Abstract
A high-affinity K+ transporter PutHKT2;1 cDNA was isolated from the salt-tolerant plant Puccinellia tenuiflora. Expression of PutHKT2;1 was induced by both 300 mM NaCl and K+-starvation stress in roots, but only slightly regulated by those stresses in shoots. PutHKT2;1 transcript levels in 300 mM NaCl were doubled by the depletion of potassium. Yeast transformed with PutHKT2;1, like those transformed with PhaHKT2;1 from salt-tolerant reed plants (Phragmites australis), (i) were able to take up K+ in low K+ concentration medium or in the presence of NaCl, and (ii) were permeable to Na+. This suggests that PutHKT2;1 has a high affinity K+-Na+ symport function in yeast. Arabidopsis over-expressing PutHKT2;1 showed increased sensitivities to Na+, K+, and Li+, while Arabidopsis over-expressing OsHKT2;1 from rice (Oryza sativa) showed increased sensitivity only to Na+. In contrast to OsHKT2;1, which functions in Na+-uptake at low external K+ concentrations, PutHKT2;1 functions in Na+-uptake at higher external K+ concentrations. These results show that the modes of action of PutHKT2;1 in transgenic yeast and Arabidopsis differ from the mode of action of the closely related OsHKT2;1 transporter.
Keywords: Potassium transporter, Puccinellia tenuiflora, PutHKT2;1, salt stress, salt tolerance
Introduction
Soil salinity is one of the most important abiotic stresses for plants and is a substantial constraint to agricultural production worldwide. Understanding the molecular mechanism of salt stress tolerance and subsequently developing salt-tolerant crops are now essential for solving the current problem of salt stress. High salinity causes both osmotic and ionic stress and can lead to growth inhibition and plant death. For many plants, such as graminaceous crops, sodium disequilibrium is the primary consequence of ionic stress and often leads to adverse effects on plant nutrition, enzyme activities, photosynthesis, and metabolism (Tester and Davenport, 2003; Mahajan and Tuteja, 2005; Munns and Tester, 2008).
In saline environments, high external Na+ favours Na+ entry down the electrochemical gradient across the plasma membrane. Outright exclusion of Na+ and vacuolar compartmentalization of internal Na+ are some of the mechanisms by which plants acquire salt tolerance. Plasma membrane Na+/H+ antiporters, also known as SOS1 or NHA-type transporters, are responsible for extruding Na+ out of the salt-stressed cell, while vacuolar membrane Na+/H+ antiporters, also known as NHX-type antiporters, are responsible for the sequestration of Na+ from the cytosol into vacuoles under salt stress (Hasegawa et al., 2000; Zhu, 2003; Munns and Tester, 2008; Mahajan et al., 2008).
A third mechanism for improving salt tolerance is gaining control over Na+ influx. Toxic Na+ influx into roots under high external Na+ concentrations has been suggested to be mediated by non-selective cation channels (NSC) or voltage-independent channels (VIC) (Amtmann and Sanders, 1999; Schachtman and Liu, 1999; Blumwald et al., 2000; Maathuis and Sanders, 2001; Demidchik and Tester, 2002). However, the detailed molecular identities of NSC/VIC remain unknown. Finally, high-affinity K+ transporters (HKT-type transporters) could mediate Na+ entry into plant cells.
HKT-type transporters have been characterized in several plant species (Schachtman and Schroeder, 1994; Rubio et al., 1995; Fairbairn et al., 2000; Uozumi et al., 2000; Horie et al., 2001; Garciadeblas et al., 2003; Su et al., 2003; Ren et al., 2005), and the known plant HKTs have been shown to perform diverse functions. For example, TaHKT2;1 of wheat showed a dual mode of action in a heterologous expression system. That is, it acted as a Na+-K+-symporter at low [NaExt+] but switched to a Na+-uniporter at high [NaExt+] (Rubio et al., 1995; Gassmann et al., 1996). Genetic knock-down of TaHKT2;1 in wheat reduced the Na+ content of root exudates and enhanced the growth of transgenic plants under salinity (Laurie et al., 2002) showing that TaHKT2;1 can function as a Na+-uptake pathway in wheat roots. AtHKT1;1 from Arabidopsis and OsHKT2;1 from rice were also characterized as Na+-uniporters in heterologous expression systems (Uozumi et al., 2000; Horie et al., 2001; Garciadeblas et al., 2003). Further functional characterization in planta by genetic mutations revealed that AtHKT1;1 does not mediate Na+-influx into Arabidopsis roots (Maser et al., 2002a; Berthomieu et al., 2003; Horie et al., 2006), while OsHKT2;1 mediates Na+-uptake into K+-starved roots (Horie et al., 2007).
Because most of the economically important crops are monocotyledonous, understanding the salt-tolerance mechanisms of monocotyledonous halophytes will aid in improving the salt tolerance of cereals. HKT genes have been isolated from several halophytes (Liu et al., 2001; Takahashi et al., 2007; Shao et al., 2008), but so far only one of them, the reed plant Phragmites australis, is a monocotyledon. Puccinellia tenuiflora (Griseb.) Scrib. et Merr. is a monocotyledonous plant found in saline-alkali soil in China, and is highly tolerant to salinity stress (Peng et al., 2004; Wang et al., 2004, 2007; Zhang et al., 2008). The salt tolerance of P. tenuiflora is thought to be due to its ability to restrict the unidirectional Na+ influx to the root cells which leads to a large Na+ concentration gradient between roots and shoot (Wang et al., 2009). The endodermal barrier to Na+ in P. tenuiflora roots was proposed to restrict Na+ uptake as the uptake of K+ is maintained (Peng et al., 2004). Although high K+/Na+ selective transporter(s) were suggested to contribute in the restriction of unidirectional Na+ influx (Wang et al., 2009), the molecular identities of these channels/transporters have not been reported.
In this study, a cDNA encoding an HKT-type potassium transporter was isolated from P. tenuiflora (PutHKT2;1) and its expressions has been analysed under K+-starvation and salt stress conditions. The function of PutHKT2;1 was investigated in transgenic yeast and Arabidopsis and compared with closely related OsHKT2;1 of rice and PhaHKT2;1 (previously named PhaHKT1, Takahashi et al., 2007) of the reed plant.
Materials and methods
Plant materials, growth conditions, and stress treatments
Seeds of P. tenuiflora were collected in an alkaline soil area located in North-East China. Plants were germinated in tap water for 14 d and then transferred to nutrient solution and allowed to grow for another 14 d before stress treatments. The nutrient solution contained 0.75 mM NH4NO3, 0.5 mM KCl, 0.25 mM NaH2PO4, 0.75 mM CaCl2, 0.5 mM MgCl2, 0.075 μM ZnSO4, 0.032 μM CuSO4, and 2 mg l−1 Fe-EDTA. The temperatures of the growth chamber were maintained at 28 °C during the day and 22 °C at night while the daily photoperiod of 350–400 mmol m−2 s−1 was 12 h. For the salt-stress treatment, NaCl was added to this solution at 300 mM. For the K+-starvation treatment, KCl was removed from the standard solution. Plants were harvested at 6, 12, and 24 h after stress treatments and preserved in –80 °C for further analyses.
