Abstract
Drought tolerance is a key trait for increasing and stabilizing barley productivity in dry areas worldwide. Identification of the genes responsible for drought tolerance in barley (Hordeum vulgare L.) will facilitate understanding of the molecular mechanisms of drought tolerance, and also facilitate the genetic improvement of barley through marker-assisted selection or gene transformation. To monitor the changes in gene expression at the transcriptional level in barley leaves during the reproductive stage under drought conditions, the 22K Affymetrix Barley 1 microarray was used to screen two drought-tolerant barley genotypes, Martin and Hordeum spontaneum 41-1 (HS41-1), and one drought-sensitive genotype Moroc9-75. Seventeen genes were expressed exclusively in the two drought-tolerant genotypes under drought stress, and their encoded proteins may play significant roles in enhancing drought tolerance through controlling stomatal closure via carbon metabolism (NADP malic enzyme, NADP-ME, and pyruvate dehydrogenase, PDH), synthesizing the osmoprotectant glycine-betaine (C-4 sterol methyl oxidase, CSMO), generating protectants against reactive-oxygen-species scavenging (aldehyde dehydrogenase,ALDH, ascorbate-dependent oxidoreductase, ADOR), and stabilizing membranes and proteins (heat-shock protein 17.8, HSP17.8, and dehydrin 3, DHN3). Moreover, 17 genes were abundantly expressed in Martin and HS41-1 compared with Moroc9-75 under both drought and control conditions. These genes were possibly constitutively expressed in drought-tolerant genotypes. Among them, seven known annotated genes might enhance drought tolerance through signalling [such as calcium-dependent protein kinase (CDPK) and membrane steroid binding protein (MSBP)], anti-senescence (G2 pea dark accumulated protein, GDA2), and detoxification (glutathione S-transferase, GST) pathways. In addition, 18 genes, including those encoding Δl-pyrroline-5-carboxylate synthetase (P5CS), protein phosphatase 2C-like protein (PP2C), and several chaperones, were differentially expressed in all genotypes under drought; thus they were more likely to be general drought-responsive genes in barley. These results could provide new insights into further understanding of drought-tolerance mechanisms in barley.
Keywords: Barley, drought stress, drought tolerance, microarray, reproductive stage
Introduction
Drought is one of the main environmental constraints to agricultural productivity worldwide. Many efforts have been made to elucidate the mechanisms of drought tolerance in plants through molecular and genomics approaches, and a number of genes that respond to drought stress at the transcriptional level have been reported (Bray, 1993; Seki et al., 2002; Cheong et al., 2003; Liu and Baird, 2004; Hazen et al., 2005; Talamè et al., 2007). The functions of many genes have been predicted based on their sequence similarity with proteins of known functions in different species. Some of these genes have been reported to play important roles in protecting plants from drought stress through stress perception, signal transduction, transcriptional regulatory networks in cellular responses, or tolerance to dehydration (Zhang et al., 2004; Nakashima and Yamaguchi-Shinozaki, 2006; Umezawa et al., 2006). Several stress-induced putative drought-tolerance genes have been used for improving the stress tolerance of plants through gene transformation (Xu et al., 1996; Pellegrineschi et al., 2002, 2004; Abebe et al., 2003; Umezawa et al., 2006). Although some progress has been made, the molecular basis of plant tolerance to drought stress remains to be discovered (Ingram and Bartels, 1996; Bruce et al., 2002; Vinocur and Altman, 2005; Umezawa et al., 2006; Guo et al., 2007b).
Barley is one of the most important cereal crops grown in many developing countries, where it is often subject to extreme drought stress that significantly affects production (Ceccarelli, 1994; Ceccarelli et al., 2007). Investigating the drought-tolerance mechanisms in barley could facilitate a better understanding of the genetic bases of drought tolerance, and so enable the effective use of genetic and genomic approaches to improve its drought tolerance. More recently, high-throughput screening techniques such as microarray analysis have been used to monitor the expression of genes that respond to abiotic stresses. However, few experiments have reported the use of microarrays for gene expression analysis in barley under drought or drought-related stresses (Ozturk et al., 2002; Ueda et al., 2004; Walia et al., 2006; Talamè et al., 2007). Most of these experiments were performed with a short period of dehydration shock (Ozturk et al., 2002; Ueda et al., 2004; Walia et al., 2006), and only one simulated the slow development of drought stress that occurs in field conditions (Talamè et al., 2007). Many differentially expressed genes responding to drought-related stresses were identified after short drought treatments in these studies; however, transcriptional changes responding to longer periods of stress may not have been identified, even though they may be crucial to adaptation under field conditions (Talamè et al., 2007). Moreover, a limitation in most reported gene expression experiments is the use of single genotypes, without comparing differences in transcription levels between drought-tolerant and drought-sensitive barley genotypes under drought-stress conditions. Consequently, it is impossible to separate drought-tolerance-related genes from drought-responsive genes in these studies. Therefore, many of the differentially expressed genes so far identified may not be responsible for enhancing drought tolerance. Furthermore, all previous studies were conducted on seedlings, whereas drought stress at the reproductive stage may have much more effect on grain yield than drought at the vegetative stage (Ceccarelli et al., 2004). Therefore, analysis of gene expression for drought tolerance during the reproductive stage may provide further insight into the molecular mechanisms of drought tolerance in barley.
Differences in transcription levels at the reproductive stage between drought-tolerant and drought-sensitive barley genotypes under drought conditions may, therefore, identify genes important in enhancing drought tolerance. The results of a study in which an Affymetrix Barley 1 GeneChip was used to identify barley genes that were differentially expressed between drought-stressed and normal growth conditions at the reproductive stage are reported here. Based on putative functions of the identified genes, possible mechanisms for drought tolerance in barley are elaborated.
Materials and methods
Plant materials and experimental treatment
Three barley genotypes (Martin, HS41-1, and Moroc9-75) were used for the measurement of physiological traits and gene expression. Martin is cultivated in North Africa and is well adapted to drought stress; HS41-1 is a pure line of Hordeum vulgare ssp. spontaneum and has been selected for its very good drought tolerance; and Moroc9-75 is considered to be sensitive to drought stress (Ceccarelli, 1994; Ceccarelli et al., 2004).
A pot experiment was arranged in a randomized complete-block design with two treatments (well-watered and drought-stressed) and three replications (10 pots/replication) under controlled conditions in a greenhouse at the International Centre for Agricultural Research in the Dry Areas (ICARDA) (Tel Hadya, Aleppo, Syria). Three vernalized seedlings of the same genotype were transplanted into a 3.0 l pot (15 cm in height and 16 cm in diameter) filled with 2.2 kg of sterilized field soil, which contained about 6% water. Field capacity, wilting point, and available water content (AWC) of the soil were measured at ICARDA's soil laboratory. For barley, 70% and 10% of AWC in the soil were considered to be well-watered and severe drought conditions, respectively (Doorenbos and Pruit, 1977). Each genotype was planted in 60 pots giving a total of 180 plants; all plants were grown with 16 h daylight at 28 °C and an 8 h dark period at 20 °C under controlled conditions. The drought treatment was started by withholding water at the flowering stage. The soil moisture for the pots of the well-watered and drought-stressed conditions was maintained with the required amounts of water by weighing pots and watering plants daily. The days were counted after the AWC in the soil reached 10% to allow drought measurements at precisely determined intervals. The grain yield was determined and analysed when grains were mature for both drought-stress and control conditions.
Measurement of chlorophyll content and chlorophyll fluorescence parameters
Leaf chlorophyll was determined using a chlorophyll meter (SPAD-502, Minolta, Japan). Six flag leaves for each genotype in both well-watered and drought-stressed conditions were measured after drought stress. There were three measurements at random locations in the middle of the leaf for each plant and the average used for the analysis. From each genotype, 20 leaves with incremental chlorophyll levels (determined by SPAD-502 readings) were then harvested to construct a standard curve for the quantification of chlorophyll content using the method for chlorophyll analysis described by Arnon (1949).
After drought treatment, six flag leaves from both well-watered and drought-stressed conditions for each genotype were selected to measure chlorophyll fluorescence parameters. The dark adaptation period for all measurements was about 25 min, and chlorophyll fluorescence was measured using a portable fluorescence spectrometer Handy PEA (Hansatech Instruments, Norfolk, UK) following the manufacturer's instructions. Fluorescence values recorded include: Fo which is the initial/minimal fluorescence, a measure of the stability of the light-harvesting complex; Fv/Fm represents the maximum quantum yield of PSII, which, in turn, is highly correlated with the quantum yield of net photosynthesis. Where Fm is the maximal fluorescence value, and Fv is variable fluorescence, Fv=Fm–Fo.
RNA isolation, target preparation, and processing for GeneChip analysis
Seven flag leaves of a replication for each genotype were harvested at 0, 1, 3, and 5 d after reaching 10% of AWC in soil to constitute a single biological replicate. These flag leaves were used for RNA isolation by using Trizol reagent following the manufacturer's protocol (Invitrogen, Karlsruhe, Germany). The RNA was further purified using the RNeasy Kit (Qiagen, Hilden, Germany). RNA yield and quality were determined by using an Agilent 2100 Bioanalyser (Agilent Technologies, Boblingen, Germany).
