n = 151 | |
| GG | 140 (0.44) | 63 (0.40) | 64 (0.42) | 61 (0.40) | |
| GA | 133 (0.42) | 74 (0.47) | 68 (0.45) | 62 (0.41) | |
| AA | 44 (0.15) | 21 (0.13) | 19 (0.13) | 28 (0.19) | |
| G | 413 (0.65) | 200 (0.63) | 196 (0.65) | 184 (0.61) | |
| A | 227 (0.35) | 116 (0.37) | 106 (0.35) | 118 (0.39) | |
| A55V | n = 321 | n = 156 | n = 151 | n = 156 | |
| AA | 110 (0.34) | 52 (0.33) | 50 (0.33) | 52 (0.33) | |
| AV | 169 (0.53) | 82 (0.53) | 77 (0.51) | 75 (0.48) | |
| VV | 42 (0.13) | 22 (0.14) | 24 (0.16) | 29 (0.19) | |
| A | 389 (0.61) | 186 (0.60) | 177 (0.59) | 179 (0.57) | |
| V | 253 (0.39) | 126 (0.40) | 125 (0.41) | 133 (0.43) | |
| 3′UTR 45bp insertion | n = 325 | n = 159 | n = 155 | n = 157 | |
| del/del | 177 (0.54) | 85 (0.53) | 81 (0.53) | 78 (0.50) | |
| del/ins | 135 (0.42) | 64 (0.40) | 60 (0.39) | 69 (0.44) | |
| ins/ins | 13 (0.04) | 10 (0.06) | 12 (0.08) | 10 (0.06) | |
| del | 489 (0.75) | 234 (0.74) | 222 (0.73) | 225 (0.72) | |
| ins | 161 (0.25) | 84 (0.26) | 84 (0.27) | 89 (0.28) | |
Genotype and allele frequencies are in parenthesis. Because of rounding, not all genotype and allele frequencies sum to 1.
Table 2.
Homozygous genotype frequency comparisons in NTD cases, NTD mothers and controls.
| Marker | Genotype | Frequency | OR | 95% CI | p-value |
|---|---|---|---|---|---|
| UCP2 −866G>A | Control AA | 0.15 | |||
| NTD Case AA | 0.13 | 0.89 | 0.51-1.55 | 0.681 | |
| NTD Mother AA | 0.13 | 0.84 | 0.47-1.48 | 0.540 | |
| UCP2 −866G>A | Control GG | 0.44 | |||
| NTD Case GG | 0.40 | 0.85 | 0.58—1.26 | 0.420 | |
| NTD Mother GG | 0.42 | 0.95 | 0.64-1.40 | 0.780 | |
| UCP2 A55V | Control VV | 0.13 | |||
| NTD Case VV | 0.14 | 1.26 | 0.73-2.16 | 0.412 | |
| NTD Mother VV | 0.16 | 1.09 | 0.63-1.90 | 0.760 | |
| UCP2 A55V | Control AA | 0.34 | |||
| NTD Case AA | 0.33 | 0.96 | 0.64-1.44 | 0.840 | |
| NTD Mother AA | 0.33 | 0.95 | 0.63-1.43 | 0.805 | |
| UCP2 3′UTR indel | Control ins/ins | 0.04 | |||
| NTD Case ins/ins | 0.05 | 1.61 | 0.69-3.76 | 0.270 | |
| NTD Mother ins/ins | 0.08 | 2.04 | 0.91-4.59 | 0.084 | |
| UCP2 3′UTR indel | Control del/del | 0.54 | |||
| NTD Case del/del | 0.53 | 0.96 | 0.66-1.41 | 0.835 | |
| NTD Mother del/del | 0.52 | 0.92 | 0.62-1.34 | 0.651 | |
| UCP2 A55V, 3′UTR indel | Control VV, del/del | 0.02 | |||
| NTD Case VV, del/del | 0.01 | 0.58 | 0.12-2.80 | 0.493 | |
| NTD Mother VV, del/del | 0.03 | 1.20 | 0.35-4.17 | 0.771 | |
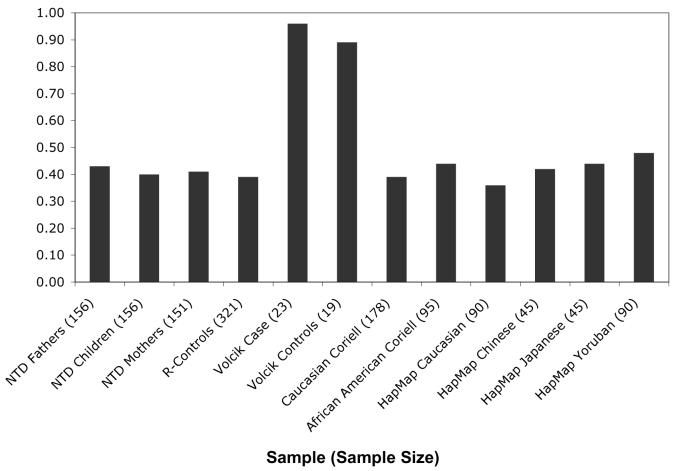
We also genotyped DNA samples from unaffected Caucasian and African Americans. UCP2 A55V allele frequencies in these groups were similar to those observed in the Irish population and consistent with the allele frequencies reported in HapMap (Figure 1). Allele frequencies were also similar for the other UCP2 markers; the −866G>A minor allele frequency ranged from 0.35-0.44 and the 3′UTR 45bp insertion/deletion minor allele frequency ranged from 0.18-0.27 in the Irish, Caucasian and African American populations.
