Abstract
The Escherichia coli heat-labile enterotoxin B subunit (LT-B) has been used as a model antigen for the production of plant-derived high-valued proteins in maize. LT-B with its native signal peptide (BSP) has been shown to accumulate in starch granules of transgenic maize kernels. To elucidate the targeting properties of the bacterial LT-B protein and BSP in plant systems, the subcellular localization of visual marker green fluorescent protein (GFP) fused to LT-B and various combinations of signal peptides was examined in Arabidopsis protoplasts and transgenic maize. Biochemical analysis indicates that the LT-B::GFP fusion proteins can assemble and fold properly retaining both the antigenicity of LT-B and the fluorescing properties of GFP. Maize kernel fractionation revealed that transgenic lines carrying BSP result in recombinant protein association with fibre and starch fractions. Confocal microscopy analysis indicates that the fusion proteins accumulate in the endomembrane system of plant cells in a signal peptide-dependent fashion. This is the first report providing evidence of the ability of a bacterial signal peptide to target proteins to the plant secretory pathway. The results provide important insights for further understanding the heterologous protein trafficking mechanisms and for developing effective strategies in molecular farming.
Keywords: GFP, LT-B, recombinant proteins, secretory pathway, signal peptide, targeting, transgenic plants
Introduction
Production of recombinant proteins such as high-value pharmaceutical or industrial products in plants has many advantages, including safety and scalability of plant production systems (Streatfield, 2007). However, achieving high levels of recombinant proteins in a heterologous system is an ongoing challenge. Transgenic protein expression is driven mostly by the selection of strong and/or tissue-specific promoters and specific targeting signals for proper compartmentalization within the cell. This allows for different levels of regulation and modifications that result in variable levels of recombinant protein expression and accumulation (Hood, 2004). The subcellular localization of a protein accounts greatly for its proper maturation and function within the cell, hence it affects the degree to which functional protein is accumulated.
Among the plant systems used for recombinant protein expression, high-yielding crops have received a lot of attention in the past decade. Maize has been demonstrated as an effective expression system for functional proteins of prokaryotic, viral, and eukaryotic origins (Murry et al., 1993; Hood et al., 1997; Kusnadi et al., 1998; Streatfield et al., 2001, 2002). The commercialization of corn-produced β-glucuronidase (Witcher et al., 1998), aprotinin (Zhong et al., 1999), and avidin (Hood et al., 1997) demonstrated the viability of this expression system at the commercial level. Maize has also been used for the production of edible vaccines against deadly diseases such as traveller's diarrhea through expression of the antigenic subunit B of E. coli heat-labile enterotoxin (Chikwamba et al., 2002a, b; Streatfield et al., 2003). Despite concerns over using grain crops for the production of recombinant proteins, both the scientific and economic advantages of maize-based approaches are undeniable (Ramessar et al., 2008).
Maize seed is known for its large storage capacity and stability of proteins and starches, hence it is considered as an ideal organ for manufacturing recombinant proteins (Ramessar et al., 2008). While there are extensive studies on maize native seed storage proteins (Shewry and Tatham, 1990; Shewry and Halford, 2002), knowledge related to heterologous protein production and storage are limited. A number of studies report the subcellular localization of the recombinant protein in the transgenic maize (Hood et al., 1997; Witcher et al., 1998) and the relationship between subcellular localization and recombinant protein yield (Streatfield et al., 2003). These reports highlighted the importance of proper compartmentalization for optimum expression and accumulation of functional protein in plants. Expression of the E. coli heat-labile enterotoxin subunit B (LT-B) gene in maize with its N-terminus native bacterial signal peptide or with a native 27 kDa γ-zein signal peptide resulted in the accumulation of LT-B in the starch granules of the transgenic kernels (Chikwamba et al., 2003). This observation has raised a question on how the bacterial protein and its signal peptide behave in the plant cellular machinery. Considerable interest has also arisen in the possible trafficking pathway of recombinant proteins in plants.
Protein translocation pathways in bacteria (Papanikou et al., 2007; Driessen and Nouwen, 2008) and plants (Raikhel and Chrispeels, 2000; Hanton et al., 2007; Hormann et al., 2007; Brown and Baker, 2008; Rojo and Denecke, 2008) have been the focus of many studies and continue to be of great interest in today's research. In Gram-positive bacteria, secreted proteins only need to cross the plasma membrane, while in Gram-negative bacteria, secreted proteins are translocated through the plasma membrane into the periplasm and through the outer membrane. Most secreted proteins carry an N-terminal signal sequence that directs them to the inner membrane protein translocation machinery to be internalized into the membrane, translocated to the periplasm or secreted (Saier, 2006; Saier et al., 2008).
In plants, however, the sorting of proteins in the cell is somewhat more complex due to the presence of different organelles, and hence, different types of membranes. Finely orchestrated gene regulation and tissue gene expression along with subcellular compartmentalization of plant proteins allows cells to become specialized and differentiated to fulfil their role in tissue specificity, organ identity, and organism performance. Proteins carrying an N-terminal signal peptide are often directed to the secretory pathway, transported into the endoplasmic reticulum (ER) lumen, and sorted thereafter to the Golgi, cell wall, vacuole or plasma membrane, or retained in the ER. Proteins lacking a signal sequence are translated in the cytosol, and if carrying appropriate targeting signals, they are then translocated into other organelles such as the plastids (Inaba and Schnell, 2008; Paul, 2008) or mitochondria (Attardi and Schatz, 1988). Two cases have been described in which proteins are targeted to the plastid through the secretory pathway (Villarejo et al., 2005; Nanjo et al., 2006). Two native plant proteins, α-carbonic anhydrase from Arabidopsis (Villarejo et al., 2005) and rice plastidial N-glycosylated nucleotide pyrophosphatase/phosphodiesterase (Nanjo et al., 2006) were shown to localize to the plastids after being processed in the ER, describing a novel pathway for protein trafficking into the plastids. Most recently, a novel mechanism for targeting into plant mitochondria has also been reported; successful targeting of Arabidopsis thaliana mitochondrial-targeting signal 1 (MITS1) protein to mitochondria is influenced by an N-terminal extension serving as a targeting peptide as well as by domains in the full-length protein (Chatre et al., 2009). The MITS1 N-terminal extension was shown to contain three regions that co-ordinate the mitochondrial targeting signal, including a cryptic signal for protein targeting to the secretory pathway (Chatre et al., 2009).
Although much has been learned about the translocation machineries of bacteria and plants, less is known about how plants utilize their molecular translocation machineries to sort bacterial proteins when expressed in transgenic plants. LT-B has a native signal peptide that directs the protein for secretion in bacteria. In E. coli, LT-B is exported to the periplasm in a Sec-dependent fashion via the general secretory pathway. Once the signal peptide is cleaved, the protein assembles into a functional pentamer and associates with the A subunit (LT-A) to form the holotoxin before being secreted from the cell via a type II protein secretion pathway (Tauschek et al., 2002). LT-B has been expressed in several plant species such as potato (Mason et al., 1998; Tacket et al., 1998), tobacco (Kang et al., 2003), maize (Chikwamba et al., 2002b; Streatfield et al., 2003) and soybean (Moravec et al., 2007). Although several of these studies describe the targeting of LT-B to different cellular compartments by the inclusion of targeting signals within their constructs (Streatfield et al., 2003), only two studies show the detailed subcellular localization of LT-B in the transgenic material (Chikwamba et al., 2003; Moravec et al., 2007). Streatfield and colleagues (Streatfield et al., 2003), for example, used the signal sequences from barley α-amylase and aleurin to target LT-B to the cell surface and vacuole, respectively, and the maize granule-bound glycogen synthase to target LT-B to the plastids. In the Chikwamba work (Chikwamba et al., 2003) the functional LT-B was found in the starch granules of transgenic maize kernels expressing the LT-B with its native bacterial signal peptide. In the Moravec work (Moravec et al., 2007) the replacement of bacterial signal peptide with an Arabidopsis basic chitinase signal peptide at the N-terminus of LT-B resulted in the localization of LT-B in protein bodies of transgenic soybean seeds.
The focus of this study is to examine the subcellular trafficking of the bacterial LT-B protein and its native signal peptide in plant systems using the green fluorescent protein (GFP) as a reporter. Using the functional LT-B::GFP fusion proteins it is shown that the LT-B signal peptide, not LT-B protein itself, can direct cargo proteins to the secretory pathway in Arabidopsis thaliana and maize. In addition, in maize seed, the bacterial signal peptide leads to a strong association of the cargo proteins with the starch fraction, although most of the soluble fusion protein is found in the fibre fraction. The results provide insights for further understanding the processing of a bacterial protein in the plant cells and future design of a high level production system for recombinant proteins in plants.
Materials and methods
DNA constructs
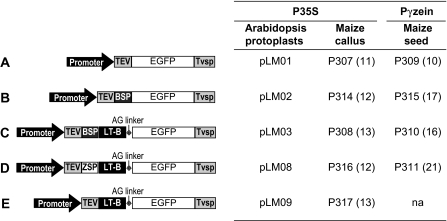
A schematic representation of the constructs used in this study is presented in Fig. 1. The enhanced green fluorescence protein (EGFP) sequence in pLM01, pLM02, pLM03, pLM08, and pLM09 was cloned from p27zn-signal (Shepherd and Scott, 2009) using standard molecular biology techniques for restriction enzyme-based cloning. Construct pTH210 containing the CaMV 35S promoter (P35S), tobacco etch virus translational enhancer (TEV) and LT-B was used as a cloning vector (Mason et al., 1998) for some of the constructs. For the generation of plasmids pLM01, pLM02, and pLM03 the EGFP sequence was cloned into pTH210 at the NcoI–SacI sites, SacI–SacI sites, and KpnI–SacI sites, respectively. Plasmid pLM01 was used as a backbone for the generation of pLM08 and pLM09. Plasmid pRC5, a pUC19-based vector carrying the maize 27 kDa γ-zein promoter (Pγzein), TEV, the maize 27 kDa γ-zein signal peptide (ZSP) fused to LT-B and the soybean vegetative storage protein terminator (Tvsp), was used as a source of LT-B fused to the maize 27 kDa γ-zein signal peptide (R Chikwamba, unpublished results). The ZSP-LT-B fragment was amplified by the polymerase chain reaction (PCR) method and the digested product was cloned into the NcoI–BstXI site of pLM01. An NcoI–EcoRI fragment of pLM08 was inserted into the NcoI–EcoRI backbone of pLM01 to generate pLM09. A simple alanine-glycine linker (AG linker) consisting of six amino acids with three AG repeats (AGAGAG) was added between LT-B and GFP using oligonucleotide extensions in the PCR primers in plasmids pLM03, pLM08, and pLM09. Expression cassettes (Fig. 1) were cloned and recombined into a Gateway version of pTF101.1 (Paz et al., 2004), pTF101.1gw1, for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plasmid pRC4 (Chikwamba et al., 2002b) was used as a donor of the Pγzein, and derived plasmids.