Cloning of HKT cDNA from P. tenuiflora (PutHKT2;1)
Total RNA was extracted from the shoots of P. tenuiflora using RNeasy plant extraction Mini Kit (Qiagen, Germany) according to the manufacturer's instructions. Total RNA was reverse transcribed by using an oligo dT primer and MMLV-reverse transcriptase. The partial cDNA fragment was amplified by PCR, using a degenerate forward primer (5′-TCCATATCAAGCTGCATAGC-3′) and a reverse primer (5′-AGCCTTCCATAGAGCATGAC-3′) deduced from the conserved regions of HKT, and reverse transcription product as a template. Full-length cDNA was obtained using 5′- and 3′-RACE techniques. The full-length cDNA of PutHKT2;1 was amplified using a forward primer (5′-ATATAACATCATGGGTCGGGTGAAG-3′) and a reverse primer (5′-GCAAAGGCAAGTGTACCAAATTGGT-3′). The PCR product was subcloned into pBluescript vector and sequenced.
Extraction of genomic DNA and Southern hybridization
Genomic DNA of P. tenuiflora was isolated from 4-week-old plants using the CTAB method. The genomic DNAs (20 μg lane−1) were digested with restriction enzymes, BamHI and HindIII, separated on a 0.8% (w/v) agarose gel, and transferred to a Hybond-N+ nylon membrane (GE Healthcare Life Sciences, UK). The membrane was hybridized overnight at 45 °C with a digoxigenin (DIG)-labelled PutHKT2;1 cDNA probe that was amplified by PCR with a forward primer (5′-AGGAGAACACGAAAGGCAAGAGATG-3′) and a reverse primer (5′-TCCATTGCAAGCGCAACACGTATAT-3’) using PCR DIG Labeling Mix (Roche Diagnostics, Switzerland). The signals were detected with CDP-Star detection reagent (GE Healthcare Life Sciences, UK) using a LAS-1000 plus image analyser (Fuji Film, Japan).
RNA extraction and cDNA synthesis for real-time RT-PCR
For real-time RT-PCR, total RNA was extracted from the roots and shoots of P. tenuiflora subjected to 300 mM NaCl, K+-starvation, or both stresses using the RNeasy plant extraction Mini Kit (Qiagen, Germany) according to the manufacturer's instructions. Two micrograms of DNase-treated RNA was reverse transcribed to cDNA using the High Capacity RNA-to-cDNA Kit (Applied Biosystems, USA) following the manufacturer's instructions. The cDNA was diluted 10 times and 1 μl of the diluted cDNA was used as the template in each well for quantitative real-time PCR analysis. The cDNA was amplified using Power SYBR Green PCR Master Mix (Applied Biosystems, USA) on the ABI 7300 thermocycler (Applied Biosystems, USA). A tubulin gene from P. tenuiflora was cloned and used as an internal standard to normalize the expression data for the PutHKT2;1 gene. The following primers were used: 5′-TTCCATCGACTGCTCACTCA-3′ and 5′-CCATCACTGGGTGGTGCAAA-3′ for PutHKT2;1, 5′-GCTGACCACACCTAGCTTCGGGG-3′ and 5′-GACCAGGGAACCTCAGGCAGC-3′ for tubulin. The PCR was performed as follows: 50 °C for 2 min and 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s and 60 °C for 1 min. A dissociation kinetic analysis was performed to check the specificity of the annealing. Three replications were performed for each sample. Standard determination curves were generated using serial dilutions of 1000, 500, 250, 125, and 62.5 ng μl−1 cDNA. The results were analysed according to Muller et al. (2002).
Subcellular localization of PutHKT2;1::GFP fusion protein
A pBluscript plasmid containing a green fluorescent protein (GFP) gene, sGFP(S65T), was digested by KpnI and SalI. The PutHKT2;1 open reading frame (ORF) containing KpnI and SalI sites was amplified by PCR, digested, and fused in-frame with the GFP. Transient expression of GFP fusion constructs was performed via bombardment of onion epidermal cells using a Biolistic PDS/He system (Bio-Rad, USA) following standard protocols. After bombardment, the cells were recovered on plates for 10 h at 28 °C in the dark before observation using an Olympus BX51 microscope (Olympus, USA).
Functional characterization of PutHKT2;1 in yeast
The ORF of PutHKT2;1, OsHKT2;1, and PhaHKT2;1 was inserted into the protein expression vector pAUR123 (Takara, Japan), and introduced into Saccharomyces cerevisiae strain 9.3 (ATCC 201409), in which the original potassium transporters (TRK1, 2) and P-type ATPase involved in Na+ extrusion (ENA1–4) were deleted. The ion uptake experiment was performed as described previously (Takahashi et al., 2007). For the yeast growth test experiment, transformed yeasts were cultured overnight at 30 °C in SC/-His medium containing 0.5 μg/ml Aureobasidin until the OD600 reached 0.8, and 10-fold serial diluted cultures were incubated on SC/-His plates containing the indicated concentrations of K+ and Na+. The plates were incubated at 30oC for 5 d. For both ion uptake and yeast growth test experiments, control experiment were performed with the yeast 9.3 strain transformed with plasmid pAUR123 without an insert.
Generation of transgenic Arabidopsis plants over-expressing the PutHKT2;1 and OsHKT2;1 genes
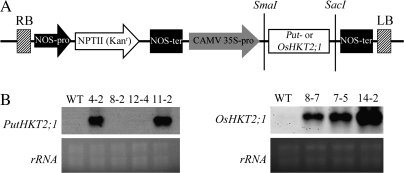
The coding regions of PutHKT2;1 and OsHKT2;1 were subcloned into the plant transformation binary vector pBI121 (Clontech, Japan) under the control of CaMV 35S promoters with nptII as the selectable marker for kanamycin resistance (Fig. 1A). The constructs were introduced into Agrobacterium tumefaciens strain EHA 105. Arabidopsis (ecotype Colombia) was transformed by the floral dip method (Clough and Bent, 1998). A total of 14 PutHKT2;1 and 16 OsHKT2;1 independent T0 transgenic plants were obtained by screening kanamycin-resistant regenerated Arabidopsis plants. An RNA gel blot analysis of the T0 transgenic plants showed the introduced gene transcript in 10 out of 14 lines of PutHKT2;1 and in 13 out of 16 lines of OsHKT2;1 with varied expression levels among the lines (data not shown). Expression of the transgene was confirmed in the T2 generation lines by an RNA gel blot analysis (Fig. 1B). Lines 4-2 and 11-2 of PutHKT2;1 and lines 7-5 and 14-2 of OsHKT2;1 transgenic plants were used for root length assays and ion content measurements. All of the lines used in these experiments were homozygous, except for line 4-2 of PutHKT2;1. This heterozygous PutHKT2;1 no. 4-2 was used based on the PutHKT2;1 expression level. The germination rate of PutHKT2;1 no. 4-2 seeds were more than 80% on the kanamycin selection medium. The seedlings that survived on the selection medium consisted of both homozygous and heterozygous plants. However, no difference was observed in the phenotype of the seedlings. In addition, the phenotype of PutHKT2;1 no. 4-2 seedlings were indistinguishable from those of the homozygous PutHKT2;1 no. 11-2 seedlings, both under normal or stress conditions. These results suggest that the introduced gene action is dominant.
Fig. 1.
(A) Construction of plasmid pBI121 that contains PutHKT2;1 or OsHKT2;1 open reading frame driven by the CaMV 35S promoter and nptII. RB, right border; LB, left border. (B) Expression of PutHKT2;1 and OsHKT2;1 in wild-type (WT) and T2 generation transgenic lines. Total RNA (4.5 μg) extracted from 4-week-old seedlings were analysed by RNA gel blot.
