Abstract
Neutral/alkaline invertases are a subgroup, confined to plants and cyanobacteria, of a diverse family of enzymes. A family of seven closely-related genes, LjINV1–LjINV7, is described here and their expression in the model legume, Lotus japonicus, is examined. LjINV1 previously identified as encoding a nodule-enhanced isoform is the predominant isoform present in all parts of the plant. Mutants for two isoforms, LjINV1 and LjINV2, were isolated using TILLING. A premature stop codon allele of LjINV2 had no effect on enzyme activity nor did it show a visible phenotype. For LjINV1, premature stop codon and missense mutations were obtained and the phenotype of the mutants examined. Recovery of homozygous mutants was problematic, but their phenotype showed a severe reduction in growth of the root and the shoot, a change in cellular development, and impaired flowering. The cellular organization of both roots and leaves was altered; leaves were smaller and thicker with extra layers of cells and roots showed an extended and broader zone of cell division. Moreover, anthers contained no pollen. Both heterozygotes and homozygous mutants showed decreased amounts of enzyme activity in nodules and shoot tips. Shoot tips also contained up to a 9-fold increased level of sucrose. However, mutants were capable of forming functional root nodules. LjINV1 is therefore crucial to whole plant development, but is clearly not essential for nodule formation or function.
Keywords: Cellular development, legume, Lotus japonicus, neutral/alkaline invertase, mutants, plant development, sucrose metabolism, TILLING
Introduction
The major carbohydrate transported in plants, sucrose, is exquisitely partitioned between different plant organs to optimize its use. A product of photosynthesis in source leaves, sucrose is transported throughout the plant via the phloem to sink tissues. Plant growth and development relies on the breakdown of this disaccharide to hexoses within these sink tissues before it can be utilized by metabolism, but there are only two routes by which this can be achieved, namely (i) conversion by sucrose (Suc) synthase (EC 2.4.1.13) to UDP-glucose and fructose, and (ii) cleavage by invertase (INV; EC 3.2.1.26) to glucose and fructose. The former is considered to be a reversible reaction (Geigenberger and Stitt, 1993), whereas invertase catalyses an irreversible breakdown to hexoses (Avigad, 1982). Sucrose synthase is believed to be important in polymer synthesis (e.g. cellulose) and to be the dominant pathway under conditions of low oxygen availability (Koch, 2004), which presumably underlies its importance in root nodules where it maintains nitrogen fixation and assimilation (Craig et al., 1999; Baier et al., 2007; Horst et al., 2007). By contrast, invertases are critical for turgor control, sugar signalling, and for apoplastic-required unloading during pollen and seed development (Koch, 2004). There is also evidence for their role in defence mechanisms (Koch, 2004). Both enzymes exist as a number of isoforms encoded by small gene families that permit both spatial and temporal controls. Whereas Suc synthase is located in the cytoplasm, three known locations have been described for invertases: the cell wall, vacuole, and cytoplasm (Roitsch and Gonzalez, 2004). Both cell wall and vacuolar invertase are defined as acidic by their pH optima, whereas the cytoplasmic forms are neutral to alkaline in their pH optima. Although there is a considerable literature on the roles of acidic invertases, very little information is available on the role of neutral/alkaline (N/A) invertases (Bonfig et al., 2007), since it was generally believed that they had low and unstable activity (Roitsch and Gonzalez, 2004). They have also been considered ‘maintenance’ enzymes involved in Suc degradation when the activities of acid invertase and Suc synthase are low (Winter and Huber, 2000).
Over the past few years, genome analyses have expanded our knowledge of N/A invertases and small gene families have been described in cyanobacteria (Vargas et al., 2003), in Arabidopsis and rice (Ji et al., 2005), and in poplar (Bocock et al., 2008). In the last species, the group is larger than in most others due to multiple gene duplication and selective retention events that have increased the N/A invertase family in particular. In rice it was proposed from a comparison of the sequences at their N-termini that subgroups of the family were located in mitochondria and plastids (Ji et al., 2005) and experimental proof has been obtained for both locations (Murayama and Handa, 2007; Szarka et al., 2008; Vargas et al., 2008). In comparison to acidic invertases, there has been a paucity of information on the function of the group until quite recently. In Arabidopsis, the major isoform, AtCYTINV1 (aka AtCINV1) has been found to be important in stress-inhibited root growth (Lou et al., 2007; Qi et al., 2007). Very recently, Jia et al. (2008) identified a gene by map-based cloning in rice in which a missense mutation caused a short root phenotype. The protein encoded by this gene, named OsCYT-INV1 (aka OsNIN8), was the homologue of AtCYT-INV1, and was needed for converting sucrose to hexoses in the root. Both the main root and the lateral roots in this mutant were decreased in length due to a reduction in length of the cells, but its seed set was also reduced due to the low fertility of its pollen. It is becoming clear, therefore, that at least one isoform of N/A invertase has a role in controlling the hexose needed for root growth in plants.
In Lotus japonicus, Flemetakis et al. (2006) described two cDNAs isolated from EST libraries that represented two isoforms, LjINV1 and LjINV2, of N/A invertases. One of the invertases, LjINV1, was up-regulated upon inoculation with rhizobia and they speculated, therefore, that it may have a role in nodulation. The increase in the expression was much less, however, than that for the main isoform of Suc synthase, the alternative route for sucrose metabolism. This isoform was recently shown to be required for nodule function (Horst et al., 2007). The N/A invertase family in L. japonicus is described here and evidence is presented that LjINV1 is the major isoform in the plant and plays a crucial role during plant establishment and subsequent development, but not in nitrogen fixation.
Materials and methods
Plant material
Lotus japonicus plants for the alkaline invertase assays were grown in either a greenhouse with supplementary lighting at 20/18 °C (day/night) or in a controlled environment room at a constant 20 °C. Seeds were germinated in compost and grown for 8–10 weeks. To generate nodules, the roots were truncated to stimulate lateral root formation, the plants transferred to perlite:vermiculite (1:1, v/v) and at the same time inoculated with Mesorhizobium loti strain Tono for nodulation. Plants were fed with Hornum solution (Handberg and Stougaard, 1992) twice a week. Plants for RNA extracts were grown in the greenhouse on a Terragreen® (Oil-Dri UK Ltd, Wisbech, UK) and sand mix (1:1, v/v).
Seedlings for invertase assays and RNA extraction were grown under aseptic conditions in a growth chamber at 25 °C with a 16 h daylength. L. japonicus ‘Gifu’ seeds were scarified and sterilized with 10% Parazone domestic bleach (Jeyes Ltd, Thetford, UK) for 10 min, then rinsed thoroughly with sterile water and left to imbibe for 5 h in water. Seeds were then plated onto 1% agarose. Seedlings were harvested at 2, 5, 7, and 9 d after sterilization. The testae were removed before tissue homogenization.
Data mining and phylogenetic analysis
Full-length invertase coding sequences, TC8973 and TC16464, had previously been identified and characterized by searching EST databases (Flemetakis et al., 2006). These sequences were used to search the Kazusa database; genomic sequences corresponding to the TCs were identified and aligned fully. These sequences were designated LjINV1 and LjINV2, respectively. Genomic sequence corresponding to another isoform was identified by a BLASTN database searches using TC8973 and TC16464. A sequence with high similarity was identified in EMBL accession no. AP006378 and designated LjINV3. Coding sequences of LjINV1 to LjINV3 were used to search the Kazusa TAC/BAC and whole-genome shotgun assembly databases for other similar sequences. The following databases hold information on the Lotus INV genes that are presented in Table 1: Kazusa: http://www.kazusa.or.jp/lotus/index.html; EMBL: www.ebi.ac.uk.
