Abstract
During phase II metabolism, a substrate is rendered more hydrophilic through the covalent attachment of an endogenous molecule. The cytosolic sulfotransferase (SULT) and UDP-glucuronosyltransferase (UGT) families of enzymes account for the majority of phase II metabolism in humans and animals. In general, phase II metabolism is considered to be a detoxication process, as sulfate and glucuronide conjugates are more amenable to excretion and elimination than are the parent substrates. However, certain products of phase II metabolism (e.g., unstable sulfate conjugates) are genotoxic. Members of the nuclear receptor superfamily are particularly important regulators of SULT and UGT gene transcription. In metabolically active tissues, increasing evidence supports a major role for lipid-sensing transcription factors, such as peroxisome proliferator-activated receptors (PPARs), in the regulation of rodent and human SULT and UGT gene expression. This review summarizes current information regarding the regulation of these two major classes of phase II metabolizing enzyme by PPARs.
1. Introduction
Phase II, or conjugative, metabolism is defined as the covalent attachment of an endogenous molecule to a functional group on a substrate molecule. Although a substrate containing a suitable functional group can directly undergo phase II metabolism, conjugation often occurs subsequent to a phase I reaction (e.g., catalyzed by a cytochrome P450), during which the functional group is added to the substrate. The conjugating moiety is most often a sulfonate or glucuronate group, although other conjugating moieties include glutathione, glycine, acetate, and the methyl group. Phase II metabolism usually increases the hydrophilicity of the substrate molecule, which facilitates transport and elimination of the product. Phase II sulfonation and glucuronidation reactions are catalyzed by the cytosolic sulfotransferase (SULT) and the UDP-glucuronosyltransferase (UGT) families of enzymes, respectively, (Figure 1). The SULT and UGT enzymes represent a highly responsive defense system against the mutagenicity of carcinogenic environmental chemicals and the toxicity of xenobiotics and endogenous metabolic intermediates. Members of the nuclear receptor superfamily are particularly important regulators of UGT and SULT gene transcription. In metabolically active tissues, increasing evidence supports a major role for lipid-sensing transcription factors, such as peroxisome proliferator-activated receptors (PPARs), in the regulation of rodent and human SULT and UGT gene expression. This review summarizes current information regarding the regulation of these two major classes of phase II metabolizing enzyme by PPARs.
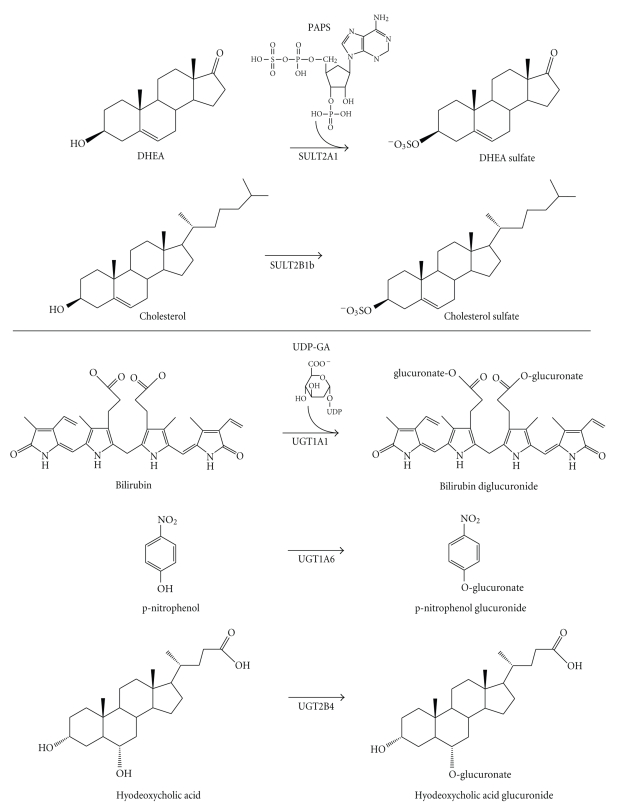
Figure 1.
