Abstract
Ceramide is an important bioactive lipid, intimately involved in many cellular functions, including the regulation of cell death, and in cancer and chemotherapy. Ceramide is synthesized de novo from sphinganine and acyl CoA via a family of 6 ceramide synthase enzymes, each having a unique preference for different fatty acyl CoA substrates and a unique tissue distribution. However, little is known regarding the regulation of these important enzymes. In this study we focus on ceramide synthase 1 (CerS1) which is the most structurally and functionally distinct of the enzymes, and describe a regulatory mechanism that specifically controls the level of CerS1 via ubiquitination and proteasome dependent protein turnover. We show that both endogenous and ectopically expressed CerS1 have rapid basal turnover and that diverse stresses including chemotherapeutic drugs, UV light and DTT can induce CerS1 turnover. The turnover requires CerS1 activity and is regulated by the opposing actions of p38 MAP kinase and protein kinase C (PKC). p38 MAP kinase is a positive regulator of turnover, while PKC is a negative regulator of turnover. CerS1 is phosphorylated in vivo and activation of PKC increases the phosphorylation of the protein. This study reveals a novel and highly specific mechanism by which CerS1 protein levels are regulated and which directly impacts ceramide homeostasis.
1. Introduction
Ceramide is a major bioactive lipid in eukaryotic cells. In addition to its structural role as a membrane component [1], it is involved in a variety of cellular functions including the regulation of cell growth, differentiation and viability [2–4]. Ceramide homeostasis in turn is dependent on the de novo synthesis of ceramide from sphinganine and acyl CoA, which is catalyzed by a family of 6 ceramide synthases [5, 6]. These enzymes, named CerS1-6, are each the product of a different gene and preferentially use different fatty acyl CoA substrates containing fatty acid chains of different length, thereby producing ceramides with different acyl chains [7–11]. Thus, the regulation of these enzymes is of central importance to cell function.
Despite this rapid increase in our understanding of the enzymes and pathways involved with regulating ceramide homeostasis, we know relatively little about the distinct roles of each of these pathways in normal cell physiology and in pathology. Several studies have reported differential tissue expression patterns of the CerS genes [10–13] and it has been reported that certain ceramides have different roles in cancer and chemotherapy [14–16]. We previously showed that ectopic expression of the CerS1, CerS4 or CerS5 genes in human embryonic kidney cells had unique effects on controlling the sensitivity of the cells to different drugs used in cancer chemotherapy [17]. Thus, while CerS1 sensitized cells to a wide range of drugs including cisplatin, carboplatin, doxorubicin and vincristine, CerS5 only sensitized cells to doxorubicin and vincristine, and CerS4 did not affect sensitivity to any of the tested drugs. Paralleling these findings, it was shown that the specific effect of CerS1 was mediated through the activation of the MAP kinase p38 [17]. In other studies it was shown that the level of C18 ceramide is linked to head and neck cancer [18, 19], and CerS1 increased sensitivity to imatinib in cultured chronic myeloid leukemia cells [20].
Underlying our lack of understanding of the specific roles of the CerS enzymes is our lack of understanding of the levels of regulation of the individual enzymes. The differential tissue distribution suggests that there is a basic level of transcriptional control, but nothing is known about possible regulation by post-translational mechanisms, or about how drugs and other forms of stress affect these enzymes. In this report we demonstrate that CerS1 is a protein with a short half-life, and is turned over by ubiquitination and rapid proteasomal degradation. A wide variety of cellular stresses including drugs used in cancer chemotherapy, cause increased turnover of CerS1. Furthermore, we demonstrate that CerS1 turnover is regulated by the opposing functions of p38 MAP kinase and protein kinase C (PKC).
2. Materials and Methods
2.1 Materials
Cell culture reagents and Lipofectamine 2000 were from Invitrogen Corp., Carlsbad, CA. Fetal bovine serum (FBS) was from Atlanta Biologicals, Lawrenceville, GA. Restriction enzymes were from Promega Corp., Madison, WI. Monoclonal mouse anti-FLAG M2 antibody, polyclonal rabbit anti-FLAG antibody, cisplatin, doxorubicin, dithiothreitol (DTT), epoxomicin and lactacystin were from Sigma-Aldrich, St. Louis, MO. Protein A agarose beads were from Gibco, Basel, Switzerland. MG132 was from Calbiochem, San Diego, CA. 12-O-tetradecanoyl-phorbol-13 acetate (TPA) and anti-ubiquitin (P4D1) antibodies were from Cell Signaling, Danvers, MA. SB203580 was from BIOMOL, Plymouth Meeting, PA, and bis-indolylmaleimide (BIM) was from LC Laboratories, Woburn, MA. Goat anti-mouse and goat anti-rabbit horseradish peroxidase conjugated secondary antibody and chemiluminescent detection kits were from Pierce Biotechnology, Rockford, IL. Optitran BA-S 85 nitrocellulose membrane was from Whatman Schleicher and Schuell, Dassel, Germany.
2.2. Cell culture and transfection
Human embryonic kidney (HEK 293) cells were cultured in Dulbecco’s modified Eagle’s medium (MEM) supplemented with 10% fetal bovine serum, 2 mM L-glutamine, non-essential amino acids, 100 IU/ml penicillin and 100 µg/ml streptomycin in a 5% CO2 incubator at 37°C. Stable cell lines were generated by transfecting cells at 70–80% confluence in 6 well plates with 4 µg purified plasmid DNA using Lipofectamine 2000 according to manufacturer’s instructions. After 48 h the cells were transferred to a 75 cm2 culture flask and were selected in the presence of G418 sulfate, gradually increasing from 100 to 500 µg/ml [17]. G418 resistant cell lines were assayed for CerS1 expression by immunoblotting using monoclonal mouse anti-FLAG antibody, directed against the FLAG epitope. The cell lines were stored frozen in FBS containing 10% DMSO under liquid nitrogen. The human glioblastoma cell line U373 and the human lung cancer cell line A549 were used to establish the regulation of endogenous CerS1 protein.
2.3. Constructs
N-terminal 3xFLAG-tagged mouse CerS1 or CerS5 fusion proteins were generated by cloning the CerS1 or CerS5 gene into the mammalian expression vector pCMV-3tag-1A (Stratagene) with kanamycin selection. A mutant FLAG-CerS1 in which histidines 182 and 183 were replaced with alanines (H182A, H183A) was obtained by amplifying two separate fragments: a 5' fragment of the gene using forward primer (a) 5’- aaggatccatggctgctgccgcggc and reverse primer (b) 5’-gaggagcagggtgaccacCGCGGCcaccagcatgaccaccg-3’, and a 3' fragment of the gene, using forward primer (c) 5'-cggtggtcatgctggtgGCCGCGgtggtcaccctgctcctc-3’ and reverse primer (d) 5'-aaagaattcttatcagctagcgtaatctggaacatcgtatgggtagaagagcttgtccttcacc (mutation sites are in uppercase letters). The two fragments were then used as templates to amplify the full length mutated gene, using primers (a) and (d). The resultant PCR product was cloned as a BamHI-EcoRI fragment. The expression, enzyme activity and endoplasmic reticulum localization of the N-terminally tagged proteins were the same as previously reported [17].
2.4. Drug/stress treatment
Most experiments were performed with stable cell lines to minimize differences in expression levels between experiments. HEK 293 or HeLa cells expressing the FLAG-CerS1 or FLAG-CerS5 protein, U373 or A549 cells, at 90% confluence, were treated with the indicated amounts of drugs for the indicated times. Cisplatin and doxorubicin were prepared in Pt-buffer (3 mM NaCl/1 mM Na-PO4, pH 6.4), and DTT in water. Solvent alone was added to control untreated wells. For UV treatment, cells were exposed to UV-B for the indicated doses followed by 6 h incubation, prior to harvesting. UV fluence was measured using a Black Ray meter. The output of the lamp was determined to be 1.6 Joules/sec/ meter2, and the doses in the figures are indicated as Joules/meter2. In the case of drug (inhibitor/activator) co-treatment, cells were pre-treated with indicated amounts of the second drug for 1 h prior to the application of stress. The stresses are used at the lowest dose for which it is possible to see a cellular response in a reasonable amount of time and which is consistent with many published reports (see [21–23]. These cytotoxic agents all function in a dose- and time-dependent manner.
2.5. Inhibitors and activators
Inhibitors and activators were used at the following concentrations: SB203580, 2.5 µM; BIM, 2.5 µM; TPA, 100nM; MG132, 20 µM; epoxomicin, 1µM; and lactacystin, 10 µM. Control samples received an equivalent volume of the solvent.