Figure 1. UCP2 A55V Allele Frequencies Among Multiple Ethnic Groups.
T (Valine) allele distributions for UCP2 A55V (rs660339) observed in different populations. The frequencies for Volcik cases and controls were previously reported (2003). Genotyping data for this SNP in the HapMap populations is publicly available (ss69320500). The remaining allele distributions were determined in the current study.
DISCUSSION
Our evaluation of three UCP2 polymorphisms, −866G>A, A55V, and the 3′UTR 45bp insertion/deletion in exon 8, suggests common variants in this gene do not influence NTD risk in the Irish population. Allele and genotype frequencies were not statistically different between NTD mothers, affected children, and controls. These data contrast with those of Volcik et al. (2003), who detected an association between a compound homozygous UCP2 genotype and spina bifida among a population of 24 spina bifida case infants and 20 control infants. They observed a more than threefold increase in spina bifida risk for subjects who were homozygous for the 55VV and 3′UTR 45bp del/del genotypes (OR = 3.6, 95% CI = 1.0-13.1). We genotyped both of these variants in our cohort and we also genotyped a third variant in the promoter region (−866G>A) that has been shown to influence expression levels (Esterbauer et al., 2001; Krempler et al., 2002; Wang et al., 2004).
The frequency of the previously reported risk double homozygous genotype (55VV, 3/UTR 45bp del/del) was an order of magnitude smaller in our study compared to that reported by Volcik et al., and displayed no association with NTDs. Discrepancies between frequencies of this double homozygous genotype in the two studies might be attributed to ethnic stratification; unfortunately the ethnic background of their sample population was not specified. While this precludes comparing our results directly to that of Volcik et al. (2003), our finding of similar allele and haplotype frequencies in European and African American samples suggests this region exhibits little ethnic specific LD block structure stratification. It is likely that the allele frequencies observed by Volcik et al. (2003) are an artifact of a small sample size and not representative of any population.
As obesity is a major, and increasingly more important, risk factor for NTDs, UCP2 was a logical candidate to study for NTD risk. In this report, by far the largest of its kind to date, we found no relationship between NTDs and any of the variants examined in UCP2. However, we cannot rule out the possibility that these variants do influence NTD risk in other populations. It is also possible that one or more of these variants in conjunction with other variants increases risk for NTDs. As with most complex phenotypes, we expect that individual genetic variants will not have a powerful influence on NTD risk. More studies are needed to elucidate the function of UCP2 and the possible role variants in this gene may play in obesity, type-2 diabetes, and NTDs.