Fig. 1.
Gene cassettes, plasmids, and transgenic lines for studying localization of LT-B using GFP as a reporter. Gene cassettes A–E were used in transient assays using Arabidopsis leaf and root protoplasts, and in stable transformation of maize callus and endosperm tissues. Constitutive expression in Arabidopsis protoplasts and in maize callus was driven by the double CaMV 35S promoter (P35S promoter). The maize 27 kDa γ-zein promoter (Pγzein promoter) was used to drive expression of gene cassettes A–E in maize endosperm. The number of independent transgenic events for each line is presented in parenthesis. TEV, tobacco etch virus translational enhancer leader sequence; EGFP, enhanced green fluorescent protein; Tvsp, the soybean vegetative storage protein terminator; BSP, the LT-B bacterial signal peptide; LT-B, the B subunit of E. coli heat-labile enterotoxin; ZSP, 27 kDa γ-zein signal peptide; AG linker, alanine-glycine linker; na, not available.
The nuclear marker VirD2::RFP was a kind gift of Dr Stanton Gelvin. The ER marker plasmid designated here as ER cherry (Nelson et al., 2007) was obtained from the Arabidopsis Biological Resource Center (ABRC Stock No. CD3-959, http://arabidopsis.org).
Stable transformation of maize
Maize transformation was carried out at the Center for Plant Transformation at Iowa State University as described previously (Frame et al., 2000). Briefly, DNA constructs (Fig. 1) were co-bombarded with a selectable marker gene that confers resistance to the herbicide bialaphos. Herbicide-resistant calli were analysed using PCR for presence of the GFP and LT-B genes. For constructs driven by P35S, the calli of transgenic events were tested for GFP fluorescence and imaged as described in the ‘Fluorescence imaging and laser scanning confocal microscopy’ section of the Materials and methods. Transgenic calli were regenerated and plants brought to maturity in the greenhouse.
Protein extraction from transgenic tissue
Endosperm powder samples were collected from transgenic kernels using a hand-held drill as described by Sangtong et al. (2002). Plant materials were incubated with the following protein extraction buffer at a rate of 10 μl buffer mg−1 maize powder: 25 mM sodium phosphate (pH 6.6), 100 mM NaCl, 0.1% Triton X-100 (v/v), 1 mM EDTA, 10 μg ml−1 leupeptin, and 0.1 mM serine protease inhibitor Perfabloc SC (Fluka), for 2 h at 37 °C. Total aqueous extractable protein (TAEP) was determined using the Bradford assay (Bradford, 1976).
LT-B detection by GM1 capture ELISA
Quantification of LT-B in the samples was carried on using a modification of the monosialoganglioside-dependent enzyme-linked immunosorbent assay (ELISA) described previously (Chikwamba et al., 2003). The described protocol was modified as follows: monosialoganglioside, GM1, from bovine brain (G7641, Sigma, St Louis, MO, USA) was used at a 10 μg ml−1 concentration, 50 μl per well. Streptavidin–horseradish peroxidase conjugate (554066, BD Biosciences, San Jose, CA, USA) was used at a dilution of 1:1000 in 1% dry milk (DM) (w/v) in phosphate buffered saline [PBS; 0.01 M Na2HPO4, 0.003 M KH2PO4, 0.1 M NaCl (pH 7.2)]. Horseradish peroxidase substrate [ABTS; 0.5 mM 2,2-azino-bis(3-ethylbenzothiazoline-6-sulphonic acid) diammonium salt, 0.1 M citric acid, pH 4.35] was activated prior to use, by adding 5.5 μl 30% H2O2 to 5.5 ml ABTS solution. Activated horseradish peroxidase substrate was added to the plate and incubated in the dark at room temperature for 30 min. Absorbance was measured spectrophotometrically at 405 nm at the end of the reaction. Sample wells were blanked against non-transgenic maize protein extracts and all measurements were performed in duplicate. Raw ELISA data was converted to per cent LT-B of total soluble protein by reference to an ELISA standard curve constructed using purified bacterial LT-B (kindly provided by Dr John Clements, Tulane University, LA, USA).
Western blotting
An aliquot containing 50–100 μg of total aqueous extractable protein (TAEP) from maize kernels was boiled for 5 min and loaded onto a 15% polyacrylamide SDS-PAGE (Laemmli, 1970). The separated proteins were transferred to a 0.45 μm nitrocellulose membrane using the Bio-Rad Semidry Transblot apparatus according to the manufacturer's instructions. Membranes were blocked with 5% DM in PBST [PBS–0.05% Tween-20 (v/v)] for 3 h at room temperature. The presence of GFP and LT-B::GFP fusions was evaluated by overnight incubation of the membranes in goat anti-GFP (Cat no. 600-101-215, Rockland Immunochemicals, Inc., Gilbertsville, PA, USA) diluted 1:8000 in 1% DM in PBST at room temperature. Rabbit anti-LT-B (RECO-55G, Immunology Consultants Laboratory, Inc., Newberg, OR, USA) was also used at a 1:1000 dilution in 1% DM in PBST and incubated for 1 h at room temperature. Maize waxy and γ-zein proteins were probed using rabbit anti-waxy (1:2000; S Wessler, University of Georgia, Athens, GA, USA) and rabbit anti-zein (1:3000; P Scott, USDA/ARS, Ames, IA, USA) antibodies, respectively. Membranes were washed four times with PBST. A horseradish peroxidase conjugated rabbit anti-goat IgG (1:3000; A4174, Sigma, St Louis, MO, USA) or goat anti-rabbit (1:2000 for LT-B or 1:5000 for waxy and zeins; A0545, Sigma, St Louis, MO, USA) in PBST was used as secondary antibody. Coloured bands were revealed by incubation with horseradish peroxidase substrate, 3,3′,5,5′-tetramethylbenzidine (T0565, Sigma, St Louis, MO, USA).
Immunoprecipitation
For immunoprecipitation, 500 μg of TAEP were incubated with 10 μl of rabbit anti-LT-B (RECO-55G, Immunology Consultants Laboratory, Inc., Newberg, OR, USA) overnight in the cold room (9–11 °C). Protein G beads (IP50-1KT, Sigma, St Louis, MO, USA) were used to recover the immunoprecipitation (IP) complex, following the supplier's recommendations. Ten μl of recovered IP complex were analysed by Western blotting using goat anti-GFP antibody (Cat no. 600-101-215, Rockland Immunochemicals, Inc., Gilbertsville, PA, USA) as described in the ‘Western blotting’ section of the Materials and methods.
Small-scale starch isolation from transgenic maize kernels
A modified protocol based on (Chikwamba et al., 2003) was used for small-scale starch isolation. Five to ten dry transgenic maize kernels were imbibed in sterile water at 37 °C overnight. Alternatively, 5–10 developing transgenic kernels of ears harvested 15–20 d after pollination were directly dissected from the cob. The pericarp and embryo were removed from the kernels using a small bent forcep, and the dissected endosperms were placed in 10 ml of fresh sterile water in a conical 50 ml Falcon tube. The kernels were then homogenized using a Polytron homogenizer. Samples were then filtered using a 30 μm Nylon Filter (146506, Spectrum Laboratories, Houston, TX, USA). The original tube was rinsed three times with water and all fractions were collected into a clean Falcon tube. The filtrate was centrifuged at 3000 rpm for 30 min, and the material on the filter was saved as the fibre fraction. After aspiration of the supernatant, the starch slurry was transferred to an Eppendorf tube and centrifuged for 5 min at 5000 rpm. The starch pellet was washed four times each with water, 70% ethanol, 95% ethanol, and 75% ethanol/3% mercaptoethanol, and then spun to dryness in a Speed Vac. The fibre was placed directly into a 50 ml Falcon tube and lyophilized overnight. This fraction was designated as the fibre fraction.
Thermolysin treatment of starch samples
Thermolysin (3097-ZN, R&D Syetem, Inc., Minneapolis, MN, USA) was diluted to 10 μg ml−1 with 5 mM CaCl2 solution. Ten milligrams of starch were incubated with 100 μl thermolysin solution at 37 °C for 2 h. The reactions were terminated by the addition of EDTA to a final concentration of 20 mM. Samples were subsequently washed five times with 1 ml distilled water. After the last wash, 100 μl 1× SDS sample buffer was added to each sample and boiled for 5 min. Twenty μl were loaded onto an SDS-PAGE gel for Western blot analysis, as described in the ‘Western blotting’ section of the Materials and methods.
Arabidopsis thaliana mesophyll and root culture protoplast isolation for transient transformation assays
Arabidopsis thaliana (ecotype Columbia) seeds were vernalized in water at 4 °C for 48 h before sowing in prewetted LC1 Sunshine Mix. Flats were placed in a growth chamber at 21 °C and 16 h photoperiod covered with humidomes for 2–3 d. One to two weeks after germination, plants were thinned and transplanted to individual pots for further growth. Isolation of mesophyll protoplasts was carried on as described by Sheen (2001).
Arabidopsis root protoplasts were isolated from a root culture that was maintained on Lindsmaier–Skoog media (LSP003, Caisson Laboratories, North Logan, UT, USA) supplemented with 20 g l−1 sucrose, 1 mg l−1 naphthaleneacetic acid (NAA), and 0.05 mg l−1 kinetin (Contento et al., 2005), kindly provided by Dr D Bassham at Iowa State University. For the isolation of root culture protoplasts, 10–20 ml of root cell culture was pelleted in a 50 ml Falcon tube by centrifuging for 4 min at 50 g, or resting on the bench. The supernatant was removed and replaced by 40 ml of fresh enzyme solution [0.5× artificial sea water (ASW, 1.7 M NaCl, 9.4 mM MgSO4, 3.4 mM CaCl2, 5 mM MES, 3.45 mM KCl, 8.35 mM MgCl2·6H2O, 0.875 mM NaHCO3, pH 6.0), 0.3 M mannitol, 0.31% Cellulase Onozuka R10, 0.15% Macerozyme R10, pH 5.7, filter sterilized]. Cells were transferred to a deep Petri dish and were vacuum infiltrated for 5 min. The Petri plate was covered with aluminium foil and placed on an orbital shaker for 3.5 h at 50 rpm. The digested cell solution was filtered through a 40 μm mesh into a 50 ml conical Falcon tube, and the plate was washed once with cold W5 solution (154 mM NaCl, 125 mM CaCl2, 5 mM KCl and 2 mM MES, pH 5.7, filter sterilized). Cells were spun at 50 g for 4 min. The supernatant was removed and cells were washed twice with W5. The cells were carefully resuspended in fresh W5 and incubated on ice for at least 30 min prior to transformation with polyethylene glycol (PEG). Leaf and root protoplasts were transformed using PEG as described in Sheen (2001).