Sample processing, hybridization, and scanning of Affymetrix Barley 1 GeneChip with 22 792 probe sets were performed in the Microarray Facility at the KFB (Kompetenzzentrum für Fluoreszente Bioanalytik) Regensburg, Germany, following the standard Affymetrix protocol (Affymetrix GeneChip® Expression Analysis Technical Manual). Briefly, 5 μg of high quality total RNA was reverse transcribed using the One-Cycle cDNA Synthesis Kit (Affymetrix, Santa Clara, CA, USA). The resulting double-stranded cDNA was used as a template to generate biotin-tagged cRNA from an in vitro transcription (IVT) reaction, using the Affymetrix IVT Labeling Kit. The resulting biotin-tagged cRNA was fragmented to strands of 35–200 bases in length, and 15 μg of fragmented cRNA was used for each hybridization. Barley 1 GeneChips were hybridized for 16 h at 45 °C with rotation in a GeneChip® Hybridization Oven 640 (Affymetrix, Santa Clara, CA, USA). The arrays were washed and stained with R-phycoerythrin streptavidin in a Fluidics Station, and subsequently scanned with an Affymetrix GeneChip® Scanner 3000.
GeneChip data processing and analysis
All scanned data from Barley 1 GeneChips were processed first by robust multiarray average (RMA; Irizarry et al., 2003) using ArrayAssist software version 3.4 (Stratagene, La Jolla, CA, USA). Normalized expression values were computed from raw CEL files by first applying the RMA model of probe-specific correction of perfect match probes. The algorithm consisted of three steps: a model-based background correction stage neutralized the effects of background noise and the processing artefacts, a subsequent quantile normalization stage aligned expression values to a common scale, and, finally, an iterative median polishing procedure summarized the data and generated a single expression value for each probe set. The resulting RMA expression values were log2-transformed. Average log signal intensity values of three biological replicates for each sample were then computed and used for further analysis. The baseline files were generated by using the intensity data of 0 d under drought stress for each genotype, and the intensity data of Moroc9-75 for comparison of drought-tolerant and drought-sensitive genotypes. All detailed protocols and data can be accessed online at http://www.ncbi.nlm.nih.gov/projects/geo/query/acc.cgi?acc=GSE15970. Differentially expressed genes were identified using significance analysis by unpaired t test. Differentially expressed genes were explored through pairwise analysis of control versus multiple treatment comparison (0 d as the reference for the detection of differentially expressed genes responding to drought stress, and Moroc9-75 as the reference for the identification of constitutively expressed genes in drought-tolerant genotypes) with parameters asymptotic for P-value computation and Benjamini–Hochberg for correction type. Based on the differential expression report of ArrayAssist, genes whose time-specific differences varied significantly (P <0.0001, |log2-fold| value ≥2) across time points were identified as differentially expressed for each genotype. Significantly expressed genes were then hierarchically clustered with average linkage and Euclidean distance as a measurement of similarity using Genesis version 1.5 (Graz University of Technology, http://www.genome.tugraz.at). The probe sets that showed differential expression under drought stress were annotated using Affymetrix new release (July 2008) annotation data for Barley 1 GeneChip and current plant databases of NCBI and DFCI-Compbio.
Quantitative real-time PCR
To validate the results from the microarray experiment, 12 genes, which represented up-regulated, unchanged, and down-regulated genes identified through microarray analysis, were analysed using quantitative real-time PCR as described by Guo et al. (2007a). RNAs used for real-time PCR were the aliquots of RNA samples used for the hybridization of Barley 1 GeneChip and included 0 d RNAs for all three genotypes, 1 d RNAs for genotypes HS41-1 and Martin, 3 d RNAs for genotypes Martin and Moroc9-75, and 5 d RNAs for genotypes Martin and HS41-1. In general, specific primers (see Supplementary Table S3 at JXB online) for selected genes were designed for a 100 bp amplicon with Tm at 58–59 °C. First-strand cDNA was synthesized from 2.0 μg of total RNA using SuperScript II RNase H– Reverse Transcriptase (Invitrogen, Carlsbad, CA). Real-time RT PCR was performed in an ABI Prism 7000 DNA analyser (Applied Biosystems, Foster, CA) using the QuantiTect SYBR green PCR kit (Qiagen, Valencia, CA) for signal detection. To normalize the total amount of cDNAs present in each reaction, a housekeeping gene encoding actin was co-amplified. Its expression among three genotypes was similar (log2-fold was around 0) at all time points of the experiment. The RT-PCR primers designed for all genes used in this study were evaluated for PCR amplification efficiencies by performing real-time PCR using a series dilution of each cDNA at rates of 1/2, 1/5, 1/10, 1/20, 1/50, and 1/100. The efficiency test showed that all the primers amplified the genes with approximately the same efficiency as that of the normalizer actin. The ΔΔCT method of relative gene quantification recommended by Applied Biosystems was used to calculate the expression level of three genotypes under drought-stressed conditions relative to genotypes under control conditions, respectively.
Results
Effect of drought stress on chlorophyll content, maximum quantum yield of PSII (Fv/Fm) and grain yield
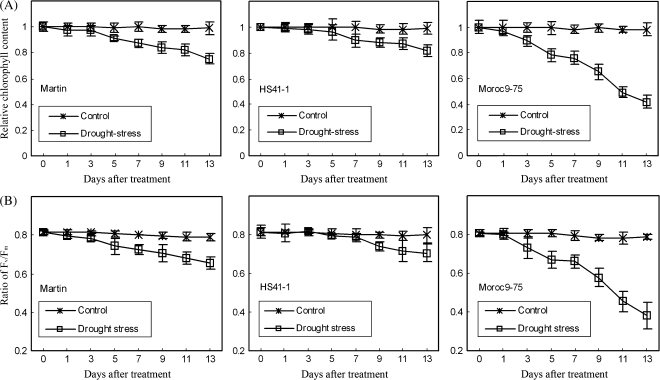
Although chlorophyll content decreased in all three genotypes under post-anthesis drought conditions (Fig. 1a), there were differential responses between drought-tolerant and drought-sensitive genotypes. A visible decline in chlorophyll content started 3 d after drought stress in the drought-sensitive Moroc9-75 and at 5 d in drought-tolerant Martin and HS41-1. After 13 d of drought stress, the relative reduction in chlorophyll content was at least double in Moroc9-75 (41.8%) compared with Martin (24.9%) and HS41-1 (19.4%).
Fig. 1.
Chlorophyll contents and maximum quantum yield of PSII (Fv/Fm) of three genotypes under well-watered conditions (70% available water in the soil) and drought stress (10% available water in the soil). Results are presented as mean ±SD of six individual measurements. (A) and (B) represent relative chlorophyll content and ratio of Fv/Fm, respectively, for three genotypes (Martin, HS41-1, and Moroc9-75).
Fv/Fm, which represents the maximum quantum yield of PSII, did not show any significant difference among the three genotypes under well-watered control conditions and declined in all three genotypes under drought stress during the 13 d of the experiment (Fig. 1b). The change in Fv/Fm during the 13 d of drought stress followed a similar pattern to chlorophyll content, where Moroc9-75 showed a much quicker decline in Fv/Fm after 5 d of drought stress than for Martin and HS41-1.
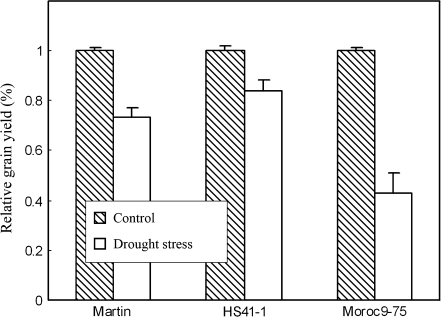
Under post-anthesis drought conditions, grain yield reduction was significant in all three genotypes but with different reduction rates (Fig. 2); the reduction was 56.8% for drought-sensitive genotype Moroc9-75, and 26.6% and 16.3% for drought-tolerant Martin and HS41-1, respectively. Based on chlorophyll content, Fv/Fm and yield reduction rate, the genotype Moroc9-75 was more sensitive to drought stress than Martin and HS41-1.
Fig. 2.
Relative grain yields of three genotypes (Martin, HS41-1, and Moroc9-75) under two moisture regimes in the soil at the post-anthesis stage. Control indicates well-watered conditions, 70% available water in the soil; drought stress, 10% available water in the soil. Values are the means ±SD.
Exploration of differentially expressed genes in response to drought stress
A total of 144, 66, and 53 genes were differentially expressed between drought-stressed and control plants of Martin, HS41-1 and Moroc9-75, respectively, in at least one of the three time points (Table 1; see Supplementary Table S1 at JXB online, and GSE15970). Among them, 96, 58, and 42 genes were up-regulated in Martin, HS41-1, and Moroc9-75, respectively, after drought stress (Table 1). All these differentially expressed genes were selected for further analysis. After a comparison of the gene expression profiles among the three genotypes, 188 differentially expressed genes (containing 65 unknown genes) were identified between drought-treated and control plants (see Supplementary Table S1 at JXB online). Among them, 17 genes were differentially expressed in both Martin and HS41-1, 20 were differentially expressed only in Martin and Moroc9-75, one was differentially expressed only in HS41-1 and Moroc9-75, and 18 were differentially expressed in all three genotypes (Table 2; Fig. 3).
Table 1.