ACKNOWLEDGEMENTS
These studies would not be possible without the participation of the affected families, and their recruitment by the Irish Association of Spina Bifida and Hydrocephalus and the Irish Public Health Nurses. The authors acknowledge research support from the intramural research programs of the National Human Genome Research Institute, the National Institute of Child Health and Human Development and the Health Research Board, Ireland. FP was supported by a Pharmacology Research Associate Fellowship from the National Institute of General Medical Sciences, National Institutes of Health.
This research was supported by the intramural research programs of the National Human Genome Research Institute, the National Institute of Child Health and Human Development, and the Health Research Board, Ireland.
LITERATURE CITED
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Becerra JE, Khoury MJ, Cordero JF, Erickson JD. Diabetes mellitus during pregnancy and the risks for specific birth defects: a population-based case-control study. Pediatrics. 1990;85(1):1–9. [PubMed] [Google Scholar]
- Bulotta A, Ludovico O, Coco A, Di Paola R, Quattrone A, Carella M, Pellegrini F, Prudente S, Trischitta V. The common −866G/A polymorphism in the promoter region of the UCP-2 gene is associated with reduced risk of type 2 diabetes in Caucasians from Italy. J Clin Endocrinol Metab. 2005;90(2):1176–1180. doi: 10.1210/jc.2004-1072. [DOI] [PubMed] [Google Scholar]
- D'Adamo M, Perego L, Cardellini M, Marini MA, Frontoni S, Andreozzi F, Sciacqua A, Lauro D, Sbraccia P, Federici M, Paganelli M, Pontiroli AE, Lauro R, Perticone F, Folli F, Sesti G. The −866A/A genotype in the promoter of the human uncoupling protein 2 gene is associated with insulin resistance and increased risk of type 2 diabetes. Diabetes. 2004;53(7):1905–1910. doi: 10.2337/diabetes.53.7.1905. [DOI] [PubMed] [Google Scholar]
- Dalgaard LT, Andersen G, Larsen LH, Sorensen TI, Andersen T, Drivsholm T, Borch-Johnsen K, Fleckner J, Hansen T, Din N, Pedersen O. Mutational analysis of the UCP2 core promoter and relationships of variants with obesity. Obes Res. 2003;11(11):1420–1427. doi: 10.1038/oby.2003.191. [DOI] [PubMed] [Google Scholar]
- Esterbauer H, Schneitler C, Oberkofler H, Ebenbichler C, Paulweber B, Sandhofer F, Ladurner G, Hell E, Strosberg AD, Patsch JR, Krempler F, Patsch W. A common polymorphism in the promoter of UCP2 is associated with decreased risk of obesity in middle-aged humans. Nat Genet. 2001;28(2):178–183. doi: 10.1038/88911. [DOI] [PubMed] [Google Scholar]
- Gable DR, Stephens JW, Dhamrait SS, Hawe E, Humphries SE. European differences in the association between the UCP2 −866G > A common gene variant and markers of body mass and fasting plasma insulin. Diabetes Obes Metab. 2007;9(1):130–131. doi: 10.1111/j.1463-1326.2006.00573.x. [DOI] [PubMed] [Google Scholar]
- Humphries SE, Cooper JA, Talmud PJ, Miller GJ. Candidate gene genotypes, along with conventional risk factor assessment, improve estimation of coronary heart disease risk in healthy UK men. Clin Chem. 2007;53(1):8–16. doi: 10.1373/clinchem.2006.074591. [DOI] [PubMed] [Google Scholar]
- Janssen PA, Rothman I, Schwartz SM. Congenital malformations in newborns of women with established and gestational diabetes in Washington State, 1984-91. Paediatr Perinat Epidemiol. 1996;10(1):52–63. doi: 10.1111/j.1365-3016.1996.tb00026.x. [DOI] [PubMed] [Google Scholar]