2.6. Immunoblotting and immunoprecipitation
For immunoblotting, following drug/stress treatment the cells were quickly rinsed in ice-cold PBS and lysed by adding HEPES lysis buffer (20 mM HEPES buffer, pH 7.4, 25 mM KCl, 250 mM sucrose, 10 mM N-ethyl malemide (NEM), 1% protease inhibitor cocktail (2.5 mM PMSF, 0.01 µg/ml pepstatin A, 0.001 µg/ml leupeptin in methanol) containing 1% digitonin directly to the dish. The lysates were scraped from the plates, collected in 1.5 ml tubes and centrifuged at 12,000 rpm in a Sorvall desktop centrifuge for 1 min to remove insoluble material. Total protein in the supernatant was quantified using the Pierce BCA protein assay reagent according to manufacturer’s instructions. Protein concentrations between plates within an experiment were very close and varied by less that 5%. Equal amounts of protein were brought to the same volume and mixed with an equal volume of 2X urea sample buffer (50 mM Tris HCl pH 6.8, 1.6% SDS, 7% glycerol, 8 M urea, 4% β-mercaptoethanol, 0.016% bromophenol blue). Samples were separated by SDS-PAGE on 15% gels and transferred onto nitrocellulose membranes by standard techniques. CerS proteins were detected with primary antibody (see below) in 0.5% casein blocking buffer, and HRP conjugated secondary antibody, and blots were developed with a chemiluminescent detection kit. The blots were scanned at high resolution using an Epson Expression 1680 scanner. Ectopically expressed Flag-CerS1 or FLAG-CerS5 were detected with anti-FLAG mAb. Ubiquitinated CerS1 was detected with mouse anti-ubiquitin antibody (P4D1). Endogenous CerS1 was detected with anti-CerS1 rabbit antibody that was made against a synthetic peptide, encompassing amino acids 319–337 of the human CerS1 (DLREYDTAEAQSLKPSKAE), conjugated to KLH. Antibodies were obtained after an initial course of three injections, and subsequent boosts, and were subjected to ammonium sulfate enrichment and purification (see [24] for general methods for producing anti-peptide antibodies). The antibodies were tested against the cognate peptide in ELISA assays, and by immunoblotting lysates of HEK 293 cells expressing the recombinant CerS1 protein, and was specifically reactive in both cases. For loading control, blots were stripped in stripping buffer (62.5 mM Tris HCl pH 7.0, 2% SDS, 0.78% β-mercaptoethanol) at 60°C for 1 h and probed with mouse monoclonal anti-β tubulin antibody.
For immunoprecipitation of reduced and denatured protein, 3xFLAG-CerS1 overexpressing HEK293 cells, treated or untreated with either 50 µM cisplatin or 2.5 µM doxorubicin, in the presence or absence of pretreatment with 20 µM MG132, were lysed in a buffer containing 10 mM Tris (pH 8.0), 150 mM NaCl, 2% SDS and 10 mM NEM. The lysates were sonicated (20 pulses) and centrifuged at 10,000 rpm for 10 mins to remove insoluble material. The lysates were prepared for immunoprecipitation by dilution in 4 volumes of a buffer containing 10 mM Tris (pH 8.0), 150 mM NaCl and 1% Triton-X-100 (PBST). DTT was added to a final concentration of 1 mM. The lysates were precleared with protein A agarose beads at 4°C for 2 h, followed by incubation with rabbit anti-FLAG antibody overnight at 4°C. FLAG-CerS1-antibody complexes were pulled down by incubation with protein A agarose beads for 2 h at 4°C. The beads were washed once with 0.5 M LiCl and 3x with 0.1% Tween in PBS. Proteins were eluted from the beads by boiling for 5 min with urea sample buffer. The eluted proteins were separated by SDS-PAGE (15 % gel) and western blotting was done as described above with mouse anti-ubiquitin P4D1 antibody. All immunoprecipitation and/or immunoblotting experiments were done in their entirety multiple times (noted in each figure legend) and similar results were obtained.
2.7. In vivo phosphorylation
HEK 293 cells stably expressing N-terminal FLAG tagged CerS1 or vector control were cultured in 6-well plates, at exactly the same cell density. The culture medium was replaced with phosphate-free DMEM containing dialyzed FBS (Invitrogen, Carlsbad, CA) 1 h before adding 0.3 mCi [32P] orthophosphate (MP Biomedical, Irvine, CA; 285 Ci/mg) per well. After 30 minutes incubation with the labeled phosphate, 20 µM MG132 and/or 200 nM TPA in DMSO were added to the cells and incubated for 5 h. Three replicate wells of cells were used for each treatment. The cells were lysed directly in the wells in HEPES lysis buffer containing digitonin, protease inhibitor cocktail (described above) and phosphatase inhibitors (15 mM sodium fluoride, 1 mM orthovanadate). The lysates from the 3 replicate wells were pooled, and the lysates were spun at 13000 rpm at 4°C for 5 minutes and each of the supernatants were precleared with agarose beads. Each of the entire cleared supernatants were immunoprecipitated with mouse anti-FLAG mAb and protein A agarose. The beads are washed once with 0.5 M LiCl and 3x with 0.1% Tween in PBS. After washing, the immunoprecipitated proteins were eluted with 2x urea sample buffer and the entire eluate was separated by SDS-PAGE (15 %). The gel was dried under vacuum and scanned in a phosphorimager to detect phosphorylated CerS1.
2.8. Ceramide synthase assay
In vitro ceramide synthase assay was performed as described [17, 25]. Briefly, pellets were resuspended in 1 ml homogenization buffer and homogenized as described above, and protein concentration was determined using the Bradford reagent. Homogenates (50–250 µg protein in 0.25 ml final volume) were incubated with 0.25 µCi of [4,5-3H] sphinganine/15 µM sphinganine/20 µM defatted BSA, and 50 µM fatty acyl CoA (C16 for CerS5, and C18 for CerS1) for 20 min at 37°C. Lipids were extracted, and the level of dihydroceramide synthesis was analyzed by TLC, using chloroform/methanol/2 M ammonium hydroxide (40:10:1; v/v/v) as the developing solvent and using N-palmitoyl-sphingosine, and N-stearoyl-sphingosine as authentic standards. Lipids were visualized using a phosphorimager (Fuji), recovered from the TLC plates by scraping the silica directly into scintillation vials, and quantified by liquid scintillation counting. Specific enzyme activity is presented as picomoles of dihydroceramide synthesized per minute per milligram of protein.
3. Results
3.1. CerS1 protein undergoes turnover in response to a variety of stresses
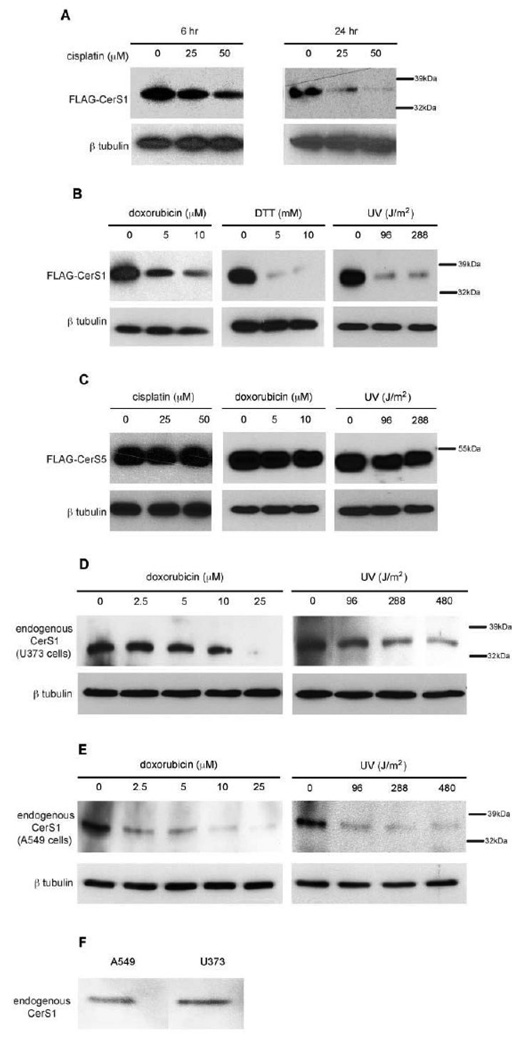
CerS1 is structurally and functionally distinct from the other CerS enzymes in terms of phylogeny, substrate specificity, tissue distribution, and cellular function including the response to stress (see [5] and Discussion). However, nothing is known about the regulation of this enzyme at the biochemical level. To explore this, we followed the fate of CerS1 in response to a variety of stress stimuli. Figure 1A shows that in HEK 293 cells stably expressing FLAG-CerS1, the level of FLAG-CerS1 protein decreases following exposure to cisplatin in a dose- and time-dependent manner. Moreover, this response was not unique for cisplatin since treatment of the cells with diverse stresses such as doxorubicin, dithiothreitol, or UV light resulted in a similar or greater reduction in CerS1 protein level (Figure 1B). In contrast, in HEK 293 cells stably expressing FLAG-CerS5, the level of FLAG-CerS5 remains constant after exposure to cisplatin, doxorubicin, or UV light (Figure 1C), and shows no stress-induced turnover. Turnover of endogenous CerS1 was demonstrated in the human glioblastoma cell line U373 and the human lung cancer cell line A549. Figure 1D shows that in the U373 glioblastoma cell line, following treatment with either doxorubicin or UV light, the endogenous CerS1 protein level goes down as well. Figure 1E shows the decrease in endogenous CerS1 in human A549 lung cancer cells following treatment with doxorubicin or UV light. This is interesting because there are mixed reports regarding CerS1 expression in lung cells [8, 11]. In this regard, Figure 1F shows a direct comparison (on the same gel) of equal amounts of U373 and A549 cells, and it can be seen that these cell lines express approximately equal levels of CerS1 protein. It should be noted that CerS1 is also expressed in head and neck carcinoma cells [18, 19]. Taken together, these experiments indicate that the level of both ectopically expressed and endogenous CerS1 protein is reduced in response to a wide variety of stresses.