Functional comparison of PutHKT2;1 and OsHKT2;1 in transgenic Arabidopsis
Seeds were surface-sterilized with 70% (v/v) ethanol for 2 min and 1% (v/v) NaClO solution for 15 min. The seeds were rinsed three times with water, and sown on MS medium containing 1.5% (w/v) sucrose, 0.8% (w/v) agar, and 50 mg l−1 kanamycin. Seven-day-old seedlings were transferred from germination medium to minimal medium (Maser et al., 2002a), supplemented with the indicated amounts of NaCl, KCl, and CaCl2. The standard minimal medium contains 1.75 mM K+, thus for medium containing more or less K+ the KNO3 and KH2PO4 were replaced with NH4NO3 and NaH2PO4.2H2O, respectively, and KCl was added as indicated. For the determination of shoot and root ion contents, plant material (15 plants in each pool) was dried in an oven at 68 °C for 7 d. The dried material was pulverized with Micro Smash MS-100 bead beater (TOMY, Japan). Ions were extracted with 1 M HCl, and were measured with an atomic absorption spectrophotometer AA670-G (Shimadzu, Japan).
Results
Isolation and characterization of PutHKT2;1 cDNA
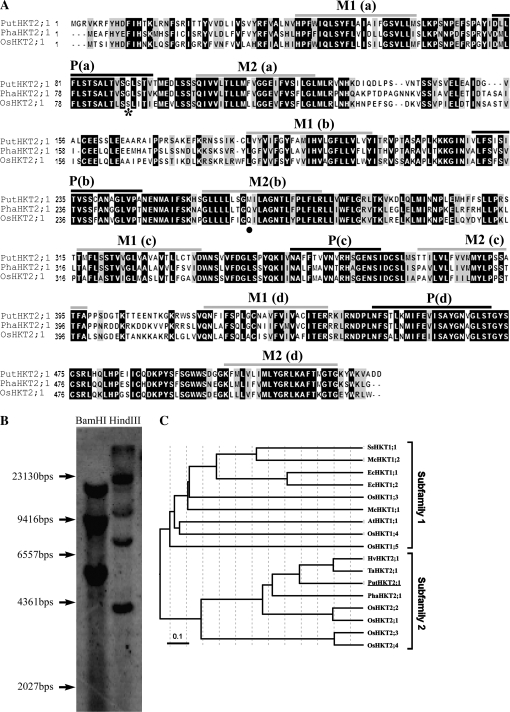
By using degenerate primers deduced from several HKT sequences and standard reverse transcription (RT)-PCR methods, a cDNA homologue of an HKT high-affinity K+ transporter from P. tenuiflora was cloned. The length of the amplified cDNA fragment was about 1500 bp, and the translated amino acid sequence showed high homology to TaHKT2;1 (Schachtman and Schroeder, 1994) and HvHKT2;1 (Haro et al., 2005). The full-length cDNA was obtained by 5′- and 3′-RACE and was designated as PutHKT2;1 (Accession no. FJ716169) which is 1778 bp long and contains an ORF of 1593 bp encoding 531 amino acid residues. PutHKT2;1 belongs to subfamily 2 in a phylogenetic tree proposed by Platten et al. (2006) (Fig. 2C) and shares high homology with the other members of the subfamily (80, 77, 67, and 64% identities at the amino acid level with TaHKT2;1, HvHKT2;1, PhaHKT2;1, and OsHKT2;1, respectively). Analysis of the PutHKT2;1 sequence by the TMpred program predicted eight transmembrane-spanning domains (Fig. 2A) which result from sequential membrane-pore-membrane (MPM) motifs as has been previously suggested in HKT structural models (Durell and Guy, 1999; Durell et al., 1999; Kato et al., 2001; Maser et al., 2002b). A glycine residue at the filter position in the first P-region is conserved in the PutHKT2;1 sequence (Fig. 2A), suggesting that PutHKT2;1 is a K+-Na+-type rather than a Na+-Na+-type transporter (Maser et al., 2002b).
Fig. 2.
(A) Amino acid sequence alignment of PutHKT2;1 with PhaHKT2;1 (Phragmites australis, BAE44385) and OsHKT2;1 (Oryza sativa, BAB61789). The sequences were aligned by the program ClustalW. Black and grey backgrounds indicate identical residues and similar residues, respectively. Grey and black bars above the sequences indicate the putative transmembrane (M) and pore (P) domains, respectively, by the TMpred program. The conserved Gly residue is indicated by an asterisk, while the Gln270 of TaHKT2;1 which is conserved in PhaHKT2;1 and OsHKT2;1 is indicated by a black dot. Putative M1PM2 motifs (a–d) are marked. (B) Genomic Southern hybridization of PutHKT2;1. Genomic DNA (20 μg) was digested by BamHI and HindIII. Hybridization was performed overnight at 45 °C, with a digoxigenin (DIG)-labelled PutHKT2;1 cDNA probe. (C) Unrooted minimum-evolution tree of the HKT transporters. The translated sequences of known HKT genes were aligned by the ClustalW program and the tree was constructed using the MEGA 4 program (Kumar et al., 2008). Scale bar indicates 0.1 substitutions per site.
While AtHKT1;1 was reported to be a single copy gene in the Arabidopsis genome (Uozumi et al., 2000), reed plants and rice possess several HKTs (Takahashi et al., 2007; Garciadeblas et al., 2003). Genomic Southern-hybridization using DIG-labelled PutHKT2;1 cDNA as a probe revealed a number of PutHKT2;1 hybridizing bands, suggesting that PutHKT2;1 belongs to a small gene family (Fig. 2B).
Subcellular localization of PutHKT2;1::GFP fusion protein
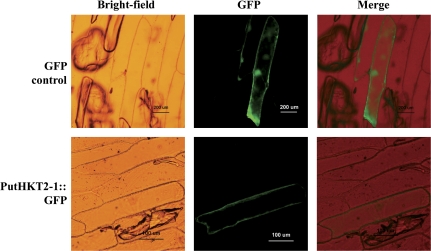
To determine the exact subcellular localization of the PutHKT2;1 protein within plant cells, the GFP gene was fused in-frame to the C-terminus of PutHKT2;1. Transient expression analysis was performed by introducing the fusion protein into onion epidermal cells by particle bombardment (Fig. 3). A bright fluorescence bordering the cell was found in cells expressing PutHKT2;1::GFP. The fluorescence derived from GFP in control experiments was distributed throughout the cell, including the nucleus. The transient expression analysis confirmed that PutHKT2;1 localizes at the plasma membrane of plant cells. The plasma membrane was also the site of other HKT proteins, including McHKT1;1 (Su et al., 2003), AtHKT1;1 (Sunarpi et al., 2005), and OsHKT2;1 (Horie et al., 2007).
Fig. 3.
Subcellular localization of PutHKT2;1::GFP fusions. GFP fusion constructs of PutHKT2;1::GFP or control GFP were introduced into onion epidermal cells by particle bombardment, and the fluorescent signals were examined 10 h after bombardment.
