Abstract
Single-stranded RNA viruses encompass broad classes of infectious agents and cause the common cold, cancer, AIDS, and other serious health threats. Viral replication is regulated at many levels, including using conserved genomic RNA structures. Most potential regulatory elements within viral RNA genomes are uncharacterized. Here we report the structure of an entire HIV-1 genome at single nucleotide resolution using SHAPE, a high-throughput RNA analysis technology. The genome encodes protein structure at two levels. In addition to the correspondence between RNA and protein primary sequences, a correlation exists between high levels of RNA structure and sequences that encode inter-domain loops in HIV proteins. This correlation suggests RNA structure modulates ribosome elongation to promote native protein folding. Some simple genome elements previously shown to be important, including the ribosomal gag-pol frameshift stem-loop, are components of larger RNA motifs. We also identify organizational principles for unstructured RNA regions. Highly used splice acceptors lie in unstructured motifs and hypervariable regions are sequestered from flanking genome regions by stable insulator helices. These results emphasize that the HIV-1 genome and, potentially, many coding RNAs are punctuated by numerous previously unrecognized regulatory motifs and that extensive RNA structure may constitute an additional level of the genetic code.
Genomes of all single stranded RNA viruses contain internal structures fundamental to viral replication and host defense evasion. Important viral RNA structures include internal ribosome entry sites, packaging signals, pseudoknots, tRNA mimics, ribosomal frameshift motifs, and cis-regulatory elements1,2. In the human immunodeficiency virus (HIV), RNA structures activate transcription, initiate reverse transcription, facilitate genomic dimerization, direct HIV packaging, manipulate reading frames, regulate RNA nuclear export, signal polyadenylation, and interact with viral and host proteins2–6. These RNA regulatory motifs have been identified by focusing on the 5′ and 3′ untranslated regions plus a few internal sequences. Most potential regulatory structures within viral RNA genomes, including in ~85% of the HIV-1 genome, are uncharacterized. This raises the possibility that new categories of RNA structure-mediated regulation remain to be identified.
The HIV-1 genome is primarily a coding RNA and contains nine open reading frames which produce 15 proteins2,3. The Gag polyprotein precursor is proteolytically processed to generate the matrix (MA), capsid (CA), nucleocapsid (NC), and p6 proteins. The Gag-Pol polyprotein adds protease (PR), reverse transcriptase (RT), and integrase (IN). The env gene encodes a 30 amino acid signal peptide (SP), gp120 and gp41. Additional sequences encode auxiliary proteins (gray boxes, Fig. 1a). Inside virions, HIV genomic RNA is found as a non-covalent dimer, is 5′ capped and 3′ polyadenylated, and is annealed to a host tRNALys3 molecule2. Viral proteins, especially nucleocapsid, chaperone folding of HIV RNA7.
Figure 1.
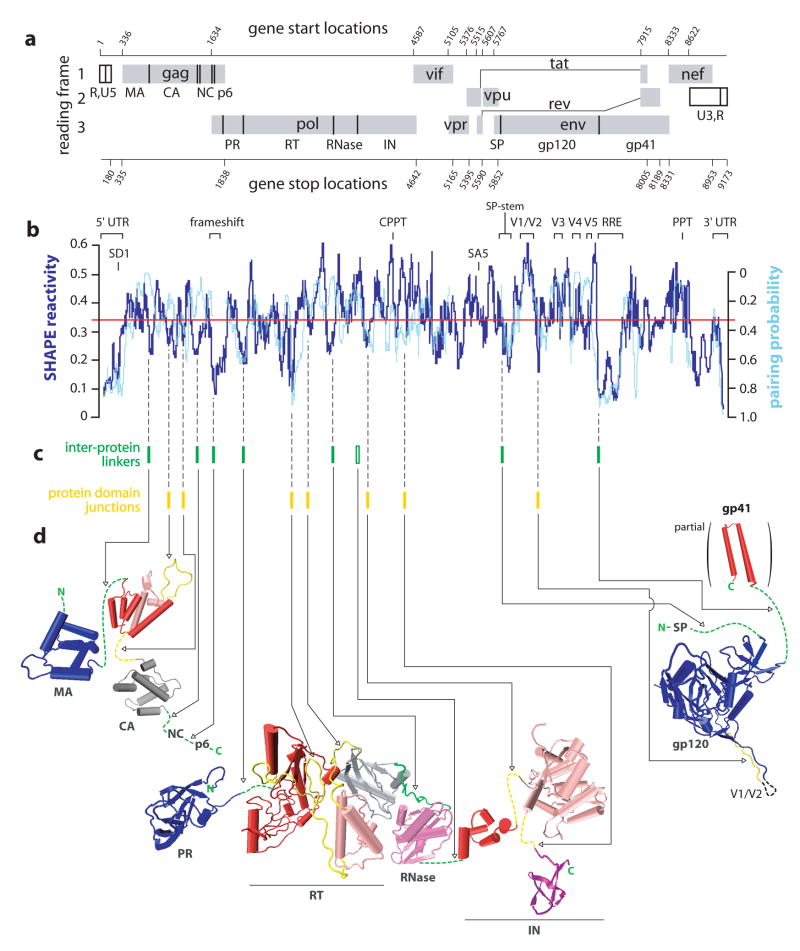
Organization, extent of RNA structure, and relationship to protein structure for an HIV-1 genome. (a) HIV-1 genome organization. Protein coding regions are shown as gray boxes; polyprotein domain junctions are depicted as solid vertical lines. Gene start and end sites are numbered according to NL4-3. (b) Comparison of median SHAPE reactivities (thick blue line) and evolutionary pairing probabilities (cyan line). Medians are calculated using a 75 nt window. The global median (= 0.34) is depicted as a red line. Pairing probability is not reported for regions encoding overlapping reading frames. (c) Inter-protein linkers in polyprotein precursors and the unstructured peptide loops that link protein domains are indicated with green and yellow bars, respectively. The single inter-protein linker that is not encoded by a region of highly structured RNA (at the RNase H-IN junction) is shown with an open green bar. (d) Comparison of domain structures for the large HIV proteins with the structure of the encoding RNA. Polyprotein linkers are green; inter-domain loops are yellow; folded protein domains are blue, red, light magenta, purple, and gray (Table S2).
Whole-genome structure analysis
To develop an accurate view of RNA structure in the full-length genome, we analyzed authentic genomic RNA extracted from HIV-1 virions. Our gentle purification maintained both previously characterized secondary structures and the few known RNA tertiary structures. For example, the host tRNALys3 was bound to the genome2 and a pseudoknot in the 5′ untranslated region (UTR)6,8 remained stably formed. The RNA was sufficiently intact to serve as a template for primer extension reactions spanning the entire genome (Table S1 and Supporting Methods).
High-throughput selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE)6,9–11 was used to chemically interrogate local nucleotide flexibility at 99.4% of the 9,173 nts in the NL4-3 HIV-1 RNA genome. 1-methyl-7-nitroisatoic anhydride (1M7) preferentially acylates conformationally flexible nucleotides at the ribose 2′-OH position9,10. The resulting 2′-O-adducts are detected as stops to primer extension using fluorescently labeled primers and capillary electrophoresis6,10 (Fig. 3a) and quantified by whole-trace Gaussian integration11 (Fig. 3b). SHAPE measurements are reproducible between independent biological replicates (R2 = 0.75; Fig. S1). SHAPE reactivities are highly sensitive to local nucleotide flexibility and disorder, but are insensitive to solvent accessibility (Fig. S2 and refs. 9,12). SHAPE reactivities therefore provide direct model-free information regarding the overall level of structure, or architecture, for any RNA. The median SHAPE reactivity varies dramatically across the HIV-1 genome (dark blue line, Fig. 1b). Regions with median reactivities below 0.25 indicate domains with significant base-paired secondary RNA structure while median SHAPE reactivities of 0.5 and greater indicate regions of largely unstructured nucleotides.