Sample preparation for microscopy
Transformed mesophyll and root culture protoplasts of Arabidopsis thaliana were washed and resuspended in fresh W5 media. Thirty μl of protoplast solution were placed directly on the microscope slides for visualization. Transgenic maize callus and developing endosperm were mounted directly onto microscope slides using Vecta Shield mounting media with propidium iodide (H1200, Vector Laboratories, Burlingame, CA, USA).
Fluorescence imaging and laser scanning confocal microscopy
Arabidopsis protoplasts, maize callus, and maize endosperm samples were visualized using a Leica TCS/NT (Leica Microsystems Inc., Exton, PA, USA) laser scanning confocal microscope equipped with Argon (488 nm) and Krypton (568 nm) lasers and a double dichroic DD488/568 filter. Green fluorescence was detected using a combination of RSP580 and BP525/25 filters under wavelengths between 500 and 550 nm. Red fluorescence was detected using a LP590 filter. During scanning, pinhole was maintained at 1 (airy units) for all images. Images were processed and analysed using ImageJ software (Abramoff et al., 2004).
Ears from self-pollinated transgenic maize plants harvested at 10–14 d after pollination were visualized and imaged using an Olympus SHZ10 stereoscope (Leeds Precision Instruments, Inc., Minneapolis, MN, USA) coupled to a SPOT RT colour CCD camera (Diagnostic Instrument Inc., Sterling Heights, MI). Images were taken under bright field or using a band pass exciter at 460–490 nm with emission filter 510–550 nm (for GFP detection), and acquired using SPOT Advanced software.
Hydropathy plots, secondary structure, and subcellular localization prediction
Hydropathy scores for signal peptide sequences of BSP (signal peptide of the B subunit of E. coli heat-labile enterotoxin; gi:88687064), ZSP (signal peptide of the maize 27 kDa γ-zein; gi:16305109), and BiPSP (signal peptide of the maize luminal binding protein; gi:1575128) were obtained based on Kyte and Doolittle (1982) using ProtScale prediction software (Gasteiger et al, 2005). Hydrophobicity plots were generated in Excel. Secondary structure for the aforementioned signal peptides were predicted using PSIPred (Jones, 1999; McGuffin et al., 2000). Prediction of subcellular localization was done using TargetP 1.1 (Emanuelsson et al., 2000; Nielsen et al., 1997) and iPSort (Bannai et al., 2002) software.
Results
Protein fusions of heat-labile enterotoxin subunit B (LT-B) and the green fluorescent protein (GFP) assemble and fold properly in plant cells
Constructs listed in Fig. 1 were made to investigate the subcellular targeting properties of the LT-B signal peptide and the LT-B protein using GFP as a visual marker. The first two constructs have GFP without a signal peptide (Fig. 1A, control), or fused to the bacterial signal peptide (BSP) of LT-B (Fig. 1B). The last three constructs have GFP fused at the C-terminus of the full-length LT-B protein with its native BSP (Fig. 1C), a maize 27 kDa γ-zein signal peptide (ZSP) (Fig. 1D), or no signal peptide (Fig. 1E).
To facilitate rapid construct verification and protein localization in different plants, each construct set was made with two different promoters. A constitutive CaMV 35S promoter was used for transient analysis in Arabidopsis protoplasts and stable analysis in transgenic maize callus culture, while a maize seed-specific 27 kDa γ-zein promoter was used for endosperm analysis in transgenic kernels. For each construct, a transgenic line identification was designated and multiple independent transgenic events were generated (Fig. 1). All transgenic maize lines were confirmed by gene-specific PCR analysis. The transgene insertion copy numbers were estimated by Southern blot analysis (data not shown). Strong transgene expressing events were chosen for further localization and biochemical analyses.
To examine whether LT-B and GFP retain their functional properties in the fusions described, experiments were carried out to assess their correct folding and assembly in maize. In stably transformed callus, green fluorescence could easily be distinguished when the fusion constructs were present (data not shown). Similarly, it was possible to detect green fluorescent transgenic maize seeds (Fig. 2A) confirming that GFP, a molecule capable of folding and fluorescing without exogenous substrates or cofactors (Chalfie et al., 1994) is active and correctly folded.
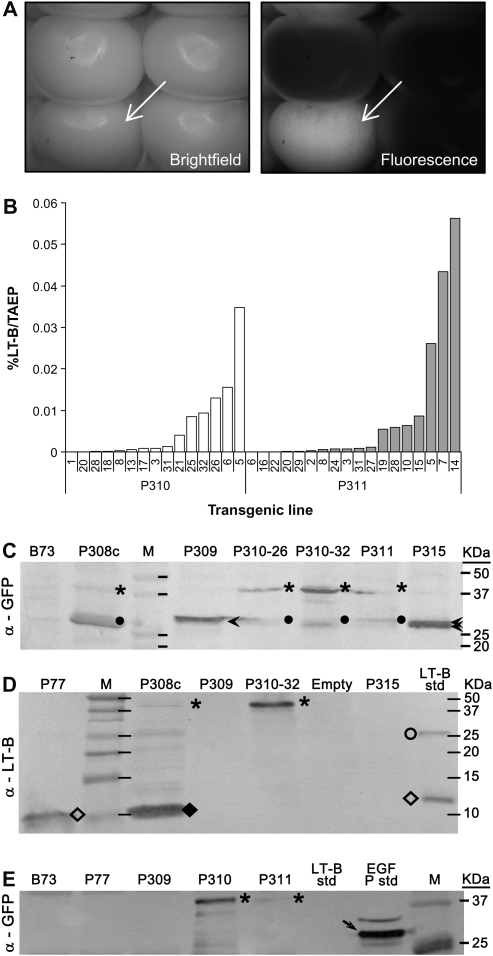
Fig. 2.
Gene expression analyses of LT-B, GFP, and LT-B::GFP fusions in transgenic maize kernels. (A) Bright field and fluorescence imaging of a representative self-pollinated ear of transgenic line P310 (Pγzein-BSP-LT-B::GFP) expressing GFP in the endosperm. A GFP-expressing kernel is marked by a white arrow. (B) LT-B levels as a percentage of total aqueous extractable protein (% LT-B/TAEP) in endosperm of P310 and P311 (Pγzein-ZSP-LT-B::GFP) kernels. Both transgenic maize carrying BSP- or ZSP-led LT-B::GFP fusion protein show the expression of functional LT-B. However, independent lines from both constructs have different levels of LT-B. (C) Western blot of TAEP extracts from transgenic callus (P308c, P35S-BSP-LT-B::GFP) and endosperms (P309, Pγzein-GFP, GFP control; P310-28, P310-32, two independent lines from P310; P311; and P315, Pγzein-BSP-GFP) using anti-GFP antibody. (D) Western blot of TAEP extracts from transgenic callus (P308c) and endosperms (P309, P310-32, and P315) using anti-LT-B antibody. (E) Western blot of immuno-precipitated samples using anti-LT-B antibody, probed with anti-GFP antibody. B73, non-transgenic maize line. P77, transgenic maize line expressing LT-B with its native bacterial signal peptide. The empty lane in (D) was a sample lost during loading. LT-B std, bacterial LT-B protein standard. EGFP std, commercial enhanced GFP standard. Arrowheads in (C), GFP. Dots in (C), possible cleavage peptides cross-react to GFP antibody. Asterisks in (C), (D), and (E), LT-B::GFP fusion. Open diamonds in (D), LT-B monomer. Closed diamond in (D), truncated LT-B::GFP fusion. Open circle in (D), LT-B multimer. Arrow in (E), commercial EGFP. Multiple EGFP bands in GFP standard may due to incomplete protein denaturation during boiling before loading.
A monosialoganglioside (GM1) capture Enzyme Linked Immunosorbent Assay (ELISA) was used to determine whether the LT-B protein assembles into functional pentamers when fused to GFP protein. To be biologically functional, the 11.7 kDa LT-B protein needs to be correctly assembled into pentameric form, which can then bind to monosialoganglioside specifically (Spangler, 1992). Figure 2B shows that multiple events of transgenic maize lines P310 and P311 accumulate pentameric LT-B at various levels.
These results combined indicate that both LT-B and GFP were able to retain their properties when fused; LT-B being in a pentameric form remains recognizable by its receptor, the GM1 receptor, and GFP retains its characteristic of self catalysis for folding and fluorescing.
Western blot analysis using anti-GFP and anti-LT-B antibodies on total aqueous extractable protein (TAEP) (Fig. 2C, D) were conducted to further confirm the presence of the fusion proteins in transgenic maize kernels. As shown in Fig. 2C, using anti-GFP antibody it was possible to detect bands around 27 kDa (arrowheads) in transgenic lines P309 (Pγzein-GFP control) and P315 (Pγzein-BSP-GFP) as predicted. In P315, two bands with similar molecular weights probably represent populations of GFP with and without cleaved signal peptide. On the other hand, in transgenic maize lines P308c (P35S-BSP-LT-B::GFP), P310-26 and -32 (Pγzein-BSP-LT-B::GFP), and P311 (Pγzein-ZSP-LT-B::GFP), the analysis also detects a number of bands that cross-reacted to anti-GFP antibody. One high molecular weight band estimated around 39 kDa (asterisks in Fig. 2C) is possibly the LT-B (11.7 kDa) and GFP (26.9 kDa) fusion product. Bands at the lower molecular weights (dots in Fig. 2C) cross-reacting to the antibody suggest possible proteolytic degradation of the fusion protein in these transgenic lines. No signal was observed for B73, a non-transgenic maize inbred line.