Expression of PutHKT2;1
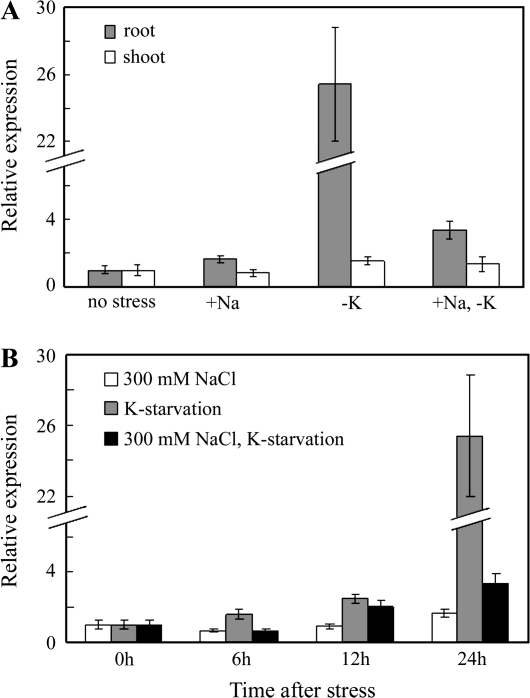
Since most HKTs were found to be regulated by K+-starvation and NaCl stress conditions (Kader et al., 2006; Takahashi et al., 2007; Shao et al., 2008), real-time RT-PCR was used to investigate the expression levels of PutHKT2;1 in shoots and roots of P. tenuiflora subjected to NaCl and K+-starvation stresses for 24 h (Fig. 4A). The expression of PutHKT2;1 was induced dramatically in roots under K+-starvation conditions (approximately 25-fold compared with the control). PutHKT2;1 expression in roots was also induced by 300 mM NaCl (1.7-fold compared with the control) or by 300 mM NaCl and K+-starvation stress (3.4-fold compared with the control), but its expression was only slightly regulated by those stresses in shoots. To gain further insight into ionic stress regulation of PutHKT2;1 expression in roots, PutHKT2;1 expression was monitored over a 24 h period (Fig. 4B). Under potassium starvation stress, PutHKT2;1 expression gradually increased. Under 300 mM NaCl, in the presence of both high and low external K+ concentration, PutHKT2;1 transcript levels decreased by approximately 30% compared to the control at 6 h. After 12 h and 24 h of 300 mM NaCl stress, the PutHKT2;1 transcript level at low external K+ concentration was twice that at high external K+ concentration.
Fig. 4.
(A) Expression analysis of PutHKT2;1 by means of real-time RT-PCR in roots and shoots of P. tenuiflora subjected to 300 mM NaCl (+Na), K+-starvation (–K), and both 300 mM NaCl and K+-starvation stresses (+Na, –K) for 24 h. (B) Time-course expression analysis of PutHKT2;1 in roots of P. tenuiflora. Results are expressed as means ±SE (n=3).
Functional comparison of PutHKT2;1, PhaHKT2;1, and OsHKT2;1 genes in yeast
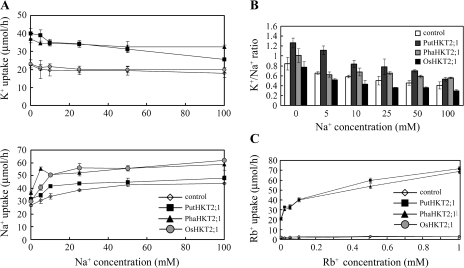
Since PutHKT2;1 had been shown to share high similarity with PhaHKT2;1 and OsHKT2;1 at the amino acid level, the function of these proteins was compared in a yeast expression system. PutHKT2;1, OsHKT2;1, and PhaHKT2;1 cDNAs were introduced and expressed in yeast strain 9.3 cells and the ion uptake characteristics of the cells were analysed. Previous functional characterization in yeasts showed that PhaHKT2;1 from the Nanpi area functions as a high affinity K+-Na+ co-transporter (Takahashi et al., 2007), while OsHKT2;1 functions as a Na+ selective uniporter (Uozumi et al., 2000). In this study, yeasts expressing PutHKT2;1 or PhaHKT2;1 grew better than the strain expressing OsHKT2;1 under micromolar K+ concentrations (10 μM K+) and were able to complement the K+ uptake deficiency phenotype, whereas the yeast strain expressing OsHKT2;1 did not (Fig. 5). Furthermore, the suppression of yeast growth by 100 mM NaCl was rescued by the addition of K+ in the yeasts expressing PutHKT2;1 and PhaHKT2;1, but not in the yeast expressing OsHKT2;1. To investigate further whether the growth promotion of yeasts expressing PutHKT2;1 and PhaHKT2;1 cDNA was due to their ability to take up K+, the ion uptake ability of those yeasts was measured. The yeasts expressing PutHKT2;1 or PhaHKT2;1 showed a high K+ uptake ability in the Rb+ uptake experiment, while the yeast expressing OsHKT2;1 did not (Fig. 6C). Similar to PhaHKT2;1, PutHKT2;1 functioned in the micromolar range of Rb+ (K+), indicating that it mediated high affinity K+ uptake. PutHKT2;1 also showed permeability to Na+ (Fig. 6A, lower panel). Increased Na+ influx with an increasing concentration of NaCl was observed in all of the transformed yeasts. However, yeasts expressing PutHKT2;1 and PhaHKT2;1 could maintain their K+ uptake ability (Fig. 6A, upper panel), and, as a result, had a higher K+/Na+ ratio than the yeast strain expressing OsHKT2;1 (Fig. 6B).
Fig. 5.
Growth of yeast cells transformed by empty vector pAUR123 (control) or with the vector containing PutHKT2;1, PhaHKT2;1, or OsHKT2;1 cDNA. Yeast cells were inoculated on SC/-His plates (except for K+-starvation treatment when the plate was a one-quarter-strength SC/-His plate) supplemented by various concentration of NaCl and KCl as indicated with serially diluted drops of yeast cells suspensions and cultured for 5 d.
Fig. 6.
(A) Concentration dependence of K+ uptake (upper panel) or Na+ influx (lower panel) in yeasts expressing HKT2;1 of P. tenuiflora, reed plant, or rice in the presence of 5, 10, 25, 50, 100 mM NaCl, and 50 μM KCl. Strain 9.3 yeast cells were transformed with empty vector pAUR123 (open diamonds) or with the vector containing PutHKT2;1 (closed squares), PhaHKT2;1 (closed triangles) or OsHKT2;1 (grey circles) cDNA. (B) K+/Na+ ratio of yeasts cells transformed with empty plasmid (open bar) or with plasmid containing PutHKT2;1 (dark-grey bar), PhaHKT2;1 (light-grey bar) or OsHKT2;1 (black bar) cDNA treated as in (A). (C) Concentration dependence of Rb+ uptake in yeasts expressing HKT2;1 of P. tenuiflora, reed plant, or rice. Yeast strains and symbol are indicated as in (A). Results are expressed as means ±SE (n=3).
Ion-specific sensitivity of PutHKT2;1 and OsHKT2;1 transgenic plants
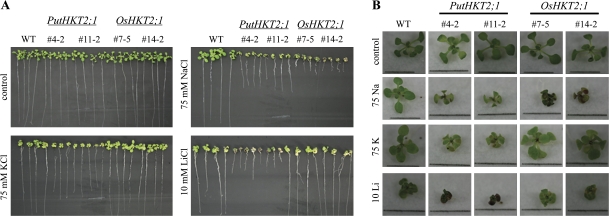
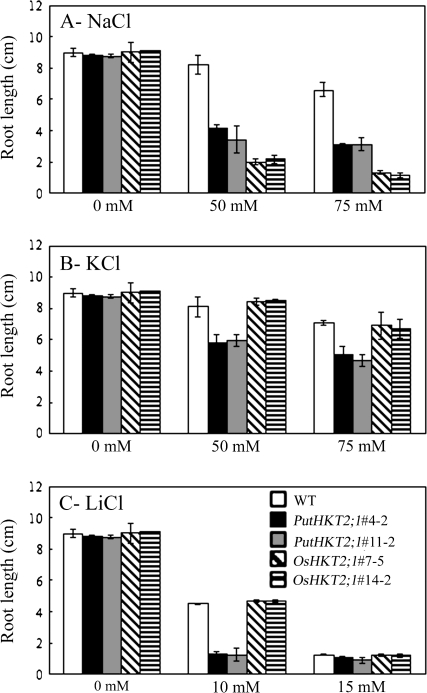
PutHKT2;1- and OsHKT2;1-expressing seedlings exhibited normal growth and were indistinguishable from WT plants on minimal medium with respect to root length (Figs 7A, 8) and shoot phenotype (Fig. 7B). Because the functional characterization of PutHKT2;1 and OsHKT2;1 in yeast showed that they have different ion uptake modes, various ionic stresses were imposed (75 mM KCl, 75 mM NaCl, and 10 mM LiCl) in order to determine the ion specificity of the two transporters. Our results showed that OsHKT2;1 over-expression leads to Na-specific sensitivity, as OsHKT2;1-expressing seedlings had shorter roots (Figs 7A, 8A) and chlorotic leaves (Fig. 7B) in the presence of elevated Na+ but not in the presence of K+ and Li+ (Figs 7, 8B, C). Interestingly, PutHKT2;1-expressing seedlings show defective root growth (Figs 7A, 8) and inhibited shoot growth (Fig. 7B) in the presence of all the cations tested, indicating that PutHKT2;1 mediates sensitivity not only to Na+ but also to K+ and Li+, cations that are physically similar to Na+. These results showed that PutHKT2;1 and OsHKT2;1 have different ion specificity.