The genes that were differentially expressed (at a minimum of one time point) between drought-treated and control plants of three barley genotypes
| Genotype | No. of drought–induced genes |
||||
| Total | Up–regulated |
Down–regulated |
|||
| Knowna | Unknownb | Knowna | Unknownb | ||
| Martin | 144 | 63 | 33 | 33 | 15 |
| HS41-1 | 66 | 37 | 21 | 7 | 1 |
| Moroc9-75 | 53 | 23 | 19 | 10 | 1 |
| Genes differentially expressed in all three genotypes | 18 | 10 | 7 | – | 1 |
| Genes differentially expressed in Martin and HS41-1 | 17 | 10 | 7 | – | – |
| Genes differentially expressed in Martin and Moroc9-75 | 20 | 7 | 8 | 5 | – |
| Genes differentially expressed in HS41–1 and Moroc9-75 | 1 | 1 | – | – | – |
‘Known’ represents genes with functional annotation based on the annotation of Barley 1 GeneChip released in July 2008.
‘Unknown’ represents no protein name or function based on the annotation of Barley 1 GeneChip released in July 2008.
Table 2.
Affymetrix probe set ID, accession number, E value, and annotation of genes in Barley 1 GeneChip that were differentially expressed in three barley genotypes between control and drought-stress conditions
| Contig IDa | Accession no.b | E-valuec | Annotation | Presence of maximum value of transcriptional changes |
|||
| Genotype | Time point | FDR corrected value | Log2 ratio | ||||
| Genes differentially expressed in all three genotypes | |||||||
| Contig4430_at | Unknownd | HS41-1 | 5 d | 2.3E-05 | 3.03 | ||
| Contig13656_at | Unknown | Martin | 3 d | 9.2E-07 | 3.88 | ||
| Contig24415_at | Unknown | Martin | 3 d | 1.2E-06 | 5.66 | ||
| HY09M19u_s_at | Unknown | HS41-1 | 1 d | 3.8E-05 | 6.30 | ||
| Contig8340_at | NP_030664.1 | 9E-94 | Unknown | Moroc9-75 | 5 d | 5.6E-05 | 3.61 |
| Contig15276_at | BAB62547.1 | 1E-45 | Unknown | HS41-1 | 3 d | 2.5E-05 | 3.46 |
| Contig16113_at | AAM53278.1 | 4E-11 | Unknown | HS41-1 | 3 d | 2.8E-05 | 4.91 |
| Contig21426_at | NP_201140.1 | 5E-06 | Unknown | Moroc9-75 | 5 d | 3.8E-05 | 4.16 |
| Contig8961_at | T02663 | 9E-17 | Abscisic acid- and stress-induced protein | Martin | 3 d | 4.2E-06 | 3.69 |
| rbah48h06_s_at | AAL92880.1 | 6E-84 | Fructosyltransferase | Moroc9-75 | 1 d | 2.5E-05 | 5.89 |
| Contig2012_s_at | S72544 | 8E-57 | Heat shock protein 17.9 | HS41-1 | 5 d | 4.6E-05 | 4.52 |
| Contig873_s_at | AAB99745.1 | 1E-115 | HSP70 | Moroc9-75 | 3 d | 2.8E-05 | 3.33 |
| Contig11041_at | AAD09209.1 | 0.0004 | Late embryogenesis abundant protein | Martin | 3 d | 9.2E-07 | 4.37 |
| rbasd16a13_s_at | S28872 | 9E-09 | Non-specific lipid transfer protein | Moroc9-75 | 5 d | 6.4E-05 | 3.95 |
| Contig21613_at | BAB89059.1 | 7E-35 | PDI-like protein | Martin | 5 d | 3.1E-06 | 4.30 |
| Contig13161_at | BAC05575.1 | 4E-69 | Protein phosphatase 2C-like protein | Martin | 3 d | 1.7E-06 | 3.65 |
| Contig3814_at | BAB64280.1 | 1E-122 | Δl-pyrroline-5-carboxylate synthetase | Martin | 3 d | 8.4E-07 | 7.08 |
| Contig15719_at | NP_180081.1 | 5E-31 | CBL-interacting protein kinase 16 | Moroc9-75 | 5 d | 9E-05 | 3.08 |
| Genes differentially expressed in Martin and HS41-1 | |||||||
| Contig5034_at | Unknown | Martin | 3 d | 1.1E-06 | 2.96 | ||
| Contig12748_at | Unknown | Martin | 3 d | 1.8E-06 | 4.76 | ||
| HS05D20u_s_at | Unknown | HS41-1 | 5 d | 5E-05 | 2.85 | ||
| EBem09_SQ001_O02_s_at | Unknown | Martin | 3 d | 3.4E-05 | 3.26 | ||
| basd23g06_s_at | Unknown | Martin | 5 d | 6.8E-05 | 4.84 | ||
| Contig6830_at | AAD27569.1 | 2E-96 | Unknown | Martin | 3 d | 1.6E-05 | 3.11 |
| Contig7437_at | AAN05517.1 | 4E-86 | Unknown | Martin | 3 d | 4.9E-07 | 6.05 |
| Contig2924_s_at | AF323586.1 | 1.4E-72 | Aldehyde dehydrogenase | Martin | 3 d | 9.2E-07 | 5.01 |
| HVSMEa0007I03r2_at | AF527606 | 2E-47 | Iron/ascorbate-dependent oxidoreductase | Martin | 3 d | 3.1E-06 | 4.97 |
| Contig1724_s_at | AAD02255.1 | 6E-19 | Dehydrin 3 | Martin | 3 d | 2.3E-06 | 7.38 |
| Contig10029_at | AF350423.1 | 1.7E-20 | Small heat shock protein HSP17.8 | HS41-1 | 5 d | 2E-05 | 4.72 |
| Contig10522_at | NP_059065.1 | 1E-05 | γ-aminobutyric acid (GABA A) receptor | Martin | 1 d | 7.7E-08 | 4.94 |
| Contig9971_at | AAL87189.1 | 5E-82 | Putative amino acid transport protein AAP2 | Martin | 3 d | 1.4E-05 | 3.14 |
| Contig4095_s_at | AB098063.1 | 5E-47 | Spermidine synthase | Martin | 3 d | 3.7E-06 | 3.10 |
| Contig6208_at | AAK20047.1 | 3E-84 | C-4 sterol methyl oxidase | HS41-1 | 3 d | 4.7E-05 | 7.10 |
| HVSMEi0006K11r2_at | BAB91939.1 | 1E-32 | NADP dependent malic enzyme | Martin | 3 d | 8.4E-07 | 5.49 |
| Contig10726_at | AAC72195.1 | 1E-100 | Pyruvate dehydrogenase E1 α subunit | Martin | 3 d | 2.4E-05 | 4.02 |
| Genes differentially expressed in Martin and Moroc9-75 | |||||||
| rbags13d01_s_at | Unknown | Martin | 3 d | 3.1E-06 | 2.79 | ||
| Contig23817_at | Unknown | Martin | 3 d | 1.7E-06 | 2.67 | ||
| Contig26247_at | Unknown | Moroc97-5 | 1 d | 4.6E-05 | 3.74 | ||
| S0000200065C10F1_at | Unknown | Martin | 3 d | 5E-06 | 4.81 | ||
| Contig9143_at | Unknown | Martin | 1 d | 1.2E-06 | 5.43 | ||
| Contig15682_at | Unknown | Martin | 3 d | 9.5E-07 | 6.56 | ||
| Contig5609_at | CAD41089.1 | 9E-57 | Unknown | Moroc97-5 | 5 d | 6.4E-05 | 2.24 |
| Contig15773_at | NP_565890.1 | 3E-06 | Unknown | Martin | 3 d | 9.2E-07 | 5.93 |
| Contig19029_at | AAD43561.1 | 7E-62 | Bacterial-induced peroxidase precursor | Martin | 3 d | 5.8E-07 | −6.53 |
| Contig2864_at | T02128 | 5E-42 | β-glucosidase homolog F8K4.3 | Martin | 5 d | 1.6E-05 | 2.97 |
| HVSMEa0006I22r2_s_at | AAD02262.1 | 0.0001 | Dehydrin 5 | Martin | 1 d | 1.2E-06 | 6.96 |
| Contig1718_s_at | AAD02260.1 | 3E-30 | Dehydrin 9 | Martin | 1 d | 1.8E-05 | 5.60 |
| Contig8538_at | Q99090 | 2E-35 | Light-inducible protein CPRF-2 | Martin | 3 d | 5E-06 | −2.77 |
| Contig8246_at | AAK27799.1 | 1E-93 | Putative amylase | Martin | 5 d | 5.8E-07 | 4.05 |
| Contig7712_at | AAK15441.1 | 4E-97 | Putative nitrate transporter | Martin | 3 d | 4.5E-06 | −3.74 |
| Contig11696_at | BAB89728.1 | 5E-66 | Putative peptide chain release factor subunit 1 | Martin | 5 d | 9.2E-07 | 4.89 |
| Contig8641_at | NP_181401.1 | 1E-108 | Ethanolamine-phosphate cytidylyltransferase | Martin | 1 d | 4.3E-06 | 2.99 |
| Contig14687_at | CAD21000.1 | 3E-31 | Putative potassium transporter | Martin | 1 d | 1.7E-06 | 2.44 |
| Contig14224_at | AAL14615.1 | 8E-13 | Putative sugar transporter | Martin | 5 d | 6.7E-05 | −3.30 |
| Contig422_at | AAB18209.1 | 1E-132 | Chlorophyll a/b -binding protein | Martin | 3 d | 2.4E-06 | −6.01 |
| Genes differentially expressed in HS41-1 and Moroc9-75 | |||||||
| Contig2717_s_at | T06489 | 2E-44 | Peptidylprolyl isomerase FKBP77 | HS41-1 | 3 d | 1E-04 | 3.46 |
Indicates probe set in Affymetrix Barley 1 GeneChip.