Table 1.
Lotus japonicus N/A invertase genes, locations and accession numbers
| Gene | Published gene name (Kasuzaa) | Chromosome/Position (cM; Kazusaa) | Corresponding TAC/BAC contig (Kazusaa) | Corresponding genome clone (Kazusa*) | Accession number (EMBL*) |
| LjINV1 | CM1887.40.nc | – | CM1887 (DF093534) | LjT06N24 (TM1887) | AP010579 |
| LjINV2 | CM1598.160.nc | chr5 26.4 | CM1598 (DF093522) | LjB16B21 (BM1888) | AP010554 |
| LjINV3 | – | chr1 66.2 | CM0206 (DF093325) | LjT44A10 (TM0223) | AP006378 |
| LjINV4 | This study | – | – | LjT07N19 (TM2287) | AP010908 |
| CM0333.360.nc (partial) | chr4 44.1 | CM0333 (DF093390) | LjT36O22 (TM0333) | AP006434 | |
| LjINV5 | This study | – | – | LjB16K09 (BM2319) | AP010909 |
| LjT03L06.210.nd (partial) | – | – | LjT03L06 (TM1622) | AP010232 | |
| LjINV6 | LjSGA_049478.1 (partial) | – | – | – | BABK01029127 |
| LjINV7 | CM0231.20.nd | chr1 42.2 | CM0231 (DF093338) | LjT04A18 (TM1266) | AP007943 |
See Materials and Methods for web addresses.
The programs Genscan (http://genes.mit.edu/GENSCAN.html) or FGENESH (www.softberry.com) were used to predict amino acid sequences of Lotus genomic sequences. These were aligned with available N/A invertase sequences from Arabidopsis, rice, and Medicago truncatula. Alignments and phylogenetic analysis was performed using the Mega4 program (www.megasoftware.net/) including bootstrap values for 1000 datasets. The exons and introns for all sequences were identified by aligning the TC and/or predicted coding sequences with the genomic sequence using ClustalW. Gene structures were predicted using the CODDLE program as used for TILLING below.
qRT-PCR
RNA was prepared from plant tissue and both RT and qRT-PCR were performed as previously described (Horst et al., 2007) on an Opticon instrument (MJ Research, Massachusetts, USA). Isoform-specific primers were designed at the 3′ end of the coding sequence as the sequence for this region was available for all seven isoforms. The specificity of the primers was confirmed by sequencing the PCR product after RT-PCR reaction. Table 2 shows primer sets used for qRT-PCR.
Table 2.
Primers for qRT-PCR, genotyping and TILLING
| Gene family | Target gene | Primer sequence |
| (A) qRT-PCR (forward and reverse) | ||
| N/A invertase | LjINV1 | 5′-TGCTTGCATCAAAACGGGGCGAC-3′ |
| 5′-TTTGCCACCAGATAACCTGCAATAG-3′ | ||
| LjINV2 | 5′-ATTCTTGGCCCGAGTATTATGATAC-3′ | |
| 5′-CACTCTTCTTAAGCCCACAAGCAC-3′ | ||
| LjINV3 | 5′-GACACCCGCACAGGAAGATTTATT-3′ | |
| 5′-TGCCATTCTTCTTATTAAGTACACAAA-3′ | ||
| LjINV4 | 5′-ATACACGAAATGGAAGGTTTATCG-3′ | |
| 5′-GCCTCTTTGCTTAGCATGCAGAC-3′ | ||
| LjINV5 | 5′-GACACAAAGAGATCTCGAATCATTG-3′ | |
| 5′-GGTTGGCACTTATCAAGGCATTCA-3′ | ||
| LjINV6 | 5′-TCCTCATATGGCTTCTTGCTGCG-3′ | |
| 5′-GCATCATCCTAGCCACCAGGTAA-3′ | ||
| LjINV7 | 5′-TTCTTTTATGGCTTCTGTCTGCGG-3′ | |
| 5′-ACCATCATCCTTGCCACCAAGTAA-3′ | ||
| EF-1α | TC24735 | 5′-GACAGACTCGTGAGCACGGAC-3′ |
| 5′-CAGGCTTCAAGACACCAGTTTCAAC-3′ | ||
| (B) TILLING and genotyping, (forward, reverse, forward/genotyping) | ||
| N/A invertase | LjINV1 | 5′-AGCTAGGTGAAGGTGTGATGCCAGCTA-3′ |
| 5′-CATAATTCCGAACGACGCATCAATGAA-3′ | ||
| 5′-GCATTGGCCGTTTTATGATGCAAAG-3′ | ||
| LjINV2 | 5′-GTGTTGCACCTGTGGATTCAGGTTGTT-3′ | |
| 5′-CAGAGTCACGAGACTTGGGGGTTCAAT-3′ | ||
| 5′-GCCCTACTCTATAGGTATATCACT-3′ | ||
TILLING
TILLING was carried out to isolate mutants for genes LjINV1 and LjINV2 using the general (GENPOP) and pre-selected nodule mutant population (NODPOP) as previously described (Perry et al., 2003). Gene-specific primers were designed using the CODDLE program (http://www.proweb.org/coddle/) to screen a 1.5 kb fragment of genomic sequence of each isoform. Primers were labelled with IRD 700 and IRD 800 and the TILLING procedure was carried out on a Li-Cor (Cambridge, UK) 4300 DNA analyser as previously described (Horst et al., 2007). Genotyping was carried out using gene-specific primers as indicated in Table 2.
Neutral/alkaline invertase assay
Material from either the first (LjINV1-3) or second (LjINV1-1 and LjINV1-2) outcrossed material was used for analysis.
Nodules
Between five and 10 nodules were harvested into a prechilled Eppendorf tube using fine forceps and weighed. The tissue was homogenized with a mini pestle with extraction buffer (50 mM HEPES, pH 7.5, 5 mM MgCl2, 1 mM EDTA, and 2 mM DTT) at a ratio of c. 5 mg tissue per 100 μl buffer. The suspension was assayed for N/A invertase activity as previously described by Hill et al. (2003).
Shoot tips/seedlings
Organs were harvested and extracted at a ratio of 20 mg tissue per 100 μl extraction buffer (as above). In these organs, there are substantial quantities of free glucose and fructose that interfere and hence need to be removed prior to assay. The values obtained for these extracts need to be expressed on a protein basis for accuracy. In order to remove these free sugars in the extract, the homogenate was centrifuged at 15 800 g at 4 °C for 10 min. One hundred and fifty to 500 μl supernatant was loaded onto an illustra™ Sephadex G-25, NAP5 column (GE Healthcare, Little Chalfont, UK) which had previously been equilibrated with extraction buffer. If less than 500 μl were used, the sample was washed onto the column with up to 350 μl of additional buffer. The protein was eluted with 600–1000 μl buffer and the eluant collected. The protein content of the purified extract was estimated using the Bradford reagent (Sigma, Poole, UK) with BSA as a standard. The N/A invertase activity was assayed as previously described (Hill et al., 2003). Three to five biological replicates were used for each assay.