The anti-LT-B antibody (Fig. 2D) cross-reacted with bands around 39 kDa in P308c and P310 (asterisks in Fig. 2D), which are probably the LT-B::GFP fusion proteins. In P308c callus, a lower molecular weight band around 11 kDa was also detected (closed diamond in Fig. 2D). Although the size of this band is similar to the monomeric form of LT-B (open diamonds in Fig. 2D) as shown in bacterial LT-B standard and P77, a control transgenic maize line carrying LT-B with its own BSP (Chikwamba et al., 2002b), current evidence is not enough to show that it represents a processed LT-B monomer. No LT-B cross-reacting bands are seen in constructs carrying GFP alone (P309 and P315), as expected.
Figure 2E shows an anti-GFP Western blot on immunoprecipitated transgenic maize kernel samples using anti-LT-B antibody. A reacting band was detected around the 39 kDa position in maize lines P310 and P311, but no band could be seen in two control maize lines P309 (Pγzein-GFP) and P77 (Pγzein-LT-B, Chikwamba et al., 2002b) carrying GFP or LT-B alone, respectively. This result confirms that the ∼39 kDa bands observed in Fig. 2C are, in fact, fused to LT-B, as predicted. Figure 2E included a commercially EGFP standard (Cat no. 4999-100; Biovision, Mountain View, CA, USA), which shows more than one cross-reacting band when probed using goat anti-GFP antibody (arrow). This phenomenon was observed in other Western blots when using the anti-GFP antibody in our work. It is possible that the multiple GFP bands in commercial EGFP standard are the result of incomplete protein denaturation during boiling before loading.
LT-B::GFP fusion proteins are detected in starch and fibre fractions of transgenic maize kernels
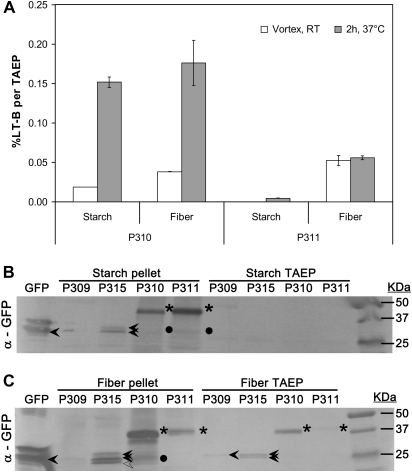
Starch and fibre fractions were analysed for functional GFP and LT-B content in transgenic lines P310 (Pγzein-BSP-LT-B::GFP) and P311 (Pγzein-ZSP-LT-B::GFP). The TAEP of all samples was obtained by two different extraction methods: simple vortex at room temperature (25 °C) and shaker-incubation at 37 °C for 2 h. These two methods of protein extraction were chosen to investigate the stability of the fusion protein by the previously described method of LT-B extraction (2 h/37 °C, Chikwamba et al., 2002b, 2003) compared to a more conservative extraction protocol (vortex/RT). As shown in Fig. 3A, the percentage of LT-B of TAEP obtained from 2 h/37 °C incubation was generally higher than that from vortex/RT extraction, suggesting that a better soluble protein recovery was achieved when using a method with higher temperature and a longer incubation time. Figure 3A also indicates that LT-B is not only associated with the starch fraction as previously reported, but also associated, at a higher percentage, to the fibre fraction obtained using the small-scale fractionation protocol. This association was also observed for the LT-B transgenic maize line P77 (Pγzein-BSP-LT-B; data not shown), the original line used in the previous work to demonstrate the LT-B association to the starch granules of transgenic maize kernels (Chikwamba et al., 2003). The observed LT-B/fibre association was not reported in the previous work due to the fact that protein recovery from non-soluble and non-starch fractions was not determined.
Fig. 3.
Association of LT-B and GFP with starch and fibre fractions of transgenic maize kernels. Transgenic maize kernels expressing constructs presented in Fig. 1 were used for small-scale fractionation. Protein extraction from starch and fibre fractions were used for LT-B and GFP determination. (A) Functional LT-B content as a percentage of TAEP. Error bars correspond to the standard deviation of two technical replicates. (B) Anti-GFP Western blot of starch soluble (TAEP) and insoluble (pellet) phases. (C) Anti-GFP Western blot of fibre soluble (TAEP) and insoluble (pellet) phases. P309, Pγzein-GFP; P310, Pγzein-BSP-LT-B::GFP; P311, Pγzein-ZSP-LT-B::GFP; P315, Pγzein-BSP-GFP; GFP, commercial EGFP standard. Arrowheads, GFP. Open arrowhead, possible truncated form of GFP. Asterisks, LT-B::GFP fusion. Dots, possible cleavage peptides cross-react to GFP antibody. Multiple EGFP bands in GFP standard may be due to incomplete protein denaturation during boiling before loading.
Western analysis using the anti-GFP antibody on proteins extracted from the aqueous phase and the insoluble pellet phase of each fraction (Fig. 3B, C) were performed to confirm the presence of the fusion proteins in starch and fibre. Interestingly, strong fusion protein bands were observed in P310 and P311 in both starch pellets (asterisks in Fig. 3B) and fibre pellets (asterisks in Fig. 3C). Weak but detectable fusion protein bands can be seen in the fibre TAEP of P310 and P311 (asterisks in Fig. 3C). However, while it was detectable by GM1 capture ELISA specific for LT-B detection, no protein bands were observed from the starch TAEP for both P310 and P311 (Fig. 3B). As for the control construct, the GFP protein from line P315 (Pγzein-BSP-GFP) behaved similar to P310 (Pγzein-BSP-LT-B::GFP) in the Western analysis. No detectable GFP band was seen for P309 (Pγzein-GFP), which carries the GFP with no signal peptide.
Thermolysin (EC 3.4.24.27) treatment based on the published protocols (Chikwamba et al., 2003) was used to verify whether the detection of the GFP and its fusion protein in starch pellets was due to internalization in starch grains of the insoluble phase, as observed previously for LT-B alone (Chikwamba et al., 2003). Any polypeptides within the starch granules should not be susceptible to hydrolysis upon treatment of intact granules with exogenous proteases such as Thermolysin (Mu-Forster and Wasserman, 1998). Starch pellets from three transgenic lines, two carrying GFP constructs (P309 and P315) and one carrying the LT-B::GFP fusion construct (P310), were treated with Thermolysin. To monitor the Thermolysin effectiveness, maize waxy protein (Mu-Forster et al., 1996) was used as an internal control for starch-bound proteins and γ-zeins (Mu-Forster and Wasserman, 1998) were used as markers for starch granular external protein contamination. The fibre fraction of P315 was used as a treatment control. Both treated and untreated samples were subjected to Western blots that cross-reacted with anti-GFP, anti-waxy, and anti-γ-zein antibodies. If the proteins are internalized in the starch granules, the reacting band should remain at the same intensity in Thermolysin-treated and untreated samples.
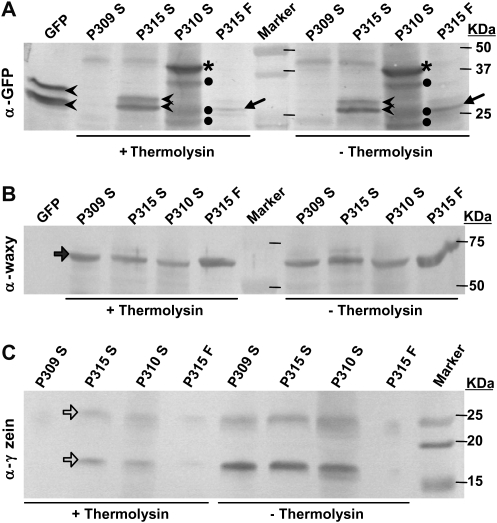
As can be seen in Fig. 4A, the GFP band in the Thermolysin-treated control sample P315F (fibre fraction) was reduced. However, the P310 fusion protein band (asterisks in Fig. 4A) and P315 GFP band (arrowheads in Fig. 4A) in the Thermolysin-treated and untreated starch samples remained at the same intensity, suggesting that these proteins were not sensitive to Thermolysin treatment, and most likely are protected by the starch granule membrane.
Fig. 4.
Western blot analysis of total proteins from starch samples treated with Thermolysin. Starch samples were treated with Thermolysin to test the susceptibility to the protease and possible internalization of fusion proteins in the starch granules of maize. SDS-loading buffer was used to extract proteins from Thermolysin-treated starch granules, and boiled for 5 min. Samples were separated on a 12% SDS-PAGE, transferred to a 0.45 μm nitrocellulose membrane, and probed with goat anti-GFP antibodies (A), rabbit anti-waxy protein antibodies (B), or rabbit anti-27 kDa γ-zein protein antibies (C), respectively. P309, Pγzein-GFP; P310, Pγzein-BSP-LT-B::GFP; P315, Pγzein-BSP-GFP; GFP, commercial EGFP. S, starch fraction; F, fibre fraction. In (A) arrowheads, GFP; asterisks, LT-B::GFP fusion; dots, possible cleavage peptides cross-react to GFP antibody; arrows, Thermolysin-sensitive GFP band from P315 fibre fraction. Block arrow in (B), waxy protein. Open block arrows in (C), zein proteins.
Because the waxy protein is strongly associated with the starch granule, Thermolysin treatment did not reduce its protein band intensity in all samples (block arrow in Fig. 4B). On the other hand, the band intensity of the 16 kDa and 27 kDa γ-zeins (open block arrow in Fig. 4C), known to accumulate in protein bodies and not inside starch granules, was notably reduced upon treatment with the protease (Fig. 4C).
The bacterial signal peptide of LT-B is sufficient for localization of GFP and LT-B::GFP fusions to the secretory system of Arabidopsis and maize
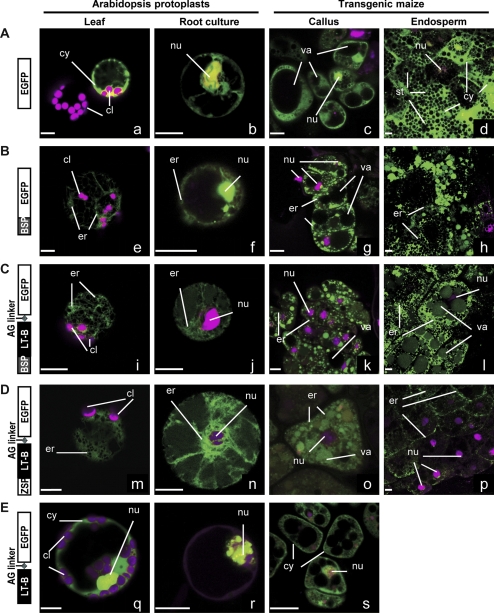
Confocal microscopy was used for localization studies in transiently transformed Arabidopsis protoplasts, and stably transformed maize callus and endosperm tissue using the constructs described in Fig. 1. The objective of such experiments was to establish the subcellular targeting properties of the bacterial signal peptide of LT-B and the LT-B protein itself. Figure 5 summarizes the results.