Gene accession no. in GenBank.
E-value in BLAST between probe set in Barley 1 GeneChip with known genes in NCBI database.
‘Unknown’ represents no protein name or function based on the annotation of Barley 1 GeneChip released in July 2008.
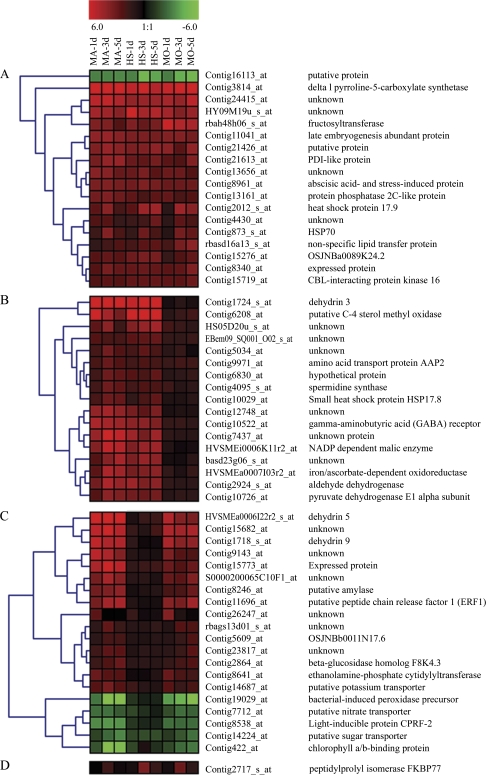
Fig. 3.
Expression changes and cluster analysis of groups of genes which were differentially expressed between control and drought stress conditions in all three genotypes (Group A), in Martin and HS41-1 (Group B), in Martin and Moroc9-75 (Group C), and in HS41-1 and Moroc9-75 (Group D). Cluster analysis for each group of genes was performed using hierarchical clustering of Genesis 1.5 with average linkage and Euclidian distance measurement. Rows represent differentially expressed genes, while columns represent the genotypes with time-course (1, 3, and 5 d) of drought treatment in which MA, HS, and MO indicate Martin, HS41-1, and Moroc9-75, respectively. Red, green, and black boxes represent genes that increased, decreased, and had equal expression levels at time points after withholding water, respectively. The contig ID and annotation of each gene are listed on the right of the figure, and the cluster numbers are listed on the left.
To validate the microarray results, 12 genes whose expression levels covered both significant and non-significant changes identified in microarray analysis were selected for real-time RT-PCR. Although the microarray log2-fold values fluctuated slightly in comparison with the corresponding values from the real-time RT-PCR, the high correlation (r2=0.85) between microarray and real-time RT-PCR data indicated that expression data from microarray analysis were in good agreement (up-regulation or down-regulation) with those obtained by real-time RT-PCR (see Supplementary Fig. S1 at JXB online).
Genes differentially expressed in at least two genotypes
To investigate the biological functions of the differentially expressed genes in response to drought stress between the barley genotypes, 56 genes that were differentially expressed in at least two genotypes (Table 2) were subjected to further analysis. The 56 differentially expressed genes were classified into four groups.
Group A consisted of 18 genes that shared a similar expression pattern at most time points in the three genotypes in response to drought stress (Fig. 3A). Of these, 12 genes were mainly up-regulated at all time points, which include genes for a Δl pyrroline-5-carboxylate synthetase (P5CS), a fructosyltransferase, a CBL (calcineurin B-like)-interacting protein kinase 16 (CIPK 16), a protein phosphatase 2C-like protein (PP2C), heat-shock proteins (HSP) HSP17.9 and HSP70, and a non-specific lipid transfer protein (nsLTP). Five genes, including genes encoding a late embryogenesis abundant (LEA) protein and a protein disulphide isomerase (PDI)-like protein, were up-regulated in all three genotypes at least at one time point under drought stress. Only one unknown gene was down-regulated at all time points in these three genotypes.
Group B contained 17 genes that were up-regulated under drought stress in the two tolerant genotypes, but not in the sensitive genotype (Fig. 3B). Among the 17 genes, 11 encoded a putative C-4 sterol methyl oxidase (CSMO), a dehydrin 3 (DHN3), a γ-aminobutyric acid (GABA) A receptor, a NADP-dependent malic enzyme (NADP-ME), an aldehyde dehydrogenase (ALDH), an ascorbate-dependent oxidoreductase (ADOR), a pyruvate dehydrogenase (PDH) E1 α subunit, and four other unknown proteins, and were consistently up-regulated in the two tolerant genotypes at all three time points. Six genes encoding an amino acid transport protein AAP2, a heat-shock protein HSP17.8, a spermidine synthase (SPDS), and three other unknown proteins were differentially expressed at 3 d and 5 d of drought stress but not at 1 d.
Group C represented 20 genes that showed similar expression patterns between Martin and Moroc9-75 (Fig. 3C). Among the 20 genes, eight were up-regulated at all time points; with five encoding two dehydrins (DHN5 and DHN9), a putative amylase, a putative potassium transporter (TRK), and a putative peptide chain release factor 1 (ERF1), and four others had unknown functions. Five genes were down-regulated at all time points and annotated as a bacterial-induced peroxidase precursor, a putative nitrate transporter (NRT), a light-inducible protein CPRF-2, a putative sugar transporter, and a chlorophyll a/b-binding protein (CABP). Four genes, with one encoding a β-glucosidase homologue and the other three of unknown function, did not show significant changes at 1 d of drought stress, but were up-regulated at least at one of the other two time points in both Martin and Moroc9-75. The remaining genes were only up-regulated at 1 d of drought stress. In Group D, only a peptidylprolyl isomerase FKBP77 gene showed similar expression changes between HS41-1 and Moroc9-75 and was significantly up-regulated only at 3 d of drought stress (Fig. 3D). The genes in Groups C and D are probably common genes responding to drought challenge between the drought-sensitive genotype Moroc9-75 and either drought-tolerant genotype Martin or HS41-1.
Abundantly expressed genes in drought-tolerant genotypes in comparison with Moroc9-75 under both conditions
In addition to the drought responsive genes identified in the three genotypes, 232 genes in either Martin or HS41-1 exhibited significant differences in transcriptional levels compared with Moroc9-75 under both well-watered and drought-stressed conditions (see Supplementary Table S2 at JXB online). Among them, 17 genes were abundantly expressed in both drought-tolerant genotypes (Table 3; see Supplementary Table S2 at JXB online). These genes with known annotations encoded a calcium-dependent protein kinase (CDPK), a G2 pea dark-accumulated protein GDA2, a membrane-related protein CP5, a putative glutathione S-transferase (GST), a putative membrane steroid binding protein (MSBP), a serine/threonine kinase-like protein (STKL), and an UVB-resistance protein UVR8. The functions of other genes are unknown. These 17 genes were not regulated by drought challenge in the three genotypes, and, therefore, they are most probably constitutively expressed genes in the drought-tolerant genotypes.
Table 3.
Constitutive genes abundantly expressed in two drought-tolerant genotypes
| Contig IDa | Accession no.b | E-valuec | Annotation |
| Contig15894_at | NP_680156.1 | 6E-37 | Similar to UVB-resistance protein UVR8 |
| Contig17366_at | NP_194051.1 | 3E-14 | Serine/threonine kinase-like protein |
| Contig17647_at | NP_195136.1 | 1E-19 | Unknown |
| Contig18339_at | AAF23901.2 | 1E-78 | Calcium-dependent protein kinase |
| Contig2458_s_at | CAD37200.1 | 7E-67 | GDA2 protein |
| Contig2488_s_at | AAG32473.1 | 6E-19 | Putative glutathione S-transferase |
| Contig25330_at | CAD39672.1 | 8E-31 | Unknownd |
| Contig3339_at | AAM91533.1 | 1E-82 | Membrane related protein CP5 |
| Contig4361_at | BAB92203.1 | E-107 | Unknown |
| Contig6026_at | Unknown | ||
| Contig6615_at | CAB53479.1 | 5E-29 | Unknown |
| Contig6926_at | AAG46109.1 | 1E-08 | Unknown |
| Contig6997_at | BAB85314.1 | 6E-32 | Unknown |
| Contig7373_at | NP_565524.1 | 6E-18 | Unknown |
| Contig8651_at | BAA83368.1 | 3E-92 | Unknown |
| HVSMEg0015I15r2_at | AAG13629.1 | 3E-11 | Putative steroid membrane binding protein |
| rbags10j11_s_at | BAB63616.1 | 7E-16 | Unknown |
Represents probe set in Affymetrix Barley 1 GeneChip.
Indicates gene accession number in GenBank.
Indicates E-value in BLAST between probe set in Barley 1 GeneChip with known genes in NCBI database.