Localization predictions
The following tools were used to predict the subcellular localization of the protein using full-length sequences of N/A invertase isoforms:
Predotar (http://urgi.versailles.inra.fr/predotar/predotar.html);
PSORT (http://wolfpsort.org/);
MitoprotII (http://ihg.gsf.de/ihg/mitoprot.html);
Targetp (http://www.cbs.dtu.dk/services/TargetP/);
ChloroP (http://www.cbs.dtu.dk/services/ChloroP/).
Cytology
Plant material from the Ljinv1-2 mutant and its corresponding wild type was cut into small pieces and placed in a solution of 2.5% (v/v) glutaraldehyde in 0.05 M sodium cacodylate, pH 7.3. The samples were then vacuum infiltrated briefly until they sank and were left overnight in fresh fixative. The fixative was removed by washing in 0.05 M sodium cacodylate and then the tissues were post-fixed in 1% (w/v) OsO4 in 0.05 M sodium cacodylate for 1 h at room temperature before washing in distilled water and dehydrating through an alcohol series. Once dehydrated, the samples were gradually infiltrated with LR White resin (London Resin Company, Reading, UK) by successive changes of resin/ethanol mixes over approximately 5 d at room temperature before being transferred to gelatine capsules in fresh resin. The resin was polymerized at 60 °C for 16 h. For light microscopy, semi-thin (0.5 μm) sections were dried onto glass slides and stained with 0.5% (w/v) Toluidine blue ‘O’ in 0.5% (w/v) borax. Digital photographs were taken on a Nikon E800 microscope and composites made and adjusted using Adobe Photoshop. Transmission electron microscopy was carried out on a FEI Tecnai 20 microscope using 0.09 μm thick sections stained with uranyl acetate and lead citrate.
Acetylene reduction assay
The ability to fix nitrogen was measured using the acetylene reduction assay as described previously (Horst et al., 2007), but using 3 ml vessels with the same acetylene concentration.
Sucrose measurements
Shoot tips were harvested, immediately frozen in liquid nitrogen and then freeze dried. Samples were then ground using tungsten beads in a Qiagen Tissuelyser. Approximately 2 mg of sample was extracted in 70% ethanol and the amount of sucrose present was measured by gas chromatography-mass spectrometry (GCMS) as previously described (Horst et al., 2007).
Results
The neutral/alkaline invertase gene family in L. japonicus
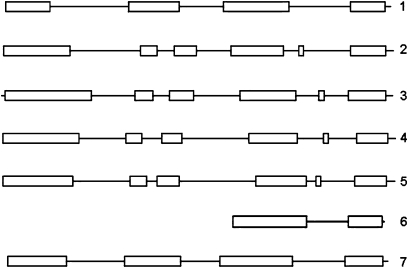
Seven genes, LjINV1 to LjINV7, encoding isoforms of N/A invertases were identified in total from data mining of the EMBL and Kazusa databases. Full-length genomic sequences were identified for six genes and a partial genomic sequence identified for one, designated LjINV6. The full accession numbers and assigned gene names are in Table 1. The genes encoding the LjINV isoforms fall into two classes based on their structures (Fig. 1): LjINV1 and LjINV7 with four exons and LjINV2, LjINV3, LjINV4, and LjINV5 with six exons. LjINV6 most likely falls in the group containing LjINV1 based on the sequence information we have to date (see below).
Fig. 1.
Exon–intron structure of the seven L. japonicus N/A invertase genes. The structure was determined using the CODDLE program (http://www.proweb.org/coddle/) and shows the same organization as that in rice (Ji et al., 2005) with six exons (boxes) for the α clade (LjINV2, LjINV3, LjINV4, LjINV5) and four for the β (LjINV1, LjINV7). The predicted structure for the partial sequence of LjINV6 places it in the β clade.
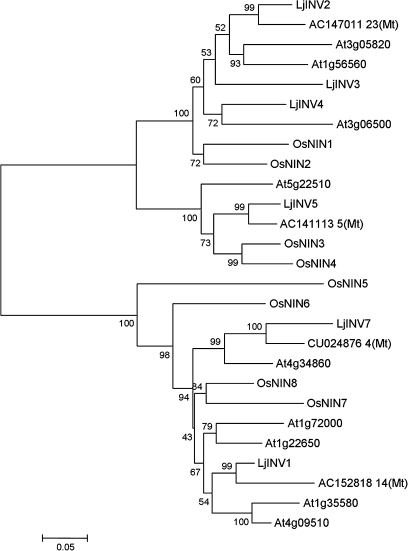
The phylogenetic relationship between the predicted sequences is shown in Fig. 2 in comparison to sequences from Arabidopsis, rice, and Medicago truncatula compiled using the Mega4 program. Since LjINV6 was only represented by a partial genomic sequence, its predicted protein sequence has not been included in this analysis. Only four INV genes could be identified in the latest release of sequences for the related model legume, Medicago truncatula (Mt2.0; www.medicago.org), however, alignment of the available sequence for LjINV6 with similar sequences in Medicago over the same region indicated that it was likely to be more similar to LjINV7 than the others. The dendrogram shows that the sequences fall into two clades, α (Fig. 2, upper) and β (Fig. 2, lower), as shown previously for rice (Ji et al., 2005). The α clade includes sequences some of which have been shown to be localized to organelles (Murayama and Handa, 2007). Following the procedures of Murayama and Handa (2007), therefore, the subcellular localization of the six full-length N/A invertases were predicted using the same five web tools. Three LjINV proteins were predicted to be localized to the mitochondria or plastids (see Supplementary Table S1 at JXB online) in accordance with their findings for the most closely related sequences in rice and Arabidopsis.
Fig. 2.
A phylogenetic dendrogram showing the relationships between the deduced amino acid sequences of N/A invertases in Lotus japonicus, Arabidopsis thaliana, Medicago truncatula, and Oryza sativa. The accession numbers for the named L. japonicus sequences are given in the text and are taken from Ji et al. (2005) for rice, the remainder are given in the figure. Bootstrap values are for 1000 datasets. Accession numbers for the L. japonicus genes are given in Table 1.
LjINV1 is the most highly expressed N/A invertase transcript
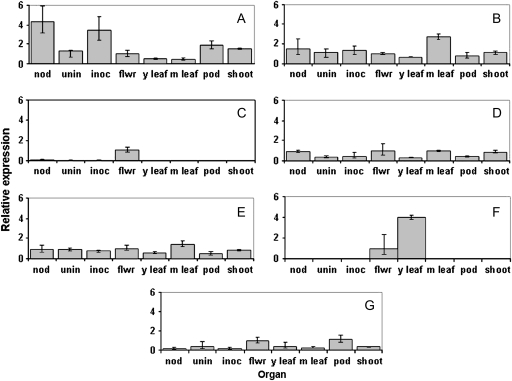
Transcript analysis was carried out using qRT-PCR for LjINV1–LjINV7 with primers specific to the 3′ region of each gene (Table 2). The predominant transcript in all organs examined except mature leaves was LjINV1. The levels of LjINV1 were 17-fold greater in inoculated and infected roots than the amount of next most abundant transcript, LjINV5. In other tissues, levels were up to 6-fold greater. Transcripts for most of the LjINV genes were present in all organs (Fig. 3) and some levels varied substantially between the organs; LjINV3 transcripts were predominantly in flowers (Fig. 3C) and LjINV6 only in flowers and young leaves (Fig. 3F). In young seedlings (2, 5, 7, or 9-d-old) the profile of transcripts was very similar to that of young leaves or uninfected roots, with LjINV1 the predominant transcript. Furthermore, the profiles did not change substantially throughout the period (see Supplementary Fig. S1 at JXB online).
