Abstract
Mapping and sequencing of the non-dormant evg mutant in peach [Prunus persica (L.) Batsch] identified six tandem-arrayed DAM (dormancy-associated MADS-box) genes as candidates for regulating growth cessation and terminal bud formation. To narrow the list of candidate genes, an attempt was made to associate bud phenology with the seasonal and environmental patterns of expression of the candidates in wild-type trees. The expression of the six peach DAM genes at the EVG locus of peach was characterized throughout an annual growing cycle in the field, and under controlled conditions in response to a long day–short day photoperiod transition. DAM1, 2, 4, 5, and 6 were responsive to a reduction in photoperiod in controlled conditions and the direction of response correlated with the seasonal timing of expression in field-grown trees. DAM3 did not respond to photoperiod and may be regulated by chilling temperatures. The DAM genes in peach appear to have at least four distinct patterns of expression. DAM1, 2, and 4 are temporally associated with seasonal elongation cessation and bud formation and are the most likely candidates for control of the evg phenotype.
Keywords: Annual, bud, chilling, perennial, photoperiod
Introduction
Autumn bud set and spring bud flush are dominant phenological events in many terrestrial ecosystems. Bud endodormancy in perennial species serves to preserve meristematic tissues in a quiescent state during potentially damaging environmental conditions. Seasonal bud endodormancy timing is a critical life history trait in perennials, balancing the opportunity cost of foregoing productive growing conditions against the risk of freezing temperatures. The phenology of local genotypes is adapted to the winter regime in which they naturally occur, complicating the introduction of superior germplasm to regions of different photoperiods and winter climate (Frewen et al., 2000; Rousi and Pusenius, 2005). Understanding the regulatory events involved in bud set and endodormancy establishment will enable the targeted adaptation of superior genotypes to alternative winter climate regions (Rohde et al., 2000). Targeted adaptation of bud endodormancy to predicted climate change regimes, particularly warmer winter conditions, could also contribute to food and timber security (Easterling, 2007; Kirilenko and Sedjo, 2007; Baldocchi and Wong, 2008). Unfortunately, a sophisticated understanding of the regulatory events between the perception of dormancy-inducing signals and growth arrest, bud formation, and entry into endodormancy are lacking.
The discovery of molecular regulators of developmental events relies heavily on the analysis of naturally occurring mutants or mutant germplasm derived from large-scale screens. For most perennial species (particularly trees) the second approach is prohibitively lengthy and expensive and, as a result, investigators have been limited to a reverse genetic strategy using candidate genes (Howe et al., 1996; Olsen and Junttila, 2002; Rohde et al., 2002; Bohlenius et al., 2006). Unfortunately naturally occurring mutants for seasonal dormancy processes are extremely rare (Thompson et al., 1985; Rodriguez et al., 1994). The evergrowing (evg) peach [P. persica (L.) Batsch] mutant is a dormancy-impaired genotype identified in Mexico (Rodriguez et al., 1994; Werner and Okie, 1998). Evergrowing (evg) peach maintains continuous apical growth and has persistent leaf production even when exposed to shortened days and low temperatures (Rodriguez et al., 1994). Lateral buds on the mutant appear to have wild-type behaviour in response to dormancy-inducing conditions suggesting the mutation specifically affects the terminal meristems. The frost hardiness of evg mutant trees is reduced relative to wild-type sibling trees (Arora et al., 1992, 1996; Rodriguez et al., 1994; Arora and Wisniewski, 1996), however, the mutant trees remain capable of some cold hardiness induction in response to cold temperatures.
Six dormancy-associated MADS-box (DAM) genes were recently identified as candidates for the evergrowing (evg) mutation in peach [Prunus persica (L.) Batsch] (Bielenberg et al., 2008). The DAM genes at the EVG locus are arranged in tandem, are highly similar in sequence and thus appear to have arisen by duplications at the EVG locus (Bielenberg et al., 2008). The peach DAM genes are members of the SVP/StMADS11 clade of MIKCC MADS-box genes and this clade appears to be expanded in perennials, or at least woody perennials (Leseberg et al., 2006). Comprehensive cataloging of MADS-box genes in annual models has revealed only two (Arabidopsis and tomato) or three (rice) members of the SVP/StMADS11 clade (Alvarez-Buylla et al., 2000; Hileman et al., 2006; Arora et al., 2007). Leseberg et al. (2006) suggest that there has been an expansion of this gene group in Populus through a combination of chromosomal and tandem duplication events resulting in eight genomic loci of the SVP/StMADS11 clade MADS-box genes. A recent analysis of the grape (Vitis vinifera) MIKCC-type MADS-box family identified five SVP/StMADS11 clade members (Diaz-Riquelme et al., 2009). In peach, tandem duplication appears to have resulted in at least six members of the SVP/StMADS11 MADS-box clade (Bielenberg et al., 2008). The expansion of this class of gene in perennial genomes relative to annual models may represent a need for additional regulatory genes for developmental events like dormancy that are not part of annual plant life histories. Altered expression of putative DAM genes during dormancy establishment or dormancy breaking have been observed in the buds of several species by transcript profiling experiments during dormancy establishment or breaking (Mazzitelli et al., 2007; Ruttink et al., 2007; Campbell et al., 2008; Horvath et al., 2008; Yamane et al., 2008; Diaz-Riquelme et al., 2009).
Expression of all six of the duplicated DAM genes are completely absent in the mutant genotype because of a deletion at the evg locus. This complicates definitive association of a specific gene or group of genes with the mutant phenotype (Bielenberg et al., 2008). The fact that all six genes are missing in plants that show the mutant phenotype raises the possibility that the six genes are functionally redundant. However, Bielenberg et al. (2008) evaluated expression of the six DAM genes in wild-type peach at four times (March, June, September, and December) during the year and found that the expression of these genes did appear to be broadly seasonal with none of the DAM genes expressed at the March sampling date. DAM1, 2, and 4 were expressed in the summer and autumn samples (Bielenberg et al., 2008). DAM3 was expressed in the summer, autumn, and winter samples, and DAM5 and 6 were expressed only in the autumn and winter samples (Bielenberg et al., 2008). These results suggest that the expression of the six DAM genes do not entirely overlap and therefore may not be redundant or functionally redundant. However, the coarse scale of sampling (three month intervals) did not allow the association of specific genes with developmental events such as bud set or bud break.