FIGURE 1. Cytotoxic stresses result in down-regulation of CerS1 in a time and concentration dependent manner.
Western blot analysis of CerS protein levels after stress stimuli. Ectopically expressed FLAG-CerS1 or FLAG-CerS5 was detected with anti-FLAG monoclonal antibody (mAb), and endogenous CerS1 was detected with rabbit anti-CerS1 antibody. The blots were stripped and probed with an anti-β-tubulin antibody as a loading control (bottom panels). A) FLAG-CerS1 levels after 6 and 24 h treatments with the indicated concentrations of cisplatin. B) FLAG-CerS1 levels after 6 h treatment with indicated doses of doxorubicin (µM), DTT (mM) or UV-B light (Joules/meter2. C) FLAG-CerS5 levels after 6 h treatment with cisplatin, doxorubicin or UV light. D) Endogenous CerS1 in glioblastoma U373 cells after 6 h treatment with doxorubicin or UV-B light. E) Endogenous CerS1 in A549 lung cancer cells after 6 h treatment with doxorubicin or UV-B light. F) Direct comparison of CerS1 levels in U373 and A549 cells. Equal amounts of cell extract were separated on the same gel and probed with anti-CerS1 antibody. The experiments were repeated 3 times with similar results. Molecular weight markers are shown.
3.2. Stress induced CerS1 protein turnover is proteasome dependent
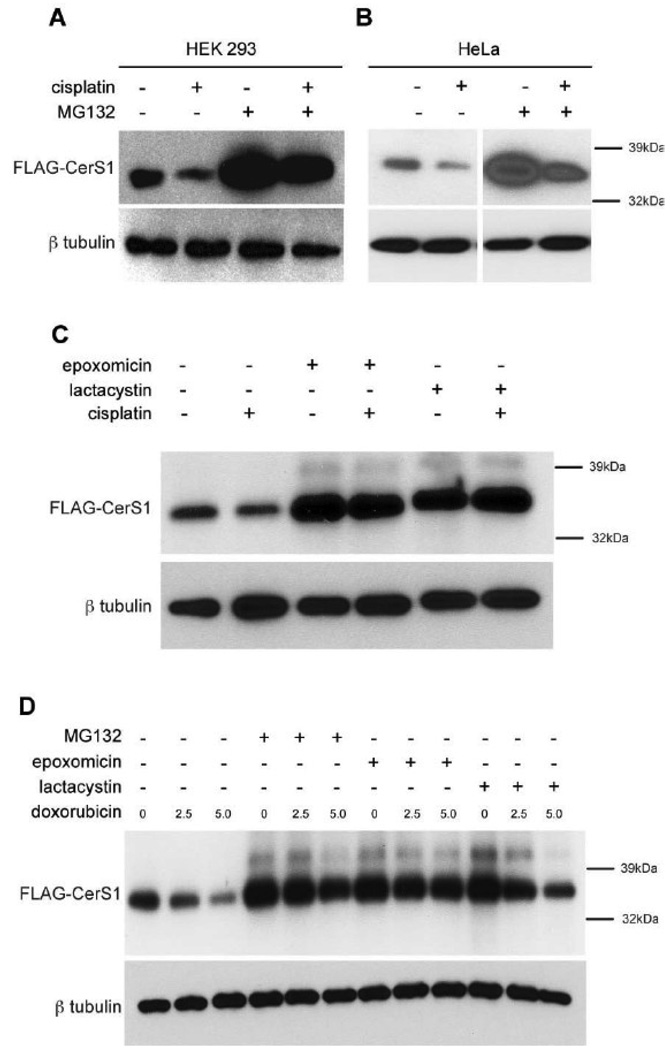
To investigate the nature of the stress induced degradation of CerS1, HEK 293 cells expressing FLAG-CerS1 were treated with the proteasomal inhibitor MG132 in the presence or absence of cisplatin (Figure 2A). As shown before, cisplatin induces turnover of CerS1 protein compared to untreated cells (lanes 1 and 2). MG132 causes a buildup of FLAG-CerS1 protein in untreated control cells (compare lanes 1 and 3), and substantially blocks the turnover of FLAG-CerS1 in cells treated with cisplatin (compare lanes 2 and 4). Quantitation of the blot shows that the MG132 inhibition of cisplatin induced turnover (lane 4) is 75% of that seen with MG132 alone (lane 3). 2B presents similar results when FLAG-CerS1 is transiently expressed in HeLa cells. Again MG132 substantially blocks the cisplatin-induced turnover of FLAG-CerS1 in HeLa cells, and shows that the effect is not cell-type specific. In addition, two other proteasome inhibitors, epoxomicin and lactacystin, block the cisplatin and doxorubicin induced turnover of FLAG-CerS1 stably expressed in HEK 293 cells (Figure 2C and D). These results indicate that under non-stress conditions, CerS1 is turned over by a proteasome dependent process, and that the increased turnover due to stress is also dependent on proteasome function.
FIGURE 2. Stress-mediated down-regulation of CerS1 is inhibited in the presence of the proteasome inhibitors.
Western blot analysis of ectopically expressed FLAG-CerS1 protein in A) stably transfected HEK 293 cells, or B) or transiently transfected HeLa cells, untreated or treated for 6 h with 50 µM cisplatin, with or without 1 h pre-treatment with 20 µM MG132 (note these samples were run on a single gel). C) Western blot analysis of ectopically expressed FLAG-CerS1 protein in stably transfected HEK 293 cells untreated or treated for 6 h with 50 µM cisplatin, with or without 1 h pre-treatment with epoxomicin or lactacystin. D) Western blot analysis of ectopically expressed FLAG-CerS1 protein in stably transfected HEK 293 cells untreated or treated for 6 h with 2.5 or 5 µM doxorubicin, with or without 1 h pre-treatment with MG132, epoxomicin or lactacystin. Bottom panels, β-tubulin control. The experiments were repeated 2 times with similar results.
3.3. CerS1 proteasomal degradation is ubiquitin mediated
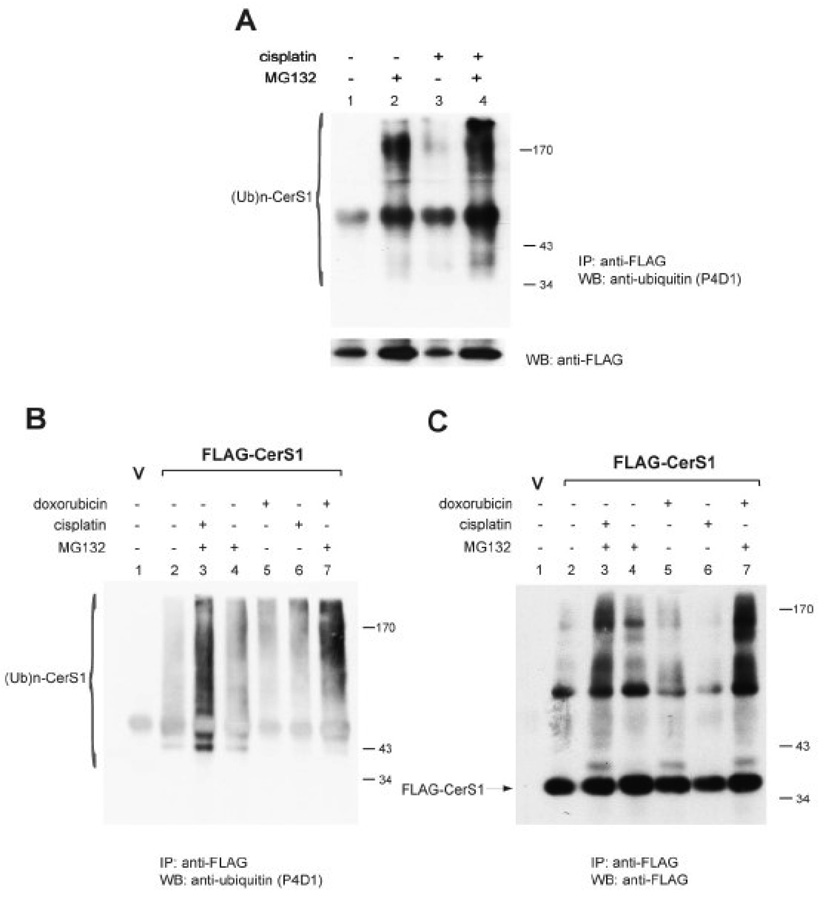
The ability of proteasome inhibitors to block CerS1 protein turnover suggested that ubiquitination is required. HEK 293 cells stably expressing FLAG-CerS1 were treated with cisplatin or buffer alone. Extracts were prepared using the standard non-denaturing buffer that is used to prepare samples for immunoblotting. The FLAG-CerS1 was immunoprecipitated using anti-FLAG antibody, and the immunoprecipitated proteins were separated by SDS-PAGE and analyzed by immunoblotting with anti-ubiquitin mAb or anti-FLAG mAb (Figure 3A). The data show that slowly migrating ubiquitinated protein accumulates in cells treated with the proteasomal inhibitor MG132 (compare lane 2 to lane 1) and that treatment with cisplatin increases the amount of the ubiquitinated protein (compare lanes 3 and 4 to lanes 1 and 2, respectively). Parallel immunoprecipitation from the same extracts with anti-FLAG mAb followed by immunoblotting with anti-FLAG mAb showed that the 39 kDa FLAG-CerS1 protein was indeed immunoprecipitated (lower panel of Figure 3A).