Unknown represents no protein name or function based on the annotation of Barley 1 GeneChip released in July 2008
Discussion
Optimized method for the identification of candidate genes related to drought tolerance
An appropriate design of drought-stress experiments and rigorous statistical analysis are critical for evaluating the drought tolerance of barley genotypes. The available water content (AWC) in the soil represents the amount of water that a plant can extract from the soil for its growth. Accurate determination of whether, when, and to what degree, a plant suffers from water stress is a key step for drought tolerance assessment (Doorenbos and Pruit, 1977). In this study, a procedure to simulate field drought conditions in a greenhouse by regulating soil AWC during the reproductive stage of barley was developed. In this procedure, the thresholds for stressed (10% AWC) and optimum water (70% AWC) conditions were determined from actual soil AWC measured from the rainfed field at Breda, one of ICARDA's drought-stress research stations; and the irrigated field at Tel Hadya, ICARDA's main experimental station for favourable growth conditions (Li et al., 2006; Guo et al., 2008).
With this procedure, several physiological and morphological traits were measured under both drought and control conditions to estimate the drought tolerance of the three barley genotypes. After 13 d of drought stress, Martin and HS41-1 had much higher chlorophyll contents and Fv/Fm ratios than Moroc9-75. The yield losses due to drought stress were lower for drought-tolerant Martin and HS41-1 than for drought-susceptible Moroc9-75 (Fig. 2), in good agreement with previous reports (Ceccarelli, 1994; Ceccarelli et al., 2004; Li et al., 2006). The results showed that the three genotypes consistently showed significant contrasts in three drought-tolerance parameters, and so were appropriate plant materials for the current study.
Genes were identified that were differentially expressed between drought-tolerant and drought-sensitive barley genotypes with high confidence, by using three genotypes that contrasted in drought tolerance, a gradual water deficit to simulate natural drought condition at the reproductive stage, and the ArrayAssist software to analyse data of the Affymetrix Barley 1 GeneChip. The differentially expressed genes found in this study should provide useful information for understanding how different barley genotypes respond to drought stress at the reproductive stage and how drought-tolerant genotypes can adapt to drought-stress conditions.
Drought responsive genes in three barley genotypes
When plants are subjected to drought stress, they try to adapt to the new environment by changing gene expression pattern after perceiving stress signals (Ozturk et al., 2002; Seki et al., 2002; Hazen et al., 2005). Therefore, the genes with altered expression are probably those involved in the pathways of plant responses to drought. By comparing gene expression patterns between drought-stressed and unstressed plants of three contrasting barley genotypes, 144, 66, and 53 differentially expressed genes (at one time point at least) were identified in Martin, HS41-1, and Moroc9-75, respectively. Among them, 18 genes showed similar expression patterns in all three genotypes (Table 2; Fig. 3A) and were divided into two groups. The first group included genes encoding for a P5CS, a fructosyltransferase, an LEA protein, an nsLTP, an HSP70, an HSP17.9, and a PDI-like protein, which have roles in membrane and protein stabilization and cellular homeostasis in several species (Amiard et al., 2003; Wang et al., 2004; Houston et al., 2005; Boudet et al., 2006; Cameron et al., 2006). The second group, including CIPK 16 and PP2C, may function as signal molecules under drought stress (Takezawa, 2003; Boominathan et al., 2004; Ok et al., 2005). For 21 differentially expressed genes in Moroc9-75 and Martin (Table 2; Fig. 3C, D), some (DHN5 and DHN9) are involved in osmotic adjustment (Taiz and Zeiger, 2002; Brini et al., 2007), others in nitrogen (NRT) and potassium metabolism (TRK), and chlorophyll synthesis/degradation (CABP) (Taiz and Zeiger, 2002). Among the 39 genes, all annotated genes (except for the genes encoding fructosyltransferase and PDI-like protein) have been reported to be drought-stress responsive genes at the seedling stage in previous barley microarray studies (Ozturk et al., 2002; Ueda et al., 2004; Walia et al., 2006; Talamè et al., 2007). Therefore, these general drought-responsive genes identified at the reproductive stage of barley are similar to those identified at the seedling stage. Since these genes were expressed in both tolerant and susceptible genotypes, they may not be directly responsible for drought tolerance.
Differentially expressed genes only in drought-tolerant barley genotypes
To identify the genes responsible for drought tolerance, genotypes with similar genetic backgrounds, but with contrasting drought tolerance, are ideal for linking candidate genes to drought tolerance. However, developing such near-isogenic lines requires several years of backcrossing and selection. One alternative is to identify common genes that are differentially expressed between drought-resistant genotypes with different genetic backgrounds and drought-sensitive genotypes under drought conditions. In this study, 17 genes were differentially expressed in both drought-tolerant genotypes under drought stress but not in the drought-sensitive genotype (Table 2; Fig. 3B), thus they might play important roles in adaptive responses to water deficit.
Among the 17 genes, one encodes an NADP-ME that is located in guard cell complexes of a C3 plant. NADP-ME facilitates lignin biosynthesis by providing NADPH, and regulates cytosolic pH through balancing the synthesis and degradation of malate (Wheeler et al., 2005). Another possible role of NADP-ME is to control stomatal closure by degrading malate during the day under water-deficit conditions, because NADP-ME expression leads to decreased stomatal aperture and increased fresh mass gained per unit water used. Therefore, manipulation of organic anion metabolism in guard cells through regulating NADP-ME expression has been proposed as an approach for drought avoidance and water conservation (Laporte et al., 2002). The pyruvate produced by NADP-ME could be further degraded and used in other pathways. Interestingly, a gene encoding PDH, which is known to be involved in the oxidative decarboxylation of pyruvate to generate acetyl-CoA for the TCA cycle, was induced under water-deficit conditions in both tolerant genotypes in this study. The transcripts of genes encoding other enzymes in the TCA cycle did not increase significantly. This phenomenon was also observed in drought-stressed loblolly pine (Watkinson et al., 2003). Since the TCA cycle provides carbon skeletons for many biosynthetic pathways, a high level of PDH provides rich carbon sources for diverse uses within a plant. Therefore, up-regulation of NADP-ME and PDH in drought-tolerant genotypes suggests that carbon metabolism may be important in acclimation to water deficit.
Polyamines such as spermidine may be pivotal in plant defence against environmental stresses; in the control of cell division, root formation, and flowering; and in slowing senescence (Tamaoki et al., 2004). A higher level of free spermidine was accumulated in drought-tolerant rice cultivars than in sensitive cultivars under water-deficit conditions and was associated with enhanced activity of SPDS that synthesizes spermidine by the addition of aminopropyl groups to putrescine (Yang et al., 2007). Spermidine may have dual functions in plant stress tolerance: as a protectant in reactive oxygen species (ROS)-scavenging and a membrane-protecting compound and as a signalling regulator in stress signalling pathways that leads to the build-up of stress-tolerant mechanisms under stress conditions (Kasukabe et al., 2004). In this study, a gene encoding SPDS was significantly induced by drought stress only in the two drought-tolerant genotypes at all time points, suggesting that increased expression of SPDS may play an important role in the drought tolerance of barley through membrane protection and/or regulating the stress signalling pathway.
Two genes encoding for a CSMO and an AAP were abundantly expressed in Martin and HS41-1 under drought stress at all time points. Castigioni et al. (2005) reported that CMSO was involved in the synthesis of glycine-betaine; many species (maize, soybean, rice, and wheat) of transgenic plants with CSMO had a significantly increased glycine-betaine content, and gained tolerance to water deficit, cold, and freezing stresses. Studies in alfalfa indicated a significant increase of glycine-betaine content in the phloem sap under drought stress and suggested that transport processes might play a role in the accumulation of compatible solutes for the adaptation to water stress (Schwacke et al., 1999). Direct transport measurements showed that AAPs were efficient transporters of glycine-betaine, proline, and the stress-induced GABA (Kwart et al., 1993; Schwacke et al., 1999). Therefore, differential expression of CSMO and AAP may enhance drought tolerance in Martin and HS41-1 by the accumulation of glycine-betaine.
Expression of genes encoding an ALDH and an ADOR was up-regulated in both tolerant genotypes under drought stress. Stress-inducible ALDH in Arabidopsis thaliana catalysed the oxidation of various toxic aldehydes, which accumulated as a result of side reactions of ROS with lipids and proteins, to protect cells against the excessive accumulation of ROS (Sunkar et al., 2003). Transgenic lines with this gene were accompanied by a decreased accumulation of lipid peroxidation-derived toxic aldehydes and showed an improvement of tolerance to dehydration, as well as to other stresses (Chaves and Oliveira, 2004). In addition, ADOR was proposed to have functions in the reduction of the ascorbate free-radical outside cells, recycling of α-tocopherol, and the reduction of lipid hydroperoxides (May, 1999). Therefore, reducing excessive ROS and reactive aldehydes may be one important mechanism of drought tolerance in barley.