The expression of the six tandem-arrayed DAM genes at the EVG locus of peach is characterized here throughout an annual growing cycle in the field and in response to a long day/short day photoperiod transition. Our specific questions were: (i) do the six genes have overlapping spatial and temporal expression patterns? (ii) do one or more of the genes have patterns of expression that coincide with seasonal bud phenology? and (iii) can seasonal expression patterns be correlated with environmental conditions?
Materials and methods
Plant material and growth conditions
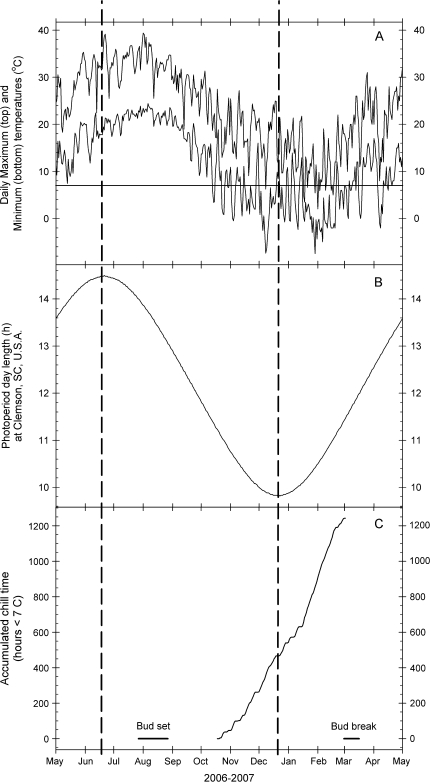
Clones of wild-type peach trees [Prunus persica (L.) Batsch] from an F2 population used to map the evg mutation (Wang et al., 2002; Bielenberg et al., 2008) were propagated by bud grafting onto peach Guardian® rootstock and planted in an irrigated nursery in 2003 at the Clemson University Musser Fruit Research Center located near Seneca, South Carolina, USA. All trees in the study were reproductively mature. Tissue samples from three randomly selected trees were collected at approximately 2-week intervals throughout the 2006–2007 growth cycle (May to May). Day length (photoperiod) values were derived from daily sunrise and sunset times for Clemson, SC as calculated by the US Naval Observatory Astronomical Applications Department. During the 2006–2007 winter season, air temperature was recorded at 10 min intervals (WatchDog™ Model 110, Spectrum Technologies, Inc., Plainfield, IL, USA). Cumulative chilling hours were calculated by the sum of 10 min intervals below 7 °C air temperature from 15 October 2006 until all the trees had broken bud and flowered in the field in early March, 2007. Day length, chilling accumulation, and daily maximum and minimum temperatures for the 12-month period of the experiment are shown in Fig. 1.
Fig. 1.
Daily maximum and minimum temperatures (A), day length (B), and cumulative chilling hours from bud set to bud break (C) at the Musser Fruit Research Center near Clemson University, SC, USA. Day length and cumulative chilling hours were calculated as described in the Materials and methods. The horizontal line in (A) is the temperature threshold below which chilling was calculated. Vertical dashed lines indicate the approximate winter or summer solstice dates. Solid bars in (C) show the intervals of terminal bud set or bud break in field-grown trees.
Field-grown trees were clonally propagated by rooting in June 2006 and grown in pots in a greenhouse at the Biosystems Research Complex, Clemson University, Clemson, SC, USA. Potted cuttings were maintained in 16 h photoperiods by supplemental lighting in the greenhouse until February 2007, when they were transferred to growth rooms maintained at a 16 h photoperiod for acclimation. Illumination in the growth rooms was 250–300 μmol m−2 s−1 photosynthetically active radiation at canopy height provided by AgroSun® Gold 1000 W sodium/halide lamps. Temperatures were maintained at 22.5 °C in the light and 18.7 °C in the dark and relative humidity was 48%/55% (light/dark). Following acclimation in the growth room environment for 3 weeks, the photoperiod was reduced to an 8 h photoperiod. Two weeks prior to the shift from long day to short day photoperiods, all plant terminal axes were marked to allow the calculation of primary axis elongation and leaf formation prior-to and following the photoperiod transition. Terminal tissues for RNA extraction and quantitative gene expression experiments were harvested from three randomly selected plants weekly for 8 weeks following the transition to a short day photoperiod. An additional two to three plants at each harvest were returned to the greenhouse from the growth room and allowed to grow in permissive conditions (16 h light period) to assess the potential for terminal meristem growth to resume.