Fig. 3.
Relative transcript levels of N/A invertases in different plant organs. Analysis by qRT-PCR of LjINV1 (A); LjINV2 (B); LjINV3 (C); LjINV4 (D); LjINV5 (E); LjINV6 (F); and LjINV7 (G) in nodules (nod), uninfected roots (unin), rhizobium-inoculated roots (inoc), flowers (flwr), young leaves (y leaf), mature leaves (m leaf), pods (pod), and shoots (shoot). Values are means ±SD from three individual plants. Transcript levels were expressed relative to the level in flowers after normalization to the levels of LjEF-1α. Note comparisons are within transcripts representing individual isoforms in different organs and the asymmetrical distribution of the SD caused by the conversion of the exponential process into a linear comparison (Livak and Schmittgen, 2001).
TILLING mutants indicate that LjINV1 is required for plant establishment and subsequent development
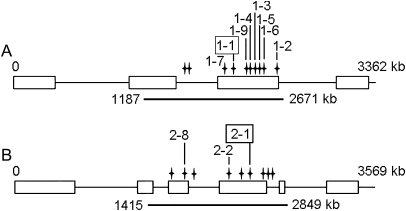
A population of EMS-mutagenized L. japonicus M2 lines (Perry et al., 2003) was used to screen for mutants in LjINV1 and LjINV2 using TILLING, the CODDLE prediction program being used to pick a suitable region of genomic sequence. Ten different alleles were detected across a c. 1.5 kb region of LjINV1 and nine for LjINV2 from a total of 12 lines as two mutations were detected in more than one line. For both genes, mutations were detected that caused premature stop codons together with a number of missense mutations with significant Position Specific Scoring Matrix (PSSM) changes according to CODDLE. The positions of the non-silent mutations are given in Fig. 4; the premature stop codon alleles were designated Ljinv1-1 and Ljinv2-1. (Full details of all the lines are given in Supplementary Tables S2 and S3 at JXB online.)
Fig. 4.
Positions of the non-silent mutations found via TILLING of LjINV1 (A); and LjINV2 (B). The underscore shows the position of the amplicon derived from the primer pair used (Table 2). The allele number bearing the mutation is given for each position and the corresponding line number can be found in Supplementary Tables 2 and 3. The two mutations producing premature stop codons are framed.
Recovery of some homozygous LjINV1 mutant lines proved problematic. The original mutations detected by TILLING were not homozygous and hence seed was sown to allow segregation of homozygous mutant plants. M3 generation plants were genotyped using allele-specific primers (Table 2), but no homozygous mutants were obtained for Ljinv1-1, Ljinv1-3, and Ljinv1-5 and only a single weak plant for Ljinv1-2 that maintained slow vegetative growth for more than a year, but never flowered and eventually died. The remaining lines segregated homozygous mutant plants. Of a total of 65 seed sown for LjINV1-1, the premature stop codon line, 43 were heterozygous and 22 homozygous wild type. At the next generation, larger numbers of seed were sown for the lines lacking homozygous mutants and the following numbers of plants were obtained (homozygous wild type/heterozygous/homozygous mutant): LjINV1-1 (78/114/4); LjINV1-2 (45/54/12); LjINV1-3 (88/131/10); LjINV1-5 (17/44/6). None of these values gives a ratio that is close to the expected ratio of 1:2:1. Of the M4 lines, none of the Ljinv1-1 mutants survived; many of the other mutant lines grew, but they did not produce seed. The Ljinv1-2 and Ljinv1-3 mutant plants had small, poorly expanded leaves and short stems. By contrast, all LjINV2 lines segregated normally and homozygous mutants were recovered in all cases. The phenotype of both seedlings and mature plants did not differ from that of the wild type (data not shown).
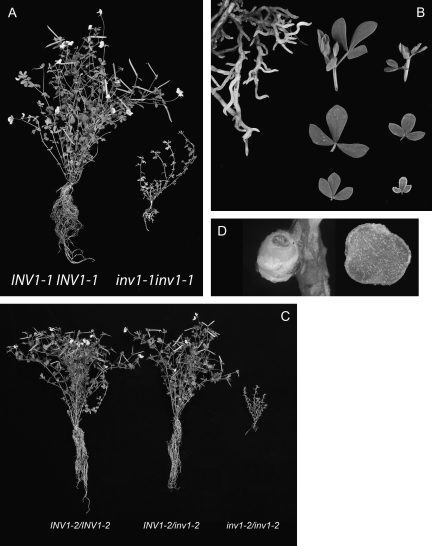
Crosses were carried out using LjINV1-1/inv1-1, LjINV1-2/inv1-2, and LjINV1-3/inv1-3 heterozygous plants to the wild type, MG20. L. japonicus MG20 is a more vigorous plant and is used as a mapping partner for L. japonicus ‘Gifu’, the background genotype of the TILLING population. Heterozygous plants were identified in the F1 population and seeds from these sown to segregate in the F2. In the F2 generation, two homozygous Ljinv1-1 mutants were identified from a population of 99 individuals, 14 homozygous Ljinv1-2 and 14 homozygous Ljinv1-3 mutants were identified from populations of 107 and 105 individuals, respectively. One of the Ljinv1-1 plants died, the phenotype at 19 weeks of the remaining one (with compost removed) is shown in Fig. 5A (and in colour in Supplementary Fig. S2 at JXB online), in comparison to its wild-type segregant. The plant had the same small poorly expanded leaves, short stems, and lack of mature flowers as M4 mutants above. Removal of the compost revealed it had a poorly developed root system with very short lateral roots (Fig. 5B). Furthermore, root hairs were not obvious as they were on wild-type plants. The leaves were much smaller and their margins chlorotic, especially in the ones closer to the apex (Fig. 5B). All Ljinv1-2 plants and Ljinv1-3 plants survived, and an example for Ljinv1-2 is shown in Fig. 5C at 16 weeks, in comparison with both wild-type and heterozygous F2 segregants. The phenotype of Ljinv1-2 and Ljinv1-3 plants was the same as that of the Ljinv1-1 mutant. This indicated that all three mutant alleles gave rise to the same heritable phenotype. Although the plants grew slowly and could be maintained for more than a year, they did not appear to flower. Subsequently, some flowers developed and these were examined microscopically (see below).
Fig. 5.
The phenotype of LjINV1 mutants. Wild type and Ljinv1-1 mutant after 19 weeks of growth (A); images (B), of the Ljinv1-1 roots (left) and shoot tips/leaflets (right); the top row shows a shoot tip of wild type (left) and mutant (right), the middle row the maximum and minimum leaf size for the wild type, and the bottom row the same for the mutant. Note the chlorosis of the smaller leaflet of the mutant. Wild type, heterozygote, and mutant plants for LjINV1-2 (C; from left to right, respectively). A nodule (D) on a Ljinv1-2 plant (left) and the same nodule cut open (bar=1 mm). A version of this figure in colour (Supplementary Fig. S2) can be found at JXB online.