A gene encoding a GABA receptor was up-regulated under drought at all time points in the two drought-tolerant genotypes. GABA is a non-protein amino acid widely distributed in many organisms. In plants, GABA rapidly accumulates in response to several abiotic stresses, such as drought, cold, heat, and mechanical injury (Mazzucotelli et al., 2006; Shelp et al., 2006). Although little is known about the physiological role of GABA in higher plants, experimental evidence suggests that GABA might be involved in pH regulation, nitrogen storage, and plant development and defence (Shelp et al., 2006). GABA might also function as an endogenous signalling molecule, because GABA receptors are found in Arabidopsis pollen tubes and may have a role in mediating the effect of GABA on pollen tube growth (Yu et al., 2006). It is therefore possible that GABA may bind to these GABA receptors and modulate the metabolic pathways to confer drought tolerance in barley.
Two genes that encode an HSP17.8 and a DHN3 were up-regulated only in Martin and HS41-1 under water deficit conditions. HSP17.8 is a cytosolic class II small HSP that mainly functions in preventing aggregation, stabilizing non-native proteins, and protecting cells from injury under stress conditions (Wang et al., 2004). Expression of another class II HSP, HSP17.9, was induced in both drought-tolerant and drought-sensitive barley genotypes by drought stress, suggesting that different members of HSP may play different roles in response to drought stress. DHN3 belongs to the family of LEA D11 proteins, a large and important class of proteins involved in protecting plants from dehydration-associated injury (Lopez et al., 2003; Buchanan et al., 2005). These proteins contribute to membrane and protein stability, metal scavenging, and suppression of ROS-induced damage in plants that are exposed to a high level of salinity or water deficit (Buchanan et al., 2005). Withholding water in barley led to over-expression of DHN3, and it was thus proposed to be associated with the adaptation to dehydration (Zhu et al., 2000).
Among the 17 genes discussed above, only three have been previously reported when seedlings of a single barley genotype were exposed to drought stress. They are those encoding for ALDH (Ozturk et al., 2002; Talamè et al., 2007), CSMO (Ozturk et al., 2002; Walia et al., 2006), and NADP-ME (Ozturk et al., 2002). The gene encoding DHN3 was reported in two drought-tolerant genotypes when exposed to drought stress at the adult stage (Zhu et al., 2000). The other six annotated genes have not previously been reported in barley, and may be unique genes responsible for drought tolerance at the reproductive stage in drought-tolerant genotypes. In addition, seven differentially expressed genes in tolerant genotypes have not been annotated to date, and their functions remain to be explored. Further investigation of their functions in drought tolerance may facilitate further understanding of drought tolerance mechanisms in barley.
Constitutive abundantly expressed genes in drought-tolerant genotypes
In the current study, 17 genes were found to be differentially expressed with high abundance in Martin and HS41-1 compared with drought-sensitive genotype Moroc9-75 under both drought-stress and control conditions, but were not expressed differentially between drought-treated plants and controls of each genotype (Table 3; see Supplementary Table S2 at JXB online). Therefore, they are possibly constitutively expressed genes for drought tolerance in barley. Among them, five were related to the regulation of downstream gene expression. One of these genes is a UV-B resistant gene UVR8, which encodes a protein with sequence similarity to the eukaryotic guanine nucleotide exchange factor RCC1 (Kliebenstein et al., 2002). Recently, UVR8 has been located in the nucleus and associated with the HY5 promoter region in vivo. UVR8 has also been related to a specific signalling pathway in plants that orchestrates the protective gene expression required for plant survival in response to adverse conditions (Brown et al., 2005). In addition, two kinase genes encoding a CDPK and a STKL may regulate the downstream gene expression. CDPKs comprise a large family of serine/threonine kinases in plants, and may be involved in environmental stress signalling (Saijo et al., 2000; Romeis et al., 2001; Taiz and Zeiger, 2002). The over-expression of CDPKs enhanced cold and salt/drought-tolerance in rice (Saijo et al., 2000). The remaining two genes are CP5 and MSBP. CP5 encodes a membrane-related protein that contains the domain of lipid-binding START (steroidogenic acute regulatory protein-related lipid transfer) in the endomembrane system. The START domain may regulate intracellular lipid signalling pathways in plants (Ponting and Aravind, 1999). MSBP is a gene encoding a membrane-steroid binding protein, and is involved in the signalling of a novel steroid or sterol. Over-expression or under-expression of MSBP in transgenic plants demonstrated MSBP to be a regulator through cell elongation-related genes (Yang et al., 2005).
The other two genes are GDA2 and GST. GDA2 is an antisenescence-related protein located in the nucleus, and over-expression of GDA2 was observed in rice during salt stress, and appeared to be involved with externally applied stimuli (Kawasaki et al., 2001; Chotikacharoensuk et al., 2006). Glutathione S-transferase encoded by GST is the enzyme that catalyses the glutathione-dependent detoxification reactions and the reduction of hydroperoxides. Higher levels of gene expression and enzymatic activity of this enzyme were observed in tolerant potato genotypes under cold and osmotic stresses (Seppänen et al., 2000). Furthermore, GSTs may act as binding proteins that sequestrate flavonoids (e.g. anthocyanins) in the vacuole for protection against environmental stresses (Tahkokorpi et al., 2007).
Recent studies indicated that the levels of transcripts in several organisms (such as humans, mice, and Arabidopsis) are heritable expression traits (e-traits) that may affect downstream expression of phenotypic traits (West et al., 2007). For example, one aluminium (Al)-induced malate transporter gene (ALMT1), abundantly expressed in Al-tolerant wheat lines under both Al-stressed and control conditions, was associated with Al tolerance in wheat (Sasaki et al., 2004; Guo et al., 2007a). For barley drought tolerance, little research has been conducted that compares transcript levels between drought-tolerant and drought-sensitive barley genotypes. Thus the genes abundantly expressed in both Martin and HS41-1 in comparison with Moroc9-75 in the present study are new findings in exploring genes related to drought tolerance in barley, and suggests that they might be critical genes for drought tolerance.
Putative mechanism of drought tolerance in barley
In the current study, 18 genes (Fig. 3A) showed a similar expression pattern across three genotypes under drought stress, therefore they are probably general drought-responsive genes as reported in several other studies (Seki et al., 2002; Amiard et al., 2003; Bray, 2004; Wang et al., 2004; Boudet et al., 2006; Cameron et al., 2006; Talamè et al., 2007). Thirty-four genes were highly expressed in only the two drought-tolerant genotypes, not in the drought-sensitive genotype, thus they most probably participated in the process of drought tolerance in barley. Based on the putative functions of annotated genes, they can be classified into two groups: as regulators in signal transduction and as functional genes that directly enhanced drought-stress tolerance. The first group includes transcription factors (UVR8 and CP5), genes for protein kinases (CDPK and STKL), and other types of signalling regulators (MSBP and SPDS). The second group includes NADP-ME and PDH in carbon metabolism for stomatal behaviour; CSMO and an AAP in the biosynthesis and translocation of glycine-betaine for osmoprotection; ADOR, ALDH, GST, and SPDS in scavenging ROS for detoxification; and HSP17.8 and DHN3 in the stability of proteins and membranes for protecting the cell from injury under drought stress. Therefore, drought-tolerant barley genotypes probably gain their tolerance under drought stress through the re-establishment of cellular homeostasis, the enhancement of functional and structural protection of proteins and membranes, and the adjustment of stomata.
Supplementary data
Supplementary data are available at JXB online.
Supplementary Table S1. Transcriptional changes in log base 2 ratio for differentally expressed genes of three barley genotypes in response to drought stress
Supplementary Table S2. Transcriptional changes in log base 2 ratio for genes differentially expressed under both control and drought conditions in Martin and HS41-1 compared with Moroc9-75
Supplementary Table S3. Oligonucleotide sequences used for real-time RT-PCR in this study
Supplementary Fig. S1: Confirmation of microarray data by real-time RT-PCR
Acknowledgments
This research was supported by a GTZ project (No. 2002.7860.6-001.00 and Contract No. 81060503) sponsored by the German Federal Ministry of Economic Cooperation and Development (BMZ), grants of the National Natural Science Foundation of China (No. 30871526), the Research Project of Returned Overseas Students and Scholars of the Chinese Educational Ministry (2007-1108), the Science and Technology Planning Project of Guangdong Province (2008B050300003), the Program of Science and Technology of Guangzhou (No. 2007J1-C0111), the Education Bureau of Guangzhou Municipality (No. 62002), and the ‘Global Center of Excellence (COE) Program’ Japan.