RNA extraction and cDNA synthesis
Plant tissue samples were frozen in liquid N2 immediately following harvest and stored at –80 °C until processed. Total RNA was isolated according to the method described by Meisel et al. (2005) and modified for use in our laboratory. Tissues were finely ground in liquid N2 and 100 mg of powdered tissue was transferred to a 2.0 ml centrifuge tube and 13 μl of β-mercaptoethanol and 600 μl of 85 °C extraction buffer (2% SDS, 1.4 M NaCl, 50 mM EDTA pH 8.0, 0.5% PVP, 0.1 M TRIS-HCl, pH 8.0) were added sequentially. The mixture was homogenized by vortexing, followed by incubation at 65 °C for 15–20 min and allowed to cool to room temperature. After the tubes cooled to room temperature, 60 μl of 5 M potassium acetate, 180 μl of cold (–20 °C) 100% ethanol, 600 μl of equilibrated phenol, and 120 μl of chloroform were added sequentially, mixing well after each addition. Tubes were vortexed and incubated for 30 min on ice. Tubes were then centrifuged at 12 000 g at 4 °C for 30 min. The supernatant was transferred to a clean tube and an equal volume of chloroform:isoamyl alcohol (24:1 v/v) was added. Tubes were mixed by vortexing and centrifuged at 12 000 g at 4 °C for 20 min. The supernatant was transferred to a new tube and precipitated with LiCl at a final concentration of 3 M at –20 °C for 3 h. The RNA was pelleted by centrifugation at 12 000 g at 4 °C for 20 min. The pellet was dissolved completely into 400 μl of DEPC-treated SS buffer (1 M NaCl, 0.5% SDS, 10 mM TRIS-HCl, pH 8.0). The sample solution was extracted with an equal volume of chloroform:isoamyl-alcohol (24:1 v/v) by vortexing. These phases were separated by centrifugation at 14 000 g for 10 min at 4 °C. The aqueous phase was transferred to a clean tube and the RNA was ethanol precipitated at –80 °C for 30 min. RNA was pelleted by centrifugation at 12 000 g for 20 min at 4 °C and the pellet was washed with 75% ethanol twice and resuspended in 20 μl of RNase free water. Total RNA was quantified using spectrophotometry, and the quality was assessed by the ratio of A260 nm:A280 nm (≥1.8) and A260 nm:A230 nm (≥2.1) followed by electrophoresis in a denaturing 1.3% agarose gel.
Two μg of DNase I-treated total RNA were reverse-transcribed with oligomer (dT)30 as a primer using the SuperScript III First Strand Synthesis System for RT-PCR (Invitrogen, Carlsbad, CA, USA) following the manufacturer's instructions. Aliquots of first strand cDNA were stored at –20 °C.
DAM expression localization
Gene-specific primers (Table 1) were designed based on the DAM cDNA sequences (GenBank accession numbers: DAM1, DQ863253; DAM2, DQ863255; DAM3, DQ863256; DAM4, DQ863250; DAM5, DQ863251; DAM6, DQ863252) using Primer3_www.cgi v0.2 (Rozen and Skaletsky, 2000), and ordered from Integrated DNA Technologies (Coralville, IA, USA). All primer pairs span one or more introns in the genomic sequence to allow for the detection of genomic DNA contamination (Table 1). Each PCR reaction consisted of one DAM gene target and the tubulin-α target; fragment sizes of the amplicons are indicated in Table 1. PCR products were cloned and sequenced to confirm identity. PCR was performed with 0.5 μl of first strand cDNA (in a volume of 25 μl, containing 1 U of Platinum™ Taq DNA polymerase (Invitrogen, Carlsbad, CA, USA), 0.2 mM dNTPs, 1.5 mM MgCl2, 0.4 μM of each primer and 1×Taq polymerase reaction buffer. An initial denaturation step of 120 s at 95 °C was followed by 39 cycles of 94 °C for 30 s, 68 °C for 30 s, and 72 °C for 60 s. The PCR products were fractioned on a 2% agarose gel and scored for the presence or absence of amplification products.
Table 1.
Primers used to detect and quantify expression of the six DAM genes from the EVG locus and a tubulin-α reference gene
| Gene | Accession number | Oligonucleotide sequence pairs used for amplification (5′–3′) | Primer position | Amplicon size (bp) |
| DAM1 | DQ863253 | F–GGGGACGATGAAAATGACGAGGGAG | 5′UTR/1 | 428 |
| R–CAATCACCCGGCCAAGGCTTGCATC | 4 | |||
| DAM2 | DQ863255 | F–CAGTCAGCCAGCAGGAGAAGCAGCC | 5′UTR | 585 |
| R– GCTCAGCTCCCTTTCTTTCAAGTGCC | 5/6 | |||
| DAM3 | DQ863256 | F–ACCAGCTAAGGCAGACGATGA | 6/7 | 166 |
| R–GAGGGAGAGAGACTGAGAGCA | 7 | |||
| DAM4 | DQ863250 | F–TGTGGCACTTGAGAAAAAGGGA | 5/6 | 419 |
| R–CAGGTTACTTTCCCCAGGCCAC | 3′UTR | |||
| DAM5 | DQ863251 | F–CCCCGAAACCCACCAACGAAGATG | 5′UTR/1 | 685 |
| R–CAGCACTGTTGCAGGTGGTG | 7 | |||
| DAM6 | DQ863252 | F–CCAACAACCAGTTAAGGCAGAAGA | 6/7 | 229 |
| R–GGAAGCCCCAGTTTGAGAGA | 7/8 | |||
| Tua5 | DY650410 | F–CAGATGCCCAGTGATGCCTCAG | – | 336 |
| R–ACCAGTACCACCACCAACAGC | – |
Primers were designed to amplify regions containing introns in genomic DNA sequences to minimize amplification resulting from contaminating DNA. Primer position in the gene is indicated by the exon number, beginning with the exon that contains the translation start signal. The presence of a ‘/’ between exon numbers indicates that the primer sequence spans an intron.