Fig. 7.
Ion specific sensitivity of PutHKT2;1 and OsHKT2;1 over-expression lines. (A) Seven-day-old WT and transgenic seedlings grown on MS were transferred to minimal-medium (Maser et al., 2002a) supplemented with 75 mM NaCl, 75 mM KCl, and 10 mM LiCl. Seedlings were grown vertically for nine additional days before being photographed. (B) Shoot phenotype of seedling grown in (A). Bar in each picture indicates 1 cm.
Fig. 8.
Ion specific sensitivity of PutHKT2;1 and OsHKT2;1 over-expression lines by means of root length. Seedlings were grown as in Fig. 7. Results are expressed as means ±SE (n=5).
PutHKT2;1 and OsHKT2;1 mediate K-dependent Na-sensitivity in different ways
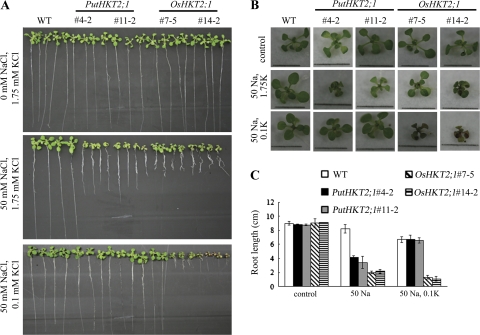
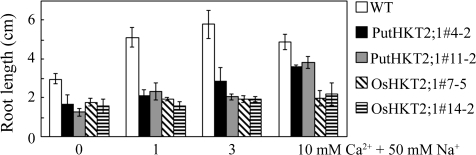
OsHKT2;1 has been reported to mediate Na+ uptake into K+-starved roots in vivo (Horie et al., 2007). Therefore, WT and transgenic seedlings were subjected to various external K+ concentrations in the presence of 50 mM NaCl to determine the K+-dependency of PutHKT2;1. As expected, the root growth and shoot phenotype of OsHKT2;1-expressing seedlings were more adversely affected as the external K+ concentration decreased (Fig. 9), confirming that low external K+ concentration induces OsHKT2;1-mediated Na+-uptake. By contrast, the root growth and shoot phenotype of PutHKT2;1-expressing seedlings were more adversely affected as the external K+ concentration increased. The root growth and shoot phenotype of PutHKT2;1-expressing seedlings were indistinguishable from those of the WT in the presence of 50 mM NaCl and 0.1 mM KCl, indicating that PutHKT2;1 mediates Na+ uptake in the higher external K+ concentration range. Furthermore, external Ca2+ addition could rescue the Na-sensitivity of the WT, but Na+-sensitivities of PutHKT2;1- and OsHKT2;1-expressing seedlings were not affected by increasing the Ca2+ concentration in the medium from 0 mM to 3 mM (Fig. 10). In the lines expressing PutHKT2;1, Ca2+ was able to rescue the effects of Na+ stress slightly but only at 10 mM [CaExt2+]. Increasing [CaExt2+] from 0 mM to 10 mM increased the root length of PutHKT2;1-expressing seedlings 2–3-fold.
Fig. 9.
Potassium-dependent Na-sensitivity of PutHKT2;1 and OsHKT2;1 over-expression lines. (A) Seven-day-old WT and transgenic seedlings grown on MS were transferred to minimal-medium supplemented with 1.75 mM or 0.1 mM KCl and 50 mM NaCl. (B) Shoot phenotype of seedlings grown in (A). Bar in each picture indicates 1 cm. (C) Root-lengths of WT and transgenic seedlings after the indicated treatments. Results are expressed as means ±SE (n=5).
Fig. 10.
Calcium regulation on Na-sensitivity of PutHKT2;1 and OsHKT2;1 over-expression lines. Root lengths of WT and transgenic seedlings after the indicated treatments. Seven-day-old WT and transgenic seedlings grown on MS were transferred to minimal-medium supplemented with 0, 1, 3, or 10 mM CaCl2 and 50 mM NaCl. Results are expressed as means ±SE (n=5).
Low tissue K+/Na+ ratio account to Na-sensitivity of PutHKT2;1 and OsHKT2;1 transgenic seedlings
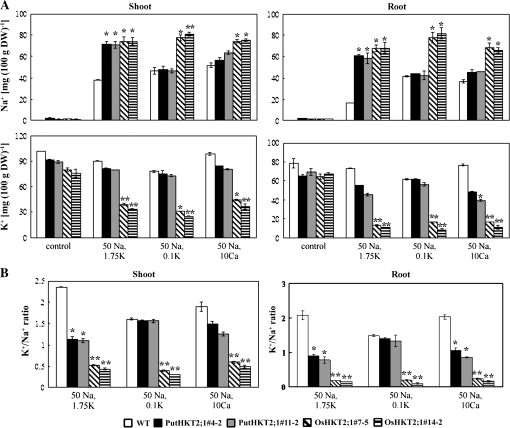
The Na+ and K+ contents of WT, PutHKT2;1, and OsHKT2;1 transgenic seedlings were determined. Figure 11 shows that the Na+ and K+ accumulation patterns of WT and transgenic seedlings were similar between shoot and root. PutHKT2;1 and OsHKT2;1 transgenic seedlings accumulated more Na+ than the WT when the growth medium contained 50 mM Na+ and 1.75 mM K+ (Fig. 11A, upper panel). Interestingly, external Na+ addition significantly reduced the K+ content of OsHKT2;1-expressing seedlings but had little effect on the K+ content of PutHKT2;1-expressing seedlings (Fig. 11A, lower panel). Furthermore, the Na+ and K+ contents of PutHKT2;1-expressing seedlings did not significantly differ from those of the WT when grown in the presence of 50 mM Na+ and 0.1 mM K+, confirming that PutHKT2;1 does not mediate Na+ uptake under low external K+ concentration. The Na+ accumulation in shoots and roots of PutHKT2;1-expressing seedlings was decreased when the [CaExt2+] was increased from 3 mM to 10 mM, but higher [CaExt2+] did not greatly affect the Na+ accumulation of OsHKT2;1-expressing seedlings. Overall, the Na-sensitivity symptoms of PutHKT2;1- and OsHKT2;1-expressing seedlings were due, primarily, to decreased K+ to Na+ ratios both in the shoot and root (Fig. 11B).
Fig. 11.