Figure 3.
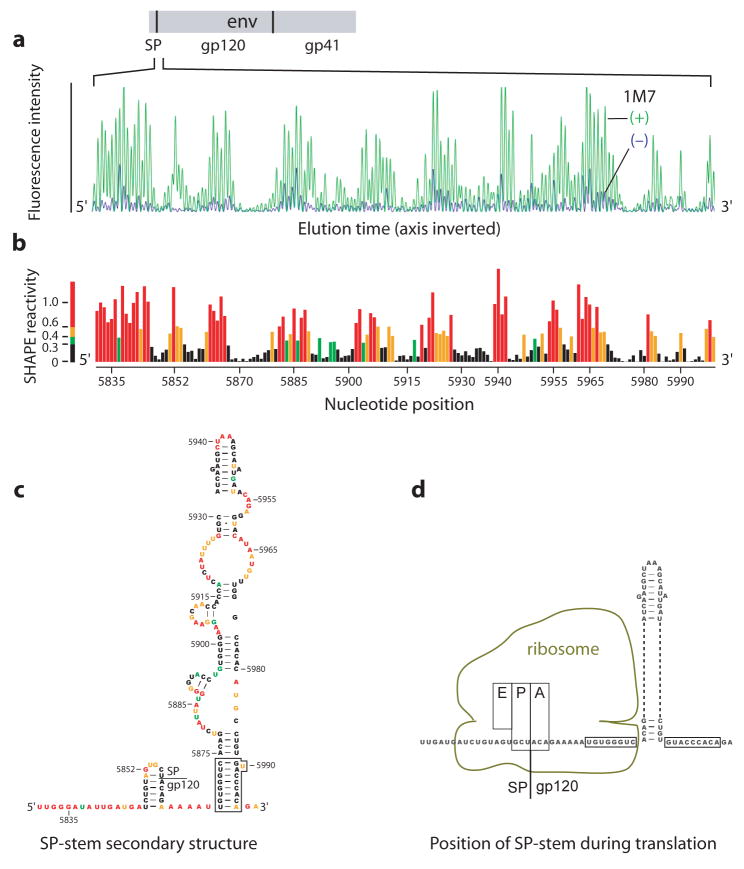
SHAPE analysis of the signal peptide (SP) gp120 region. (a) Processed capillary electrophoresis trace showing intensity versus position for the (+) and (−) reagent reactions. (b) Histogram of integrated and normalized SHAPE reactivities as a function of nucleotide position. The SHAPE reactivity scale shown here is used consistently throughout this work. (c) RNA secondary structure model for the SP pause site stem. (d) Location of the SP-stem relative to the eukaryotic ribosome at the pause site. Base pairs disrupted when the ribosome is at the pause site are boxed.
We also assessed HIV-1 genome structure by examining evolutionary information contained in nucleotide and amino acid variation to assign a pairing probability at each nucleotide13,14. This algorithm does not use chemical reactivity or thermodynamic information and thus infers RNA structure using information that is orthogonal to SHAPE.
We identify at least 10 “structured” regions that exhibit both low SHAPE reactivity and high pairing probability (compare blue and cyan traces, Fig. 1b). This group includes the 5′ UTR and the Rev Responsive Element (RRE), which are known HIV regulatory elements (labels, Fig. 1b). The majority of these highly structured and evolutionarily conserved elements have not been characterized previously. These regions include the PR-RT junction, domains in the RT, IN, and Vif open reading frames, an element 3′ of the Env signal peptide (SP), and the nef-3′ UTR region.
We also identify at least 7 “unstructured” regions, extending over 200–600 nts, with high SHAPE reactivities and low pairing probabilities. These include the RNase H coding domain, variable domains (Vx) in gp120, and the polypurine tract (PPT) (Fig 1b). On a smaller scale, the consensus sequences for the highly-used splice site acceptors are also unstructured (Fig. S3). There are ~4 regions of apparent disagreement in the level of RNA structure, having high pairing probabilities and high SHAPE reactivities (one each in the RT, RNase, IN and gp41 coding regions). This small group may reflect sequence conservation that is not accounted for by the evolutionary model13 or may form critical structures at an alternate stage of the viral replication cycle.
RNA structure encodes protein structure
We first evaluated whether global RNA genome structure is linked to protein structure. HIV-1 produces three major classes of mRNA. The 9 kb class encodes Gag and Gag-Pol and is identical to the packaged genomic RNA analyzed here except, as an mRNA, it is not dimerized at its 5′ end2. There are very few differences in the SHAPE reactivity of dimeric and monomeric RNAs at the 5′ end of the genome6. Thus, genome structures outside of the dimerization region will correlate closely to the mRNA that encodes Gag and Gag-Pol. The most abundant 4 kb env mRNA is generated by splicing nucleotide 288 (SD1, the major splice donor) to nucleotide 5522 (termed the SA5 site)15. SA5 is followed by an unstructured genome region (Figs. 1a, b). Thus, RNA structures identified in the env coding region are likely to exist in the spliced mRNA that encodes Env. Structures for the 1.8 kb class of mRNAs, which generate Tat and Rev, cannot be predicted using the genomic RNA because discontinuous segments are joined in the final mRNA.
The Gag, Gag-Pol and Env polyprotein precursors are synthesized roughly as beads on a string and the constituent proteins are liberated by proteolytic cleavage2,3 (Figs. 1a, d). Eight inter-protein peptides link the HIV proteins (green bars, Fig. 1c). The RNA sequences that encode these spacer peptide linkers in Gag (at the MA-CA, CA-NC, and NC-p6 junctions), Pol (PR-RT, RT-RNase H junctions) and Env (SP-gp120, gp120-gp41 junctions) all (except the RNase-IN junction) have SHAPE reactivities that are much lower than the median (Fig. 1b). RNA sequences that encode these inter-protein peptide linkers are more highly structured than 95.2% of randomly selected regions in the genome (Fig. S4a).
Domains within the individual HIV-1 proteins CA, RT, and IN are also linked by unstructured peptide elements and each domain junction is encoded by an RNA region of low SHAPE reactivity (compare yellow bars in Fig. 1c with dark blue trace in Fig. 1b). Protein loops encoded by RNA regions with low SHAPE reactivity include the cyclophilin loop and the linker between the N- and C-terminal domains in CA, both loops that link independently folded domains in RT, and the 8 and 9 amino acid loops linking the three domains in IN (in yellow, Fig. 1d). These protein domain junctions are more highly structured than 88.9% of randomly selected equivalent-length regions in the genome (Fig. S4b).
In contrast to the other large HIV proteins, domains in gp120 (termed inner, outer, and bridging sheet) are not structurally autonomous. The C-terminal 35 residues of gp120 weave from the outer to the inner domain and the bridging sheet is comprised of residues that are 315 positions distant16. Junctions between domains in gp120 are also not encoded by highly structured RNA, suggesting that gp120 folding is not linked to RNA structure in the same way as for other HIV proteins because its constituent domains are not structurally independent.
The recurring pattern of structure, conspicuously located near or after autonomously folding protein coding domains, is consistent with a model in which HIV protein structure is encoded in its RNA at two distinct levels. The first is the linear relationship between RNA and protein primary sequences. In the second level, higher-order RNA structure directly encodes protein tertiary structure because unstructured protein loops are derived from highly structured RNA elements. Many proteins appear to fold during translation17, highly structured RNA slows and causes ribosomal pausing during translation18,19, and changes in the extent of local RNA structure modulate protein activity20. Together, these observations suggest that attenuation of ribosome elongation by highly structured RNA at protein domain junctions facilitates native folding of HIV proteins by allowing time for domains to fold independently during translation.