FIGURE 3. CerS1 degradation is ubiquitination-mediated.
HEK 293 cells stably transfected with FLAG-CerS1 (denoted FLAG-CerS1) were treated with 50 µM cisplatin or 2.5 µM doxorubicin for 6 h, with or without pretreatment with 20 µM MG132 for 1 h. Vector control cells (V) are HEK 293 cells stably transfected with empty vector. A) Cells were extracted with non-denaturing buffer containing Digitonin (Materials and Methods), and equal amounts of extracted protein were immunoprecipitated with anti-FLAG mAb and captured with protein A agarose beads. The immunoprecipitates were then subjected to immunoblot analysis, using mouse anti-ubiquitin mAb P4D1 (upper panel), or anti-FLAG mAb to show that the 39 kDa FLAG-CerS1 protein was indeed immunoprecipitated (lower panel). B and C) The cells were extracted under strongly denaturing conditions to avoid co-Immunoprecipitation proteins. Following Immunoprecipitation with anti-FLAG mAb and protein A agarose, the beads were washed in chaotropic 0.5 M LiCl to dissociate any further associated proteins (see Materials and Methods). The immunoprecipitates were then subjected to immunoblot blot analysis, using mouse anti-ubiquitin mAb P4D1 (B), or anti-FLAG mAb (C). The experiments were repeated 2 times with similar results. (Ub)n-CerS1 – ubiquitinated CerS1, IP - immunoprecipitation, WB - Western blot.
The foregoing experiment did not preclude the possibility that the ubiquitinated proteins were associated proteins that co-immunoprecipitated with CerS1. To eliminate this possibility, the experiment was re-performed where the cell extracts were prepared in denaturing and reducing conditions to avoid co-immunoprecipitation of associated proteins, and the immunoprecipitates were washed in strongly chaotropic buffer to dissociate any further associated proteins. Samples were prepared from both cisplatin and doxorubicin treated cells to demonstrate the generality of the effect. The immunoprecipitated CerS1 protein were separated by SDS-PAGE and analyzed by immunoblotting with anti-ubiquitin mAb (Figure 3B). Again, the data show clearly that either stress, cisplatin or doxorubicin, causes a significant increase in ubiquitinated CerS1 protein. Some slowly migrating ubiquitinated protein reacts with the anti-ubiquitin antibody in control cells (compare lane 2 to the vector control in lane 1). Cells stressed with either doxorubicin (lane 5) or cisplatin (lane 6) contain more of the slowly migrating ubiquitinated CerS1 proteins than the untreated cells (lane 2). Treatment with MG132 results in the accumulation of the slowly migrating ubiquitinated proteins (compare lane 4 to lane 2) and treatment with either cisplatin or doxorubicin significantly increases the level of ubiquitinated CerS1 protein (compare lanes 3 and 7 to lane 4). Parallel, anti-FLAG immunoprecipitations from the same extracts that were analyzed by immunoblotting with anti-FLAG (Figure 3C) show that the 39 kDa FLAG-CerS protein was immunoprecipitated, and that here too, the stresses result in a significant increase in the slowly migrating proteins (compare lanes 3 and 7 to lane 4). Taken together, these studies demonstrate a basal level of CerS1 ubiquitination in untreated cells and increased level in stressed cell.
3.4. CerS1 has a short half-life
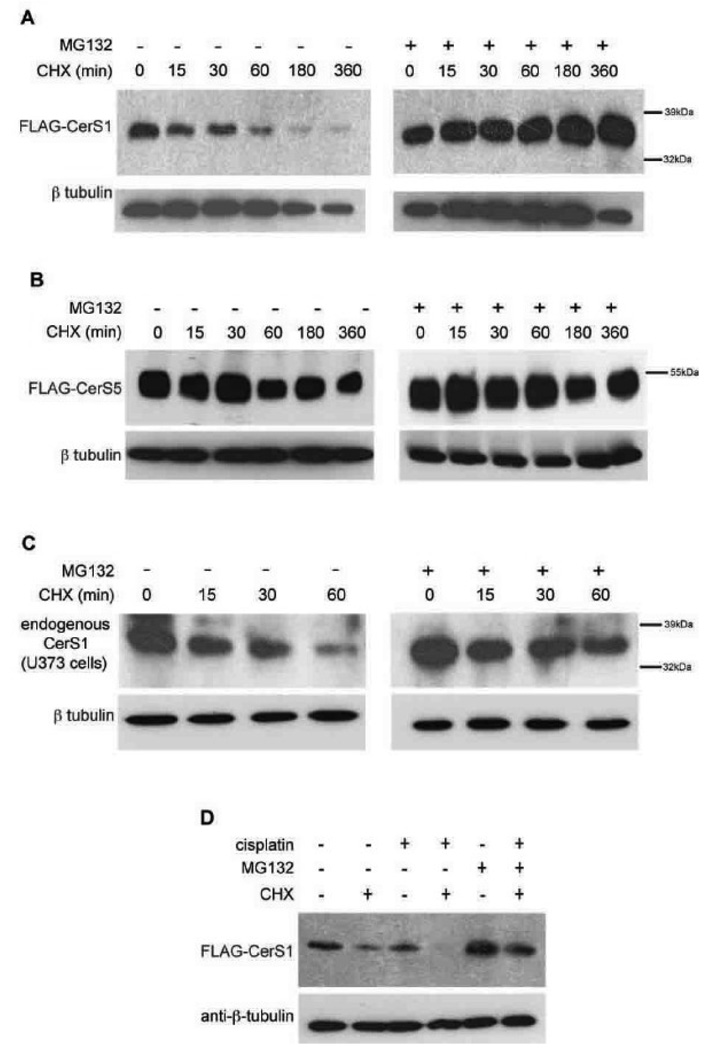
The foregoing data suggested that CerS1 protein rapidly turns over in response to stress, and this led us to ask about basal turnover. This was directly tested by monitoring the level of CerS1 protein in cells treated with cycloheximide to inhibit protein synthesis. Figure 4A shows that the 37 kDa FLAG-CerS1 stably expressed in HEK 293 cells rapidly turns over with a half-life of about 20–30 minutes and that MG132 substantially blocks this turnover. In contrast, Figure 4B shows that the level of the 50 kDa FLAG-CerS5 protein stably expressed in HEK 293 cells is stable following cycloheximide treatment, indicating that it is not being turned over. Treatment of human U373 glioblastoma cells with cycloheximide shows that the endogenous CerS1 is also turning over rapidly, and this turnover is blocked by MG132 (Figure 4C). The data in Figure 4D show that when HEK 293 cells expressing FLAG-CerS1 are treated with cisplatin in addition to cycloheximide, there is an increased level of turnover than in the presence of cycloheximide or cisplatin alone, and this is reversed by the proteasomal inhibitor MG132. These data demonstrate a high basal level of CerS1 protein turnover that can be increased with stress.
FIGURE 4. Basal turnover of CerS1 is rapid and inhibited by the proteasome-inhibitor MG132.
HEK 293 cells, stably transfected with FLAG-CerS1 (A) or FLAG-CerS5 (B) were treated with cycloheximide (CHX, 30 µg/ml) for the indicated time periods, in the presence or absence of 1 hour pre-treatment with 20 µM proteasome-inhibitor MG132. C) Glioblastoma U373 cells were treated with cycloheximide for the indicated times and endogenous CerS1 was detected. D) HEK 293 cells stably transfected with FLAG-CerS1 were treated with 30 µg/ml cycloheximide, with or without 50µM cisplatin for one hour. Addition of MG132 blocks proteasome mediated turnover. All experiments are Western analysis as described in Figure 1. The experiments were repeated 2 times with similar results.