Comparison of Na+ and K+ accumulation in shoots and roots between WT, PutHKT2;1, and OsHKT2;1 over-expression lines. Seven-day-old WT and transgenic seedlings grown on MS were transferred to minimal-medium supplemented with the indicated salts. Seedlings were grown vertically for 14 additional days before being harvested. For each treatment, the ion content was determined from a pool of 15 plants. Results are expressed as means ±SE (n=3). Asterisks indicate a significant difference from the WT (P <0.05) by Student's t test.
Discussion
PutHKT2;1 and OsHKT2;1 perform different functions in transgenic yeast and Arabidopsis
In this study, an HKT homologue, PutHKT2;1, was isolated from a graminaceous halophyte P. tenuiflora and its function was characterized and compared with two previously identified HKTs, PhaHKT2;1 and OsHKT2;1. When characterized in yeast, PutHKT2;1 shows K+-Na+ transport characteristics similar to those of PhaHKT2;1 which may be due to the conserved glycine residue in the first pore region. As previously reported, OsHKT2;1-expressing yeast could transport K+ like OsHKT2;2 but only if the serine in this position was mutated to glycine. Similarly, AtHKT1;1-expressing yeast could transport K+ only if the serine in this position was mutated to glycine. However, it is worth noting that the K+ uptake complementation rate of the AtHKT1;1 mutant is still weaker than that in yeast expressing TaHKT2;1 (Maser et al., 2002b), suggesting that some amino acids other than the well-known glycine residue are involved in determining the K+ transport properties of HKT proteins. Furthermore, other studies in yeast have shown that single-base mutations of TaHKT2;1 enable it to enhance yeast salt tolerance (Rubio et al., 1995, 1999), indicating that small changes in the HKT protein structure could be expected to alter the ion selectivity or affinity of the protein. Even though PutHKT2;1 and PhaHKT2;1 showed very similar K+ transport properties, they displayed a slight difference in Na+ uptake ability (Fig. 5). A difference in Na+ uptake ability can be expected because the amino acid sequences of PutHKT2;1 and PhaHKT2;1 differed by 37%. Among the different amino acids, one amino acid (glutamine at position 270 of TaHKT2;1) was previously shown to be responsible for the low affinity Na+ uptake of the protein (Rubio et al., 1999). Mutation of the glutamine residue to leucine resulted in reduced low affinity Na+ uptake of TaHKT2;1. This glutamine residue is conserved in PhaHKT2;1. However, PutHKT2;1 has a methionine instead of glutamine (Fig. 1). Thus, the lower Na+ uptake ability of PutHKT2;1 may be related to the glutamine-to-methionine substitution. However, the transport properties of an HKT protein may not be solely determined by single residues.
To clarify the physiological role of PutHKT2;1, PutHKT2;1 was expressed in Arabidopsis and compared with OsHKT2;1, which is a closely related HKT but functions differently in yeast. Three conclusions can be drawn from our results. First, our finding that Arabidopsis plants expressing PutHKT2;1 and OsHKT2;1 had significantly higher Na+ contents than the WT in the presence of 50 mM NaCl (which exacerbates the Na-stress effect on WT) (Fig.11) show that PutHKT2;1 and OsHKT2;1 transport Na+. PutHKT2;1 seems to be permeable to K+ and Li+ in addition to Na+, because it confers sensitivity not only to Na+ but also to K+ and Li+. This suggests that PutHKT2;1 functions as a general alkali cation transporter. Second, the Na+-transport function of PutHKT2;1 and OsHKT2;1 is dependent on [KExt+]. PutHKT2;1 transports Na+ at high [KExt+] while OsHKT2;1 transports Na+ at low [KExt+]. This different mode of action might relate to the physiological function of each protein in plants. OsHKT2;1 is responsible for a major portion of nutritional Na+ uptake and for the distribution of Na+ in K-starved roots (Horie et al., 2007). Thus OsHKT2;1 only mediates Na+ at low [KExt+]. The possible physiological functions of PutHKT2;1 are discussed below. And third, the Na+ transport function of OsHKT2;1 is independent of [CaExt+] while the Na+ transport function of PutHKT2;1 is partially dependent on [CaExt+]. This is because the addition of 10 mM Ca2+ reduced Na+ accumulation in PutHKT2;1-expressing Arabidopsis (Fig. 11). Ca2+ serves as a second messenger in plant cells during abiotic stress signalling and appears to have roles in ion channel regulation (Zhu, 2003; Cheong et al., 2007; Mahajan et al., 2008). The SOS3(CBL4)-SOS2(CIPK24) complex was first suggested to down-regulate HKT gene expression or inactivate HKT proteins during salt stress, because mutations in AtHKT1;1 suppressed the Na+-hypersensitivity of the sos3 mutant (Uozumi et al., 2000; Mahajan and Tuteja, 2005). A subsequent study showed that AtHKT1;1 mutations do not strictly repress the Na+ hypersensitivity of the sos3 mutant and that SOS3 and AtHKT1;1 are physiologically distinct major determinants of salinity tolerance (Horie et al., 2006). However, at high Ca2+, SOS3-independent mechanisms may be activated because the Arabidopsis genome encodes nine homologues of the SOS3 gene (Kolukisaoglu et al., 2004) and might, in turn, regulate (e.g. inactivate) PutHKT2;1 and thus decrease the absolute Na+ content in transgenic Arabidopsis.
PutHKT2;1 and OsHKT2;1 control K+ acquisition and nutrient status of Arabidopsis differently
This study revealed that PutHKT2;1 and OsHKT2;1 function in controlling Na+ homeostasis, which, in turn, affects K+ acquisition and the nutrient status of transgenic Arabidopsis. Our results show that OsHKT2;1 is a negative regulator of the K+-uptake system in transgenic Arabidopsis, because OsHKT2;1-expressing Arabidopsis exhibited a significantly reduced K+ content in both the roots and shoots (Fig. 11). This finding provides an additional reason for the rapid reduction of OsHKT2;1 mRNA in response to elevated levels of NaCl (Horie et al., 2007). The down-regulation of OsHKT2;1 transcripts in such conditions might also be necessary to prevent OsHKT2;1 from inhibiting the K-uptake system, rather than just to restrict Na+-influx under high [NaExt+]. By contrast, the K+ content of PutHKT2;1-expressing Arabidopsis was not greatly affected by external Na+ addition. This result implies two possibilities, that are (i) PutHKT2;1, unlike OsHKT2;1, does not negatively regulate any K+-uptake system, and (ii) PutHKT2;1 mediates a substantial K+ uptake under low [KExt+] and in the presence of NaCl.
Possible role of PutHKT2;1 in the salt tolerance of P. tenuiflora
The expression patterns of HKT transporters may provide some clues to their physiological roles. So far, the HKT transcripts from graminaceous plants such as wheat (Schachtman and Schroeder, 1994), rice (Garciadeblas et al., 2003; Kader et al., 2006), and reed plant (Takahashi et al., 2007) are mainly expressed in roots and induced under low K+ concentration conditions. Similarly, PutHKT2;1 mRNA was also expressed mainly in roots and up-regulated dramatically in response to K+ starvation (Fig. 4). In a previous study, when P. tenuiflora was subjected to K+ starvation and salt-stress conditions, K+ uptake was not influenced by Cs+, and TEA application in those conditions resulted in a decreased K+ concentration in shoots and decreased Na+ concentrations in shoots and roots (Peng et al., 2004). This result suggested that, at low external K+ availability and in the presence of Na+, P. tenuiflora had a high affinity K+ uptake system that is also permeable to Na+. PutHKT2;1 is a candidate for a high affinity K+ uptake system in view of (i) its localization in the plasma membrane (Fig. 3), (ii) the up-regulation of PutHKT2;1 transcripts in roots under K-starvation condition (Fig. 4), (iii) the evidence that PutHKT2;1 mediates K+ transport in yeast (Fig. 6C), and (iv) the fact that PutHKT2;1-expressing Arabidopsis could maintain the tissue K+ content similar to the WT under low [KExt+] and in the presence of elevated Na+ (Fig. 11). Maintaining a high K+/Na+ ratio under salt stress is important to the cellular metabolism (Zhu, 2003; Munns and Tester, 2008), and PutHKT2;1 might facilitate K+ uptake under low [KExt+] and in the presence of elevated Na+ to keep the K+/Na+ ratio of the cell at the same level. Previous studies have shown that P. tenuiflora could maintain its shoot potassium content which remained unchanged during salt stress treatment and resulted in only a slight decrease in the K+/Na+ ratio (Peng et al., 2004; Wang et al., 2004, 2009).