Example reactions catalyzed by PPAR-regulated SULT and UGT enzymes. The upper panel shows the SULT2A1- and SULT2B1b-catalyzed 3-sulfonation of the prototype substrates, dehydroepiandrosterone and cholesterol, respectively. Human SULT2A1 is transcriptionally regulated by PPARα in human hepatocytes, while SULT2B1b is regulated by PPARα, PPARδ and PPARγ in keratinocytes. The lower panel shows the glucuronidation of bilirubin, p-nitrophenol and hyodeoxycholic acid, which are prototype substrates for UGT1A1, UGT1A6, and UGT2B4, respectively. UGT1A1, UGT1A3, UGT1A4, UGT1A6, UGT1A9, and UGT2B4 have all been identified as PPAR target genes. PAPS, 3′-phosphoadenosine-5′-phosphosulfate; UDP-GA, uridine-5′-diphospho-α-D-glucuronic acid.
2. PPARs
The PPAR nuclear receptor network represents a central determinant of cellular energy balance. In heterodimeric partnership with the retinoid X receptor (RXR), PPAR forms a ligand-activated nuclear receptor transcription factor that is capable of integrating the expression of a wide spectrum of target genes involved in cellular lipid metabolism, energy homeostasis, and inflammation (Figure 2). The three known PPAR isoforms, PPARα, PPARδ (also called PPARβ) and PPARγ are products of separate genes and are well conserved across species. PPARα expression is the most prominent in the liver, kidney, and heart where it is engaged in the regulation of fatty acid oxidation [1]. By contrast, PPARγ is most widely expressed in adipose tissues where it plays a role in adipocyte differentiation and the control of insulin sensitivity [1]. PPARδ is ubiquitously expressed and has been implicated as a regulator of a range of physiological functions from the modulation of insulin resistance to embryo implantation during pregnancy [1]. Fibrates and thiazolidinediones are well-characterized ligand-activators of PPARα and PPARγ, respectively, [1]. Fatty acids represent a major source of cellular energy and are important physiological activators of PPARα [1]. Relative to fatty acids, oxidized fatty acid intermediates are more short-lived species and thus are well poised to serve as both endogenous signaling intermediates and physiological PPARγ agonists [2]. As a key integrator of cellular energy metabolism in a wide spectrum of tissues, the PPAR·RXR transcription factor network is being increasingly recognized for its potential as a therapeutic target and for its expanded role in gene regulation.
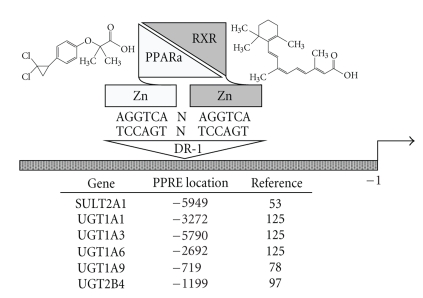
Figure 2.
Regulation of SULT and UGT gene transcription by PPARα. The PPARα and RXR nuclear receptors are each depicted as a zinc module-containing DNA-binding domain (rectangle) that is joined to a ligand-binding domain (LBD, triangle) through a hinge region. PPARα and RXR bind as a heterodimer to a peroxisome proliferator response element (PPRE) in the regulatory region of a target gene. The consensus PPRE is a nuclear receptor hexamer motif (i.e., (A/G)G(G/T)TCA) in a DR-1 configuration (direct repeat with one intervening nucleotide, N). Binding of an agonist to the LBD of PPARα (e.g., chemical structure for the potent PPARα agonist, ciprofibrate, is shown) evokes a conformational change in the receptor that results in coactivator recruitment and increased target gene transcription. In contrast to its silent role in partnership with some nuclear receptors, RXR functions as an active partner with the PPARs, whereby binding of an agonist to the LBD of RXR (e.g., chemical structure for the prototype RXR ligand, 9-cis-retinoic acid, is shown) activates target gene transcription and enhances PPAR ligand-activated transcription. The locations of functional PPREs that have been identified in the 5′-flanking regions of the human SULT2A1, UGT1A1, UGT1A3, UGT1A6, UGT1A9, and UGT2B4 genes are shown (positions are relative to the transcription start site).