Semi-quantitative gene expression of DAM genes in terminal buds
Template cDNA was synthesized from 2.5 μg of DNAse I-treated total RNA using the Invitrogen SuperScript first-strand synthesis system as above. First strand cDNAs were diluted 1:4 with nuclease-free water and aliquots of the cDNA sample were amplified using gene-specific primers (Table 1). PCR reactions were performed in a 25 μl volume containing 0.2 μM of each primer, 1 U of Platinum Taq DNA polymerase (Invitrogen), 0.2 mM dNTPs, 1.5 mM MgCl2, 0.4 μM of each primers and 1×Taq polymerase reaction buffer, 4 μl of cDNA sample (equivalent to 125 ng of the total RNA). PCR was performed as follows: denaturation at 95 °C for 60 s; 21 to 26 cycles of 94 °C for 30 s, 62 °C for 30 s, 72 °C for 40 s; followed by a final extension at 72 °C for 120 s. The appropriate number of cycles for each individual gene was determined experimentally to ensure that the amplification end-points were in a linear range. PCR products were fractionated on a 3% (w/v) agarose gel, stained with SYBR Green I (Invitrogen), and photographed using the FUJIFILM Science Lab 99 Image Gauge system. The relative intensities of the bands in each lane were quantified by imaging the gel with the Image Gauge Version 3.2 software (Fuji Photo Film, Tokyo, Japan). Target gene expression was quantified relative to the control amplification (tubulin-α) in the same lane of the gel. Two genes were tested as RT-PCR references: an 18S rRNA amplification (QuantumRNA Universal 18S Internal Standard primers with a primer:competimer ratio of 1:9, Ambion, Inc., Austin, TX, USA) and a P. persica tubulin-α EST were used (DY650410, Table 1). The QuantumRNA Universal 18S primer:competimer system significantly reduced the amplification signal into a range appropriate for use as a reference standard for our DAM genes. However, the cycle number for linear amplification of the tubulin-α gene, more closely matched the cycle number for the linear range of the DAM genes and this gene was selected to be the semi-quantitative reference (Table 1).
The stability of the tubulin-α reference throughout the experiment was verified by real-time PCR performed on an iCycler iQ system (Bio-Rad, Hercules, CA, USA) using the iQ SYBR-Green Supermix (Bio-Rad, Hercules, CA, USA). PCR was conducted on three technical replicates for each biological replicate assayed with the following programme: an initial DNA polymerase activation at 95°C for 180 s, then followed by 40 cycles of 95 °C for 30 s, 60 °C for 30 s, and 72 °C for 30 s. Finally, a melting curve was performed, and the PCR products were checked with 2% agarose gel in 1× TAE with ethidium bromide.
Fluorescence values were baseline-corrected and Ct values were calculated using LinRegPCR program (Ruijter et al., 2009). Tubulin-α showed a low variability of expression within biological replicates and a stable expression throughout the experiment with a stability index of 0.09 (calculated as in Brunner et al., 2004). This value was similar to the most stable reference genes proposed in poplar (Brunner et al., 2004) and similar to the reference genes analysed in bud tissue during dormancy transition in Norway spruce (Yakovlev et al., 2006).
Results
Organ specificity of P. persica DAM genes
Tissues from field- and greenhouse-grown trees were harvested to determine the presence or absence of expression of DAM1-6 by RT-PCR. Vegetative shoot tissues (stem, leaf, terminal and lateral buds, and growing meristems) were sampled on multiple dates throughout the growing season in the field and the greenhouse. Flower and fruit tissues were sampled throughout development and maturation in spring 2006 from field-grown trees. Flowers were divided into petals, stamens, pistils, and sepals for separate analysis, but no floral organ-specific expression was observed (expression was in all organs or none) so only the pooled results are presented. Fruit were divided into the flesh (mesocarp) and the developing seed (all tissues inside the hard outer pit) for analysis. Root samples were obtained from greenhouse-grown potted trees and were primarily young, non-suberized, white roots. Tissue-specific expression of individual genes was consistent across field and greenhouse-grown trees, except for root samples which were not obtained from field-grown trees. DAM1–6 were each expressed in all vegetative tissues (Table 2). DAM2, 3, 4, and 5 were expressed in all floral and fruit tissues and the developing seed (Table 2). DAM1 was not expressed in any floral structure but was expressed in the fruit flesh and in the developing seed (Table 2). DAM6 was expressed in all vegetative and reproductive tissues except for the developing seed (Table 2).
Table 2.
Tissue localization of peach DAM gene expression
| Gene | Organ |
|||||||
| Leaf | Root | Stem | Tip | Flower | Buds | Fruit flesh | Developing seed | |
| DAM1 | + | + | + | + | − | + | + | + |
| DAM2 | + | + | + | + | + | + | + | + |
| DAM3 | + | + | + | + | + | + | + | + |
| DAM4 | + | + | + | + | + | + | + | + |
| DAM5 | + | + | + | + | + | + | + | + |
| DAM6 | + | + | + | + | + | + | + | − |
a‘+’ Indicates detectable expression was observed in samples from at least one growing condition and sample time, but does not signify constitutive expression at all times. ‘—’ Indicates no detectable expression was observed in any growing condition or sampling date investigated.
Expression of P. persica dormancy-associated MADS genes through the annual growth cycle
Vegetative terminal growing points or terminal buds from field-grown trees were sampled for a complete growing cycle from post-bud break (May 2006) through the resumption of growth in the following spring (May 2007). In May of 2006 the peach trees were in leaf and actively growing. Growth cessation and terminal bud formation were observed visually between mid-August and early September on leading stems of the upper canopy. Following winter, the trees bloomed during the first week of March 2007. Vegetative bud break followed blooming by approximately 2 weeks (Fig. 1). A late frost event killed all fruit on these trees in April 2007 and caused some loss of new shoots.