References
- Abebe T, Guenzi AC, Martin B, Cushman JC. Tolerance of mannitol-accumulating transgenic wheat to water stress and salinity. Plant Physiology. 2003;131:1748–1755. doi: 10.1104/pp.102.003616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amiard V, Morvan-Bertrand A, Billiard JP, Huault C, Keller F, Prud'homme MP. Fructans, but not sucrosyl-galactosides, raffinose and loliose, are affected by drought stress in perennial ryegrass. Plant Physiology. 2003;132:2218–2229. doi: 10.1104/pp.103.022335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnon DI. Copper enzymes in isolated chloroplasts. Polyphenoloxidase in Beta vulgaris. Plant Physiology. 1949;24:1–15. doi: 10.1104/pp.24.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boominathan P, Shukla R, Kumar A, Manna D, Negi D, Verma PK, Chattopadhyay D. Long term transcript accumulation during the development of dehydration adaptation in Cicer arietinum. Plant Physiology. 2004;135:1608–1620. doi: 10.1104/pp.104.043141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudet J, Buitink J, Hoekstra FA, Rogniaux H, Larré C, Satour P, Leprince O. Comparative analysis of the heat stable proteome of radicles of Medicago truncatula seeds during germination identifies late embryogenesis abundant proteins associated with desiccation tolerance. Plant Physiology. 2006;140:1418–1436. doi: 10.1104/pp.105.074039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray EA. Molecular responses to water deficit. Plant Physiology. 1993;103:1035–1040. doi: 10.1104/pp.103.4.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray EA. Genes commonly regulated by water-deficit stress in Arabidopsis thaliana. Journal of Experimental Botany. 2004;55:2331–2241. doi: 10.1093/jxb/erh270. [DOI] [PubMed] [Google Scholar]
- Brini F, Hanin M, Lumbreras V, Irar S, Pages M, Masmoudi K. Overexpression of wheat dehydrin DHN-5 enhances tolerance to salt and osmotic stress in Arabidopsis thaliana. Plant Cell Reports. 2007;26:2017–2026. doi: 10.1007/s00299-007-0412-x. [DOI] [PubMed] [Google Scholar]
- Brown BA, Cloix C, Jiang GH, Kaiserli E, Herzyk P, Kliebenstein DJ, Jenkins GI. A UV-B-specific signalling component orchestrates plant UV protection. Proceedings of the National Academy of Sciences, USA. 2005;102:18225–18230. doi: 10.1073/pnas.0507187102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce WB, Edmeades GO, Barker TC. Molecular and physiological approaches to maize improvement for drought tolerance. Journal of Experimental Botany. 2002;53:13–25. [PubMed] [Google Scholar]
- Buchanan C, Lim S, Salzman RA, Kagiampakis I, Klein RR, Pratt LH, Cordonnier-Pratt M-M, Klein PE, Mullet JE. Sorghum bicolor’s transcriptome response to dehydration, high salinity and ABA. Plant Molecular Biology. 2005;58:699–720. doi: 10.1007/s11103-005-7876-2. [DOI] [PubMed] [Google Scholar]
- Cameron KD, Teece MA, Smart LB. Increased accumulation of cuticular wax and expression of lipid transfer protein in response to periodic drying events in leaves of tree tobacco. Plant Physiology. 2006;140:176–183. doi: 10.1104/pp.105.069724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castigioni P, Korte J, Bell E, Hinchey B, Bensen R, Loida P, Ahrens J. Transgenic plants with increased glycine-betaine. 2005 US Patent, 20050160500. [Google Scholar]
- Ceccarelli S, Grando S, Baum M, Udupa SM. Breeding for drought resistance in a changing climate. In: Rao S, Ryan J, editors. Challenges and strategies of dryland agriculture. 2004. CSSA Special Publication no. 32. Madison, WI: Crop Science Society of America and American Society of Agronomy, 167–190. [Google Scholar]
- Ceccarelli S. Specific adaptation and breeding for marginal conditions. Euphytica. 1994;77:205–219. [Google Scholar]
- Ceccarelli S, Grando S, Baum M. Participatory plant breeding in water-limited environments. Experimental Agriculture. 2007;43:1–25. [Google Scholar]
- Chaves MM, Oliveira MM. Mechanisms underlying plant resilience to water deficits: prospects for water-saving agriculture. Journal of Experimental Botany. 2004;55:2365–2384. doi: 10.1093/jxb/erh269. [DOI] [PubMed] [Google Scholar]
- Cheong YH, Kim KN, Pandey GK, Gupta R, Grant JJ, Luan S. CBL1, a calcium sensor that differentially regulates salt, drought, and cold responses in Arabidopsis. The Plant Cell. 2003;15:1833–1845. doi: 10.1105/tpc.012393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chotikacharoensuk T, Arteca RN, Arteca JM. Use of differential display for the identification of touch-induced genes from an ethylene-insensitive Arabidopsis mutant and partial characterization of these genes. Journal of Plant Physiology. 2006;163:1305–1320. doi: 10.1016/j.jplph.2005.12.005. [DOI] [PubMed] [Google Scholar]
- Doorenbos J, Pruit WO. Guidelines for predicting crop water requirements. 1977. FAO Irrigation and Drainage Paper no. 24. Rome: FAO. [Google Scholar]
- Guo P, Bai G, Carver B, Li R, Bernardo A, Baum M. Transcriptional analysis between two wheat near-isogenic lines contrasting in aluminum (Al) tolerance under Al stress. Molecular Genetics and Genomics. 2007a;277:1–12. doi: 10.1007/s00438-006-0169-x. [DOI] [PubMed] [Google Scholar]
- Guo P, Baum M, Li R, Grando S, Varshney RK, Graner A, Ceccarelli S, Valkoun J. Transcriptional analysis of barley genes in response to drought stress at the reproductive growth stage using Affymetrix Barley 1 genechip. Journal of Guangzhou University (Natural Science Edition) 2007b;6:32–36. [Google Scholar]
- Guo P, Baum M, Varshney R, Graner A, Grando S, Ceccarelli S. QTLs for chlorophyll and chlorophyll fluorescence parameters in barley under post-flowering drought. Euphytica. 2008;163:203–214. [Google Scholar]
- Hazen SP, Pathan MS, Sanchez A, Baxter I, Dunn M, Estes B, Chang HS, Zhu T, Kreps JA, Nguyen HT. Expression profiling of rice segregating for drought tolerance QTLs using a rice genome array. Functional and Integrative Genomics. 2005;5:104–116. doi: 10.1007/s10142-004-0126-x. [DOI] [PubMed] [Google Scholar]
- Houston NL, Fan C, Xiang JQ, Schulze JM, Jung R, Boston RS. Phylogenetic analyses identify 10 classes of the protein disulphide isomerase family in plants, including single-domain protein disulphide isomerase-related proteins. Plant Physiology. 2005;137:762–778. doi: 10.1104/pp.104.056507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingram J, Bartels D. The molecular basis of dehydration tolerance in plants. Annual Review of Plant Physiology and Plant Molecular Biology. 1996;47:377–403. doi: 10.1146/annurev.arplant.47.1.377. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs R, Collin R, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- Kasukabe Y, He L, Nada K, Misawa S, Ihara I, Tachibana S. Overexpression of spermidine synthase enhances tolerance to multiple environmental stresses and up-regulates the expression of various stress-regulated genes in transgenic Arabidopsis thaliana. Plant and Cell Physiology. 2004;45:712–722. doi: 10.1093/pcp/pch083. [DOI] [PubMed] [Google Scholar]
- Kawasaki S, Borchert C, Deyholos M, Wang H, Brazille S, Kawai K, Galbraith DW, Bohnert HJ. Gene expression profiles during the initial phase of salt stress in rice (Oryza sativa L.) The Plant Cell. 2001;13:889–906. doi: 10.1105/tpc.13.4.889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliebenstein DJ, Lim JE, Landry LG, Last RL. Arabidopsis UVR8 regulates ultraviolet-B signal transduction and tolerance and contains sequence similarity to human regulator of chromatin condensation 1. Plant Physiology. 2002;130:234–243. doi: 10.1104/pp.005041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwart M, Hirner B, Hummel S, Frommer WB. Differential expression of two related amino acid transporters with differing substrate specificity in Arabidopsis thaliana. The Plant Journal. 1993;4:993–1002. doi: 10.1046/j.1365-313x.1993.04060993.x. [DOI] [PubMed] [Google Scholar]
- Laporte MM, Shen B, Tarczynski MC. Engineering for drought avoidance: expression of maize NADP-malic enzyme in tobacco results in altered stomatal function. Journal of Experimental Botany. 2002;53:699–705. doi: 10.1093/jexbot/53.369.699. [DOI] [PubMed] [Google Scholar]
- Li R, Guo P, Baum M, Grando S, Ceccarelli S. Evaluation of chlorophyll content and fluorescence parameters as indicators of drought tolerance in barley. Agricultural Science in China. 2006;5:751–757. [Google Scholar]
- Liu X, Baird WV. Identification of a novel gene, HAABRC5, from Helianthus annuus (Asteraceae) that is upregulated in response to drought, salinity, and abscisic acid. American Journal of Botany. 2004;91:184–191. doi: 10.3732/ajb.91.2.184. [DOI] [PubMed] [Google Scholar]
- Lopez CG, Banowetz GM, Peterson CJ, Kronstad WE. Dehydrin expression and drought tolerance in seven wheat cultivars. Crop Science. 2003;43:577–582. [Google Scholar]
- May JM. Is ascorbic acid an antioxidant for the plasma membrane? FASEB Journal. 1999;13:995–1006. doi: 10.1096/fasebj.13.9.995. [DOI] [PubMed] [Google Scholar]