3. SULTs
3.1. The Role of SULTs in Metabolism
Sulfonation reactions are catalyzed by two distinct families of enzymes, the cytosolic SULTs and the membrane-bound sulfotransferases. Of these, only the SULTs participate in phase II drug metabolism; the membrane-bound sulfotransferases catalyze the sulfonation of proteins and complex carbohydrates [3]. The SULTs catalyze the transfer of a sulfonate moiety (SO3 −) from the physiological donor 3′-phosphoadenosine-5′-phosphosulfate (PAPS) to a small molecule substrate containing a nucleophilic moiety such as a susceptible hydroxyl group [4] (Figure 1). The SULTs are widely expressed in hepatic and extrahepatic tissues [5] where they represent key components of the xenobiotic defense system. They also function prominently in physiological processes by metabolizing endogenous substrates, including estrogens [6], thyroid hormones [7], bile acids [8], and neurosteroids [9–12]. In xenobiotic metabolism, sulfate conjugation is recognized as a double-edged sword. As a rule, sulfate conjugates are more polar than the parent substrate and, hence, more amenable to excretion and elimination. However, the production of unstable sulfate conjugates can lead to the focused generation of genotoxic species and carcinogen activation [13, 14].
Like other classes of xenobiotic-metabolizing enzymes, the SULTs exist as a superfamily of related proteins with each enzyme exhibiting a characteristic expression pattern and substrate specificity profile. The cytosolic SULTs are categorized into two major groups, the arylsulfotransferases (SULT1 family) and the hydroxysteroid sulfotransferases (SULT2 family) [5, 15]. The SULT1 family is divided into five subfamilies, designated SULT1A, SULT1B, SULT1C, SULT1D, and SULT1E. As a brief generalization, SULT1A subfamily enzymes metabolize phenolic substrates and function in drug metabolism. For example, SULT1A1 readily catalyzes the sulfonation of simple phenols, such as 1-naphthol and p-nitrophenol [4], and detoxifies common phenolic pharmaceuticals, such as acetaminophen [16, 17] and troglitazone [4]. Consistent with its role in drug metabolism, SULT1A1 is abundantly expressed in liver, although it is also expressed in numerous extrahepatic tissues [5]. Rodent enzymes of the SULT1B subfamily catalyze the sulfonation of 3,5,3′-triiodothyronine [18], an important step in thyroid hormone metabolism, although SULT1A1 appears to perform this function in humans [19, 20]. SULT1C enzymes are best known for their abilities to bioactivate the heterocyclic amine procarcinogen, N-hydroxy-2-acetylaminofluorene, to its ultimate carcinogenic form [21]. SULT1D enzymes, described thus far chiefly in the canine, sulfonate phenols, amines, and eicosanoids [22]. SULT1E1 is the physiologic estrogen sulfotransferase, catalyzing the 3-sulfonation of estradiol at nanomolar concentrations [5]. SULT1E1 is expressed most highly in tissues that are actively engaged in estrogen metabolism, such as breast, uterus, and placenta [23–25].
The SULT2 family is divided into two subfamilies, SULT2A and SULT2B. In general, these SULTs most effectively metabolize molecules containing a steroid or sterol nucleus (e.g., dehydroepiandrosterone (DHEA), pregnenolone, cholesterol, and bile acids) [26]. The SULT2A subfamily members, including human SULT2A1, are most highly expressed in liver, intestine, and adrenal cortex [27–30], and the prototypical SULT2A substrate is DHEA [27] (Figure 1). SULT2A-catalyzed DHEA sulfoconjugation within the adrenal cortex provides high circulating levels of DHEA sulfate, which serves as a reservoir of a precursor molecule that can be converted into potent androgens and estrogens within various tissues (e.g., prostate) [31].