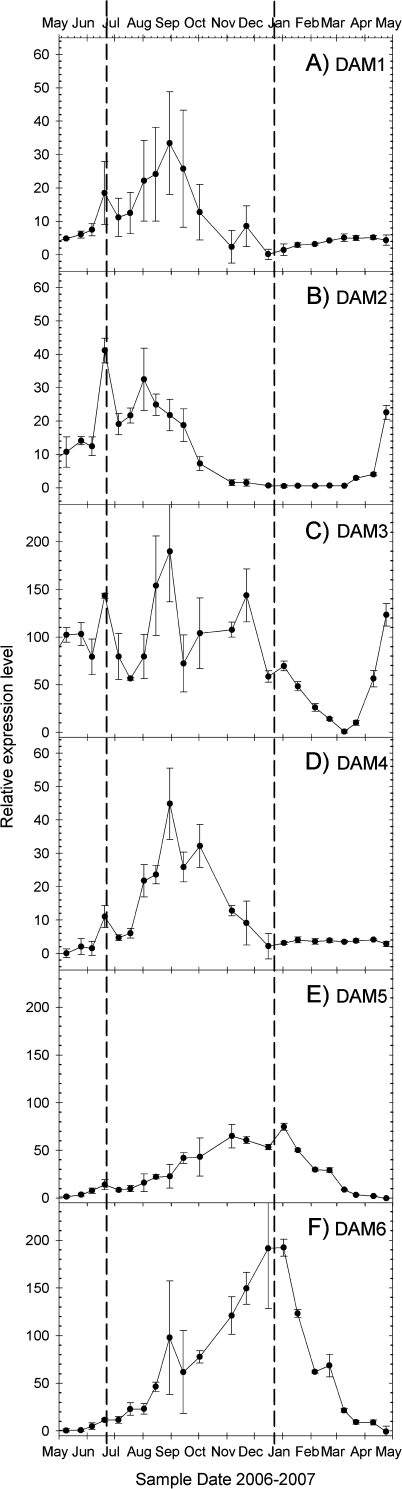
Each of the six genes had a clear seasonal pattern of gene expression (Fig. 2). DAM1 was expressed at very low levels beginning in May 2006 and gradually increased to a peak in late August (Fig. 2). DAM1 expression then declined, effectively to zero, by December and was not expressed in the winter season or the following spring through to the end of our sampling in May 2007 (Fig. 2). DAM2 expression increased steadily from May 2006 and peaked at summer solstice and again 6 weeks later, coinciding with the timing of visible bud set in the field-grown trees. Expression of DAM2 then steadily decreased until December 2006 and was not expressed again until April 2007. DAM3 was expressed at relatively high levels during the growing season and displayed a cyclic pattern of expression throughout the growing season on a 4–5 week cycle. Beginning in mid-November, DAM3 expression steadily dropped until reaching effective zero during the first week of March 2007. Following bud break in March 2007, DAM3 expression climbed rapidly through the end of our sampling period. DAM4 expression followed a pattern similar to that of DAM1 with expression peaking in late August/early September and then declining to no detectable expression in the winter and through the following spring. DAM5 and DAM6 expression followed a nearly identical pattern of steady increase through the autumn, reaching a peak coinciding with the winter solstice, followed by a rapid decline to May, by which point there was no observable expression (Fig. 2).
Fig. 2.
Semi-quantitative expression of six DAM genes in terminal tissues of field-grown peach trees throughout an annual growth cycle. Amplification products were quantified by densitometry following gel electrophoresis. A peach tubulin-α amplification was performed in each reaction tube along with the target gene specific primers as a loading control and quantitation reference. Tubulin-α band intensities within each amplification reaction were set to 100 and the target gene amplification product intensity was calculated relative to the tubulin-α control for each amplification product. Each symbol represents the average of samples from three individual trees, error bars are standard deviations. Vertical dashed lines are as in Fig. 1. DAM1, 2, and 4 are on a different y-axis scale to DAM3, 5, and 6 to improve the visualization of the expression dynamics.
Effect of short day length on expression of P. persica dormancy-associated MADS genes
Photoperiodic regulation of DAM1–6 was examined in a controlled environment experiment where LD acclimated plants were exposed to 8 weeks of SD treatment. Terminal elongation in trees exposed to the SD treatments was observed to evaluate the growth and developmental responses to a dormancy-inducing short day environment. The elongation decreased rapidly in response to a shift to SD, and no further growth was observed following 3 weeks of exposure to SD (data not shown). In addition to assessing growth cessation in a SD environment, the ability of trees to resume growth following a transfer from the SD growth chamber environment back to a LD photoperiod was observed. Peach trees had completely lost the ability to resume terminal growth in the LD photoperiod by the third week of the SD exposure (data not shown).
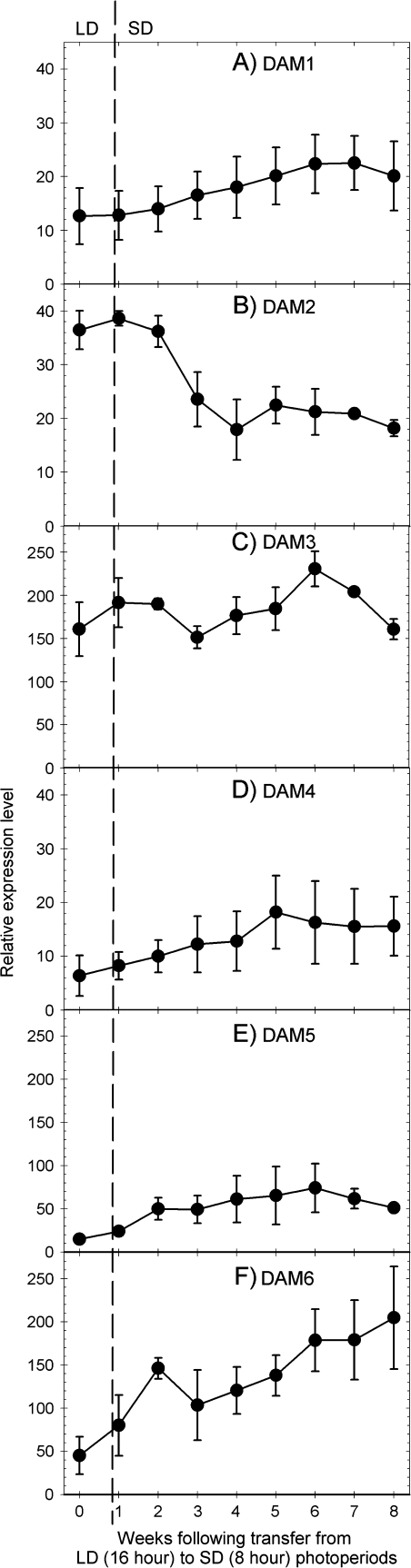
DAM1 and DAM4 were expressed at low levels in LDs and increased in response to SD treatment (Fig. 3A, D). DAM3 was expressed at higher levels in LDs than DAM1 or DAM4, but showed no clear induction or repression in response to SD exposure (Fig. 3C). DAM2 was expressed in LD conditions but decreased by approximately one-half in response to the shift to SD conditions (Fig. 3B). Both DAM5 and DAM6 increased in expression level within 1 week following the transition to SD and continued to increase until the end of the experiment (Fig. 3E, F). Within 8 weeks, expression of both genes had increased approximately 5-fold from the level observed in LDs.