Finally, this work provides a direct functional comparison of two closely related HKTs from a glycophyte (rice) and from a halophyte (P. tenuiflora). The different characteristics between PutHKT2;1 and OsHKT2;1 might contribute to the different salt tolerance of P. tenuiflora and rice. Further studies of the tissue-specific expression and the transcriptional regulation of PutHKT2;1 in response to external K+ and Na+ concentrations should elucidate the important role of PutHKT2;1 in the salt tolerance of P. tenuiflora.
Acknowledgments
The authors are grateful to Dr M Fujimoto (Laboratory of Plant Molecular Genetics, the University of Tokyo) for his help with the technique of GFP observations. This work was supported by JSPS AA Science Platform Program.
References
- Amtmann A, Sanders D. Mechanisms of Na+ uptake by plant cells. Advances in Botanical Research. 1999;29:75–112. [Google Scholar]
- Berthomieu P, Conejero G, Nublat A, et al. Functional analysis of AtHKT1 in Arabidopsis shows that Na+ recirculation by the phloem is crucial for salt tolerance. EMBO Journal. 2003;22:2004–2014. doi: 10.1093/emboj/cdg207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumwald E, Aharon GS, Apse MP. Sodium transport in plant cells. Biochimica et Biophysica Acta. 2000;1465:140–151. doi: 10.1016/s0005-2736(00)00135-8. [DOI] [PubMed] [Google Scholar]
- Cheong YH, Pandey GK, Grant JJ, Batistic O, Li L, Kim BG, lee SC, Kudla J, Luan S. Two calcineurin B-like calcium sensors, interacting with protein kinase CIPK23, regulate leaf transpiration and root potassium uptake in Arabidopsis. The Plant Journal. 2007;52:223–239. doi: 10.1111/j.1365-313X.2007.03236.x. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Demidchik V, Tester M. Sodium fluxes through nonselective cation channels in the plasma membrane of protoplast from Arabidopsis roots. Plant Physiology. 2002;128:379–387. doi: 10.1104/pp.010524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durell SR, Guy HR. Structural models of the KtrB, TrkH, and Trk1, 2 symporters based on the structure of the KcsA K+ channel. Biophysical Journal. 1999;77:789–807. doi: 10.1016/S0006-3495(99)76932-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durell SR, Hao Y, Nakamura T, Bakker EP, Guy HR. Evolutionary relationship between K+ channels and symporters. Biophysical Journal. 1999;77:775–788. doi: 10.1016/S0006-3495(99)76931-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairbairn DJ, Liu W, Schachtman DP, Gomez-Gallego S, Day SR, Teasdale RD. Characterisation of two HKT-like potassium transporters from Eucalyptus camaldulensis. Plant Molecular Biology. 2000;43:515–525. doi: 10.1023/a:1006496402463. [DOI] [PubMed] [Google Scholar]
- Garciadeblas B, Senn ME, Banuelos MA, Rodriguez-Navarro A. Sodium transport and HKT transporters: the rice model. The Plant Journal. 2003;34:788–801. doi: 10.1046/j.1365-313x.2003.01764.x. [DOI] [PubMed] [Google Scholar]
- Gassmann W, Rubio F, Schroeder JI. Alkali cation selectivity of the wheat root high-affinity potassium transporter HKT1. The Plant Journal. 1996;10:869–882. doi: 10.1046/j.1365-313x.1996.10050869.x. [DOI] [PubMed] [Google Scholar]
- Haro R, Banuelos MA, Senn ME, Barrero-Gil J, Rodriguez-Navarro A. HKT1 mediates sodium uniports in roots. Pitfalls in the expression of HKT1 in yeast. Plant Physiology. 2005;139:1495–1506. doi: 10.1104/pp.105.067553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa PM, Bressan RA, Zhu JK, Bohnert HJ. Plant cellular and molecular responses to high salinity. Annual Review of Plant Physiology and Plant Molecular Biology. 2000;51:463–469. doi: 10.1146/annurev.arplant.51.1.463. [DOI] [PubMed] [Google Scholar]
- Horie T, Costa A, Kim TH, Han MJ, Horie R, Leung HY, Miyao A, Hirochika H, An G, Schroeder JI. Rice OsHKT2;1 transporter mediates large Na+ influx component into K+-starved roots for growth. EMBO Journal. 2007;26:3003–3014. doi: 10.1038/sj.emboj.7601732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horie T, Horie R, Chan WY, Leung HY, Schroeder JI. Calcium regulation of sodium hypersensitivities of sos3 and athkt1 mutants. Plant and Cell Physiology. 2006;47:622–633. doi: 10.1093/pcp/pcj029. [DOI] [PubMed] [Google Scholar]
- Horie T, Yoshida K, Nakayama H, Yamada K, Oiki S, Shinmyo A. Two types of HKT transporters with different properties of Na+ and K+ transport in Oryza sativa. The Plant Journal. 2001;27:129–138. doi: 10.1046/j.1365-313x.2001.01077.x. [DOI] [PubMed] [Google Scholar]
- Kader MA, Seidel T, Golldack D, Lindberg S. Expressions of OsHKT1, OsHKT2, and OsVHA are differentially regulated under NaCl stress in salt-sensitive and salt tolerant rice (Oryza sativa L.) cultivars. Journal of Experimental Botany. 2006;57:4257–4268. doi: 10.1093/jxb/erl199. [DOI] [PubMed] [Google Scholar]
- Kato Y, Sakaguchi M, Mori Y, Saito K, Nakamura T, Bakker EP, Sato Y, Goshima S, Uozumi N. Evidence in support of a four transmembrane-pore-transmembrane topology model for the Arabidopsis thaliana Na+/K+ translocating AtHKT1 protein, a member of the superfamily of K+ transporters. Proceedings of the National Academy of Sciences, USA. 2001;98:6488–6493. doi: 10.1073/pnas.101556598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolukisaoglu U, Weinl S, Blazevic D, Batistic O, Kudla J. Calcium sensors and their interacting protein kinases: genomics of the Arabidopsis and rice CBL-CIPK signaling networks. Plant Physiology. 2004;134:45–58. doi: 10.1104/pp.103.033068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Nei M, Dudley J, Tamura K. MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Briefings in Bioinformatics. 2008;9:299–306. doi: 10.1093/bib/bbn017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurie S, Feeney KA, Maathuis FJM, Heard PJ, Brown SJ, Leigh RA. A role for HKT1 in sodium uptake by wheat roots. The Plant Journal. 2002;32:139–149. doi: 10.1046/j.1365-313x.2002.01410.x. [DOI] [PubMed] [Google Scholar]