This model makes the clear prediction that ribosome pause sites should occur preferentially in the highly structured regions of an HIV-1 RNA that encode protein junctions. We tested this idea using a toeprinting experiment, in which ribosome processivity is inhibited by cycloheximide and sites preferentially occupied by the ribosome are detected as stops to primer extension in an in vitro translation reaction21. Ribosome pause sites are statistically overrepresented at the MA-CA and CA-NC junctions in Gag and at the sequences encoding the cyclophilin loop in CA (Fig. S5). Conversely, ribosome pause sites are underrepresented in flanking, but unstructured, regions of the HIV RNA (p = 0.018). These experiments thus strongly support the model that mRNA structure over a region spanning 60–100 nucleotides specifically modulates ribosome processivity at protein domain junctions.
RNA secondary structure model for HIV-1
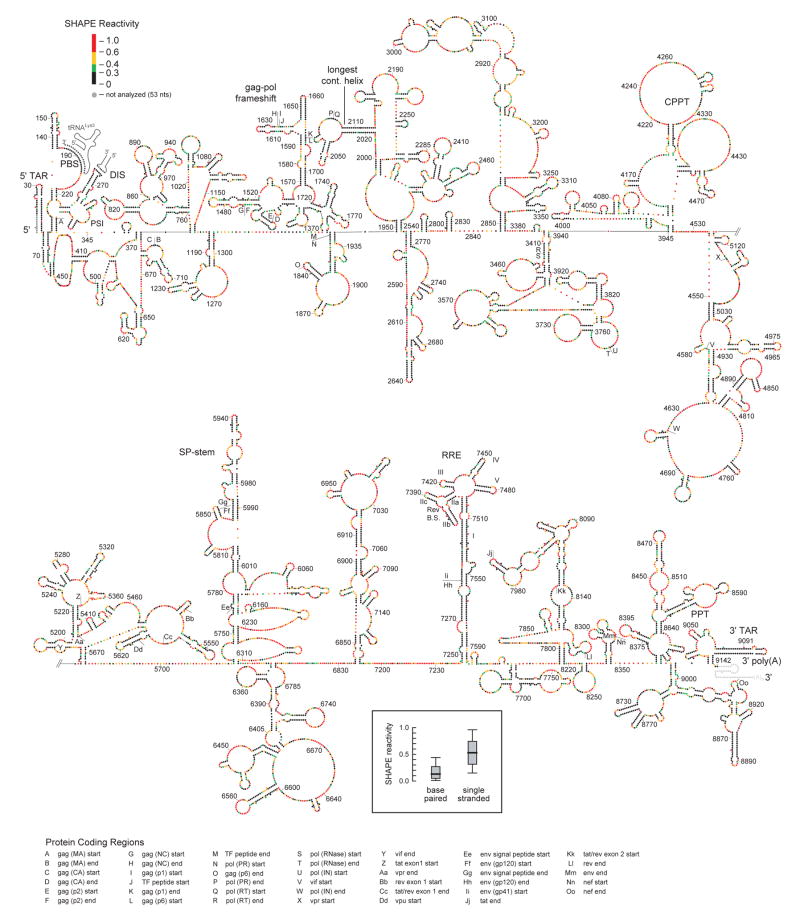
Comprehensive SHAPE reactivity information can also be used to determine a nucleotide-resolution secondary structure model for the entire NL4-3 HIV-1 genome (Fig. 2). SHAPE reactivities are converted to free energy change terms and used to constrain a thermodynamic folding algorithm22,23. The final result is a thermodynamically favored structural model highly reflective of the experimental SHAPE data, at single nucleotide resolution. For example, most nucleotides assigned to single-stranded regions are reactive towards SHAPE (red, orange, and green nucleotides; Fig. 2); whereas, base-paired nucleotides are predominantly unreactive (black nucleotides and inset; Fig. 2). (For a full discussion of SHAPE-directed RNA folding and the fundamental correctness of this model, see the Supporting Methods.)
Figure 2.
Structure of the HIV-1 NL4-3 genome. The 5′ and 3′ genome halves are shown in panels (a) and (b). Nucleotides are colored by their absolute SHAPE reactivities (see scale in panel a). Every nucleotide is shown explicitly as a sphere; base pairing is indicated by adjacent parallel orientation of the spheres. Protein domains are identified by letters. Important structural landmarks are labeled explicitly. Full nucleotide identities and pairings are provided in the supplementary information (Fig. S7). Intermolecular base pairs involving the tRNALys3 primer and the genomic dimer are shown in gray. Inset shows a box plot illustrating SHAPE reactivities for single stranded versus paired nucleotides in the model. Median reactivities are indicated by bold horizontal lines; the large box spans the central 50% of the reactivity data.
The HIV-1 genome is less structured than ribosomal RNA but, similarly, contains multiple independent RNA folding domains that extend from the overall genomic backbone. These domains include both small stem-loops plus roughly 21 large and complexly folded structures (Fig. 2). Although many genome regions are highly structured, only 7 helices span a complete turn of an 11 bp RNA duplex. The largest paired region, devoid of bulges, is the structured RNA element that bridges the coding junction between the RT and RNase H folding domains (Fig. 1). This helix is 19 bp long, contains a non-canonical G-A base pair (Fig. 2a, nts 2015–2033/2103–2121), and is thus shorter than the 30 base pair length competent to induce the interferon response24.
The HIV-1 genome structural model provides a robust starting point for identifying previously unrecognized functional elements and long-range RNA interactions. SHAPE reactivities describe a well formed stem 3′ to the signal peptide (SP) coding region in the Env protein (Fig. 3c). This stem (the SP-stem) is evolutionarily conserved (Fig. 1b), reinforcing an important biological role. The signal recognition particle (SRP) binds the nascent Env SP and translocates the cytoplasmic ribosome elongation complex to the rough endoplasmic reticulum where translation of gp120 and gp41 continue25.
RNA-induced translational pausing occurs as the ribosome unwinds highly structured RNA, typically located 6–7 nucleotides downstream of the A-site18. The SP-stem will be exactly in this conformation when the final tRNAAla from SP and the first tRNAThr of gp120 are in the P-and A-sites (boxed nucleotides, Fig. 3d). We infer that ribosomal attenuation or pausing at the SP-stem provides additional time for SRP recruitment and subsequent translocation of the elongation complex to the endoplasmic reticulum.
The SHAPE-constrained secondary structure is also informative for previously identified regulatory motifs. In HIV-1, pro and pol gene products are translated when the ribosome undergoes a –1 register shift from the gag to pol reading frames. Frameshifting occurs at a slippery sequence (U UUU UUA) and is enhanced by a downstream RNA structure. These elements are typically drawn as a single stranded slippery sequence and a 12 bp stem-loop26. Direct analysis of intact genomic RNA shows that the gag-pol frameshift signal is one component (identified here as P3) of a 3-helix structure (Figs. 2 & S6a). The slippery sequence pairs to form one of the three helices (P2). These two helices are stabilized by an anchoring helix (P1) that creates this discrete structural element (Fig. S6a). This three-helix junction structure is conserved among HIV-1 group M sequences (Fig. S6b).
Most RNA viruses require a complex pseudoknotted structure to induce ribosomal frameshifting27. The three-helix junction may function, in part, to slow translation before the ribosome encounters P3, facilitating the prerequisite pause necessary for frameshifting. The three-helix junction model may also explain why changing the slippery site to sequences which allow alternate tRNA pairing and enhance frameshifting in other RNA viruses eliminates frameshifting in HIV-128. In the SHAPE-directed model, changes to the slippery sequence compromise base pairing in the conserved P2 helix (Fig. S6).