3.5. CerS1 enzyme activity is required for its turnover
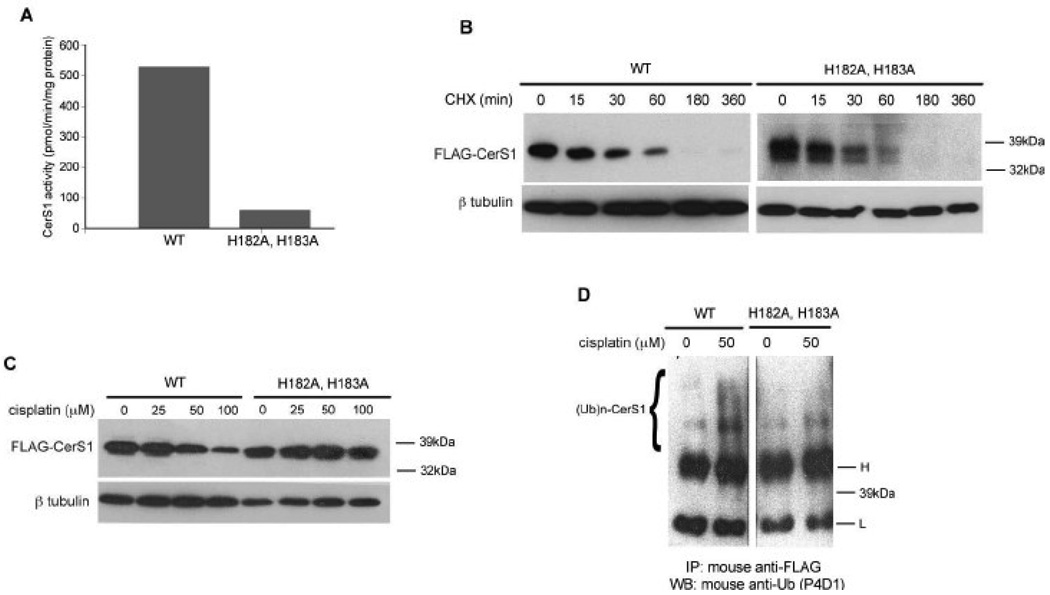
The current study has shown that upon stress there is a loss of CerS1 protein in the cells. Therefore, it was of interest to determine if the catalytic activity of CerS1 is required for its stress-induced degradation. CerS1, like all other CerS proteins, has a Lag1p motif which has been shown to be necessary for catalytic activity, and it has been shown that mutating the histidines at positions 182 and 183 to alanines results in a loss of almost all enzymatic activity [9]. This molecular genetic approach is more specific than using relatively non-specific inhibitors of ceramide synthesis such as fumonisin or myriocin which inhibit either all ceramide synthases or the upstream serine palmitoyl transferase, respectively, and the change of two amino acids is unlikely to alter the folding or membrane topology of CerS1 as previously show [7, 26]. Thus, to test whether the activity of CerS1 is necessary for stress induced degradation, we generated the H182A, H183A mutant, which showed greatly reduced activity in comparison to the parent protein (Figure 5A). We also determined the basal turnover rate of the catalytically inactive mutant protein in the presence of cycloheximide (Figure 5B). The mutant protein has a rapid basal turnover rate similar to the wild type (Figure 2), which indicates that it can turnover, and that basal turnover is not dependent on catalytic activity. In contrast, the mutant FLAG-CerS1, which lacks enzyme activity, shows no turnover (Figure 5C) in response to cisplatin even at very high concentrations while the wild-type protein is turned over. Figure 5D shows that ubiquitination of the inactive mutant FLAG-CerS1 enzyme does not increase much beyond the level seen in the absence of stress, in contrast to ubiquitination of wild-type FLAG-CerS1 which is increased in the cisplatin-treated cells. Taken together, these data indicate that CerS1 is mechanistically involved in regulation of its own level in response to stress.
FIGURE 5. Catalytically inactive CerS1 mutant does not undergo stress-induced degradation.
A) In vitro ceramide synthase enzyme activity of FLAG-CerS1 and mutant FLAG-CerS1( H182A,H183A). Equal amounts of protein extracts were assayed for in vitro activity, using 3H-sphinganine and C18 acyl CoA confirming published results [9]. B) Basal turnover of mutant FLAG-CerS1(H182A,H183A). HEK 293 cells stably expressing FLAG-CerS1 or FLAG-CerS1(H182A,H183A) were treated with cycloheximide and analyzed by Western analysis as described in Figure 1. C) The response of mutant FLAG-CerS1(H182A,H183A) to stress. HEK 293 cells stably expressing FLAG-CerS1 or FLAG-CerS1(H182A,H183A) were treated with indicated amounts of cisplatin for 6 h, and subjected to Western analysis. D) Ubiquitination of mutant FLAG-CerS1 is not increased following stress. Equal concentrations of extracts from HEK 293 cells stably expressing FLAG-CerS1 or FLAG-CerS1(H182A,H183A), treated with 50 µM cisplatin for 6 h, were immunoprecipitated with mouse anti-FLAG mAb. There is a clear increase in ubiquitination after cisplatin treatment of wild-type cells that is absent in the mutant. (Note that this analysis was done in the absence of MG132 which results in the stabilization of the ubiquitinated CerS1 (Figure 3)). Western blotting was done with mouse anti-ubiquitin (P4D1) antibody. H-immunoglobulin heavy chain; L-immunoglobulin light chain; (Ub)n-CerS1- ubiquitinated CerS1. The experiment was repeated 2 times with similar results.
3.6. The p38 MAP kinase is a positive regulator of CerS1 protein turnover
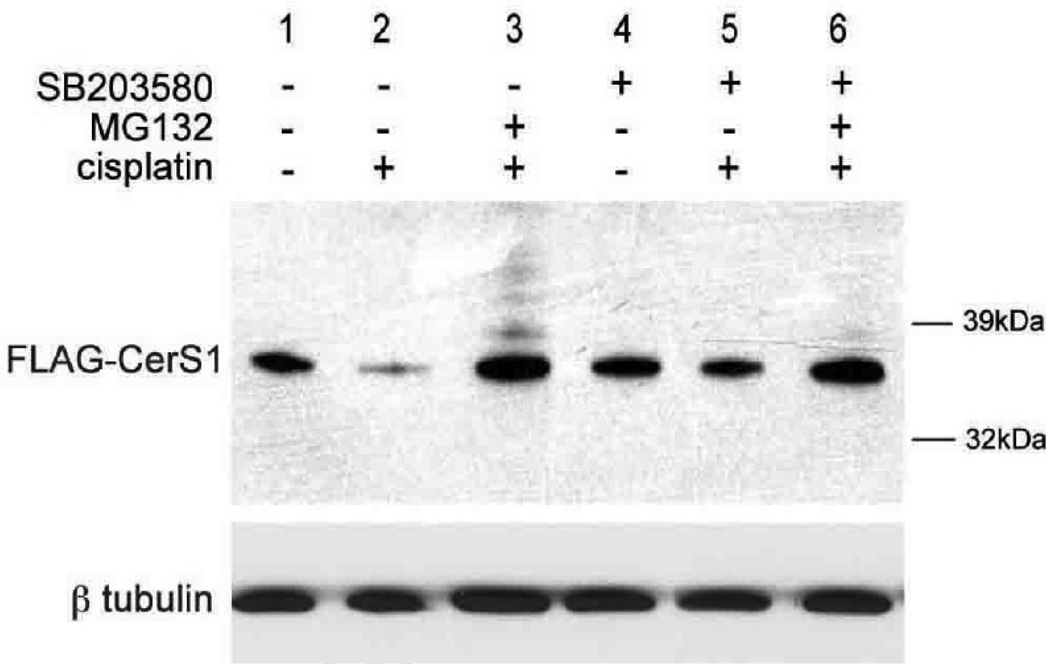
We previously showed that the p38 MAP kinase was an essential component of the signaling mechanism mediating how ceramide controls cellular sensitivity to drugs [17, 27, 28]. Moreover, it has been reported that p38 regulates the activities of ubiquitin ligases [29]. Together this suggested that the rapid turnover of CerS1 might reflect the action of p38 on turnover of the protein. To this end we used the specific p38 inhibitor SB203580 to see if the turnover of FLAG-CerS1 protein was also p38-dependent. The data in Figure 6 show that SB203580 has no effect itself on untreated control HEK 293 cells expressing FLAG-CerS1 (compare lanes 4 and 1). However, SB203580 inhibits the cisplatin induced turnover of FLAG-CerS1 (compare lanes 5 and 2). Treatment of the SB203580/cisplatin treated cells with the proteasomal inhibitor MG132 slightly increases the level of FLAG-CerS1 (compare lane 6 to lane 5) as it did in cells treated with cisplatin alone (lane 3). Thus, p38 has a novel role in positively regulating the level of CerS1 protein.
FIGURE 6. p38 MAP kinase is a positive regulator of CerS1 protein turnover.
Western blot showing the inhibition of cisplatin induced CerS1 protein turnover in HEK 293 cells stably expressing FLAG-CerS1 by the p38 MAP kinase inhibitor SB203580 (compare lane 3 to lane 2). The inhibitor (2.5 µM) was added one hour before the addition of cisplatin and the samples were harvested 6 hours later. Western blotting was as described in Figure 1. Addition of MG132 blocks turnover, confirming the results in Figure 2. The experiment was repeated 2 times with similar results.