Fig. 5.
Confocal images of transiently and stably transformed Arabidopsis and maize cells expressing GFP or LT-B::GFP fusion proteins. Constructs A–E are described in Fig. 1. Transiently transformed Arabidopsis leaf (a, e, i, m, q) and root (b, f, j, n, r) protoplasts using the constitutive P35S promoter constructs were imaged 24–48 h after transformation. Stably transformed maize callus (c, g, k, o, s) also used the P35S promoter constructs. Fresh immature endosperm (12–26 d after pollination) from transgenic maize seed carrying the Pγzein promoter constructs were excised and imaged (d, h, l, p). Images are presented as merged green and red channels (presented in magenta color) for all samples. Green signal in all images corresponds to GFP. Red signal in leaf protoplasts is the autofluorescence of chlorophyll in chloroplasts. Red signal in root protoplasts corresponds to the expression of a VirD2::RFP construct, a nuclear marker. Red signal in maize callus and endosperm samples is propidium iodide used as a counter stain that labels nucleic acids. Organelle labelling: chloroplasts (cl), cytosol (cy), nucleus (nu), endoplasmic reticulum (er), vacuole (va), starch (st). Bars=10 μm.
Figure 5A is the GFP control construct in which the gfp gene is under the control of either P35S or Pγzein promoters with no signal peptide. The transient expression of GFP in Arabidopsis leaf (Fig. 5Aa) and root (Fig. 5Ab) protoplasts, and stable expression of GFP in maize callus (Fig. 5Ac) and endosperm (Fig. 5Ad) resulted in localization of GFP in the cytoplasm and nucleus; this observation has been reported and reviewed extensively (Hanson and Kohler, 2001; Berg et al., 2008).
The fusion of GFP to the C-terminus of the LT-B signal peptide BSP (Fig. 5B) resulted in localization of the GFP signal in the endoplasmic reticulum (ER) in all types of transformed cells tested in this study (Fig. 5Be–h).
To understand whether the mature LT-B protein also plays a role in subcellular localization in plant cells, the LT-B::GFP fusion construct linked with either bacterial BSP (Fig. 5C), plant ZSP (Fig. 5D) or no signal peptide (Fig. 5E) was assessed in various transiently or stably transformed plant cells. The results show that the GFP signals were accumulated in the secretory systems for either BSP (Fig. 5Ci–l) or ZSP (Fig. 5Dm–p) led LT-B::GFP fusion proteins in both Arabidopsis and maize, similar to the observations presented Fig. 5Be–h. On the other hand, when the signal peptide (Fig. 5E) was removed, the GFP signal returned to the cytosol and nucleus, and no signal was observed in the secretory pathway (Fig. 5Eq–s).
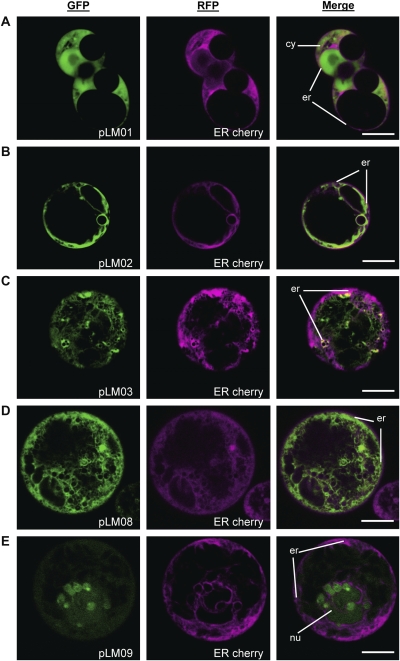
Figure 6 shows the co-localization experiments for the confirmation of subcellular localization using a known ER marker (Nelson et al., 2007). This marker (ER cherry) contains a red fluorescent protein fused to the signal peptide of the Arabidopsis thaliana wall-associated kinase 2 (He et al., 1999) and an HDEL motif for retention in the ER (Munro and Pelham, 1987). Arabidopsis root protoplasts were co-transformed with the ER cherry marker construct and the various constructs listed in Fig. 1. The results show that the ER-targeted RFP signals co-localized with the GFP signals from the constructs that contained either BSP (Fig. 6B, C) or ZSP (Fig. 6D). By contrast, GFP signals from constructs carrying GFP alone (Fig. 6A) or LT-B::GFP fusion (Fig. 6E) with no signal peptide were constantly detected in separate subcellular compartments than ER-targeted RFP signal.
Fig. 6.
Co-localization experiments in Arabidopsis root protoplasts. Protoplasts were co-transformed using the constructs presented in Fig. 1 and an ER marker protein fused to RFP, ER cherry (Nelson et al., 2007). (A) pLM01 (GFP control). (B) pLM02 (BSP-GFP). (C) pLM03, (BSP-LT-B::GFP). (D) pLM08 (ZSP-LT-B::GFP). (E) pLM09 (LT-B::GFP). Green channel corresponds to GFP signal. Red channels (presented in magenta color) corresponds to ER-cherry signal. Merged images are also presented. Organelle labelling: cytosol (cy), nucleus (nu), endoplasmic reticulum (er). Bars=10 μm.
Discussion
The aim of this study was to determine the targeting properties of the B-subunit of E. coli heat-labile enterotoxin (LT-B) and its signal peptide (BSP) in plant cells using GFP as a visual marker. The main findings of this study are as follows: (i) BSP directs LT-B and GFP to the secretory pathway of plant cells; this is the first report of a bacterial signal peptide carrying this function in plants; (ii) LT-B protein alone has no obvious targeting properties; (iii) GFP fused to BSP, and LT-B::GFP fusions carrying BSP or a maize zein signal peptide (ZSP) can be found in starch granules of transgenic maize kernels, but are mostly strongly associated with the fibre fraction; and (iv) it is possible to generate successful functional translational fusions of LT-B (11.7 kDa) with GFP (∼27 kDa); this property of LT-B has great potential applications in the biotechnology industry. Our results indicate that subcellular localization of LT-B in plant cells is determined by the presence (or absence) of a signal sequence such as BSP or ZSP. The findings also indicate that BSP plays a targeting role in plants similar to its role in bacteria, directing proteins (GFP and LT-B) to the secretory pathway. Finally, the findings also indicate that LT-B can be fused to proteins as large as 27 kDa and maintain functionality, a property highly desirable for the potential use of LT-B as a carrier molecule.
BSP acts as a functional signal peptide in plants
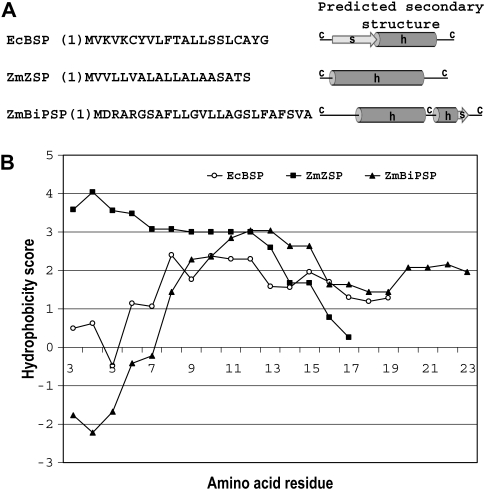
Signal peptides control the entry of all proteins to the prokaryotic and eukaryoic secretory pathways (Nielsen et al., 1997). The predicted secondary structure (PSIPred; Jones, 1999; McGuffin et al., 2000) of the signal peptide of LT-B (BSP) was compared with two signal peptides from maize proteins, 27 kDa γ-zein and luminal binding protein (Fig. 7A). It can be seen that all three signal peptides have common coil–helix–coil motifs, and that all three of them follow the (–3, –1) rule: the residues at positions –-3 and –1 from the cleavage site must be small and neutral (Nielsen et al., 1997). Figure 7B also shows the hydropathy plot for these signal peptides. A clear hydrophobic region comprising residues 7 to 17 is consistent with the predicted helical motif. Furthermore, subcellular localization prediction softwares TargetP 1.1 (Nielsen et al., 1987; Emanuelsson et al., 2000) and iPSort (Bannai et al., 2002) identified the sequences used in constructs B–D (Fig. 1) as having a signal peptide that would target the fusion proteins to the secretory pathway. By contrast, constructs lacking a signal peptide were predicted to accumulate in the cytosol.
Fig. 7.
Amino acid sequence, predicted secondary structure, and hydropathy plots of bacterial and plant signal peptides. (A) Amino acid sequence and predicted secondary structure. (B) Hydropathy plots. EcBSP (signal peptide of the B subunit of E. coli heat-labile enterotoxin), ZmZSP (signal peptide of the maize 27 kDa γ-zein), and ZmBiPSP (signal peptide of the maize luminal binding protein). c, coil; h, helix; s, strand. Hydropathy scores were obtained based on Kyte and Doolittle (1982) using ProtScale prediction software (Gasteiger et al., 2005). Subcellular localization prediction was performed using TargetP 1.1 (Emanuelsson et al., 2000; Nielsen et al., 1997) and iPSort (Bannai et al., 2002) software.
In E. coli enterotoxigenic strains, BSP is the signal peptide that directs LT-B protein to be translocated to the periplasm where the BSP is cleaved off, and the LT-B protein assembles into functional pentamers before being secreted in a folded state (Tauschek et al., 2002). The results presented in Figs 5 and 6 show that BSP plays the role of a signal peptide in plants, and results in the localization of the fusion proteins in the secretory pathway of Arabidopsis and maize. It is also clear that the mature LT-B protein has no targeting ability for the secretory system, and that the signal peptide alone, is sufficient for targeting cargo proteins to the ER. All the constructs used lack an ER retention motif, which raises interesting questions regarding the ER retention observed. Proteins carrying a signal peptide are usually transported further downstream in the secretory pathway unless they carry an ER retention motif, interact strongly with ER resident proteins, or fail to fold properly (Pagny et al., 1999). Therefore, in the case of our constructs carrying a BSP or ZSP, they were expected to be secreted from the cell. It has been reported that secreted forms of GFP reaching the apoplast showed none to low fluorescence levels due to the effect of pH on GFP folding (Batoko et al., 2000; Zheng et al., 2004). Hence, it cannot be ruled out that some of the GFP from BSP- or ZSP- carrying constructs were secreted but not visualized due to unfavourable experimental conditions (e.g. different pH). At the same time, it is also possible that the LT-B::GFP fusions, in particular, might require strong interaction with ER resident chaperones and protein isomerases for proper folding and assembly, thereby resulting in accumulation in the ER lumen, as has been shown for other proteins (Saito et al., 2009).