Unstructured motifs and insulator helices
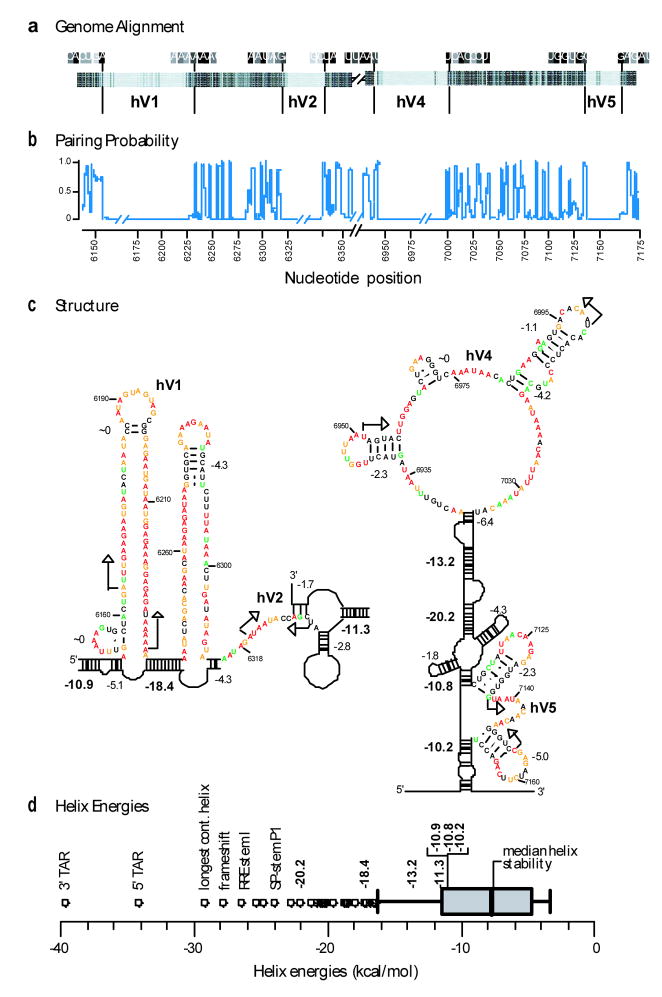
Analysis of the HIV-1 genome structure supports a role for RNA structures in sequestering unstructured regions. Five variable domains (V1–V5; see Fig. 1a,b) in the Env surface protein, gp120, account for much of the genetic diversity in HIV-114 and are a critical component of the viral host evasion strategy. Four of these domains are hypervariable (hV1, hV2, hV4, and hV5) and exhibit large amino acid insertions and deletions between viral isolates14.
Sequences encoding hypervariable domains are internally unstructured and are bordered by evolutionarily conserved and stable RNA structures (Figs. 4a,b). For example, hypervariable region hV1 is encoded by RNA sequences with high SHAPE reactivities and is flanked by two stable helices (with free energies of −10.9 and −18.4 kcal/mol, Fig. 4c). Similar patterns are evident in the other hypervariable regions (Fig. 4c). Some hypervariable regions, especially hV4, do contain internal helices with non-trivial free energies; however, these helices are not evolutionarily conserved (Fig. 4b) and are much less stable than the flanking helices whose stabilities are in the 10–20 kcal/mol range (Fig. 4c). These helices are also highly stable relative to the distribution of duplex stabilities over the entire genome (Fig. 4d).
Figure 4.
RNA structure in Env hypervariable regions. (a) Schematic sequence alignment for group M reference sequences14 at the Env hypervariable regions (hV1, hV2, hV4 & hV5). Nucleotides are represented as vertical bars; light gray and black indicate low versus universal conservation, respectively. (b) Evolutionary pairing probabilities. Breaks indicate extensive nucleotide insertions and deletions among the group M consensus sequences. (c) RNA structures at the hypervariable coding regions hV1, hV2, hV4, and hV5. Calculated free energies are shown for each helix (in kcal/mol); energies for anchoring helices proposed to function as structural insulators are emphasized in bold. (d) Distribution of helix stabilities in the HIV genome shown in a box blot representation. Whiskers illustrate 1.5 times the interquartile range and circles emphasize helices of exceptionally high stability. Free energy changes for proposed insulating helices are in bold; other significant helices are labeled.
Collectively, these data suggest RNA sequences encoding length polymorphisms in env are segregated from the rest of the genome by stable helices that function as structural insulators. The observed organization of hypervariable regions is thus well suited, first, to accommodate extensive substitutions, insertions or deletions and, second, to prevent these regions from forming non-functional base-pairing interactions with adjacent regulatory motifs, which include the 3′ splice site acceptors and the RRE.
Perspective
Structural analysis of a complete HIV-1 genome reveals that this RNA is punctuated by multiple, previously unrecognized but readily identifiable and evolutionarily conserved, RNA structures. Most genome regions with low SHAPE reactivities are associated with a regulatory function (Fig. 1). SHAPE may be generally useful for identifying new regulatory elements in large RNAs. All of these elements represent hypotheses and starting points that we hope will stimulate further detailed examination. Our discovery that the peptide loops that link independently folded protein domains are encoded by highly structured RNA indicates that these and likely other mRNAs encode protein structure at a second level beyond specifying the amino acid sequence. In this view, high-order RNA structure directly encodes protein structure, especially at domain junctions. The extraordinary density of information encoded in the structure of large RNA molecules (Figs. 1, 2 and 4d) represents another level of the genetic code, one about which we currently know the least. This work makes clear that there is much to be discovered via broad structural analyses of RNA genomes and intact messenger RNAs.
Methods Summary
Full length HIV-1 genomic RNA was gently purified from NL4-3 virions (Genbank AF324493). The RNA was equilibrated in a native buffer [50 mM Hepes (pH 8.0), 200 mM potassium acetate (pH 8.0), 3 mM MgCl2] at 37 °C for 15 min and treated with 1M710. Sites of 2′-hydroxyl modification were identified over read lengths spanning several hundred nucleotides using 31 primer extension reactions resolved by fluorescence-detected capillary electrophoresis6,11. Pairing probabilities were determined using RNA-Decoder13 and secondary structure models were developed by incorporating SHAPE reactivities as a pseudo-free energy change term, in conjunction with nearest-neighbor parameters, in an accurate thermodynamics-based prediction algorithm22,23.
Methods
Virus production
HIV-1 strain NL4-3 (group M, subtype B) was used to infect a non-Hodgkin’s T cell lymphoma cell line (a modified version of the SupT1 cell line, which was a gift from J. Hoxie, U. Pennsylvania)29. The virus-containing inoculum for infecting SupT1 cells was generated by CaPO4/DNA coprecipitation 30 and subsequent transfection of pNL43 (NIH AIDS Research and Reference Reagent Program, c/n 114; GenBank AF324493) into 293T cells31. HIV-1 virions were purified as described32 except cells were removed using a Millipore Opticap XL-5.0 micron filter. The total protein and CAp24 yields were 20.7 mg and 2.3 mg, based on total protein (BioRad DC protein assay) and HPLC with subsequent amino acid analysis assays, respectively.
Virions were purified from cellular debris by subtilisin treatment and centrifugation through a sucrose cushion. Concentrated virions (in 19 mL, corresponding to 19 liters of infected cell culture supernatant) were digested with subtilisin [1 mg/ml, in 20 mM Tris (pH 8.0), 1 mM CaC12; 37 °C, 18 hr; stopped by the addition of 5 μg/ml of phenylmethylsulfonyl fluoride33]. The resulting solution contained digested cellular proteins and viral particles free of surface proteins. The sample was centrifuged through a cushion of 20% (wt/vol) sucrose in phosphate buffered saline (PBS) (Beckman SW41 rotor, 37k rpm, 2.0 h, 4 °C); supernatant was carefully removed; and residual sucrose in the pellet was removed by overlaying PBS and repeating the centrifugation step (1 h at 4 °C).