3.7. Protein kinase C (PKC) is a negative regulator of CerS1 protein turnover
PKC is known to be involved in regulating the turnover of other proteins via phosphorylation [29–31], and examination of the sequence of the CerS1 protein indicates that there are several putative phosphorylation sites. Therefore, we looked at whether modulating PKC activity could in turn modulate the turnover of CerS1 protein.
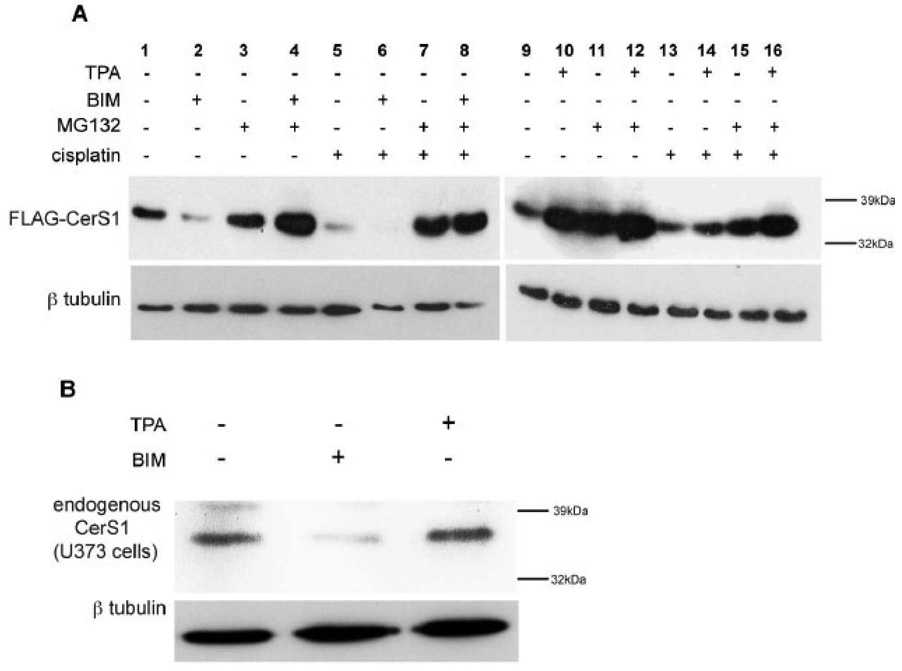
Figure 7A shows that when FLAG-CerS1 expressing cells are treated with the PKC inhibitor BIM, there is a dramatic increase in FLAG-CerS1 protein turnover (lane 2) compared to the untreated control cells (lane 1). Treatment with MG132 blocks the BIM induced turnover (lane 4). When BIM and cisplatin are added together (lane 6), there is greater FLAG-CerS1 turnover compared to BIM or cisplatin alone (lanes 2 or 5, respectively). MG132 reverses the effects of cisplatin alone (lane 7) or BIM and cisplatin together (lane 8).
FIGURE 7. PKC is a negative regulator of CerS1 protein turnover.
A) HEK 293 cells stably expressing FLAG-CerS1 were treated with the PKC inhibitor BIM, or the PKC activator TPA. BIM causes CerS1 turnover (compare lane 2 to lane1) and increases the cisplatin induced CerS1 turnover (lane 6). In contrast, TPA, inhibits the turnover of FLAG-CerS1 (compare lane 10 to lane 9) and reverses the cisplatin induced turnover of FLAG-CerS1 (lane 14). MG132 blocks the turnover of FLAG-CerS1 as shown in Figure 2. All treatments were done together and samples 1–8 and 9–16 were run on parallel gels, which were blotted and processed together. B) Human glioblastoma U737 cells were treated with BIM and TPA and endogenous CerS1 was detected. The experiments were repeated 2 times with similar results.
In contrast, when cells are treated with the PKC activator TPA, there is a decrease in FLAG-CerS1 turnover (compare lane 10 to lane 9). TPA reduced cisplatin-induced FLAG-CerS1 turnover (compare lane 14 to lane 13). TPA and MG132 together prevent cisplatin-induced FLAG-CerS1 turnover to a greater extent than TPA or MG132 alone (compare lane 16 to lanes 14 and 15, respectively). In the U373 glioblastoma cells, BIM induces turnover of endogenous CerS1 and TPA appears to have some stabilizing effect on endogenous CerS1 (Figure 7B). Therefore, activation or inhibition of PKC have opposite effects on the proteasomal mediated turnover of CerS1, and demonstrate a negative regulatory role for PKC on CerS1 protein turnover.
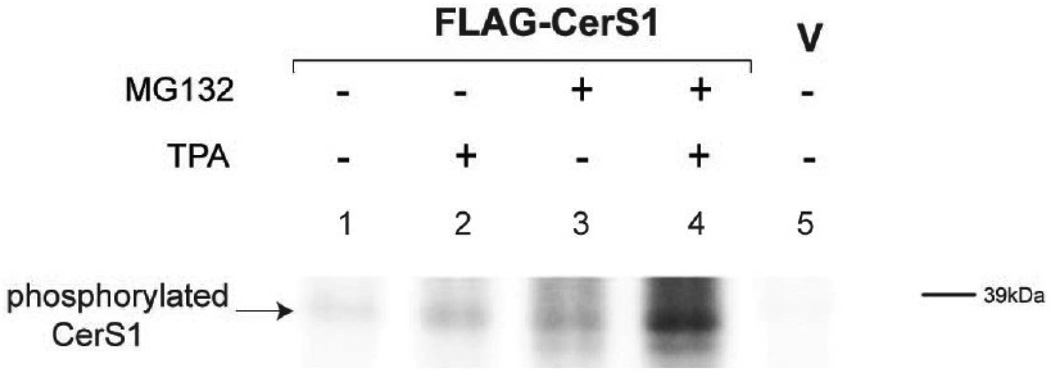
These data, and the presence of putative phosphorylation sites in CerS1, suggested that PKC acts on CerS1 by phosphorylation. We tested this by in vivo phosphorylation of HEK cells expressing FLAG-CerS1, by labeling cells with [32P] orthophosphate in the presence and absence of TPA and/or MG132. FLAG-CerS1 protein was immunoprecipitated with anti-FLAG mAb and separated by SDS-PAGE. The data in Figure 8 show that there is a basal level of phosphorylation of FLAG-CerS1 (lane 1), and TPA increases the level of phosphorylated FLAG-CerS1 (lane 2).
FIGURE 8. In vivo phosphorylation of CerS1.
HEK 293 cells stably expressing FLAG-CerS1 were metabolically labeled with [32P] orthophosphate (see Materials in Methods) in presence or absence of the PKC activator TPA and MG132 to prevent proteasome mediated turnover. The entire radiolabeled extracts were immunoprecipitated with anti-FLAG mAb. The immunoprecipitated labeled FLAG-CerS1 samples were electrophoresed on the same gel and the radioactivity was detected using a Fuji Phosphorimager. The experiments were repeated 2 times with similar results.
The proteasome inhibitor MG132 resulted in only a minor increase in phosphorylated FLAG-CerS1 due to the decreased protein turnover (lane 3), while the further addition of TPA (in addition to MG132) substantially increased the level of phosphorylated FLAG-CerS1 (lane 4) although TPA does not decrease turnover beyond the effect of MG132 alone. This is the first demonstration that CerS1 is phosphorylated. This supports the idea that TPA activation of PKC results in an increase in phosphorylation of FLAG-CerS1, rather than simply by the inhibition of CerS1 turnover.
4. Discussion
Six ceramide synthase enzymes are responsible for the de novo synthesis of all the ceramide in mammalian cells [5]. Although highly homologous, the CerS1 enzyme is structurally, catalytically, and functionally unique compared to the other CerS enzymes. CerS1 is phylogenetically distinct, lacks a HOX-like domain [5], preferentially uses C18 acyl CoA as a substrate [13], is expressed (mRNA) most abundantly in the brain and heart [10], increases sensitivity to specific chemotherapeutic agents [17, 20], and is associated with the alteration of C18 ceramide levels in carcinomas of the head and neck [18, 19]. However, nothing was known about the regulation of the CerS1 enzyme.
In this study we directly tested the idea that CerS1 is differentially regulated post-translationally, thereby providing a sensitive and rapid level of modulation beyond that controlled by transcription. The data show that both endogenous and ectopically expressed CerS1 are regulated at the level of protein turnover. We demonstrated that 1) the basal level of CerS1 is rapidly turned over by ubiquitination and proteasomal mediated degradation; 2) stresses including cisplatin, doxorubicin, DTT and UV light induce ubiquitin/proteasomal mediated CerS1 turnover; 3) CerS1 catalytic activity is necessary for turnover, thus identifying a novel negative feedback mechanism for CerS1 function; 4) the p38 MAP kinase, which has been previously shown to be specifically up-regulated in cells expressing CerS1, is also a positive regulator of the turnover of CerS1; 5) that PKC is a negative regulator of CerS1 turnover; and 6) that CerS1 is phosphorylated and that PKC activation increases CerS1 phosphorylation.