- Kirke PN, Molloy AM, Daly LE, Burke H, Weir DG, Scott JM. Maternal plasma folate and vitamin B12 are independent risk factors for neural tube defects. Q J Med. 1993;86(11):703–708. [PubMed] [Google Scholar]
- Kovacs P, Ma L, Hanson RL, Franks P, Stumvoll M, Bogardus C, Baier LJ. Genetic variation in UCP2 (uncoupling protein-2) is associated with energy metabolism in Pima Indians. Diabetologia. 2005;48(11):2292–2295. doi: 10.1007/s00125-005-1934-9. [DOI] [PubMed] [Google Scholar]
- Krauss S, Zhang CY, Lowell BB. The mitochondrial uncoupling-protein homologues. Nat Rev Mol Cell Biol. 2005;6(3):248–261. doi: 10.1038/nrm1592. [DOI] [PubMed] [Google Scholar]
- Krempler F, Esterbauer H, Weitgasser R, Ebenbichler C, Patsch JR, Miller K, Xie M, Linnemayr V, Oberkofler H, Patsch W. A functional polymorphism in the promoter of UCP2 enhances obesity risk but reduces type 2 diabetes risk in obese middle-aged humans. Diabetes. 2002;51(11):3331–3335. doi: 10.2337/diabetes.51.11.3331. [DOI] [PubMed] [Google Scholar]
- Lentes KU, Tu N, Chen H, Winnikes U, Reinert I, Marmann G, Pirke KM. Genomic organization and mutational analysis of the human UCP2 gene, a prime candidate gene for human obesity. J Recept Signal Transduct Res. 1999;19(1-4):229–244. doi: 10.3109/10799899909036648. [DOI] [PubMed] [Google Scholar]
- Liu YJ, Liu PY, Long J, Lu Y, Elze L, Recker RR, Deng HW. Linkage and association analyses of the UCP3 gene with obesity phenotypes in Caucasian families. Physiol Genomics. 2005;22(2):197–203. doi: 10.1152/physiolgenomics.00031.2005. [DOI] [PubMed] [Google Scholar]
- Marti A, Corbalan MS, Forga L, Martinez-Gonzalez MA, Martinez JA. Higher obesity risk associated with the exon-8 insertion of the UCP2 gene in a Spanish case-control study. Nutrition. 2004;20(6):498–501. doi: 10.1016/j.nut.2004.03.019. [DOI] [PubMed] [Google Scholar]
- Ochoa MC, Santos JL, Azcona C, Moreno-Aliaga MJ, Martinez-Gonzalez MA, Martinez JA, Marti A. Association between obesity and insulin resistance with UCP2-UCP3 gene variants in Spanish children and adolescents. Mol Genet Metab. 2007;92(4):351–358. doi: 10.1016/j.ymgme.2007.07.011. [DOI] [PubMed] [Google Scholar]
- Otaegui D, Saenz A, Ruiz-Martinez J, Olaskoaga J, Lopez de Munain A. UCP2 and mitochondrial haplogroups as a multiple sclerosis risk factor. Mult Scler. 2007;13(4):454–458. doi: 10.1177/1352458506070454. [DOI] [PubMed] [Google Scholar]
- Rudofsky G, Jr., Schroedter A, Schlotterer A, Voron'ko OE, Schlimme M, Tafel J, Isermann BH, Humpert PM, Morcos M, Bierhaus A, Nawroth PP, Hamann A. Functional polymorphisms of UCP2 and UCP3 are associated with a reduced prevalence of diabetic neuropathy in patients with type 1 diabetes. Diabetes Care. 2006;29(1):89–94. doi: 10.2337/diacare.29.01.06.dc05-0757. [DOI] [PubMed] [Google Scholar]
- Scialli AR. Teratology public affairs committee position paper: maternal obesity and pregnancy. Birth Defects Res A Clin Mol Teratol. 2006;76(2):73–77. doi: 10.1002/bdra.20236. [DOI] [PubMed] [Google Scholar]
- Sesti G, Cardellini M, Marini MA, Frontoni S, D'Adamo M, Del Guerra S, Lauro D, De Nicolais P, Sbraccia P, Del Prato S, Gambardella S, Federici M, Marchetti P, Lauro R. A common polymorphism in the promoter of UCP2 contributes to the variation in insulin secretion in glucose-tolerant subjects. Diabetes. 2003;52(5):1280–1283. doi: 10.2337/diabetes.52.5.1280. [DOI] [PubMed] [Google Scholar]