- Liu W, Fairbairn DJ, Reid RJ, Schachtman DP. Characterization of two HKT1 homologues from Eucalyptus camadulensis that display intrinsic osmosensing capability. Plant Physiology. 2001;127:283–294. doi: 10.1104/pp.127.1.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maathuis FJ, Sanders D. Sodium uptake in Arabidopsis root is regulated by cyclic nucleotides. Plant Physiology. 2001;127:1617–1625. [PMC free article] [PubMed] [Google Scholar]
- Mahajan S, Pandey GK, Tuteja N. Calcium- and salt-stress signaling in plants: Shedding light on SOS pathway. Archives of Biochemistry and Biophysics. 2008;471:146–158. doi: 10.1016/j.abb.2008.01.010. [DOI] [PubMed] [Google Scholar]
- Mahajan S, Tuteja N. Cold, salinity and drought stresses: an overview. Archives of Biochemistry and Biophysics. 2005;444:139–158. doi: 10.1016/j.abb.2005.10.018. [DOI] [PubMed] [Google Scholar]
- Maser P, Eckelman B, Vaidyanathan R, et al. Altered shoot/root Na+ distribution and bifurcating salt sensitivity in Arabidopsis by genetic disruption of the Na+ transporter AtHKT1. FEBS Letters. 2002a;531:157–161. doi: 10.1016/s0014-5793(02)03488-9. [DOI] [PubMed] [Google Scholar]
- Maser P, Hosoo Y, Goshima S, et al. Glycine residues in potassium channel-like selectivity filters determine potassium selectivity in four-loop-per-subunit HKT transporters from plants. Proceedings of the National Academy of Sciences, USA. 2002b;99:6428–6433. doi: 10.1073/pnas.082123799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller PY, Janovjak H, Miserez AR, Dobbie Z. Processing of gene expression data generated by quantitative real-time RT-PCR. BioTechniques. 2002;32:1372–1379. [PubMed] [Google Scholar]
- Munns R, Tester M. Mechanisms of salinity tolerance. Annual Reviews of Plant Biology. 2008;59:651–681. doi: 10.1146/annurev.arplant.59.032607.092911. [DOI] [PubMed] [Google Scholar]
- Peng YH, Zhu TF, Mao YQ, Wang SM, Su WA, Tang ZC. Alkali grass resists salt stress through high [K+] and an endodermis barrier to Na+ Journal of Experimental Botany. 2004;55:939–949. doi: 10.1093/jxb/erh071. [DOI] [PubMed] [Google Scholar]
- Platten JD, Cotsaftis O, Berthomieu P, et al. Nomenclature for HKT transporters, key determinants of plant salinity tolerance. Trends in Plant Science. 2006;11:372–374. doi: 10.1016/j.tplants.2006.06.001. [DOI] [PubMed] [Google Scholar]
- Ren ZH, Gao JP, Li LG, Cai XL, Huang W, Chao DY, Shu MZ, Wang ZY, Luan S, Lin HX. A rice quantitative trait locus for salt tolerance encodes a sodium transporter. Nature Genetics. 2005;37:1141–1146. doi: 10.1038/ng1643. [DOI] [PubMed] [Google Scholar]
- Rubio F, Gassmann W, Schroeder JI. Sodium-driven potassium uptake by the plant potassium transporter HKT1 and mutations conferring salt tolerance. Science. 1995;270:1660–1663. doi: 10.1126/science.270.5242.1660. [DOI] [PubMed] [Google Scholar]
- Rubio F, Schwarz M, Gassmann W, Schroeder JI. Genetic selection of mutations in the high affinity K+ transporter HKT1 that define functions of a loop site for reduced Na+ permeability and increased Na+ tolerance. Journal of Biological Chemistry. 1999;274:6839–6847. doi: 10.1074/jbc.274.11.6839. [DOI] [PubMed] [Google Scholar]
- Schachtman DP, Liu W. Molecular pieces to the puzzle of the interaction between potassium and sodium uptake in plants. Trends in Plant Science. 1999;4:281–287. doi: 10.1016/s1360-1385(99)01428-4. [DOI] [PubMed] [Google Scholar]
- Schachtman DP, Shroeder JI. Structure and transport mechanism of a high-affinity potassium uptake transporter from higher plants. Nature. 1994;370:655–658. doi: 10.1038/370655a0. [DOI] [PubMed] [Google Scholar]
- Shao Q, Zhao C, Han N, Wang B-S. Cloning and expression pattern of SsHKT1 encoding a putative cation transporter from halophyte Suaeda salsa. DNA Sequence. 2008;19:106–114. doi: 10.1080/10425170701447465. [DOI] [PubMed] [Google Scholar]
- Su H, Balderas E, Vera-Estrella R, Golldack D, Quigley F, Zhao C, Pantoja O, Bohnert HJ. Expression of the cation transporter McHKT1 in halophyte. Plant Molecular Biology. 2003;52:967–980. doi: 10.1023/a:1025445612244. [DOI] [PubMed] [Google Scholar]
- Sunarpi Horie T, Motoda J, et al. Enhanced salt tolerance mediated by AtHKT1 transporter-induced Na+ unloading from xylem vessels to xylem parenchyma cells. The Plant Journal. 2005;44:928–938. doi: 10.1111/j.1365-313X.2005.02595.x. [DOI] [PubMed] [Google Scholar]
- Takahashi R, Liu S, Takano T. Cloning and functional comparison of a high-affinity K+ transporter gene PhaHKT1 of salt-tolerant and salt-sensitive reed plants. Journal of Experimental Botany. 2007;58:4387–4395. doi: 10.1093/jxb/erm306. [DOI] [PubMed] [Google Scholar]
- Tester M, Davenport R. Na+ tolerance and Na+ transport in higher plants. Annals of Botany. 2003;91:503–527. doi: 10.1093/aob/mcg058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uozumi N, Kim EJ, Rubio F, Yamaguchi T, Muto S, Tsuboi A, Bakker EP, Nakamura T, Schroeder JI. The Arabidopsis HKT1 gene homolog mediates inward Na+ currents in Xenopus laevis oocytes and Na+ uptake in Saccharomyces cerevisiae. Plant Physiology. 2000;122:1240–1259. doi: 10.1104/pp.122.4.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CM, Zhang JL, Liu XS, Li Z, Wu GQ, Cai JY, Flowers TJ, Wang SM. 2009 Puccinellia tenuiflora maintains a low Na+ level under salinity by limiting unidirectional Na+ influx resulting in a high selectivity for K+ over Na+. PMID: 19183292. [Google Scholar]
- Wang SM, Zhao GQ, Gao YS, Tang ZC, Zhang CL. Puccinellia tenuiflora exhibits stronger selectivity for K+ over Na+ than wheat. Journal of Plant Nutrition. 2004;27:1841–1857. [Google Scholar]
- Wang YC, Chu YG, Liu GF, Wang MH, Jiang J, Hou YJ, Qu GZ, Yang CP. Identification of expressed sequence tags in alkali grass (Puccinellia tenuiflora) cDNA library. Journal of Plant Physiology. 2007;164:78–79. doi: 10.1016/j.jplph.2005.12.006. [DOI] [PubMed] [Google Scholar]
- Zhang CQ, Nishiuchi S, Liu S, Takano T. Characterization of two plasma membrane protein 3 genes (PutPMP3) from alkali grass, Puccinellia tenuiflora, and functional comparison of the rice homologues, OsLti6a/b from rice. BMB Reports. 2008;41:448–454. doi: 10.5483/bmbrep.2008.41.6.448. [DOI] [PubMed] [Google Scholar]
- Zhu J-K. Regulation of ion homeostasis under salt stress. Current Opinion in Plant Biology. 2003;6:441–445. doi: 10.1016/s1369-5266(03)00085-2. [DOI] [PubMed] [Google Scholar]