The observation of the ER localization properties of the LT-B signal peptide may explain why a great proportion of LT-B can be detected in the fibre fraction during kernel fractionation. The fibre fraction generated in the small-scale starch preparation process contains most maize cell components except for the starch, after removal of germ and pericarp by manual excision. The results do not explain, however, why a fraction of GFP and LT-B::GFP proteins fused to BSP was also found in the starch granules of maize seeds. Starch granules are inside the amyloplasts, specialized starch storage plastids found in cells of the maize endosperm. Proteins targeted to the ER have an N-terminal signal peptide that guides its insertion and translocation into the ER lumen, where the signal peptide is cleaved (Vitale and Boston, 2008). The ER is the point of entry for the secretory pathway, which includes ER, Golgi, plasma membrane, vacuole, cell wall or any body derived from any of these (Robinson et al., 2007). With few reported exceptions, nuclear proteins targeted to the plastids are translated in free ribosomes in the cytosol, and are then translocated using the organelle's specialized import machinery in a transit peptide-dependent fashion.
The BSP-dependent dual localization observed in this work has some similarity to plant signal peptide-dependent dual localization reported for rice amylases αAmy3 (Chen et al., 2004) and αAmyI-1 (Asatsuma et al., 2005) which accumulate in plastids and cell wall/extracellular space in plant cells. It remains to be determined whether the dual localization property of LT-B involves one or more trafficking routes, reaching the plastids via an ER-independent pathway. Recently, two groups described a novel pathway for protein trafficking into the plastids via the secretory pathway. Villarejo and colleagues (2005) showed that α carbonic anhydrase (α-CA) accumulates in the chloroplast stroma after being processed within the secretory system (Villarejo et al., 2005). In a different study it was shown that nucleotide pyrophosphatase/phosphodiesterase (NPP1) of rice and barley is glycosylated in the secretory pathway (Nanjo et al., 2006) before final accumulation in the plastids through a vesicular transport pathway from the ER and Golgi (Nanjo et al., 2006).
It is also possible that the default localization of the BSP-driven proteins is the ER, as was observed in Arabidopsis and maize, but additional fates occur in maize endosperm upon saturation of the secretory system. Overexpression of glycinin in soybean resulted in increased formation of ER-derived protein bodies carrying intermediates destined for the protein storage vacuole (Kinney et al., 2001). It is proposed that this response is most likely due to the formation of insoluble protein aggregates in the ER (Kinney et al., 2001). Because a large proportion of the fusion proteins in this study are found in the insoluble fraction of the endosperm, it cannot be ruled out that the dual localization can be a result of overexpression.
Unexpected patterns of recombinant protein deposition in endosperm have been reported for other species such as wheat (Arcalis et al., 2004) and rice (Drakakaki et al., 2006). In wheat, KDEL-tagged recombinant serum albumin was shown to accumulate in the ER lumen in leaves but was detected in prolamin aggregates inside vacules in the endosperm, as was observed with recombinant phytase targeted for secretion (Arcalis et al., 2004). In the same wheat study, recombinant legumin targeted to the vacuole resulted in the accumulation of the protein in globulin inclusion bodies around the prolamin bodies (Arcalis et al., 2004). Further effects of tissue type on recombinant protein subcellular fate was studied in rice by the expression in leaves and endosperm of recombinant phytase targeted for secretion. While, in leaves the phytase was successfully secreted, in endosperm it was retained in the endoplasmic reticulum-derived protein bodies and protein storage vacuoles (Drakakaki et al., 2006).
Because fractionation experiments were conducted specifically with transgenic maize kernels and not with Arabidopsis tissue, at this time it is difficult to establish if the observations presented for BSP-driven proteins are restricted to maize endosperm. Fractionation experiments combined with GFP localization suggest that the majority of the protein fusion population carrying a signal peptide is targeted to the ER. Using confocal microscopy, it was not possible to detect any GFP signal in plastids of either Arabidopsis or maize, but biochemical evidence suggests that a small fraction of the population was indeed associated with the starch fraction in maize kernels. In maize, the native γ-zein proteins are retained in the ER-lumen and accumulate in ER-derived protein bodies as a result of protein–protein interactions with other zeins and chaperones (Kim et al., 2002). Unlike the zein protein, the LT-B::GFP mature protein may or may not have the ability to interact with other zeins and ER resident proteins to be retained effectively in the ER or its derived systems.
BSP driven GFP and LT-B::GFP fusions associate with fibre and starch
Small-scale (5–10 kernels) starch isolation of maize kernels using a modification of the previously described protocol (Chikwamba et al., 2003) gives two main fractions: starch and fibre. As in other wet milling fractionation procedures (Johnson, 2000), this protocol allows separation and recovery of starch, fibre, germ, steep water, and pericarp. For practical purposes in this study, pericarp, embryo, and steep water, were discarded, and the endosperm was used for further separation of starch and fibre. While the starch fraction derives mostly from the amyloplasts, less is known about the cellular components of the fibre fraction.
The results presented in Fig. 3 indicate that fusions carrying BSP or ZSP are strongly associated with the fibre fraction of transgenic maize kernels, and to a lesser degree, with the starch fraction as well. While accumulation of LT-B in starch granules had been reported before (Chikwamba et al., 2003) the strong association with the fibre fraction is a novel observation, consistent with the ER-localization determined by confocal microscopy (Figs 5, 6). The strong association of LT-B with the fibre fraction may provide unique opportunities for effective downstream processing of a potential edible vaccine product, adding utility to the low value fibre fraction that results from corn fractionation (L Johnson, personal communication). The low association of LT-B with starch also presents potential advantages for the industry, as it might allow for utilization of the starch fraction for traditional applications such as biorefinery and non-food, non-feed processing.
LT-B can be used as a carrier molecule for peptide with size as large as 27 kDa
Expression of recombinant proteins in plants continues to be an area of great interest in the pharmaceutical, industrial, and vaccine industries. In this study, GFP is used as a visual marker to study the subcellular localization of the LT-B protein and its bacterial signal peptide. The data indicate that the C-terminus fusion of LT-B with GFP (using a simple alanine-glycine linker) results in retention of properties and assembly for both proteins. Because the pentameric LT-B protein is a potent antigen (Nashar et al., 1998) and binds specifically to the GM1 receptor of the epithelial cell surface, the results presented here suggest that LT-B has the potential to be used as a carrier molecule to deliver proteins larger than itself.
The use of LT-B as a carrier molecule for vaccination purposes has been extensively studied in bacterial systems (Buddenborg et al., 2008; Spangler, 1992). However, reports of plant-derived LT-B fusions are limited. Two studies report the fusion of small molecules (5–6 kDa) to the carboxy terminus of LT-B in Arabidopsis (Rigano et al., 2004) and in tobacco (Rosales-Mendoza et al., 2009). In both of these cases, the peptides used in the fusion represent about 1/5 of the size of the GFP protein (27 kDa) used in this study. Our work presented here is an important contribution to the vaccine research, because it is demonstrated that the LT-B can be fused with a protein as large as GFP and both proteins can retain their native conformations and functionalities.
The success of using LT-B as a carrier molecule would rely on the integral delivery of both proteins. In this study, it is observed that the ratio of fusion protein degradation is markedly different in lines P308c (P35S-BSP-LT-B::GFP) and P310 (Pγzein-BSP-LT-B::GFP) (Fig. 2D). A higher ratio of fused protein is detected in line P310 compared to line P308c. The main difference between the two constructs is the promoter; P308c has the constitutive P35S promoter and P311 has the seed specific γ-zein promoter. As the consequence of promoter specificity, the TAEP of P308c was extracted from the stably transformed callus culture while the TAEP of P310 was from the endosperm of transgenic seed.
The presence of degraded forms of the fusion proteins is relevant from two different perspectives: (i) for localization and visualization purposes, as the truncated forms may show different localization as the uncleaved protein; and (ii) for potential implications in the use of LT-B as a carrier molecule. Because only limited numbers of callus and seed lines were analysed, it could not be determined whether this difference was due to different fusion protein processing in different tissues or due to the protein extraction process itself. More likely, however, the enhanced instability of the fusion protein is a result of stress in callus cultures, as these are not specialized for storage of proteins. Presence of a cryptic vacuolar sorting signal in the GFP sequence can result in partial degradation (daSilva et al., 2005) of fusions of mGFP5 (Haseloff et al., 1997) with a plant signal peptide. It has also been shown that certain forms of GFP can be truncated upon secretion, resulting in a lack of fluorescence (Zheng et al., 2004).
On the other hand, endosperm tissue is designed for storage and possesses all the necessary cellular machinery for correct folding, processing, and storage of proteins to high levels. Taking into account the markedly lower degree of degradation of the fusion protein in trangenic maize kernels, the results confirm that maize endosperm is an adequate tissue for recombinant protein production and accumulation.
In conclusion, this study shows that BSP, a bacterial signal peptide, can be used to direct cargo proteins to the endoplasmic reticulum of plant cells. The LT-B protein can be fused to proteins as large as 27 kDa while retaining proper folding and pentamer assembly. The fact that high fusion protein accumulation can be achieved in maize endosperm and enriched in the fibre fraction of the corn seed provide valuable information for future design strategies of specialty maize for molecular farming.
Acknowledgments
We thank François Torney, David Jackson, Martha James, Harry Horner, Gregory Phillips, and Paul Scott for helpful and stimulating discussions, Diane Bassham for a critical review of the manuscript and valuable suggestions, Margie Carter at the ISU Confocal and Image Analysis Facility and Jessica Zimmer for technical assistance, and the staff at the Maize Transformation Facility at ISU for generating transgenic material. Partial financial support for this project was provided by the United States Department of Agriculture (USDA special grant IOW05082) and the Biopharmaceutical Initiative of the Plant Sciences Institute at Iowa State University.