- Shaw GM, Quach T, Nelson V, Carmichael SL, Schaffer DM, Selvin S, Yang W. Neural tube defects associated with maternal periconceptional dietary intake of simple sugars and glycemic index. Am J Clin Nutr. 2003;78(5):972–978. doi: 10.1093/ajcn/78.5.972. [DOI] [PubMed] [Google Scholar]
- Vogler S, Goedde R, Miterski B, Gold R, Kroner A, Koczan D, Zettl UK, Rieckmann P, Epplen JT, Ibrahim SM. Association of a common polymorphism in the promoter of UCP2 with susceptibility to multiple sclerosis. J Mol Med. 2005;83(10):806–811. doi: 10.1007/s00109-005-0661-5. [DOI] [PubMed] [Google Scholar]
- Volcik KA, Shaw GM, Zhu H, Lammer EJ, Finnell RH. Risk factors for neural tube defects: associations between uncoupling protein 2 polymorphisms and spina bifida. Birth Defects Res A Clin Mol Teratol. 2003;67(3):158–161. doi: 10.1002/bdra.10019. [DOI] [PubMed] [Google Scholar]
- Waller DK, Mills JL, Simpson JL, Cunningham GC, Conley MR, Lassman MR, Rhoads GG. Are obese women at higher risk for producing malformed offspring? Am J Obstet Gynecol. 1994;170(2):541–548. doi: 10.1016/s0002-9378(94)70224-1. [DOI] [PubMed] [Google Scholar]
- Wang H, Chu WS, Lu T, Hasstedt SJ, Kern PA, Elbein SC. Uncoupling protein-2 polymorphisms in type 2 diabetes, obesity, and insulin secretion. Am J Physiol Endocrinol Metab. 2004;286(1):E1–7. doi: 10.1152/ajpendo.00231.2003. [DOI] [PubMed] [Google Scholar]
- Weinberg CR, Wilcox AJ, Lie RT. A log-linear approach to case-parent-triad data: assessing effects of disease genes that act either directly or through maternal effects and that may be subject to parental imprinting. Am J Hum Genet. 1998;62(4):969–978. doi: 10.1086/301802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki H, Sasaki H, Ogawa K, Shono T, Tamura S, Doi A, Sasahara M, Kawashima H, Nakao T, Furuta H, Nishi M, Nanjo K. Uncoupling protein 2 promoter polymorphism −866G/A affects peripheral nerve dysfunction in Japanese type 2 diabetic patients. Diabetes Care. 2006;29(4):888–894. doi: 10.2337/diacare.29.04.06.dc05-1984. [DOI] [PubMed] [Google Scholar]
- Yanovski JA, Diament AL, Sovik KN, Nguyen TT, Li H, Sebring NG, Warden CH. Associations between uncoupling protein 2, body composition, and resting energy expenditure in lean and obese African American, white, and Asian children. Am J Clin Nutr. 2000;71(6):1405–1420. doi: 10.1093/ajcn/71.6.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuno K, Ando S, Misumi S, Makino S, Kulski JK, Muratake T, Kaneko N, Amagane H, Someya T, Inoko H, Suga H, Kanemoto K, Tamiya G. Synergistic association of mitochondrial uncoupling protein (UCP) genes with schizophrenia. Am J Med Genet B Neuropsychiatr Genet. 2007;144(2):250–253. doi: 10.1002/ajmg.b.30443. [DOI] [PubMed] [Google Scholar]
- Zhang CY, Baffy G, Perret P, Krauss S, Peroni O, Grujic D, Hagen T, Vidal-Puig AJ, Boss O, Kim YB, Zheng XX, Wheeler MB, Shulman GI, Chan CB, Lowell BB. Uncoupling protein-2 negatively regulates insulin secretion and is a major link between obesity, beta cell dysfunction, and type 2 diabetes. Cell. 2001;105(6):745–755. doi: 10.1016/s0092-8674(01)00378-6. [DOI] [PubMed] [Google Scholar]