Since the mutants did not produce mature, fertile flowers and since survival of Ljinv1-1 plants appeared to be improved following outcrossing to MG20, LjINV1-1 and LjINV1-2 heterozygous plants from the first outcross to MG20 were crossed to confirm that the phenotype was due to the mutations at the LjINV1 locus and not due to the background variation following EMS mutagenesis. Seed (51) from six successful crosses were sown (45 germinated) and the plants genotyped. This resulted in 14 wild type, 22 heterozygous for a single mutation and 9 (20%) heterozygous for both mutations. The mutant plants heterozygous for both mutations, i.e. possessing the two different, but both mutations, displayed the mutant phenotype and thus were non-complementing. These data combined with the heritable phenotype in the three different alleles, indicate that the observed phenotypes of both lines were due to mutations in the LjINV1 gene and not due to any background mutations following EMS mutagenesis.
The level of LjINV expression was examined in the shoot tips of the Ljinv1-1 and Ljinv1-2 mutants, their wild types and the heterozygotes segregating from the cross with MG20. The shoot tip was considered to be the most comparable organ and was the only one where sufficient material could be obtained for analysis without destroying the few plants available. LjINV1 transcript levels in the premature stop-codon mutant, Ljinv1-1 and the heterozygote were greatly reduced (31.6% and 59.9% of the wild type, respectively) indicating nonsense-mediated decay of this aberrant transcript whereas the expression in the Ljinv1-2 mutant, a missense mutant, was not significantly different in segregants. Moreover, this change in LjINV gene expression was the only one observed in the Ljinv1-1 mutant; no compensation by an increase in the expression of the genes encoding the other N/A invertase isoforms was observed (see Supplementary Fig. S3 at JXB online).
Neutral/alkaline invertase activity is severely decreased by mutant alleles of Ljinv1-1, Ljinv1-2, and Ljinv1-3, but not Ljinv2-1
To investigate further the basis of the phenotypes in the N/A invertase mutants, enzyme activities were measured. As a prelude to an analysis of segregating populations, activities (Table 3) in different organs of the wild types, Gifu and MG20 were assessed, and it was shown that there was little difference between the two ecotypes. Activity in whole seedlings reflected the expression levels and showed little change over a period of a week (see Supplementary Fig. S1 at JXB online).
Table 3.
N/A Invertase enzyme activities in wild-type Lotus japonicus plants 28 d post-inoculation
| Nodulea | Root tipsa | Shoot tipsb | |
| Gifu | 0.35±0.05 | 0.39±0.03 | 1.34±0.13 |
| MG20 | 0.22±0.08 | 0.33±0.03 | 1.48±0.14 |
Values are means ±SE of five replicates.
μmol hexose min−1 g−1 fresh weight.
μmol hexose h−1 mg−1 protein.
In initial experiments, homozygous mutant material was not available for most of the lines and so activity was measured in the nodules of wild types and heterozygotes. For the mutant lines with the most severe phenotypes, Ljinv1-1, Ljinv1-2, and Ljinv1-3, there was a decrease in enzyme activity in the heterozygotes (data not shown). Activity was also measured in the lines that gave homozygous mutants, Ljinv1-4, Ljinv1-6, and Ljinv1-7, following outcrossing. No significant difference was found for Ljinv1-6 and Ljinv1-7 mutants and a small difference was found for Ljinv1-4 (data not shown).
Outcrossings of the mutants, Ljinv1-1, Ljinv1-2, and Ljinv1-3 to the MG20 wild type was performed using heterozygotes since the mutants were infertile, and enzyme activities measured in F2 segregating populations in which the genotypes were determined by sequencing. Table 4 shows the enzyme activities in nodules and shoot tips of the wild type, heterozygote, and mutant line. As in the M4 generation, all three alleles showed reduced activity as heterozygotes in both nodules and shoot tips showing the trait was heritable. Shoot tips from the mutants showed a further reduction in activity, but not a complete lack of activity. The effect of the decrease in invertase activity in the shoots, however, was confirmed by GCMS analysis of shoot tips. Material from all three mutant lines examined showed an increase in sucrose content (Table 5). In normal growing conditions it proved very difficult to obtain sufficient mutant material for nodule assays. Hence, it was tested whether glucose (1%) would restore root growth as it does in rice and Arabidopsis (Qi et al., 2007; Jia et al., 2008) and found that root growth (but not shoot growth) was restored in Ljinv1 mutants sufficiently to increase nodulation (data not shown). Using nodules from one set of treated plants, it was found that N/A invertase activity was reduced to almost zero in the mutant (Table 4).
Table 4.
N/A invertase activities in nodules and shoot tips of Ljinv1 mutants
| Line | WT | HET | MUT |
| Nodule (μmol hexose min−1 g−1 fresh weight) | |||
| LjINV1-1 | 0.63±0.07 | 0.35±0.05 | n.t. |
| LjINV1-2 | 0.78±0.01 | 0.17±0.03 | n.t. |
| LjINV1-3 | 0.69±0.03 | 0.29±0.03 | 0.030±0.001 |
| Shoot tip (μmol h−1 mg−1 protein) | |||
| LjINV1-1 | 1.64±0.19 | 1.24±0.11 | 0.70±0.02 |
| LjINV1-2 | 1.96±0.28 | 1.33±0.03 | 1.12±0.20 |
| LjINV1-3 | 2.12±0.21 | 1.94±0.07 | 1.35±0.06 |
Values are means ±SE for 3–5 replicates of outcrossed lines. WT, wild-type segregant; HET, heterozygous segregant; MUT, homozygous mutant segregant; n.t., not tested.
Table 5.
Sucrose levels in shoot tips of Ljinv1 mutants
| Line | Genotype | Sucrose |
| LjINV1-1 | WT | 22.5±5.1 |
| HET | 31.7±2.6 | |
| MUT | 203.0±15.3 | |
| LjINV1-2 | WT | 27.9±6.1 |
| HET | 37.0±8.9 | |
| MUT | 147.9±13.8 | |
| LjINV1-3 | WT | 30.1±6.4 |
| HET | 43.8±9.2 | |
| MUT | 194.5±28.2 |
Values (μg mg−1 dry weight) are means ±SE for five replicates. WT, wild-type segregant; HET, heterozygous segregant; MUT, homozygous mutant segregant.
By contrast, N/A invertase activity was not significantly decreased in mature leaves of the Ljinv2-1 mutant line (0.37 μmol h−1 mg−1 protein) when compared to the wild type (0.39 μmol h−1 mg−1 protein) despite these tissues showing the highest transcript levels. This was most likely to be due to the expression of both LjINV1 and LjINV5 at comparable levels to LjINV2 in this organ.