References
- Abramoff MD, Magelhaes PJ, Ram SJ. Image processing with ImageJ. Biophotonics International. 2004;11:36–42. [Google Scholar]
- Arcalis E, Marcel S, Altmann F, Kolarich D, Drakakaki G, Fischer R, Christou P, Stoger E. Unexpected deposition patterns of recombinant proteins in post-endoplasmic reticulum compartments of wheat endosperm. Plant Physiology. 2004;136:3457–3466. doi: 10.1104/pp.104.050153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asatsuma S, Sawada C, Itoh K, Okito M, Kitajima A, Mitsui T. Involvement of α-amylase I-1 in starch degradation in rice chloroplasts. Plant and Cell Physiology. 2005;46:858–869. doi: 10.1093/pcp/pci091. [DOI] [PubMed] [Google Scholar]
- Attardi G, Schatz G. Biogenesis of mitochondria. Annual Review of Cell Biology. 1988;4:289–331. doi: 10.1146/annurev.cb.04.110188.001445. [DOI] [PubMed] [Google Scholar]
- Bannai H, Tamada Y, Maruyama O, Nakai K, Miyano S. Extensive feature detection of N-terminal protein sorting signals. Bioinformatics. 2002;18:298–305. doi: 10.1093/bioinformatics/18.2.298. [DOI] [PubMed] [Google Scholar]
- Batoko H, Zheng HQ, Hawes C, Moore I. A Rab1 GTPase is required for transport between the endoplasmic reticulum and Golgi apparatus and for normal Golgi movement in plants. The Plant Cell. 2000;12:2201–2218. doi: 10.1105/tpc.12.11.2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg RH, Beachy RN, Kevin FS. Methods in cell biology. Vol. 85. New York: Academic Press; 2008. Fluorescent protein applications in plants; pp. 153–177. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein–dye binding. Analytical Biochemistry. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Brown L-A, Baker A. Shuttles and cycles: transport of proteins into the peroxisome matrix (review) Molecular Membrane Biology. 2008;25:363–375. doi: 10.1080/09687680802130583. [DOI] [PubMed] [Google Scholar]
- Buddenborg C, Daudel D, Liebrecht S, Greune L, Humberg V, Schmidt MA. Development of a tripartite vector system for live oral immunization using a Gram-negative probiotic carrier. International Journal of Medical Microbiology. 2008;298:105–114. doi: 10.1016/j.ijmm.2007.08.008. [DOI] [PubMed] [Google Scholar]
- Chalfie M, Tu Y, Euskirchen G, Ward WW, Prasher DC. Green fluorescent protein as a marker for gene expression. Science. 1994;263:802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- Chatre L, Matheson LA, Jack AS, Hanton SL, Brandizzi F. Efficient mitochondrial targeting relies on co-operation of multiple protein signals in plants. Journal of Experimental Botany. 2009;60:741–749. doi: 10.1093/jxb/ern319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M-H, Huang L-F, H-m Li, Chen Y-R, Yu S-M. Signal peptide-dependent targeting of a rice α-amylase and cargo proteins to plastids and extracellular compartments of plant cells. Plant Physiology. 2004;135:1367–1377. doi: 10.1104/pp.104.042184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chikwamba R, Cunnick J, Hathaway D, McMurray J, Mason H, Wang K. A functional antigen in a practical crop: LT-B producing maize protects mice against Escherichia coli heat-labile enterotoxin (LT) and cholera toxin (CT) Transgenic Research. 2002a;11:479–493. doi: 10.1023/a:1020393426750. [DOI] [PubMed] [Google Scholar]
- Chikwamba R, McMurray J, Shou H, Frame B, Pegg SE, Scott P, Mason H, Wang K. Expression of a synthetic E. coli heat-labile enterotoxin B sub-unit (LT-B) in maize. Molecular Breeding. 2002b;10:253–265. [Google Scholar]
- Chikwamba RK, Scott MP, Mejia LB, Mason HS, Wang K. Localization of a bacterial protein in starch granules of transgenic maize kernels. Proceedings of the National Academy of Sciences, USA. 2003;100:11127–11132. doi: 10.1073/pnas.1836901100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contento AL, Xiong Y, Bassham DC. Visualization of autophagy in Arabidopsis using the fluorescent dye monodansylcadaverine and a GFP-atATG8E fusion protein. The Plant Journal. 2005;42:598–608. doi: 10.1111/j.1365-313X.2005.02396.x. [DOI] [PubMed] [Google Scholar]
- daSilva LLP, Taylor JP, Hadlington JL, Hanton SL, Snowden CJ, Fox SJ, Foresti O, Brandizzi F, Denecke J. Receptor salvage from the prevacuolar compartment is essential for efficient vacuolar protein targeting. The Plant Cell. 2005;17:132–148. doi: 10.1105/tpc.104.026351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drakakaki G, Marcel S, Arcalis E, Altmann F, Gonzalez-Melendi P, Fischer R, Christou P, Stoger E. The intracellular fate of a recombinant protein is tissue dependent. Plant Physiology. 2006;141:578–586. doi: 10.1104/pp.106.076661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driessen AJM, Nouwen N. Protein translocation across the bacterial cytoplasmic membrane. Annual Review of Biochemistry. 2008;77:643–667. doi: 10.1146/annurev.biochem.77.061606.160747. [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, von Heijne G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. Journal of Molecular Biology. 2000;300:1005–1016. doi: 10.1006/jmbi.2000.3903. [DOI] [PubMed] [Google Scholar]
- Frame BR, Zhang H, Cocciolone SM, Sidorenko LV, Dietrich CR, Pegg SE, Zhen S, Schnable PS, Wang K. Production of transgenic maize from bombarded type II callus: effect of gold particle size and callus morphology on transformation efficiency. In Vitro Cellular and Developmental Biology Plant. 2000;36:21–29. [Google Scholar]
- Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, Bairoch A. Protein identification and analysis tools on the ExPASy server. In: Walker JM, editor. The proteomics protocols handbook. Humana Press; 2005. pp. 571–607. [Google Scholar]
- Haseloff J, Siemering KR, Prasher DC, Hodge S. Removal of a cryptic intron and subcellular localization of green fluorescent protein are required to mark transgenic Arabidopsis plants brightly. Proceedings of the National Academy of Sciences, USA. 1997;94:2122–2127. doi: 10.1073/pnas.94.6.2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson MR, Kohler RH. GFP imaging: methodology and application to investigate cellular compartmentation in plants. Journal of Experimental Botany. 2001;52:529–539. [PubMed] [Google Scholar]
- Hanton SL, Matheson LA, Chatre L, Rossi M, Brandizzi F. Post-Golgi protein traffic in the plant secretory system. Plant Cell Reports. 2007;26:1431–1438. doi: 10.1007/s00299-007-0390-z. [DOI] [PubMed] [Google Scholar]
- He Z-H, Cheeseman I, He D, Kohorn BD. A cluster of five cell wall-associated receptor kinase genes, wak1-5, are expressed in specific organs or Arabidopsis. Plant Molecular Biology. 1999;39:1189–1196. doi: 10.1023/a:1006197318246. [DOI] [PubMed] [Google Scholar]
- Hood EE. Where, oh where has my protein gone? Trends in Biotechnology. 2004;22:53–55. doi: 10.1016/j.tibtech.2003.11.006. [DOI] [PubMed] [Google Scholar]
- Hood EE, Witcher DR, Maddock S, et al. Commercial production of avidin from transgenic maize: characterization of transformant, production, processing, extraction and purification. Molecular Breeding. 1997;3:291–306. [Google Scholar]
- Hormann F, Soll J, Bolter B. The chloroplast protein import machinery: a review. Methods in Molecular Biology. 2007;390:179–193. doi: 10.1007/978-1-59745-466-7_12. [DOI] [PubMed] [Google Scholar]
- Inaba T, Schnell DJ. Protein trafficking to plastids: one theme, many variations. Biochemistry Journal. 2008;413:15–28. doi: 10.1042/BJ20080490. [DOI] [PubMed] [Google Scholar]
- Johnson LA. Corn: the major cereal of the Americas. In: Kulp K, Ponte JG Jr, editors. Handbook of cereal science and technology. 2nd edn. New York, NY, USA: Marcel Dekker Inc; 2000. pp. 31–80. [Google Scholar]
- Jones DT. Protein secondary structure prediction based on position-specific scoring matrices. Journal of Molecular Biology. 1999;292:195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- Kang T-J, Loc N-H, Jang M-O, Jang Y-S, Kim Y-S, Seo J-E, Yang M-S. Expression of the B subunit of E. coli heat-labile enterotoxin in the chloroplasts of plants and its characterization. Transgenic Research. 2003;12:683–691. doi: 10.1023/B:TRAG.0000005114.23991.bc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim CS, Y-m Woo, Clore AM, Burnett RJ, Carneiro NP, Larkins BA. Zein protein interactions, rather than the asymmetric distribution of zein mrnas on endoplasmic reticulum membranes, influence protein body formation in maize endosperm. The Plant Cell. 2002;14:655–672. doi: 10.1105/tpc.010431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinney AJ, Jung R, Herman EM. Cosuppression of the alpha subunits of beta-conglycinin in transgenic soybean seeds induces the formation of endoplasmic reticulum-derived protein bodies. The Plant Cell. 2001;13:1165–1178. doi: 10.1105/tpc.13.5.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusnadi AR, Hood EE, Witcher DR, Howard JA, Nikolov ZL. Production and purification of two recombinant proteins from transgenic corn. Biotechnoly Progress. 1998;14:149–155. doi: 10.1021/bp970138u. [DOI] [PubMed] [Google Scholar]
- Kyte J, Doolittle R. A simple method for displaying the hydropathic character of a protein. Journal of Molecular Biology. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Mason HS, Haq TA, Clements JD, Arntzen CJ. Edible vaccine protects mice against Escherichia coli heat-labile enterotoxin (LT): potatoes expressing a synthetic LT-B gene. Vaccine. 1998;16:1336–1343. doi: 10.1016/s0264-410x(98)80020-0. [DOI] [PubMed] [Google Scholar]
- McGuffin LJ, Bryson K, Jones DT. The PSIPRED protein structure prediction server. Bioinformatics. 2000;16:404–405. doi: 10.1093/bioinformatics/16.4.404. [DOI] [PubMed] [Google Scholar]