- Mazzucotelli E, Tartari A, Cattivelli L, Forlani G. Metabolism of gamma-aminobutyric acid during cold acclimation and freezing and its relationship to frost tolerance in barley and wheat. Journal of Experimental Botany. 2006;57:3755–3766. doi: 10.1093/jxb/erl141. [DOI] [PubMed] [Google Scholar]
- Nakashima K, Yamaguchi-Shinozaki K. Regulons involved in osmotic stress-responsive and cold stress-responsive gene expression in plants. Physiologia Plantarum. 2006;126:62–71. [Google Scholar]
- Ok SH, Jeong HJ, Bae JM, Shin JS, Luan S, Kim KN. Novel CIPK1-associated proteins in Arabidopsis contain an evolutionarily conserved C-terminal region that mediates nuclear localization. Plant Physiology. 2005;139:138–150. doi: 10.1104/pp.105.065649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozturk ZN, Talame V, Deyholos M, Michalowski CB, Galbraith DW, Gozukirmizi N, Tuberosa R, Bohnert HJ. Monitoring large-scale changes in transcript abundance in drought- and salt-stressed barley. Plant Molecular Biology. 2002;48:551–573. doi: 10.1023/a:1014875215580. [DOI] [PubMed] [Google Scholar]
- Pellegrineschi A, Noguera LM, Skovmand B, Brito RM, Velazquez L, Salgado MM, Hernandez R, Warburton M, Hoisington D. Identification of highly transformable wheat genotypes for mass production of fertile transgenic plants. Genome. 2002;45:421–430. doi: 10.1139/g01-154. [DOI] [PubMed] [Google Scholar]
- Pellegrineschi A, Reynolds M, Pacheco M, Brito RM, Almeraya R, Yamaguchi-Shinozaki K, Hoisington D. Stress-induced expression in wheat of the Arabidopsis thaliana DREB1A gene delays water stress symptoms under greenhouse conditions. Genome. 2004;47:493–500. doi: 10.1139/g03-140. [DOI] [PubMed] [Google Scholar]
- Ponting CP, Aravind L. START: a lipid-binding domain in StAR, HD-ZIP and signalling proteins. Trends in Biochemical Sciences. 1999;24:130–132. doi: 10.1016/s0968-0004(99)01362-6. [DOI] [PubMed] [Google Scholar]
- Romeis T, Ludwig A, Martin R, Jones JDG. Calcium-dependent protein kinases play an essential role in a plant defence response. EMBO Journal. 2001;20:5556–5567. doi: 10.1093/emboj/20.20.5556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki T, Yamamoto Y, Ezaki B, Katsuhara M, Ahn SJ, Ryan PR, Delhaize E, Matsumoto H. A wheat gene encoding an aluminum-activated malate transporter. The Plant Journal. 2004;37:645–653. doi: 10.1111/j.1365-313x.2003.01991.x. [DOI] [PubMed] [Google Scholar]
- Saijo Y, Hata S, Kyozuka J, Shimamoto K, Izui K. Over-expression of a single Ca2+-dependent protein kinase confers both cold and salt/drought tolerance on rice plants. The Plant Journal. 2000;23:319–327. doi: 10.1046/j.1365-313x.2000.00787.x. [DOI] [PubMed] [Google Scholar]
- Schwacke R, Grallath S, Breitkreuz KE, Stransky E, Stransky H, Frommer WB, Rentsch D. LeProT1, a transporter for proline, glycine-betaine, and gamma-amino butyric acid in tomato pollen. The Plant Cell. 1999;11:377–392. doi: 10.1105/tpc.11.3.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki M, Narusaka M, Ishida J, et al. Monitoring the expression profiles of 7,000 Arabidopsis genes under drought, cold and high-salinity stresses using a full-length cDNA microarray. The Plant Journal. 2002;31:279–292. doi: 10.1046/j.1365-313x.2002.01359.x. [DOI] [PubMed] [Google Scholar]
- Seppänen MM, Cardi T, Hyökki MB, Pehu E. Characterization and expression of cold-induced glutathione S-transferase in freezing tolerant Solanum commersonii, sensitive S. tuberosum and their interspecific somatic hybrids. Plant Science. 2000;153:125–133. doi: 10.1016/s0168-9452(99)00252-6. [DOI] [PubMed] [Google Scholar]
- Shelp BJ, Bown AW, Faure D. Extracellular gamma-aminobutyrate mediates communication between plants and other organisms. Plant Physiology. 2006;142:1350–1352. doi: 10.1104/pp.106.088955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkar R, Bartels D, Kirch HH. Overexpression of a stress-inducible aldehyde dehydrogenase gene from Arabidopsis thaliana in transgenic plants improves stress tolerance. The Plant Journal. 2003;35:452–464. doi: 10.1046/j.1365-313x.2003.01819.x. [DOI] [PubMed] [Google Scholar]
- Tahkokorpi M, Taulavuori K, Laine K, Taulavuori E. After-effects of drought-related winter stress in previous and current year stems of Vaccinium myrtillus L. Environmental and Experimental Botany. 2007;61:85–93. [Google Scholar]
- Taiz L, Zeiger E. Plant physiology. 3rd edn. Sunderland, MA: Sinauer Associates Publishers; 2002. [Google Scholar]
- Takezawa D. Characterization of a novel plant PP2C-like protein ser/thr phosphatase as a calmodulin-binding protein. Journal of Biological Chemistry. 2003;278:38076–38083. doi: 10.1074/jbc.M301369200. [DOI] [PubMed] [Google Scholar]
- Talamè V, Ozturk NZ, Bohnert HJ, Tuberosa R. Barley transcript profiles under dehydration shock and drought stress treatments: a comparative analysis. Journal of Experimental Botany. 2007;58:229–240. doi: 10.1093/jxb/erl163. [DOI] [PubMed] [Google Scholar]
- Tamaoki M, Saji H, Shirano Y, Kato T, Hayashi H, Shibata D, Tabata S, Komeda Y, Takahashi T. Spermidine synthase genes are essential for survival of Arabidopsis. Plant Physiology. 2004;135:1565–1573. doi: 10.1104/pp.104.041699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda A, Kathiresan A, Inada M, Narita Y, Nakamura T, Shi W, Takabe T, Bennett J. Osmotic stress in barley regulates expression of a different set of genes than salt stress does. Journal of Experimental Botany. 2004;55:2213–2218. doi: 10.1093/jxb/erh242. [DOI] [PubMed] [Google Scholar]
- Umezawa T, Fujita M, Fujita Y, Yamaguchi-Shinozaki K, Shinozaki K. Engineering drought tolerance in plants: discovering and tailoring genes to unlock the future. Current Opinion in Biotechnology. 2006;17:113–122. doi: 10.1016/j.copbio.2006.02.002. [DOI] [PubMed] [Google Scholar]
- Vinocur B, Altman A. Recent advances in engineering plant tolerance to abiotic stress: achievements and limitations. Current Opinion in Biotechnology. 2005;16:1–10. doi: 10.1016/j.copbio.2005.02.001. [DOI] [PubMed] [Google Scholar]
- Walia H, Wilson C, Wahid A, Condamine P, Cui X, Close TJ. Expression analysis of barley (Hordeum vulgare L.) during salinity stress. Functional and Integrative Genomics. 2006;6:143–145. doi: 10.1007/s10142-005-0013-0. [DOI] [PubMed] [Google Scholar]
- Wang W, Vinocur B, Shoseyov O, Altman A. Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response. Trends in Plant Science. 2004;9:244–252. doi: 10.1016/j.tplants.2004.03.006. [DOI] [PubMed] [Google Scholar]
- Watkinson JI, Sioson AA, Vasquez-Robinet C, et al. Photosynthetic acclimation is reflected in specific patterns of gene expression in drought-stressed loblolly pine. Plant Physiology. 2003;133:1702–1716. doi: 10.1104/pp.103.026914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West MAL, Kim K, Kliebenstein DJ, van Leeuwen H, Michelmore RW, Doerge RW, Clair DAS. Global eQTL mapping reveals the complex genetic architecture of transcript level variation in Arabidopsis. Genetics. 2007;175:1441–1450. doi: 10.1534/genetics.106.064972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler MC, Tronconi MA, Drincovich MF, Andreo CS, Flugge UI, Maurino VG. A comprehensive analysis of the NADP-malic enzyme gene family of Arabidopsis. Plant Physiology. 2005;139:39–51. doi: 10.1104/pp.105.065953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Duan X, Wang B, Hong B, Ho T, Wu R. Expression of a late embryogenesis abundant protein gene, hva1, from barley confers tolerance to water deficit and salt stress in transgenic rice. Plant Physiology. 1996;110:249–257. doi: 10.1104/pp.110.1.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Zhang J, Liu K, Wang Z, Liu L. Involvement of polyamines in the drought resistance of rice. Journal of Experimental Botany. 2007;58:1545–1555. doi: 10.1093/jxb/erm032. [DOI] [PubMed] [Google Scholar]
- Yang XH, Xu ZH, Xue HW. Arabidopsis Membrane Steroid-Binding Protein 1 is involved in inhibition of cell elongation. The Plant Cell. 2005;17:116–131. doi: 10.1105/tpc.104.028381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G, Liang J, He Z, Sun M. Quantum dot-mediated detection of γ-aminobutyric acid binding sites on the surface of living pollen protoplasts in tobacco. Chemistry and Biology. 2006;13:723–731. doi: 10.1016/j.chembiol.2006.05.007. [DOI] [PubMed] [Google Scholar]
- Zhang JZ, Creelman RA, Zhu JK. From laboratory to field. Using information from Arabidopsis to engineer salt, cold, and drought tolerance in crops. Plant Physiology. 2004;135:615–621. doi: 10.1104/pp.104.040295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu B, Choi DW, Fenton R, Close TJ. Expression of the barley dehydrin multigene family and the development of freezing tolerance. Molecular and General Genetics. 2000;264:145–153. doi: 10.1007/s004380000299. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.