RNA extraction
The key features of this protocol are that genomic RNA was gently extracted from purified virions in the presence of buffers containing monovalent and divalent ions consistent with maintaining RNA secondary and tertiary structure. The HIV genomic RNA was not denatured by heat, chemical denaturants, magnesium chelation, or removal of monovalent cations during this process. Subtilisin-treated virions were suspended in virion lysis buffer [VLB; 50 mM Hepes (pH 8.0), 200 mM NaCl, and 3 mM MgCl2] and lysed with 1% (wt/vol) SDS and 20 mg/ml proteinase K (~22 °C, 30 min). The digest was extracted three times with phenol/chloroform/isoamyl alcohol (25:24:1, pre-equilibrated with VLB) followed by two extractions with pure chloroform. Quantitative reverse transcriptase PCR was used to quantify viral RNA yields against a standard curve34–36. The total yield from 19 liters of infected cells was 97.2 pmol HIV-1 genomic RNA. The aqueous layer (3.6 mL) was brought to 300 mM NaCl and precipitated with 2.5 vol ethanol. Retroviral genomes commonly contain single stranded breaks2. Approximately 30% of our genomic RNA was intact, as judged by visualization in agarose/formaldehyde gels; nicks in the remaining 70% appeared to be roughly randomly distributed based both on direct visualization of the genomic RNA and from the continuity of our primer extension reactions (see Table S1).
RNA modification
The RNA pellet containing 97.2 pmols of HIV-1 genomic RNA was dissolved in 880 μL of modification buffer [MB; 50 mM Hepes (pH 8.0), 200 mM potassium acetate (pH 8.0), 3 mM MgCl2] and incubated at 37 °C for 15 min. 405 μL of the solution was added to either 45 μL pre-warmed (37 °C) 1M7 (1-methyl-7-nitroisatoic anhydride, in DMSO)10 or DMSO. After 4 min, 45 μL of 50 mM EDTA (pH 8.0) were added to each tube. The reactions were divided into 11 μL aliquots and precipitated with ethanol.
Primer synthesis
Primers were designed with the aid of OligoWalk, part of the RNAstructure software package22 (available for download at: http://rna.urmc.rochester.edu/) (Table S1). Primers were required to be 20–22 nts in length, have high melting temperatures and low self-annealing energies, and preferably end with a 3′ G or C. Only 2 of 31 primers required redesign, giving OligoWalk a 94% success rate. Primers were synthesized to contain a 5′ six carbon linker terminating in a primary amine (IDT). The amine-tethered DNA primers (1 μL; 25 μg/ml) were labeled with one of four fluorophores (5-FAM, 6-JOE, 6-TAMARA, 5-ROX; AnaSpec) using N-hydroxysuccinimide chemistry [3 μL NHS-coupled dye (20 mg/ml in DMSO) in 0.1 M NaBO3-HCl (pH 8.5); ~22 °C, 3 hr]. Labeled primers were precipitated with ethanol, purified on a denaturing gel (20% 29:1 acryalmide/bis-acrylamide, 7 M urea, 1× TBE), recovered by passive elution in water, precipitated (300 mM NaCl, 2.5 vol ethanol, 1 vol isopropanol), pelleted, and dissolved in water. Spectrophotometric measurements indicated labeling was ~90–95% efficient as determined by the [dye]/[DNA] ratio.
Primer extension
RNA pellets (1 pmol) were dissolved in 10 μL 0.5× TE [5 mM Tris (pH 8.0), 0.5 mM EDTA] and mixed with 3.0 μL of 0.4 μM primer. The (+) and (−) 1M7 reagent reactions were labeled with JOE and FAM, respectively. Primers were annealed to the RNA by heating to 65 °C for 5 min and 45 °C for 2 min, and then placed on ice. 6 μL of RT mix37 (SuperScript III, 5× buffer, DTT, dNTPs; Invitrogen) was added to each tube and incubated for 10 sec at 45 °C, 5 min at 52 °C, 5 min at 65 °C, and cooled to 4 °C. Sodium acetate (pH 5.2; 2.0 μL at 3 M) was added to each tube; (+) and (−) 1M7 tubes were combined; and 120 μL of ethanol was added to precipitate the cDNA products. The reactions were pelleted, washed with 70% ethanol, and dissolved in 10 μL deionized formamide.
Sequencing
Dideoxy sequencing reactions (GenomeLab Methods Development Kit; Beckman), were performed using plasmids pDR0 and pDR25 (containing partial NL4-3 sequences) and primers labeled with TAMARA and ROX. Primer sequences were identical to those in Table S1 except primer 31, whose sequence (5′-CTGCAACCTC TACCTCCTGG GTGCTAGAG) annealed to the plasmid rather than to the poly(A) RNA sequence in the genomic RNA.
Capillary electrophoresis
cDNA fragments were resolved by capillary electrophoresis6,10 (Applied Biosystems AB3130 instrument). Samples were injected at 1.2 kV for 16 sec into a 36 cm capillary containing POP7 (ABI) and subjected to electrophoresis for 25 min at 15 kV. The fluorescence detector was initially calibrated with 5-FAM, 6-JOE, 6-TAMARA, and 5-ROX using fluorescent markers with fragment lengths of 242 (5-FAM), 206 (6-JOE), 188 (6-TAMARA), and 155 (5-ROX) nts. Fragments were generated by linear amplification of Hind III-digested plasmid pUC18 using primers with the sequences 5′-CAGAGCAGAT TGTACTGAGA G; 5′-GTGAAATACC GCACAGATGC; 5′-GCGTAAGGAG AAAATACCGC ATC; and 5′-CGCCATTCAG GCTGCGCAAC TG labeled with 5-FAM, 6-JOE, 6-TAMARA and 5-ROX, respectively. Fluorescent spectral overlap based on this DNA ladder was calibrated using AB3130 software.
Data processing
Raw electropherograms, containing fluorescence intensity versus elution time information, were converted to normalized SHAPE reactivities using ShapeFinder6,11,23 (available for download at http://bioinfo.unc.edu). The ShapeFinder software aligns the (+) and (−) reagent traces to the two dideoxy nucleotide sequencing ladders, corrects for signal decay38, and performs a whole-channel Gaussian integration11 to quantify all individual peak areas (see Fig. 3a). Only 11 of the 9,173 nts in the NL4-3 RNA genome had high background and were therefore excluded from analysis. Datasets were normalized to a scale such that 1.0 represents the average intensity of highly reactive nucleotide positions6,23. On this scale, ~95% of integrated intensities for the HIV-1 genome fall between 0 and 1 (see histogram in Fig. 3b). Each primer extension reaction was processed individually. The resulting intensities in regions with overlapping data from different primers correlated closely: reactivity differences were typically less than 0.1 SHAPE unit. Regions with overlapping data accounted for ~25% of the total nucleotide positions and were averaged to generate the final dataset spanning the entire NL4-3 genome.
Toeprinting ribosome pause sites at the MA-CA and CA-NC junctions
A double-stranded DNA template to direct synthesis of an mRNA spanning NL4-3 Gag nts 1 to 1795 was generated by PCR. This region encompasses the entire 5′ UTR and most of the gag coding region and ends after the three-stem frameshift element. The 5′ primer included a T7 promoter sequence (5′ TAATACGACT CACTAATGGT CTCTCTGGTT AGACCA) and the 3′ primer (5′ GCTAAAGGTT ACAGTTCCTT GTC) encoded a stop codon at position 1787. The RNA transcript was capped and polyadenylated (mSCRIPT, Epicentre) and in vitro translation was carried out in rabbit reticulocyte extract (Ambion) using ~60 μg of the capped, polyadenylated transcript, 1 μl 1.25 mM L-methionine, 1 μL [32S]-methionine (PerkinElmer), 17 μl reticulocyte extract, and 1.25 μl 20× ’medium-salt’ translation buffer (Ambion) in a total volume of 26 μL at 37 °C. Cycloheximide was added at 0, 7, 15, or 45 min to arrest translation21. Translation reaction aliquots were separated on an 8–16% SDS-PAGE gel (Invitrogen) to confirm production of a protein of the correct length. Sites of ribosome pausing were detected by adding the following to 25 μL of the in vitro translation mixture: 1.35 μl 10 mM each dNTP, 2 μl 4.0 μM fluorescently-labeled primer (primer 4 or 6 for interrogating the MA-CA and CA-NC regions, respectively), 1 μl 200 mM MgCl2, and 2 μl Superscript III (Invitrogen). The translation reaction that was pre-quenched with cycloheximide served as a background and was resolved using a JOE-labeled primer. The 7, 15, and 45 min time points were resolved using FAM-labeled primers. Primer extension reactions were incubated at 37 °C for 30 min and stopped by addition of 1 μl 0.5 M EDTA and 400 μl water. The reaction was extracted with phenol:chloroform:isoamyl alcohol (25:24:1, 2×) and chloroform (1×). 4 μl of this solution, 1 μl of a cDNA sequencing ladder, and 15 μl of formamide were combined, heated to 105 °C for 5 min, and resolved by capillary electrophoresis. Toeprinting traces were processed with ShapeFinder11 and normalized to a scale in which 1.0 is equal to the mean intensity of the most reactive positions, identically as described above for SHAPE experiments.