The stress-induced turnover of both endogenous and ectopically expressed CerS1 appears to be very general and independent of the type of stress. Thus, cytotoxic drugs, inducers of the unfolded protein response and UV-light, all of which operate by different mechanisms of action, ultimately result in CerS1 protein turnover, confirming the importance of this enzyme in regulating ceramide homeostasis. It is important to note in this context that all of these diverse cellular stresses have been associated with eliciting an increase in reactive oxygen species (ROS; [32], and future work should test whether ROS are mediators of CerS1 turnover. For example, inhibitors of ROS could be tested for their ability to abrogate stress induced CerS1 turnover. In this regard, it has been noted that changes in ROS appear to follow changes in cellular ceramide levels [33].
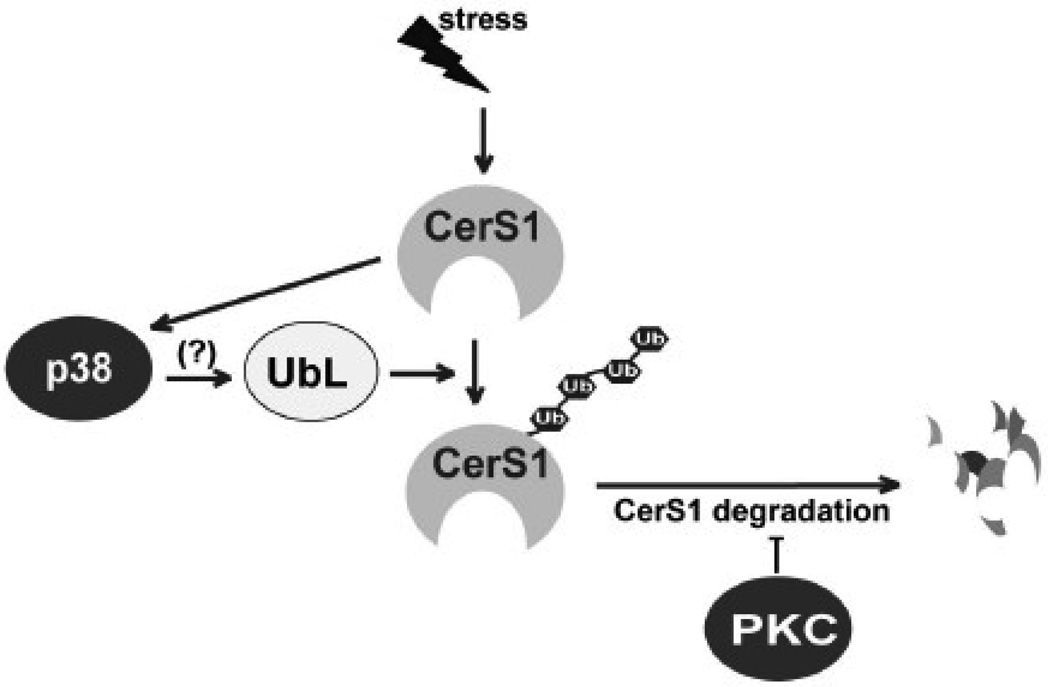
Figure 9 presents a model summarizing the results of this study. Both basal and stress-induced CerS1 turnover are mediated by its ubiquitination and proteasomal degradation. The p38 MAP kinase is a positive regulator of CerS1 protein turnover. We have previously shown that cisplatin or doxorubicin treatment of CerS1 expressing cells results in a robust activation of p38 enzyme activity [17], which is consistent with the increase in CerS1 protein turnover in stressed cells. Moreover, the demonstration that CerS1 catalytic activity is necessary for protein turnover in response to stress suggests the presence of an autoregulatory feed back loop involving ubiquitination of CerS1, but the precise molecular details will require additional investigation.
FIGURE 9. The regulation of CerS1 protein turnover.
Model showing the opposing roles for p38 MAP kinase and PKC in regulating the turnover of CerS1 in stressed and unstressed cells. Previous work showed that stress activates p38 MAP kinase in a CerS1 dependent manner [17]. There are multiple mechanisms by which PKC can be exerting its negative influence on CerS1 turnover, and they are discussed in the text. UbL-ubiquitin ligase.
The precise role of PKC in CerS1 turnover will require additional experimentation. There are multiple isoforms of PKC, which have been shown to regulate different cellular functions. Of the 8 isoforms, 5 (α,β,γ,δ,ε) are inhibited by BIM. Other inhibitors with different patterns of inhibition ultimately can be used to identify which of these is specifically involved in regulating CerS1 turnover. Mechanistically, PKC might be acting on CerS1 directly by specific phosphorylation, or indirectly via an intermediary signaling protein. Indeed, CerS1 has 5 putative serine or threonine phosphorylation sites that can be mutated to determine their role in CerS1 turnover. In vitro experiments testing the ability of PKC to phosphorylate CerS1 are needed to distinguish between these mechanisms. Nevertheless, this is the first demonstration that CerS1 is phosphorylated in vivo. Other studies have shown that PKC is a negative regulator of GMFβ, an enhancer of p38 MAP kinase [34]. In this context, inhibition of PKC could result in the upregulation of GMFβ, or another intermediate signaling molecule, which in turn would activate p38, subsequently causing the turnover of CerS1. PKC has also been shown to have an effect on p38 via ceramide generation through the activation of sphingomyelinase [35, 36], and it will be important to reconcile these studies with the current work on CerS1 turnover. Overall, these studies demonstrate a novel mechanism for the modulation of CerS1 enzyme levels in response to stress.
Acknowledgements
This work was supported by NIH grant GM53929 (SA/HA), Israel Science Foundation Grant 1404/07 (AHF), a U.S.-Israel Binational Science Foundation Grant (SA/AHF), and NIH Grant AT003899 (MH). SA acknowledges the support of a Morris Belkin Visiting Professorship at the Weizmann Institute of Science, and AHF is the Joseph Meyerhoff Professor of Biochemistry at the Weizmann Institute of Science. This work partially fulfills the requirements for the Ph.D. degree at the University of Missouri (PS). We thank the members of the Alexander and Futerman labs for support and discussions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.van Meer G, Voelker DR, Feigenson GW. Membrane lipids: Where they are and how they behave. Nat Rev Mol Cell Biol. 2008;9:112–124. doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hannun YA, Obeid LM. The ceramide-centric universe of lipid-mediated cell regulation: Stress encounters of the lipid kind. J. Biol. Chem. 2002;277:25847–25850. doi: 10.1074/jbc.R200008200. [DOI] [PubMed] [Google Scholar]
- 3.Hannun YA, Obeid LM. Principles of bioactive lipid signalling: Lessons from sphingolipids. Nat Rev Mol Cell Biol. 2008;9:139–150. doi: 10.1038/nrm2329. [DOI] [PubMed] [Google Scholar]
- 4.Morales A, Lee H, Goni FM, Kolesnick R, Fernandez-Checa JC. Sphingolipids and cell death. Apoptosis. 2007;12:923–939. doi: 10.1007/s10495-007-0721-0. [DOI] [PubMed] [Google Scholar]
- 5.Pewzner-Jung Y, Ben-Dor S, Futerman AH. When do lasses (longevity assurance genes) become cers (ceramide synthases)?: Insights into the regulation of ceramide synthesis. J Biol Chem. 2006;281:25001–25005. doi: 10.1074/jbc.R600010200. [DOI] [PubMed] [Google Scholar]
- 6.Teufel A, Maass T, Galle PR, Malik N. The longevity assurance homologue of yeast lag1 (lass) gene family (review) Int J Mol Med. 2009;23:135–140. [PubMed] [Google Scholar]
- 7.Lahiri S, Futerman AH. Lass5 is a bona fide dihydroceramide synthase that selectively utilizes palmitoyl-coa as acyl donor. J Biol Chem. 2005;280:33735–33738. doi: 10.1074/jbc.M506485200. [DOI] [PubMed] [Google Scholar]
- 8.Mizutani Y, Kihara A, Igarashi Y. Mammalian lass6 and its related family members regulate synthesis of specific ceramides. Biochem. J. 2005;390:263–271. doi: 10.1042/BJ20050291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Spassieva S, Seo JG, Jiang JC, Bielawski J, Alvarez-Vasquez F, Jazwinski SM, Hannun YA, Obeid LM. Necessary role for the lag1p motif in (dihydro)ceramide synthase activity. J Biol Chem. 2006;281:33931–33938. doi: 10.1074/jbc.M608092200. [DOI] [PubMed] [Google Scholar]
- 10.Laviad EL, Albee L, Pankova-Kholmyansky I, Epstein S, Park H, Merrill AH, Futerman AH., Jr Characterization of ceramide synthase 2: Tissue distribution, substrate specificity, and inhibition by sphingosine 1-phosphate. J Biol Chem. 2008;283:5677–5684. doi: 10.1074/jbc.M707386200. [DOI] [PubMed] [Google Scholar]
- 11.Riebeling C, Allegood JC, Wang E, Merrill AH, Futerman AH., Jr Two mammalian longevity assurance gene (lag1) family members, trh1 and trh4, regulate dihydroceramide synthesis using different fatty acyl-coa donors. J. Biol. Chem. 2003;278:43452–43459. doi: 10.1074/jbc.M307104200. [DOI] [PubMed] [Google Scholar]