Fig. 3.
Semi-quantitative expression of six dormancy-associated MADS genes from peach in response to transfer from LD (16/8 h light/dark) to SD (8/16 h light/dark) conditions in a controlled environment. Photoperiod was reduced immediately following plant sampling at Week 0. Expression levels were calculated relative to a peach tubulin-α reference as in Fig. 2. Each symbol represents the average of a sample from three individual trees, error bars are standard deviations. DAM1, 2, and 4 are on a different y-axis scale to DAM3, 5, and 6 to improve the visualization of the expression dynamics.
Discussion
Six tandemly-duplicated genes at the EVG locus of peach that are candidates for the non-dormant evg phenotype had previously been identified. All six of these genes are expressed in wild-type tissues, but are not expressed in mutant trees (Bielenberg et al., 2008). The continued expression of these six genes with apparently full-length reading frames argues that these genes have not become pseudogenes. The retention of functional copies of highly similar genes may indicate selective pressure against the accumulation of deleterious gene copies. Previous studies have indicated that these highly similar genes may have differing seasonal expression profiles and therefore differing regulatory functions in peach (Bielenberg et al., 2008).
Localization of peach DAM expression
Expression of the DAM genes in peach is not limited to the terminal tissues or tissues preparing to undergo developmental changes associated with the formation of dormant structures such as vegetative and floral buds (Table 2). Each of the genes has ubiquitous expression in peach with the exception of DAM1 and DAM6, a situation commonly found in the MADS-box gene family and the SVP/StMADS11 clade in particular (Hileman et al., 2006; Diaz-Riquelme et al., 2009).
Seasonal trends in peach DAM expression
Except for DAM3, the DAM genes are expressed in distinct phases throughout the annual cycle. DAM2 expression peaks at the summer solstice. DAM1 and DAM4 peak approximately at the date of terminal bud set. DAM5 and DAM6 peak in expression at the winter solstice. The probable photoperiodic regulation of these five genes was confirmed in controlled environment conditions. DAM2 is down-regulated, while DAM1, 4, 5, and 6 are up-regulated, in response to a shortened day length.
The expression timing of DAM1, DAM2, and DAM4 coincides most closely with the cessation of terminal growth and bud set in our experiments. These three genes are the most likely candidates for the cause of the non-dormant bud set phenotype seen in the evg mutant trees. The progressive expression of the summer/late summer genes (DAM2 decreasing while DAM1 and DAM4 are increasing) allows for the possibility that these genes are members of a single pathway leading to bud formation. Several recent studies have identified DAM-like genes from the SVP/StMADS11 clade of MIKCC MADS-box genes as responding to dormancy-inducing conditions in bud tissues of perennial species by differential display or global transcriptome analysis methods (Ruttink et al., 2007; Campbell et al., 2008; Horvath et al., 2008; Diaz-Riquelme et al., 2009). Our results add to this growing body of work that suggests that involvement of SVP clade MADS-box genes in the regulation/development of bud structures is a common mechanism across diverse perennial species.
The seasonal timing of DAM3, DAM5, and DAM6 do not correspond with the timing of bud set in wild-type plants in our experiment. The periods of greatest changes in these genes are in the winter subsequent to bud set in the late summer/early autumn. If these genes play a role in bud formation and winter dormancy, the role would appear to be limited to the establishment and/or maintenance of an endodormant state once established, as opposed to growth cessation and bud formation. An apparent homologue of DAM6 was found to be suppressed in response to cold exposure of endodormant lateral buds of Japanese apricot (Prunus mume). Similarly, a gene with similarity to DAM6 in raspberry was identified as down-regulated during the fulfilment of the chilling requirement (Mazzitelli et al., 2007). DAM5 was isolated as up-regulated in peach fruit in response to cold storage and chilling injury and, therefore, may also be responding to a cold stimulus (Ogundiwin et al., 2008). These studies provide evidence that the expression levels of DAM5 and 6 in our study were regulated, in part, by cold exposure. However, each of these experiments involved the placement of the experimental tissues (excised buds or fruit) or whole plants in complete darkness in cold storage chambers for the cold treatments. The transition to a completely dark environment coincident with the application of a cold treatment complicates the interpretation of the observed responses (Mazzitelli et al., 2007; Ogundiwin et al., 2008; Yamane et al., 2008).
The expression pattern of DAM3 during the growing season does not appear to be regulated by photoperiodic changes during the year. DAM3 steadily decreases throughout the winter (Fig. 2C), but under controlled conditions DAM3 is not affected by a decrease in photoperiod (Fig. 3C). The decrease in expression of DAM3 throughout the winter period when chilling hours are being accumulated raises the possibility that DAM3 is suppressed by cold exposure or chilling accumulation. Furthermore, DAM3 expression reaches zero at the approximate date that bud burst occurred in our experiment. One interpretation of this pattern is that DAM3 may be functioning in a growth-repressing pathway that is down-regulated by exposure to chilling temperatures. However, this hypothesis does not account for the high expression of DAM3 throughout the active growing season. Interestingly, Prassinos and Han (2006) identified a sweet cherry (Prunus avium) gene with strong similarity to DAM3 as up-regulated in a dwarfing genotype relative to a non-dwarf genotype. Expression of the sweet cherry DAM3-like gene coincided with the cessation of elongation growth in both the dwarf and non-dwarf genotypes (Prassinos and Han, 2006). Although it was expressed, an increase in expression in DAM3 associated with the cessation of elongation growth was not seen in our controlled environment study and, therefore, the correlation observed by Prassinos and Han (2006) may not be a direct causal link. It may be that DAM3 expression is a necessary, but not sufficient, component of the down-regulation of elongation growth prior to dormancy entrance. The potential role of DAM3 as a requirement for the suppression of elongation growth and the close association of the loss of DAM3 expression with bud break in this study will have to be explored in more detail with future targeted experiments.