RNA secondary structure model
The entire NL4-3 sequence – 9,173 nts plus 20 3′ adenosines [representing the poly(A) tail] – was folded using the thermodynamics-based algorithm in RNAstructure22,23. SHAPE information was used to constrain secondary structure calculations by incorporating SHAPE reactivities as pseudo free-energy change terms6,23 using slope and intercept values of 30 and −6 respectively. The maximum distance allowed between any two paired positions was 600 nucleotides. The slope and intercept values are derived from prior parameterization on long RNAs and the 600 nt cutoff reflects that 99% of all base pairs in ribosomal RNA occur between nucleotides less than this distance apart23. The genome was initially folded as a single (9,193 nts) unit; folding was then fine tuned by dividing the RNA into five independent folding regions, separated by long stretches of reactive nucleotides that were calculated to be unpaired when the entire genome was folded with SHAPE constraints (NL4-3 residues 1-2844, 2836-5722, 5676-6832, 6807-7791, and 7779-9193). Dividing the genome in this way facilitated model building and prevented formation of a few poorly supported long-distance pairings between domains. Highly reactive nucleotides at the termini of each region were prohibited from forming base pairs in these region-specific calculations. Helices consisting of a single base pair were removed from the final model and unreactive nucleotides in the primer binding site (183–199) were taken to reflect hybridization with the tRNA primer. The current version of our algorithm does not allow pseudoknots and therefore our HIV-1 structure model (Fig. 2) includes only one, heuristically predicted6,8, pseudoknot.
Quality of SHAPE-directed model of the entire HIV-1 genome
The algorithm by which SHAPE information is used to create an RNA secondary structure model does not make any specific assumptions regarding the magnitude of SHAPE reactivity that corresponds to single stranded versus based paired regions. Instead, SHAPE reactivities are converted to free energy change terms and used to constrain a thermodynamic folding algorithm22,23. SHAPE information is essential for generating this secondary structure model. Folding the genome by free energy minimization alone, using a best-of-class algorithm22,39, results in a structure that is very different from the experimentally supported model. Only 47% of the base pairs in the SHAPE-directed model also occur in the lowest free energy thermodynamics-only model. The unconstrained thermodynamics-only model is readily shown to be incorrect because many regions with high SHAPE reactivities are assigned as paired in the unconstrained model, while many regions with low SHAPE reactivities are assigned as single stranded.
Multiple lines of evidence support fundamental correctness of our working SHAPE-directed HIV-1 genome structural model (Fig. 2). First, SHAPE-directed folding is well validated and predicts the known structures of large RNAs, including 16S ribosomal RNA, with high accuracies (>90%)10,23. Second, most nucleotides assigned to single-stranded regions are reactive by SHAPE (red, orange, and green nucleotides; Fig. 2). Conversely, base-paired nucleotides are generally unreactive (black nucleotides and inset; Fig. 2). Thus, the structural modeling faithfully incorporates the experimental data. Third, many single nucleotide bulges are predicted as single reactive positions imbedded in helices whose remaining nucleotides are unreactive towards SHAPE, which speaks to the accuracy at the single nucleotide resolution level (for select examples see positions 1725, 3376, 4891, 5990, 7431, 7568, 9091; Fig. 2). Fourth, previously characterized HIV RNA structures including the 5′ TAR element, the DIS component of the packaging signal, and the five-stem RRE serve as positive controls and form structures consistent with previous work4,40 (Fig. 2). In the case of the gag-pol frameshift structure, we note that SHAPE data do not support common alternative proposals for this specific structure, including either a longer bulged stem or a pseudoknot.
The majority of structures in our current HIV-1 genome model, especially in regions with multiple closely spaced helices, are extremely well determined as evidenced by the strong correlation between SHAPE values and base pairing. This correlation is also consistent with benchmarking studies showing the SHAPE reactivities strongly discriminate between base paired and single stranded nucleotides (Fig. S2)41 and with the extent of local nucleotide disorder12. In contrast, some of the larger loop regions in our model may reflect regions that interconvert between multiple structures38,42. Elements that may require future refinement include the precise termini of helices at some multi-helix junctions and along the central backbone of the genome structure and the identification of additional pseudoknot and long-range interactions.
Calculation of evolutionary base pairing probabilities
RNA-Decoder13 was used to identify regions in the HIV-1 genome in which the ability to form base pairs is evolutionarily conserved. The program takes a set of grammar parameters, a multiple-sequence alignment, and a phylogenetic tree as input. The output is a pairing probability for each position in the genome, given the phylogenetic tree, alignment, and the grammar structural model. The pairing probability for position i in alignment D is the sum over all stem structural labels (k) of P(πi = k | M) P(D |π,T,M), where π is the structure, M is the grammar model parameters, and P (πi = k | M) is the posterior probability that position i has the specific structural label k, given the grammar43, and is calculated via the inside-outside algorithm44. In Bayesian terms, P (πi = k | M) is the prior probability of structure π and P (D | π, T, M) is the alignment probability, calculated using the Felsenstein algorithm45. Pairing predictions were made using an alignment of non-recombinant group M subtype reference sequences obtained from the Los Alamos HIV database14, with minor manual editing [and excluding subtype G, which is now considered a circulating recombinant form46]. Codon positions in genome regions encoding more than one protein in overlapping reading frames were defined according to the first open reading frame in the following pairs: gag-pro, pol-vif, vpr-vif, vpr-tat, rev-tat, env-vpu, env-tat2, env-rev2. Due to differences in nucleotide content and evolution rate within different genes in the HIV genome, the genome was scanned in two sections, upstream and downstream, that overlapped in the vif gene. This allowed use of separate phylogenetic trees for each scan, with branch lengths calculated according to the rates of evolution in each genome region. The phylogenetic tree for the 5′ half was built using the third codon position for the gag, pol, and vif genes, and the 5′ non-coding region; the tree for the 3′ half was built on the third positions of vif, vpr, rev, vpu, env, and nef genes, and the 3′ non-coding region.
Pairing probabilities were assessed across the entire genome. To accommodate as many pairing interactions as possible, we used a large window size (1300 nts), and spaced the scans at 300-nucleotide intervals. Pairing probabilities for each scan were combined using the statistical program R47 taking the maximum pairing probability in overlapping windows. It is important to note that high pairing probabilities identify regions experiencing evolutionary pressure to retain a specific, defined, secondary structure. A low pairing probability, while suggestive of a lack of structure, can also reflect (i) that an additional evolutionary constraint exists that is not accounted for by the evolutionary model or (ii) that natural selection favors folding in general, but not a precise pattern of folding.
Bootstrap Analysis of SHAPE reactivities in Inter-Protein Linkers and Protein-Domain Junctions
A bootstrap procedure was used to compare the SHAPE reactivities of particular collections of genome elements to the expectation for random genome regions of the same size. For a comparison to a collection of n genome elements, we generated 100,000 bootstrapped samples by randomly choosing n locations from the relevant portion of the genome, and randomly assigning the lengths of the actual genome elements to these n locations. For comparison to the protein domain junctions, locations were drawn randomly from the entire coding portion of the genome (bases 336–8621). We specified a length of 60 nts for each region.