- 12.Mizutani Y, Kihara A, Igarashi Y. Lass3 (longevity assurance homologue 3) is a mainly testis-specific (dihydro)ceramide synthase with relatively broad substrate specificity. Biochem J. 2006;398:531–538. doi: 10.1042/BJ20060379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Venkataraman K, Riebeling C, Bodennec J, Riezman H, Allegood JC, Sullards MC, Merrill AH, Futerman AH., Jr Upstream of growth and differentiation factor 1 (uog1), a mammalian homolog of the yeast longevity assurance gene 1 (lag1), regulates n-stearoyl-sphinganine (c18-(dihydro)ceramide) synthesis in a fumonisin B1-independent manner in mammalian cells. J. Biol. Chem. 2002;277:35642–35649. doi: 10.1074/jbc.M205211200. [DOI] [PubMed] [Google Scholar]
- 14.Huwiler A, Zangemeister-Wittke U. Targeting the conversion of ceramide to sphingosine 1-phosphate as a novel strategy for cancer therapy. Crit Rev Oncol Hematol. 2007;63:150–159. doi: 10.1016/j.critrevonc.2007.04.010. [DOI] [PubMed] [Google Scholar]
- 15.Modrak DE, Gold DV, Goldenberg DM. Sphingolipid targets in cancer therapy. Mol Cancer Ther. 2006;5:200–208. doi: 10.1158/1535-7163.MCT-05-0420. [DOI] [PubMed] [Google Scholar]
- 16.Saddoughi SA, Song P, Ogretmen B. Roles of bioactive sphingolipids in cancer biology and therapeutics Subcell. Biochem. 2008;49:413–440. doi: 10.1007/978-1-4020-8831-5_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Min J, Mesika A, Sivaguru M, Van Veldhoven PP, Alexander H, Futerman AH, Alexander S. (dihydro)ceramide synthase 1 regulated sensitivity to cisplatin is associated with the activation of p38 mitogen-activated protein kinase and is abrogated by sphingosine kinase 1. Mol Cancer Res. 2007;5:801–812. doi: 10.1158/1541-7786.MCR-07-0100. [DOI] [PubMed] [Google Scholar]
- 18.Koybasi S, Senkal CE, Sundararaj K, Spassieva S, Bielawski J, Osta W, Day TA, Jiang JC, Jazwinski SM, Hannun YA, Obeid LM, Ogretmen B. Defects in cell growth regulation by c18:0-ceramide and longevity assurance gene 1 in human head and neck squamous cell carcinomas. J. Biol. Chem. 2004;279:44311–44319. doi: 10.1074/jbc.M406920200. [DOI] [PubMed] [Google Scholar]
- 19.Senkal CE, Ponnusamy S, Rossi MJ, Bialewski J, Sinha D, Jiang JC, Jazwinski SM, Hannun YA, Ogretmen B. Role of human longevity assurance gene 1 and c18-ceramide in chemotherapy-induced cell death in human head and neck squamous cell carcinomas. Mol Cancer Ther. 2007;6:712–722. doi: 10.1158/1535-7163.MCT-06-0558. [DOI] [PubMed] [Google Scholar]
- 20.Baran Y, Salas A, Senkal CE, Gunduz U, Bielawski J, Obeid LM, Ogretmen B. Alterations of ceramide/sphingosine 1-phosphate rheostat involved in the regulation of resistance to imatinib-induced apoptosis in k562 human chronic myeloid leukemia cells. J Biol Chem. 2007;282:10922–10934. doi: 10.1074/jbc.M610157200. [DOI] [PubMed] [Google Scholar]
- 21.Wang X, Martindale JL, Holbrook NJ. Requirement for erk activation in cisplatin-induced apoptosis. J Biol Chem. 2000;275:39435–39443. doi: 10.1074/jbc.M004583200. [DOI] [PubMed] [Google Scholar]
- 22.Knudsen KE, Booth D, Naderi S, Sever-Chroneos Z, Fribourg AF, Hunton IC, Feramisco JR, Wang JY, Knudsen ES. Rb-dependent s-phase response to DNA damage. Mol Cell Biol. 2000;20:7751–7763. doi: 10.1128/mcb.20.20.7751-7763.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seger R. Map kinase signalling protocols. Humana Press; 2004. [Google Scholar]
- 24.Green N, Alexander H, Olson A, Alexander S, Shinnick TM, Sutcliffe JG, Lerner RA. Immunogenic structure of the influenza virus hemagglutinin. Cell. 1982;28:477–487. doi: 10.1016/0092-8674(82)90202-1. [DOI] [PubMed] [Google Scholar]
- 25.Lahiri S, Lee H, Mesicek J, Fuks Z, Haimovitz-Friedman A, Kolesnick RN, Futerman AH. Kinetic characterization of mammalian ceramide synthases: Determination of k(m) values towards sphinganine. FEBS Lett. 2007;581:5289–5294. doi: 10.1016/j.febslet.2007.10.018. [DOI] [PubMed] [Google Scholar]
- 26.Kageyama-Yahara N, Riezman H. Transmembrane topology of ceramide synthase in yeast. Biochem J. 2006;398:585–593. doi: 10.1042/BJ20060697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cuvillier O, Pirianov G, Kleuser B, Vanek PG, Coso OA, Gutkind S, Spiegel S. Suppression of ceramide-mediated programmed cell death by sphingosine-1-phosphate. Nature. 1996;381:800–803. doi: 10.1038/381800a0. [DOI] [PubMed] [Google Scholar]
- 28.Losa JH, Cobo CP, Viniegra JG, Sanchez-Arevalo Lobo VJ, Ramon y Cajal S, Sanchez-Prieto R. Role of the p38 mapk pathway in cisplatin-based therapy. Oncogene. 2003;22:3998–4006. doi: 10.1038/sj.onc.1206608. [DOI] [PubMed] [Google Scholar]
- 29.Ivanova IA, D'Souza SJ, Dagnino L. E2f1 stability is regulated by a novel-pkc/p38beta map kinase signaling pathway during keratinocyte differentiation. Oncogene. 2006;25:430–437. doi: 10.1038/sj.onc.1208999. [DOI] [PubMed] [Google Scholar]
- 30.Choi BH, Hur EM, Lee JH, Jun DJ, Kim KT. Protein kinase cdelta-mediated proteasomal degradation of map kinase phosphatase-1 contributes to glutamate-induced neuronal cell death. J Cell Sci. 2006;119:1329–1340. doi: 10.1242/jcs.02837. [DOI] [PubMed] [Google Scholar]
- 31.Shi RX, Ong CN, Shen HM. Protein kinase c inhibition and x-linked inhibitor of apoptosis protein degradation contribute to the sensitization effect of luteolin on tumor necrosis factor-related apoptosis-inducing ligand-induced apoptosis in cancer cells. Cancer Res. 2005;65:7815–7823. doi: 10.1158/0008-5472.CAN-04-3875. [DOI] [PubMed] [Google Scholar]
- 32.Conklin KA. Chemotherapy-associated oxidative stress: Impact on chemotherapeutic effectiveness. Integr Cancer Ther. 2004;3:294–300. doi: 10.1177/1534735404270335. [DOI] [PubMed] [Google Scholar]
- 33.Shi L, Bielawski J, Mu J, Dong H, Teng C, Zhang J, Yang X, Tomishige N, Hanada K, Hannun YA, Zuo J. Involvement of sphingoid bases in mediating reactive oxygen intermediate production and programmed cell death in arabidopsis. Cell Res. 2007;17:1030–1040. doi: 10.1038/cr.2007.100. [DOI] [PubMed] [Google Scholar]
- 34.Baldwin RM, Garratt-Lalonde M, Parolin DA, Krzyzanowski PM, Andrade MA, Lorimer A. Protection of glioblastoma cells from cisplatin cytotoxicity via protein kinase ciota-mediated attenuation of p38 map kinase signaling. Oncogene. 2006;25:2909–2919. doi: 10.1038/sj.onc.1209312. [DOI] [PubMed] [Google Scholar]
- 35.Kitatani K, Idkowiak-Baldys J, Bielawski J, Taha TA, Jenkins RW, Senkal CE, Ogretmen B, Obeid LM, Hannun YA. Protein kinase c-induced activation of a ceramide/protein phosphatase 1 pathway leading to dephosphorylation of p38 mapk. J Biol Chem. 2006;281:36793–36802. doi: 10.1074/jbc.M608137200. [DOI] [PubMed] [Google Scholar]
- 36.Zeidan YH, Wu BX, Jenkins RW, Obeid LM, Hannun YA. A novel role for protein kinase cdelta-mediated phosphorylation of acid sphingomyelinase in uv light-induced mitochondrial injury. FASEB J. 2008;22:183–193. doi: 10.1096/fj.07-8967com. [DOI] [PubMed] [Google Scholar]