Duplicated genes are a challenge for functional analysis, particularly when arranged in tandem (Jander and Barth, 2007). The environmentally regulated expression of the six peach DAM genes separates them into four distinct patterns. Four patterns of expression suggest that the six genes no longer fulfil an identical (ancestral?) regulatory function in peach and all six are, therefore, not redundant to each other. The four temporally distinct expression patterns indicate a possible sub- or neo-functionalization of the DAM genes following the duplication events that appear to have occurred in the locus. Two pairs of genes do appear to have highly similar expression patterns: DAM1 and 4 and DAM5 and 6. The expression similarity suggests that each member of a pair has the potential to be redundant to the other in the pair, although differences in tissue localization (Table 2) also argue for potential functional divergence within each pair. Future expression ‘knock-down’ experiments, performed to test the biological function of these genes, will have to account for the potential redundancy of the two gene pairs.
Acknowledgments
We would like to thank the staff of the Musser Fruit Research Center for their valuable assistance with the maintenance of our experimental plantings. The help of Dr S Jiménez-Tarodo was invaluable in the validation of the reference genes used in this work. This research was supported by the National Research Initiative of the USDA Cooperative State Research, Education and Extension Service, grant number 2007-35304-17896 to DGB and USDA CSREES Special Research Grant AC 2005-06137 ‘Peach Tree Short Life in South Carolina’ to GLR. This is Technical Contribution No. 5532 of the Clemson University Experiment Station.
References
- Alvarez-Buylla ER, Liljegren SJ, Pelaz S, Gold SE, Burgeff C, Ditta GS, Vergara-Silva F, Yanofsky MF. MADS-box gene evolution beyond flowers: expression in pollen, endosperm, guard cells, roots, and trichomes. The Plant Journal. 2000;24:457–466. doi: 10.1046/j.1365-313x.2000.00891.x. [DOI] [PubMed] [Google Scholar]
- Arora R, Agarwal P, Ray S, Singh AK, Singh VP, Tyagi AK, Kapoor S. MADS-box gene family in rice: genome-wide identification, organization and expression profiling during reproductive development and stress. BMC Genomics. 2007;8:21. doi: 10.1186/1471-2164-8-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora R, Wisniewski M. Accumulation of a 60-kD dehydrin protein in peach xylem tissues and its relationship to cold acclimation. HortScience. 1996;31:923–925. [Google Scholar]
- Arora R, Wisniewski M, Rowland LJ. Cold acclimation and alterations in dehydrin-like and bark storage proteins in the leaves of sibling deciduous and evergreen peach. Journal of the American Society for Horticultural Science. 1996;121:915–919. [Google Scholar]
- Arora R, Wisniewski ME, Scorza R. Cold-acclimation in genetically related (sibling) deciduous and evergreen peach (Prunus persica L. Batsch). 1. Seasonal changes in cold hardiness and polypeptides of bark and xylem tissues. Plant Physiology. 1992;99:1562–1568. doi: 10.1104/pp.99.4.1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldocchi D, Wong S. Accumulated winter chill is decreasing in the fruit growing regions of California. Climatic Change. 2008;87:S153–S166. [Google Scholar]
- Bielenberg DG, Wang Y, Li Z, Zhebentyayeva T, Fan S, Reighard GL, Scorza R, Abbott AG. Sequencing and annotation of the evergrowing locus in peach [Prunus persica (L.) Batsch] reveals a cluster of six MADS-box transcription factors as candidate genes for regulation of terminal bud formation. Tree Genetics and Genomes. 2008;4:495–507. [Google Scholar]
- Bohlenius H, Huang T, Charbonnel-Campaa L, Brunner AM, Jansson S, Strauss SH, Nilsson O. CO/FT regulatory module controls timing of flowering and seasonal growth cessation in trees. Science. 2006;312:1040–1043. doi: 10.1126/science.1126038. [DOI] [PubMed] [Google Scholar]
- Brunner AM, Yakovlev IA, Strauss SH. Validating internal controls for quantitative plant gene expression studies. BMC Plant Biology. 2004;4:14. doi: 10.1186/1471-2229-4-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell M, Segear E, Beers L, Knauber D, Suttle J. Dormancy in potato tuber meristems: chemically induced cessation in dormancy matches the natural process based on transcript profiles. Functional and Integrative Genomics. 2008;8:317–328. doi: 10.1007/s10142-008-0079-6. [DOI] [PubMed] [Google Scholar]
- Diaz-Riquelme J, Lijavetzky D, Martinez-Zapater JM, Carmona MJ. Genome-wide analysis of MIKCC-type MADS box genes in grapevine. Plant Physiology. 2009;149:354–369. doi: 10.1104/pp.108.131052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easterling WE. Climate change and the adequacy of food and timber in the 21st century. Proceedings of the National Academy of Sciences, USA. 2007;104:19679. doi: 10.1073/pnas.0710388104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frewen BE, Chen THH, Howe GT, Davis J, Rohde A, Boerjan W, Bradshaw HD. Quantitative trait loci and candidate gene mapping of bud set and bud flush in Populus. Genetics. 2000;154:837–845. doi: 10.1093/genetics/154.2.837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hileman LC, Sundstrom JF, Litt A, Chen MQ, Shumba T, Irish VF. Molecular and phylogenetic analyses of the MADS-Box gene family in tomato. Molecular Biology and Evolution. 2006;23:2245–2258. doi: 10.1093/molbev/msl095. [DOI] [PubMed] [Google Scholar]