A lack of LjINV1 activity leads to changes in tissue organization
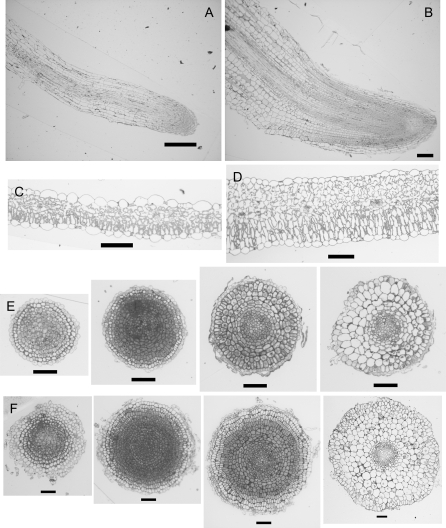
The dramatic phenotype of the Ljinv1-1 and Ljinv1-2 mutant plants led us to examine their tissue structures. Root tip and shoot material from wild-type and Ljinv1-2 segregants were fixed and embedded in plastic. Since the plants were very different in stature, leaf samples from just beneath the apical region were chosen together with the root tip as these were considered to be the most comparable. Sections stained in Toluidine Blue are shown in Fig. 6. As anticipated, the short stumpy roots of the mutant plants (Fig. 5) had a very different morphology to those of the wild type. Not only were the roots shorter and broader than the wild type (Fig. 6A, B), but the cells within them were increased in number (Fig. 6E, F). The number of cell files was also increased and the calculated cross-sectional area of the roots at its maximum was more than 20-fold greater in the mutant roots. Transverse sections through the root from the tip confirmed these changes, with extra cell layers in each zone of the root (Fig. 6E, F). The leaves were similar to the roots in that there were extra cell layers (e.g. three to four palisade layers instead of one) and enlarged cells (Fig. 6C, D).
Fig. 6.
Tissue morphologies of Ljinv1-2 mutant plants. Longitudinal sections through wild-type (A) and mutant roots (B). Cross-section of leaves of wild type (C) and mutant (D). Transverse sections at different positions along roots of wild type (E) and mutant (F) to show cell structure and organization. Bar=100 μm (A–D), 50 μm (E, F).
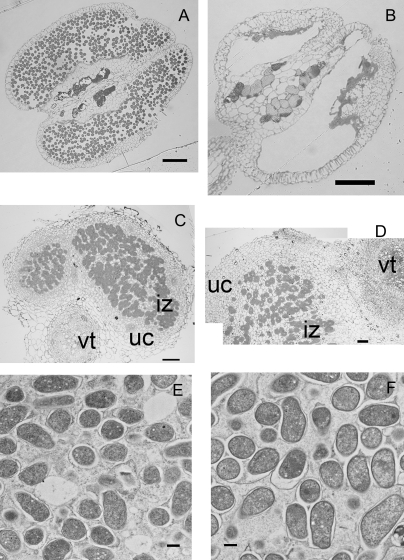
Since Jia et al (2008) noted a decrease in seed yield of the rice Oscyt-inv1 mutant that they proposed was due to low pollen viability, we also examined the few flowers that developed on our mutants. Figure 7A and B show anthers from wild-type and Ljinv1-1 mutant segregants from an outcross. It is clear from these images that there is a total absence of pollen in the mutant.
Fig. 7.
Structure of anthers and root nodule in Ljinv1 mutants. Sections through anthers of wild type (A) and mutant (B) showing a complete lack of pollen grains in the mutant. Sections through nodules of the wild type (B) and mutant (C; reconstructed from two overlapping images) showing infected central zone (iz) with bacteroids heavily stained surrounded by outer zone of uninfected cells (uc). In the top right for the mutant and bottom left for wild type the vascular tissue (vt) is shown. Sections (0.5 μm) were from LR white resin-embedded material and stained with Toluidine Blue (A–D). TEM thin sections (E, F; 0.09 μm) through wild type (E) and Ljinv1-2 mutants (F) showing bacteroids in an infected cortical cell. Bar=100 μm (A, B), 50 μm (C, D), and 0.5 μm (E, F).
A lack of LjINV1 activity does not prevent nodule formation or function
Since Suc synthase mutants that cannot assimilate nitrogen (Horst et al., 2007) can still form nodules, the question arose as to whether invertase supplies the carbon required to build a nodule. The Ljinv1 mutants and their wild types, therefore, were transferred to a vermiculite/perlite mixture, inoculated, and allowed to grow on for a further 3 weeks supplied with nutrient medium. Plants were then checked visually for signs of nodulation. A reduced number of mature nodules (Fig. 5D) was found on each mutant plant compared to the wild type or heterozygote (3±1 compared to 20±2 and 22±2, respectively). These displayed the normal wild-type structure when cut open including the red coloration indicating the presence of active leghaemoglobin (Fig. S2D). The reduction in number of nodules may have been an indirect consequence of the modified root growth as root hairs were not obvious on the mutant roots as mentioned earlier. Sections through the nodules of the wild type and Ljinv1-2 mutant showed that both types had the normal organization with a heavily stained infected zone interspersed with uninfected, vacuolate cells with little staining, and surrounded by uninfected cells of the outer cortex (Fig. 7C, D). Both the infected and uninfected mutant cells, however, were larger and more vacuolated than the corresponding cells of the wild type, as were the cells of the vascular trace. There also appeared to be a greater number of non-infected cells in the mutant although this may have been an artefact of the increase in number and volume of these cells. Both nodules accumulated starch grains that were similar in number. Examination at high magnification showed that the bacteroids were unaffected in the mutant (Fig. 7F) and they were as capable of fixing nitrogen as the wild type or heterozygote when measured by the acetylene reduction assay (3.52±0.55 compared to 4.04±1.26 and 4.44±0.4 nmol ethylene h−1 mg−1 fresh weight).
Discussion
N/A invertases are quite distinct from their acidic counterparts, so much so that they cannot be included in the same phylogeny. Their commonality resides only in their ability to breakdown sucrose to hexoses. It has been shown that L. japonicus possesses a small gene family of seven N/A invertases, LjINV1 to LjINV7, smaller than either rice (Ji et al., 2005), Arabidopsis (Qi et al., 2007) or poplar (Bocock et al., 2008). Only a partial sequence was obtained for LjINV6, but from our alignments it is most likely to be similar to LjINV7. This would yield three α-types and four β-types (Ji et al., 2005) indicating that one family member may be missing from the available Lotus genome sequence although, based on the close relationship between some pairs of genes, selective duplication appears to have occurred in different species. The situation regarding organellar localization was much less clear than in rice (Murayama and Handa, 2007; Vargas et al., 2008) for all isoforms, especially LjINV5. It was not predicted to be targeted to an organelle since it received quite low scores in all programs, despite being closely related to the recently discovered chloroplastic isoform of Arabidopsis (At-N/A-InvE, At5g22510; Vargas et al., 2008) and OsNIN3, also predicted to be plastidial (Murayama and Handa, 2007). Experimental confirmation of location, therefore, will be needed in Lotus.
Like the closely related rice sequence, OsNIN8 (Ji et al., 2005), LjINV1, was expressed more highly than the other genes in most tissues and was expressed in all organs of the plant (Fig. 3). Flemetakis et al. (2006) detected only two types of N/A invertase ESTs, representing LjINV1 and LjINV2, in cDNA libraries of L. japonicus, but only LjINV1 was detected in nodule libraries. The relatively high expression of LjINV1 in all organs shown here including the highest expression in the nodule and inoculated roots, explains the ease of its detection in their EST libraries. Similarly, the low expression of LjINV2 in the nodule would explain its lack of detection in this organ. It can also be confirmed that LjINV1 shows nodule-enhanced expression as found by Flemetakis et al. (2006).