For comparison to the intra-domain loops, locations were drawn randomly from within the domains where loops occur and assigned lengths that reflected loop sizes within the same domain (for example, for the CA domain, one element of 45 base pairs was drawn from within bases 732–1427). Bootstrap samples that contained overlapping genome regions were thrown out. The mean SHAPE reactivities for each bootstrap sample were used to generate a frequency distribution that describes the expectation for equally sized but randomly located collections of genome elements in HIV coding regions. We obtained a p-value by determining the percentage of the bootstrapped means that were lower than the mean SHAPE reactivity for the collection of genome elements. This p-value is equivalent to the probability that the low SHAPE reactivity in the actual collection of genome elements occurred by chance. P-values for inter-protein linkers and protein domain junctions were 0.0482 and 0.0777, respectively. The RT-RNase H junction functions as both an inter-protein linker and as a protein domain junction because it is cleaved one-half of the time. For this analysis, the RT-RNase H junction was counted as an inter-protein linker.
Statistical analysis of ribosome pause sites
Toeprinting data spanned 748 nts (positions 670–1018 and 1243–1652) (Fig. S5). Within these 2 reads, there were 220 nts that fell within 30 nt of the MA-CA, CA-NC, or NC-p6 junctions or in the cyclophilin loop. We evaluated whether ribosomes pause preferentially near protein junctions using the binomial distribution. A total of 36 base pairs yielded toeprint signals with an intensity of 1.0 or greater. A signal of 1.0 corresponds approximately to 1.5 standard deviations above the mean; 17 of these occurred within 30 nt of a protein junction. The probability of observing this distribution by chance is p = 0.018. This analysis was insensitive to the choice of high signal threshold. Similar p-values were obtained for toeprint thresholds between 0.6 and 1.6.
Consensus structure
The gag-pro-pol consensus structure (Fig. S6b) was generated by aligning the 37 reference group M HIV-1 sequences14 using CLUSTALW48. Regions of covariation were identified using a sequence logo49.
Helix energies
Helix free energy changes (Figs. 4c,d) were calculated using the RNAstructure program22 as the sum of the base pair stacking nearest neighbor parameters50,51. Duplex regions containing single nucleotide bulges were taken to be a single helix. The helix free energy changes do not include penalties for terminal AU or GU pairs because these are, by convention in RNAstructure, associated with the loop formation free energy changes.
RNA and protein structure display
RNA secondary structures were composed using xrna (http://rna.ucsc.edu/rnacenter/xrna); HIV protein images (Fig. 1d) were created using Visual Molecular Dynamics52.
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Supplementary Material
Acknowledgments
This project was supported by the U.S. National Institutes of Health (AI068462 to K.M.W.) and by the National Cancer Institute, under contracts N01-CO-12400 and HHSN261200800001E (to R.J.G. and J.W.B.). J.M.W. was supported as a Fellow of the UNC Lineberger Cancer Center and an NIH Kirschstein Postdoctoral Fellowship. R.S. and K.K.D. were supported by NIH grants AI44667 and T32 AI07419, respectively. We are indebted to David Mathews (U. of Rochester) and Justin Low for assistance with the RNAstructure program and genome secondary structure analysis, respectively.
Footnotes
Author Contributions. J.M.W., R.J.G. and K.M.W. conceived of and designed the HIV-1 genome structure analysis project. J.M.W. and K.M.W. analyzed and interpreted the HIV SHAPE structure information. K.K.D., R.S. and C.L.B. designed and performed the bioinformatic pairing probability analysis. J.M.W., R.J.G. and C.W.L. performed the experiments; J.M.W., C.L.B. and K.M.W. performed the statistical analyses; J.W.B. produced and purified HIV-1 virions. J.M.W. and K.M.W. wrote the manuscript with contributions from all authors.
References
- 1.Cann AJ. Principles of Molecular Virology. Elsevier Academic Press; Amsterdam: 2005. [Google Scholar]
- 2.Coffin JM, Hughes SH, Varmus HE. Retroviruses. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1997. [PubMed] [Google Scholar]
- 3.Frankel AD, Young JA. HIV-1: Fifteen proteins and an RNA. Annu Rev Biochem. 1998;67:1–25. doi: 10.1146/annurev.biochem.67.1.1. [DOI] [PubMed] [Google Scholar]
- 4.Damgaard CK, Andersen ES, Knudsen B, Gorodkin J, Kjems J. RNA interactions in the 5′ region of the HIV-1 genome. J Mol Biol. 2004;336:369–379. doi: 10.1016/j.jmb.2003.12.010. [DOI] [PubMed] [Google Scholar]
- 5.Goff SP. Host factors exploited by retroviruses. Nature Rev Microbiol. 2007;5:253–263. doi: 10.1038/nrmicro1541. [DOI] [PubMed] [Google Scholar]
- 6.Wilkinson KA, et al. High-throughput SHAPE analysis reveals structures in HIV-1 genomic RNA strongly conserved across distinct biological states. PLoS Biol. 2008;6:e96. doi: 10.1371/journal.pbio.0060096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Levin JG, Guo J, Rouzina I, Musier-Forsyth K. Nucleic acid chaperone activity of HIV-1 nucleocapsid protein: critical role in reverse transcription and molecular mechanism. Prog Nucleic Acid Res Mol Biol. 2005;80:217–286. doi: 10.1016/S0079-6603(05)80006-6. [DOI] [PubMed] [Google Scholar]
- 8.Paillart JC, Skripkin E, Ehresmann B, Ehresmann C, Marquet R. In vitro evidence for a long range pseudoknot in the 5′-untranslated and matrix coding regions of HIV-1 genomic RNA. J Biol Chem. 2002;277:5995–6004. doi: 10.1074/jbc.M108972200. [DOI] [PubMed] [Google Scholar]
- 9.Merino EJ, Wilkinson KA, Coughlan JL, Weeks KM. RNA structure analysis at single nucleotide resolution by selective 2′-hydroxyl acylation and primer extension (SHAPE) J Am Chem Soc. 2005;127:4223–4231. doi: 10.1021/ja043822v. [DOI] [PubMed] [Google Scholar]
- 10.Mortimer SA, Weeks KM. A fast-acting reagent for accurate analysis of RNA secondary and tertiary structure by SHAPE chemistry. J Am Chem Soc. 2007;129:4144–4145. doi: 10.1021/ja0704028. [DOI] [PubMed] [Google Scholar]
- 11.Vasa SM, Guex N, Wilkinson KA, Weeks KM, Giddings MC. ShapeFinder: A software system for high-throughput quantitative analysis of nucleic acid reactivity information resolved by capillary electrophoresis. RNA. 2008;14:1979–1990. doi: 10.1261/rna.1166808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gherghe CM, Shajani Z, Wilkinson KA, Varani G, Weeks KM. Strong correlation between SHAPE chemistry and the generalized NMR order parameter (S2) in RNA. J Am Chem Soc. 2008;130:12244–12245. doi: 10.1021/ja804541s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pedersen JS, Meyer IM, Forsberg R, Simmonds P, Hein J. A comparative method for finding and folding RNA secondary structures within protein-coding regions. Nucleic Acids Res. 2004;32:4925–4936. doi: 10.1093/nar/gkh839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leitner T, Foley B, Hahn B, Marx P, McCutchan F, Mellors J, Wolinsky S, Korber B. HIV Sequence Compendium, (Theoretical Biology and Biophysics Group, LA-UR 06-0680) Los Alamos National Laboratory; NM: 2005. [Google Scholar]