- Horvath DP, Chao WS, Suttle JC, Thimmapuram J, Anderson JV. Transcriptome analysis identifies novel responses and potential regulatory genes involved in seasonal dormancy transitions of leafy spurge (Euphorbia esula L.) BMC Genomics. 2008;9:17. doi: 10.1186/1471-2164-9-536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe GT, Gardner G, Hackett WP, Furnier GR. Phytochrome control of short-day-induced bud set in black cottonwood. Physiologia Plantarum. 1996;97:95–103. [Google Scholar]
- Jander G, Barth C. Tandem gene arrays: a challenge for functional genomics. Trends in Plant Science. 2007;12:203–210. doi: 10.1016/j.tplants.2007.03.008. [DOI] [PubMed] [Google Scholar]
- Kirilenko AP, Sedjo RA. Climate change impacts on forestry. Proceedings of the National Academy of Sciences, USA. 2007;104:19697–10702. doi: 10.1073/pnas.0701424104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leseberg CH, Li AL, Kang H, Duvall M, Mao L. Genome-wide analysis of the MADS-box gene family in Populus trichocarpa. Gene. 2006;378:84–94. doi: 10.1016/j.gene.2006.05.022. [DOI] [PubMed] [Google Scholar]
- Mazzitelli L, Hancock RD, Haupt S, et al. Co-ordinated gene expression during phases of dormancy release in raspberry (Rubus idaeus L.) buds. Journal of Experimental Botany. 2007;58:1035–1045. doi: 10.1093/jxb/erl266. [DOI] [PubMed] [Google Scholar]
- Meisel L, Fonseca B, Gonzalez S, Baeza-Yates R, Cambiazo V, Campos R, Gonzalez M, Orellana A, Retamales J, Silva H. A rapid and efficient method for purifying high quality total RNA from peaches (Prunus persica) for functional genomics analyses. Biological Research. 2005;38:83–88. doi: 10.4067/s0716-97602005000100010. [DOI] [PubMed] [Google Scholar]
- Ogundiwin EA, Martí C, Forment J, Pons C, Granell A, Gradziel TM, Peace CP, Crisosto CH. Development of ChillPeach genomic tools and identification of cold-responsive genes in peach fruit. Plant Molecular Biology. 2008;68:379–397. doi: 10.1007/s11103-008-9378-5. [DOI] [PubMed] [Google Scholar]
- Olsen JE, Junttila O. Far red end-of-day treatment restores wild type-like plant length in hybrid aspen overexpressing phytochrome A. Physiologia Plantarum. 2002;115:448–457. doi: 10.1034/j.1399-3054.2002.1150315.x. [DOI] [PubMed] [Google Scholar]
- Prassinos C, Han K-H. American Society of Plant Biologists Annual Meeting. Boston, MA: USA; 2006. Characterization of a StMADS11 family gene from sweet cherry (Prunus avium) [Google Scholar]
- Rodriguez J, Sherman WB, Scorza R, Wisniewski M, Okie WR. Evergreen peach, its inheritance and dormant behavior. Journal of the American Society for Horticultural Science. 1994;119:789–792. [Google Scholar]
- Rohde A, Howe GT, Olsen JE, Moritz T, Van Montagu M, Junttila O, Boerjan W. Molecular aspects of bud dormancy in trees. In: Jain SM, Minocha SC, editors. Molecular biology of woody plants. Dordrecht: Kluwer Academic Publishers; 2000. pp. 89–134. [Google Scholar]
- Rohde A, Prinsen E, De Rycke R, Engler G, Van Montagu M, Boerjan W. PtABI3 impinges on the growth and differentiation of embryonic leaves during bud set in poplar. The Plant Cell. 2002;14:1885–1901. doi: 10.1105/tpc.003186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousi M, Pusenius J. Variations in phenology and growth of European white birch (Betula pendula) clones. Tree Physiology. 2005;25:201–210. doi: 10.1093/treephys/25.2.201. [DOI] [PubMed] [Google Scholar]
- Rozen S, Skaletsky HJ. Primer3 on the www for general users and for biologist programmers. In: Krawetz S, Misener S, editors. Bioinformatics methods and protocols: methods in molecular biology. Totowa, NJ: Humana Press; 2000. pp. 365–386. [DOI] [PubMed] [Google Scholar]
- Ruijter JM, Ramakers C, Hoogaars WMH, Karlen Y, Bakker O, van den Hoff MJB, Moorman AFM. Amplification efficiency: linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Research. 2009;37:12. doi: 10.1093/nar/gkp045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruttink T, Arend M, Morreel K, Storme V, Rombauts S, Fromm J, Bhalerao RP, Boerjan W, Rohde A. A molecular timetable for apical bud formation and dormancy induction in poplar. The Plant Cell. 2007;19:2370–2390. doi: 10.1105/tpc.107.052811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson MM, Smith DC, Burgess JE. Non-dormant mutants in a temperate tree species, Corylus avellana L. Theoretical and Applied Genetics. 1985;70:687–692. doi: 10.1007/BF00252298. [DOI] [PubMed] [Google Scholar]
- Wang Y, Georgi LL, Reighard GL, Scorza R, Abbott AG. Genetic mapping of the evergrowing gene in peach [Prunus persica (L.) Batsch] Journal of Heredity. 2002;93:352–358. doi: 10.1093/jhered/93.5.352. [DOI] [PubMed] [Google Scholar]
- Werner DJ, Okie WR. A history and description of the Prunus persica plant introduction collection. HortScience. 1998;33:787–793. [Google Scholar]
- Yakovlev IA, Fossdal CG, Johnsen O, Junttila O, Skroppa T. Analysis of gene expression during bud burst initiation in Norway spruce via ESTs from subtracted cDNA libraries. Tree Genetics and Genomes. 2006;2:39–52. [Google Scholar]
- Yamane H, Kashiwa Y, Ooka T, Tao R, Yonemori K. Suppression subtractive hybridization and differential screening reveals endodormancy-associated expression of an SVP/AGL24-type MADS-box gene in lateral vegetative buds of japanese apricot. Journal of the American Society for Horticultural Science. 2008;133:708–716. [Google Scholar]