From the phenotype of the Ljinv1-1, Ljinv1-2, Ljinv1-3 mutant plants, it is clear that these mutations lead to a severe impairment of growth and development in the root tissues, shoot tissues, and microspores through a decrease in N/A invertase activity in the plant and a severe decrease in the nodule (Table 4). The high background level of activity in the shoot tips of mutants was most likely due to the high expression of genes encoding other isoforms relative to that of LjINV1. The phenotype of the mutants was heritable following selfing and outcrossing, and was observed in all three independent alleles. Hence it can be concluded that the phenotype was due to the mutations in LjINV1 and not due to EMS-induced background mutations remaining in the lines.
In the absence of LjINV1 activity, Lotus plants struggle to establish themselves and cannot grow to maturity although they can survive for a considerable length of time and will produce flowers, albeit infertile ones due to a lack of pollen (Fig. 7A, B). Clearly, the lack of invertase activity was preventing correct organ and cellular development, but not general cell expansion, and this most likely led to reduced organ elongation. The overall morphology of the root was similar to the rice mutant (Jia et al., 2008), but there were notable differences at the cellular level in that the rice mutant did not show an increase in root breadth or the number of cell files, but an overall reduction in cortical cell elongation and a change in cell shape. Cells were described as shrunken with their walls inlaid on each other. This particular change was not observed here. Indeed the transverse sections of the root showed that the cells remained turgid, were rich in cytoplasm and remained in division for a much greater distance from the root tip (Fig. 6E, F) than in the wild type indicating an extension of the cap and division zone. The phenotype of the L. japonicus mutants is much more reminiscent of the severe effect on seedling growth of over-expressing acid invertase inhibitors in Arabidopsis and tobacco under inducible promoters (Bonfig et al., 2007).
The severity of the phenotype of the LjINV1 mutant may reflect merely the different patterns of carbon partitioning in Lotus compared to rice or Arabidopsis, but perhaps a simpler explanation arises from the phylogeny of the genes (Fig. 2). In Arabidopsis, there are two closely related genes in the subclade, whereas in Lotus there is a singleton. Hence, any deleterious mutation in the Lotus gene is likely to have a more dramatic effect on plant growth than in Arabidopsis where the gene duplication may have given rise to some level of redundancy of function. This is borne out by the very recent work of Barratt et al. (2009) who generated a double mutant between the two corresponding invertase knockout lines (for At1g35580 and At4g09510). These mutants were recovered at low frequency as was the case in our study and they showed a similar phenotype: poor seedling establishment and weak plants. The Arabidopsis double mutants, however, did flower, albeit much later than the single mutants or the wild type, and they set some seed. The weaker phenotype of the rice mutant, OsCyt-inv1 (Ji et al., 2005) may be explained similarly; in our phylogeny (Fig. 2) the proteins encoded by OsNIN8 and OsNIN7 are closely related to each other and are the most closely related proteins to the product of LjINV1.
Previous studies have indicated that acid invertases are important in seed development both of legumes (Weber et al., 1997) and monocots (Chourey et al., 2006) to deliver hexoses in support of mitotic activity, but this is unlikely to be the case here for N/A invertases in Lotus since the observed changes in the mutants are complex, prolonging cell division and affecting both cell size and tissue organization. Unlike Arabidopsis and rice, therefore, mutants obtained by TILLING in L. japonicus indicate that LjINV1 is crucial for many aspects of plant growth and development. This was exemplified by the additional dimension afforded by the use of a legume—the root nodule. Enhanced expression of LjINV1 can be detected within 2 d of inoculation with rhizobium (Kouchi et al., 2004), but LjINV1 activity clearly is not required for nodule formation or function. Even though a loss of LjINV activity does alter the organization of the tissue, it is not a nodule-specific phenomenon as it has the same effect as observed in all the other plant tissues examined; maintaining correct cell proliferation and expansion of the ground tissues. The Lotus phenotype is somewhat surprising given the proposed roles for invertases, albeit the acidic forms. If a decrease in N/A activity is indeed promoting extra cell divisions leading to an extended zone of proliferation and extra cell layers, it may indicate a very specialized and novel role for some members of this family.
In contrast to LjINV1, the main Suc synthases in Lotus, including the nodule-enhanced isoform, are required for nodule function, but not for plant growth, as exemplified by the phenotype of the Suc synthase double mutant in the absence of nitrate (Horst et al., 2007). This mutant cannot grow unless additional nitrate is supplied, but there is no dramatic invertase-like phenotype in the presence of added nitrogen (Horst et al., 2007). Nodule-enhanced expression as shown by both LjSUS3 and LjINV1, therefore, is not a robust indicator of a specific role in the nodule. In Ljinv1 mutants other nodule N/A invertase isoforms may be supplying hexoses for this process or Suc synthase may be compensating for the lack of invertase. The converse is not the case for invertase, since invertase cannot compensate for a lack of Suc synthase activity in L. japonicus or Pisum sativum (Craig et al., 1999; Horst et al., 2007). The possibility cannot be excluded, however, that very low levels of invertase can support the nodule. Now that this study and other recent ones (Qi et al., 2007; Jia et al., 2008; Vargas et al., 2008) have uncovered roles for some N/A invertases, the challenge will be to determine how this group of enzymes interacts to influence sucrose metabolism, not only in the cytosol alongside sucrose synthases, but also in concert with their relatives in the cell wall and vacuole. In the light of the recent crystallography results on an AtcwINV1 mutant's binding to sucrose (Matrai et al., 2008), the Lotus TILLING mutants may be particularly helpful in defining some of the properties of the N/A invertase enzymes.
In conclusion, it has been shown that reduced N/A invertase activity caused by mutations in LjINV1 affects both root and aerial parts of the plant through an effect on cell proliferation and expansion, but they do not affect the ability of the plant to form functional nodules. In this respect, the phenotype of Lotus mutants is more severe than in either the Arabidopsis or rice mutants. Phylogenetic analysis indicates that the weaker phenotype is most likely due to gene duplication is these two species. Examination of further isoforms by reverse genetic means will no doubt reveal additional novel functions for this less-well studied family of sucrose-metabolizing enzymes.
Supplementary data
The following data are available at JXB online.
Supplementary Table S1. Subcellular location data for N/A invertase isoforms.
Supplementary Table S2. L. japonicus INV1 alleles.
Supplementary Table S3. L. japonicus INV2 alleles.
Supplementary Fig. S1. Expression of LjINV genes and enzyme activities during seedling growth.
Supplementary Fig. S2. Colour plate of Fig. 5.
Supplementary Fig. S3. Expression of INV genes in the Ljinv1-1 mutant.
Acknowledgments
This work was supported by the Biotechnology and Biological Sciences Research Council via a grant-in-aid to the John Innes Centre. We should like to thank Andrew Davies for photography, Sue Bunnewell for cytology, and the Horticultural Services staff for plant husbandry. We should also like to thank Cathie Martin and Alison Smith for helpful comments on the manuscript.