- 15.Purcell DF, Martin MA. Alternative splicing of human immunodeficiency virus type 1 mRNA modulates viral protein expression, replication, and infectivity. J Virol. 1993;67:6365–6378. doi: 10.1128/jvi.67.11.6365-6378.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kwong PD, et al. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Komar AA. A pause for thought along the co-translational folding pathway. Trends Biochem Sci. 2009;34:16–24. doi: 10.1016/j.tibs.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 18.Farabaugh PJ. Programmed translational frameshifting. Microbiol Rev. 1996;60:103–134. doi: 10.1128/mr.60.1.103-134.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wen JD, et al. Following translation by single ribosomes one codon at a time. Nature. 2008;452:598–603. doi: 10.1038/nature06716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nackley AG, et al. Human catechol-O-methyltransferase haplotypes modulate protein expression by altering mRNA secondary structure. Science. 2006;314:1930–1933. doi: 10.1126/science.1131262. [DOI] [PubMed] [Google Scholar]
- 21.Hartz D, McPheeters DS, Traut R, Gold L. Extension inhibition analysis of translation initiation complexes. Methods Enzymol. 1988;164:419–425. doi: 10.1016/s0076-6879(88)64058-4. [DOI] [PubMed] [Google Scholar]
- 22.Mathews DH, et al. Incorporating chemical modification constraints into a dynamic programming algorithm for prediction of RNA secondary structure. Proc Natl Acad Sci USA. 2004;101:7287–7292. doi: 10.1073/pnas.0401799101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Deigan KE, Li TW, Mathews DH, Weeks KM. Accurate SHAPE-directed RNA structure prediction. Proc Natl Acad Sci USA. 2009;106:97–102. doi: 10.1073/pnas.0806929106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim DH, et al. Synthetic dsRNA Dicer substrates enhance RNAi potency and efficacy. Nat Biotechnol. 2005;23:222–226. doi: 10.1038/nbt1051. [DOI] [PubMed] [Google Scholar]
- 25.Stein BS, Engleman EG. Intracellular processing of the gp160 HIV-1 envelope precursor. Endoproteolytic cleavage occurs in a cis or medial compartment of the Golgi complex. J Biol Chem. 1990;265:2640–2649. [PubMed] [Google Scholar]
- 26.Wilson W, et al. HIV expression strategies: ribosomal frameshifting is directed by a short sequence in both mammalian and yeast systems. Cell. 1988;55:1159–1169. doi: 10.1016/0092-8674(88)90260-7. [DOI] [PubMed] [Google Scholar]
- 27.Giedroc DP, Theimer CA, Nixon PL. Structure, stability and function of RNA pseudoknots involved in stimulating ribosomal frameshifting. J Mol Biol. 2000;298:167–185. doi: 10.1006/jmbi.2000.3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Biswas P, Jiang X, Pacchia AL, Dougherty JP, Peltz SW. The human immunodeficiency virus type 1 ribosomal frameshifting site is an invariant sequence determinant and an important target for antiviral therapy. J Virol. 2004;78:2082–2087. doi: 10.1128/JVI.78.4.2082-2087.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Means RE, et al. Ability of the V3 loop of simian immunodeficiency virus to serve as a target for antibody-mediated neutralization: correlation of neutralization sensitivity, growth in macrophages, and decreased dependence on CD4. J Virol. 2001;75:3903–3915. doi: 10.1128/JVI.75.8.3903-3915.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Graham FL, van der Eb AJ. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973;52:456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- 31.Adachi A, et al. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J Virol. 1986;59:284–291. doi: 10.1128/jvi.59.2.284-291.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chertova E, et al. Envelope glycoprotein incorporation, not shedding of surface envelope glycoprotein (gp120/SU), is the primary determinant of SU content of purified human immunodeficiency virus type 1 and simian immunodeficiency virus. J Virol. 2002;76:5315–5325. doi: 10.1128/JVI.76.11.5315-5325.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ott DE, et al. Analysis and localization of cyclophilin A found in the virions of human immunodeficiency virus type 1 MN strain. AIDS Res Hum Retroviruses. 1995;11:1003–1006. doi: 10.1089/aid.1995.11.1003. [DOI] [PubMed] [Google Scholar]
- 34.Thomas JA, et al. Human immunodeficiency virus type 1 nucleocapsid zinc-finger mutations cause defects in reverse transcription and integration. Virology. 2006;353:41–51. doi: 10.1016/j.virol.2006.05.014. [DOI] [PubMed] [Google Scholar]
- 35.Cline AN, Bess JW, Piatak M, Jr, Lifson JD. Highly sensitive SIV plasma viral load assay: practical considerations, realistic performance expectations, and application to reverse engineering of vaccines for AIDS. J Med Primatol. 2005;34:303–312. doi: 10.1111/j.1600-0684.2005.00128.x. [DOI] [PubMed] [Google Scholar]
- 36.Buckman JS, Bosche WJ, Gorelick RJ. Human immunodeficiency virus type 1 nucleocapsid Zn2+ fingers are required for efficient reverse transcription, initial integration processes, and protection of newly synthesized viral DNA. J Virol. 2003;77:1469–1480. doi: 10.1128/JVI.77.2.1469-1480.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wilkinson KA, Merino EJ, Weeks KM. Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE): quantitative RNA structure analysis at single nucleotide resolution. Nat Protoc. 2006;1:1610–1616. doi: 10.1038/nprot.2006.249. [DOI] [PubMed] [Google Scholar]
- 38.Badorrek CS, Weeks KM. Architecture of a gamma retroviral genomic RNA dimer. Biochemistry. 2006;45:12664–12672. doi: 10.1021/bi060521k. [DOI] [PubMed] [Google Scholar]
- 39.Dowell RD, Eddy SR. Evaluation of several lightweight stochastic context-free grammars for RNA secondary structure prediction. BMC Bioinformatics. 2004;5:71. doi: 10.1186/1471-2105-5-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Olsen HS, Nelbock P, Cochrane AW, Rosen CA. Secondary structure is the major determinant for interaction of HIV rev protein with RNA. Science. 1990;247:845–848. doi: 10.1126/science.2406903. [DOI] [PubMed] [Google Scholar]
- 41.Wilkinson KA, et al. Influence of nucleotide identity on ribose 2′-hydroxyl reactivity in RNA. RNA. 2009;15 doi: 10.1261/rna.1536209. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Badorrek CS, Weeks KM. RNA flexibility in the dimerization domain of a gamma retrovirus. Nat Chem Biol. 2005;1:104–111. doi: 10.1038/nchembio712. [DOI] [PubMed] [Google Scholar]
- 43.Knudsen B, Hein J. Pfold: RNA secondary structure prediction using stochastic context-free grammars. Nucleic Acids Res. 2003;31:3423–3428. doi: 10.1093/nar/gkg614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Durbin R, Eddy S. Biological sequence analysis: probabalistic models of proteins and nucleic acids. xi. Cambridge, UK; New York: 1998. p. 356. [Google Scholar]
- 45.Felsenstein J. Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol. 1981;17:368–376. doi: 10.1007/BF01734359. [DOI] [PubMed] [Google Scholar]
- 46.Abecasis AB, et al. Recombination confounds the early evolutionary history of human immunodeficiency virus type 1: subtype G is a circulating recombinant form. J Virol. 2007;81:8543–8551. doi: 10.1128/JVI.00463-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.R Development Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; Vienna, Austria: 2008. http://www.R-project.org (R Foundation for Statistical Computing, 2008) [Google Scholar]
- 48.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chang TH, Horng JT, Huang HD. RNALogo: a new approach to display structural RNA alignment. Nucleic Acids Res. 2008;36:W91–96. doi: 10.1093/nar/gkn258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xia T, et al. Thermodynamic parameters for an expanded nearest-neighbor model for formation of RNA duplexes with Watson-Crick pairs. Biochemistry. 1998;37:14719–14735. doi: 10.1021/bi9809425. [DOI] [PubMed] [Google Scholar]
- 51.Mathews DH, Sabina J, Zuker M, Turner DH. Expanded sequence dependence of thermodynamic parameters provides improved prediction of RNA secondary structure. J Mol Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- 52.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996;14(33–38):27–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.