- Moravec T, Schmidt MA, Herman EM, Woodford-Thomas T. Production of Escherichia coli heat-labile toxin (LT) B subunit in soybean seed and analysis of its immunogenicity as an oral vaccine. Vaccine. 2007;25:1647–1657. doi: 10.1016/j.vaccine.2006.11.010. [DOI] [PubMed] [Google Scholar]
- Mu-Forster C, Huang R, Powers JR, Harriman RW, Knight M, Singletary GW, Keeling PL, Wasserman BP. Physical association of starch biosynthetic enzymes with starch granules of maize endosperm. Granule-associated forms of starch synthase I and starch branching enzyme II. Plant Physiology. 1996;111:821–829. doi: 10.1104/pp.111.3.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu-Forster C, Wasserman BP. Surface localization of zein storage proteins in starch granules from maize endosperm. Proteolytic removal by thermolysin and in vitro cross-linking of granule-associated polypeptides. Plant Physiology. 1998;116:1563–1571. doi: 10.1104/pp.116.4.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro S, Pelham HR. A C-terminal signal prevents secretion of luminal ER proteins. Cell. 1987;48:899–907. doi: 10.1016/0092-8674(87)90086-9. [DOI] [PubMed] [Google Scholar]
- Murry LE, Elliott LG, Capitant SA, et al. Transgenic corn plants expressing mdmv strain b coat protein are resistant to mixed infections of maize dwarf mosaic virus and maize chlorotic mottle virus. Biotechnology. 1993;11:1559–1564. doi: 10.1038/nbt1293-1559. [DOI] [PubMed] [Google Scholar]
- Nanjo Y, Oka H, Ikarashi N, Kaneko K, Kitajima A, Mitsui T, Munoz FJ, Rodriguez-Lopez M, Baroja-Fernandez E, Pozueta-Romero J. Rice plastidial N-glycosylated nucleotide pyrophosphatase/phosphodiesterase is transported from the ER-Golgi to the chloroplast through the secretory pathway. The Plant Cell. 2006;18:2582–2592. doi: 10.1105/tpc.105.039891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nashar TO, Williams NA, Hirst TR. Importance of receptor binding in the immunogenicity, adjuvanticity and therapeutic properties of cholera toxin and Escherichia coli heat-labile enterotoxin. Medical Microbiology and Immunology. 1998;187:3–10. doi: 10.1007/s004300050068. [DOI] [PubMed] [Google Scholar]
- Nelson BK, Cai X, Nebenfuhr A. A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. The Plant Journal. 2007;51:1126–1136. doi: 10.1111/j.1365-313X.2007.03212.x. [DOI] [PubMed] [Google Scholar]
- Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Engineering. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- Pagny S, Lerouge P, Faye L, Gomord V. Signals and mechanisms for protein retention in the endoplasmic reticulum. Journal of Experimental Botany. 1999;50:157–164. [Google Scholar]
- Papanikou E, Karamanou S, Economou A. Bacterial protein secretion through the translocase nanomachine. Nature Reviews Microbiology. 2007;5:839–851. doi: 10.1038/nrmicro1771. [DOI] [PubMed] [Google Scholar]
- Paul J. Targeting of nucleus-encoded proteins to chloroplasts in plants. New Phytologist. 2008;179:257–285. doi: 10.1111/j.1469-8137.2008.02452.x. [DOI] [PubMed] [Google Scholar]
- Paz M, Shou H, Guo Z, Zhang Z, Banerjee A, Wang K. Assessment of conditions affecting Agrobacterium-mediated soybean transformation using the cotyledonary node explant. Euphytica. 2004;136:167–179. [Google Scholar]
- Raikhel N, Chrispeels MJ. Protein sorting and vesicle traffic. In: Buchanan BB, Gruissem W, Jones RL, editors. Biochemistry and molecular biology of plants. 1st. edn. Rockville, MD: American Society of Plant Physiologists; 2000. pp. 160–201. [Google Scholar]
- Ramessar K, Sabalza M, Capell T, Christou P. Maize plants: an ideal production platform for effective and safe molecular pharming. Plant Science. 2008;174:409–419. [Google Scholar]
- Rigano M, Alvarez M, Pinkhasov J, Jin Y, Sala F, Arntzen C, Walmsley A. Production of a fusion protein consisting of the enterotoxigenic Escherichia coli heat-labile toxin B subunit and a tuberculosis antigen in Arabidopsis thaliana. Plant Cell Reports. 2004;22:502–508. doi: 10.1007/s00299-003-0718-2. [DOI] [PubMed] [Google Scholar]
- Robinson DG, Herranz M-C, Bubeck J, Pepperkok R, Ritzenthaler C. Membrane dynamics in the early secretory pathway. Critical Reviews in Plant Sciences. 2007;26:199–225. [Google Scholar]
- Rojo E, Denecke J. What is moving in the secretory pathway of plants? Plant Physiology. 2008;147:1493–1503. doi: 10.1104/pp.108.124552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosales-Mendoza S, Alpuche-Solís AG, Soria-Guerra RE, Moreno-Fierros L, Martínez-González L, Herrera-Díaz A, Korban SS. Expression of an Escherichia coli antigenic fusion protein comprising the heat-labile toxin B subunit and the heat-stable toxin, and its assembly as a functional oligomer in transplastomic tobacco plants. The Plant Journal. 2009;57:45–54. doi: 10.1111/j.1365-313X.2008.03666.x. [DOI] [PubMed] [Google Scholar]
- Saier MH, Ma CH, Rodgers L, Tamang DG, Yen MR, Laskin AI, Sariaslani S, Gadd GM. Advances in applied microbiology. Vol. 65. Academic Press; 2008. Protein secretion and membrane insertion systems in bacteria and eukaryotic organelles; p. 141. [DOI] [PubMed] [Google Scholar]
- Saier MHJ. Protein secretion and membrane insertion systems in gram-negative bacteria. Journal of Membrane Biology. 2006;214:75–90. doi: 10.1007/s00232-006-0049-7. [DOI] [PubMed] [Google Scholar]
- Saito Y, Kishida K, Takata K, Takahashi H, Shimada T, Tanaka K, Morita S, Satoh S, Masumura T. A green fluorescent protein fused to rice prolamin forms protein body-like structures in transgenic rice. Journal of Experimental Botany. 2009;60:615–627. doi: 10.1093/jxb/ern311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangtong V, Moran DL, Chikwamba R, Wang K, Woodman-Clikeman W, Long MJ, Lee M, Scott MP. Expression and inheritance of the wheat Glu-1DX5 gene in transgenic maize. Theoretical and Applied Genetics. 2002;105:937–945. doi: 10.1007/s00122-002-1036-8. [DOI] [PubMed] [Google Scholar]
- Sheen J. Signal transduction in maize and Arabidopsis mesophyll protoplasts. Plant Physiology. 2001;127:1466–1475. [PMC free article] [PubMed] [Google Scholar]
- Shepherd CT, Scott MP. Construction and evaluation of a maize chimeric promoter with activity in kernel endosperm and embryo. Biotechnology and Applied Biochemistry. 2009;52:233–243. doi: 10.1042/BA20070269. [DOI] [PubMed] [Google Scholar]
- Shewry PR, Halford NG. Cereal seed storage proteins: structures, properties and role in grain utilization. Journal of Experimental Botany. 2002;53:947–958. doi: 10.1093/jexbot/53.370.947. [DOI] [PubMed] [Google Scholar]
- Shewry PR, Tatham AS. The prolamin storage proteins of cereal seeds: structure and evolution. Biochemical Journal. 1990;267:1–12. doi: 10.1042/bj2670001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spangler BD. Structure and function of cholera toxin and the related Escherichia coli heat-labile enterotoxin. Microbiological Reviews. 1992;56:622–647. doi: 10.1128/mr.56.4.622-647.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streatfield SJ. Approaches to achieve high-level heterologous protein production in plants. Plant Biotechnology Journal. 2007;5:2–15. doi: 10.1111/j.1467-7652.2006.00216.x. [DOI] [PubMed] [Google Scholar]
- Streatfield SJ, Jilka JM, Hood EE, et al. Plant-based vaccines: unique advantages. Vaccine. 2001;19:2742–2748. doi: 10.1016/S0264-410X(00)00512-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streatfield SJ, Lane JR, Brooks CA, et al. Corn as a production system for human and animal vaccines. Vaccine. 2003;21:812–815. doi: 10.1016/s0264-410x(02)00605-9. [DOI] [PubMed] [Google Scholar]
- Streatfield SJ, Mayor JM, Barker DK, et al. Development of an edible subunit vaccine in corn against enterotoxigenic strains of Escherichia coli. In Vitro Cellular and Developmental Biology–Plant. 2002;38:11–17. [Google Scholar]
- Tacket CO, Mason HS, Losonsky G, Clements JD, Levine MM, Arntzen CJ. Immunogenicity in humans of a recombinant bacterial antigen delivered in a transgenic potato. Nature Medicine. 1998;4:607–609. doi: 10.1038/nm0598-607. [DOI] [PubMed] [Google Scholar]
- Tauschek M, Gorrell RJ, Strugnell RA, Robins-Browne RM. Identification of a protein secretory pathway for the secretion of heat-labile enterotoxin by an enterotoxigenic strain of Escherichia coli. Proceedings of the National Academy of Sciences, USA. 2002;99:7066–7071. doi: 10.1073/pnas.092152899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villarejo A, Buren S, Larsson S, et al. Evidence for a protein transported through the secretory pathway en route to the higher plant chloroplast. Nature Cell Biology. 2005;7:1224–1231. doi: 10.1038/ncb1330. [DOI] [PubMed] [Google Scholar]
- Vitale A, Boston RS. Endoplasmic reticulum quality control and the unfolded protein response: Insights from plants. Traffic. 2008;9:1581–1588. doi: 10.1111/j.1600-0854.2008.00780.x. [DOI] [PubMed] [Google Scholar]
- Witcher DR, Hood EE, Peterson D, et al. Commercial production of beta-glucuronidase (gus): a method system for the production of proteins in plants. Molecular Breeding. 1998;4:301–312. [Google Scholar]
- Zheng H, Kunst L, Hawes C, Moore I. A GFP-based assay reveals a role for RHD3 in transport between the endoplasmic reticulum and Golgi apparatus. The Plant Journal. 2004;37:398–414. doi: 10.1046/j.1365-313x.2003.01969.x. [DOI] [PubMed] [Google Scholar]
- Zhong GY, Peterson D, Delaney DE, et al. Commercial production of aprotinin in transgenic maize seeds. Molecular Breeding. 1999;5:345–356. [Google Scholar]