References
- Avigad G. Sucrose and other disaccharides. In: Loewus FA, Tanner W, editors. Encyclopedia of plant physiology. Vol. 13A. Berlin: Springer-Verlag; 1982. pp. 217–347. [Google Scholar]
- Baier MC, Barsch A, Kuster H, Hohnjec N. Antisense repression of the Medicago truncatula nodule-enhanced sucrose synthase leads to a handicapped nitrogen fixation mirrored by specific alterations in the symbiotic transcriptome and metabolome. Plant Physiology. 2007;145:1600–1618. doi: 10.1104/pp.107.106955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barratt DHP, Derbyshire P, Findlay K, Pike M, Wellner N, Lunn J, Feil R, Simpson C, Maule A, Smith AM. Sucrose catabolism in Arabidopsis requires cytosolic invertase but not sucrose synthase. Proceedings of the National Academy of Sciences, USA. 2009 doi: 10.1073/pnas.0900689106. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bocock PN, Morse AM, Dervinis C, Davis JM. Evolution and diversity of invertase genes in Populus trichocarpa. Planta. 2008;227:565–576. doi: 10.1007/s00425-007-0639-3. [DOI] [PubMed] [Google Scholar]
- Bonfig KB, Berger S, Fatima T, Gonzalez MC, Roitsch T. Metabolic control of seedling development by invertases. Functional Plant Biology. 2007;34:508–516. doi: 10.1071/FP06206. [DOI] [PubMed] [Google Scholar]
- Chourey P, Jain M, Li QB, Carlson S. Genetic control of cell wall invertases in developing endosperm of maize. Planta. 2006;223:159–167. doi: 10.1007/s00425-005-0039-5. [DOI] [PubMed] [Google Scholar]
- Craig J, Barratt P, Tatge H, et al. Mutations at the rug4 locus alter the carbon and nitrogen metabolism of pea plants through an effect on sucrose synthase. The Plant Journal. 1999;17:353–362. [Google Scholar]
- Flemetakis E, Efrose RC, Ott T, Stedel C, Aivalakis G, Udvardi MK, Katinakis P. Spatial and temporal organization of sucrose metabolism in Lotus japonicus nitrogen-fixing nodules suggests a role for the elusive alkaline/neutral invertase. Plant Molecular Biology. 2006;62:53–69. doi: 10.1007/s11103-006-9003-4. [DOI] [PubMed] [Google Scholar]
- Geigenberger P, Stitt M. Sucrose synthase catalyzes a readily reversible reaction in vivo in developing potato tubers and other plant tissues. Planta. 1993;189:329–339. doi: 10.1007/BF00194429. [DOI] [PubMed] [Google Scholar]
- Handberg K, Stougaard J. Lotus japonicus, an autogamous, diploid legume species for classical and molecular genetics. The Plant Journal. 1992;2:487–496. [Google Scholar]
- Hill LM, Morley-Smith ER, Rawsthorne S. Metabolism of sugars in the endosperm of developing seeds of oilseed rape. Plant Physiology. 2003;131:228–236. doi: 10.1104/pp.010868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horst I, Welham T, Kelly S, Kaneko T, Sato S, Tabata S, Parniske M, Wang TL. TILLING mutants of Lotus japonicus reveal that nitrogen assimilation and fixation can occur in the absence of nodule-enhanced sucrose synthase. Plant Physiology. 2007;144:806–820. doi: 10.1104/pp.107.097063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji XM, van den Ende W, van Laere A, Cheng SH. Structure, evolution, and expression of the two invertase gene families of rice. Journal of Molecular Evolution. 2005;60:615–634. doi: 10.1007/s00239-004-0242-1. [DOI] [PubMed] [Google Scholar]
- Jia LQ, Zhang BT, Mao CZ, Li JH, Wu YR, Wu P, Wu ZC. OsCYT-INV1 for alkaline/neutral invertase is involved in root cell development and reproductivity in rice (Oryza sativa L.) Planta. 2008;228:51–59. doi: 10.1007/s00425-008-0718-0. [DOI] [PubMed] [Google Scholar]
- Koch K. Sucrose metabolism, regulatory mechanisms and pivotal roles in sugar sensing and plant development. Current Opinion in Plant Biology. 2004;7:235–246. doi: 10.1016/j.pbi.2004.03.014. [DOI] [PubMed] [Google Scholar]
- Kouchi H, Shimomura K, Hata S, et al. Large-scale analysis of gene expression profiles during early stages of root nodule formation in a model legume, Lotus japonicus. DNA Research. 2004;11:263–274. doi: 10.1093/dnares/11.4.263. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lou Y, Gou JY, Xue HW. PIP5K9, an Arabidopsis phosphatidylinositol monophosphate kinase, interacts with a cytosolic invertase to negatively regulate sugar-mediated root growth. The Plant Cell. 2007;19:163–181. doi: 10.1105/tpc.106.045658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matrai J, Lammens W, Jonckheer A, Le Roy K, Rabijns A, Van den Ende W, De Maeyer M. An alternate sucrose binding mode in the E203Q Arabidopsis invertase mutant: an X-ray crystallography and docking study. Proteins: Structure, Function, and Bioinformatics. 2008;71:552–564. doi: 10.1002/prot.21700. [DOI] [PubMed] [Google Scholar]
- Murayama S, Handa H. Genes for alkaline/neutral invertase in rice, alkaline/neutral invertases are located in plant mitochondria and also in plastids. Planta. 2007;225:1193–1203. doi: 10.1007/s00425-006-0430-x. [DOI] [PubMed] [Google Scholar]
- Perry JA, Wang TL, Welham TJ, Gardner S, Pike JM, Yoshida S, Parniske M. A TILLING reverse genetics tool and a web-accessible collection of mutants of the legume Lotus japonicus. Plant Physiology. 2003;131:866–871. doi: 10.1104/pp.102.017384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi XP, Wu ZC, Li JH, Mo XR, Wu SH, Chu J, Wu P. AtCYT-INV1, a neutral invertase, is involved in osmotic stress-induced inhibition on lateral root growth in Arabidopsis. Plant Molecular Biology. 2007;64:575–587. doi: 10.1007/s11103-007-9177-4. [DOI] [PubMed] [Google Scholar]
- Roitsch T, Gonzalez M-C. Function and regulation of plant invertases, sweet sensations. Trends in Plant Science. 2004;9:1360–1385. doi: 10.1016/j.tplants.2004.10.009. [DOI] [PubMed] [Google Scholar]
- Szarka A, Horemans N, Passarella S, Tarcsay A, Örsi F, Salgó A, Bánhegyi G. Demonstration of an intramitochondrial invertase activity and the corresponding sugar transporters of the inner mitochondrial membrane in Jerusalem artichoke (Helianthus tuberosus L.) tubers. Planta. 2008;228:765–775. doi: 10.1007/s00425-008-0778-1. [DOI] [PubMed] [Google Scholar]
- Vargas W, Cumino A, Salerno GL. Cyanobacterial alkaline/neutral invertases. Origin of sucrose hydrolysis in the plant cytosol? Planta. 2003;216:951–960. doi: 10.1007/s00425-002-0943-x. [DOI] [PubMed] [Google Scholar]
- Vargas W, Pontis HG, Salerno GL. New insights on sucrose metabolism: evidence for an active A/N Inv in chloroplasts uncovers a novel component of the intracellular carbon trafficking. Planta. 2008;227:795–807. doi: 10.1007/s00425-007-0657-1. [DOI] [PubMed] [Google Scholar]
- Weber H, Borisjuk L, Wobus U. Sugar import and metabolism during seed development. Trends in Plant Science. 1997;2:169–174. [Google Scholar]
- Winter H, Huber SC. Regulation of sucrose metabolism in higher plants: localization and regulation of activity of key enzymes. Critical Reviews in Plant Science. 2000;19:31–67. doi: 10.1080/10409230008984165. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.