The SULT2B subfamily consists of two gene products, SULT2B1a and SULT2B1b, that are transcribed from the same gene locus through the utilization of different promoters and incorporation of different first exons [11, 32, 33]. SULT2B1b preferentially catalyzes the sulfonation of cholesterol (Figure 1), and SULT2B1b expression has been demonstrated in skin [11, 33–35], prostate [11, 34, 36], placenta [34, 36], lung [34, 37], intestine [11, 33, 36], endometrium [36, 38], breast [39], ovary [36], platelets [40], kidney [33], and muscle [11]. SULT2B1b protein has not been detected in liver [34]. SULT2B1b has been detected in both the cytosol and nuclei of human cells [34, 39, 41].
By comparison to SULT2B1b, SULT2B1a has minimal activity toward cholesterol but readily catalyzes the sulfonation of pregnenolone [42]. Pregnenolone sulfate is a neurosteroid that is synthesized in glial cells [43]. It is, therefore, noteworthy that SULT2B1a mRNA expression has been detected in brain [11] as well as in rat C6 glioma cells [44]. However, to date SULT2B1a protein has not been detected in any human tissues or cell lines [41].
In addition to the major SULT1 and SULT2 enzymes, SULT3A1, SULT3A2, SULT4A1, SULT5A1, and SULT6B1 have been described [45–48] but are not highly characterized. Of these, only SULT4A1 and SULT6B1 have been identified in humans [48–50].
3.2. Regulation of SULTs by PPAR
3.2.1. Regulation of Mouse Liver SULT Expression by PPARα Agonists
A comprehensive survey of SULT regulation by PPARα-activating treatments has been performed by the Klaassen research group, which evaluated the sex-dependent regulation of hepatic SULT expression following in vivo treatment of mice with a panel of prototypical nuclear receptor activators, including three PPARα agonists [51]. Hepatic transcript levels were evaluated for murine SULTs 1a1, 1b1, 1c1, 1c2, 1d1, 1e1, 2a1/2a2, 2b1, and the lesser characterized SULT family members 3a1, 4a1, and 5a1 [51]. In addition, treatment effects on the mRNA expression of both forms of PAPS synthase (PAPSs1 and PAPSs2), the enzymes responsible for synthesizing the PAPS cofactor for sulfate conjugation, were examined [51]. Male and female 8 week old C57BL/6 mice were treated for 4 days with either corn oil vehicle or one of the prototypical PPARα ligands, clofibrate (500 mg/kg IP), ciprofibrate (40 mg/kg IP), or diethylhexylphthalate (1000 mg/kg IP), and euthanized for hepatic SULT mRNA content analysis [51]. Overall, the effects of PPARα activation on murine hepatic SULT expression were not striking [51]. SULT expression in male mouse liver was not appreciably perturbed in response to in vivo treatment with PPARα agonists [51]. In the rat, SULT1E1, which is more abundantly expressed in male relative to female liver, was previously reported to decrease substantially following in vivo treatment with any of the PPARα agonists, WY-14,643, gemfibrozil or di-n-butylphthalate [52]. In Klaassen's study, the mRNA levels of several SULTs were suppressed in female mouse liver following PPARα agonist treatment, including SULTs 1c1, 1c2, 1e1, 2a1/2a2, 3a1, and 5a1 [51]. Treatment with clofibrate increased PAPSs2 mRNA content in male mouse liver [51]. However, such induction was not produced by ciprofibrate or diethylhexylphthalate treatment, suggesting that regulation of murine PAPSs2 expression is not an effect common to PPARα agonists [51]. These studies demonstrate that PPARα activation produces gene- and sex-dependent effects on the hepatic expression of SULT enzymes in the mouse.
3.2.2. Transactivation of Human Hepatic SULT2A1 Transcription by PPARα
The above-described data suggest that PPARα does not function as a positive regulator of murine hepatic SULT expression. However, our laboratory has demonstrated that human hepatic SULT2A1 expression is increased by PPARα activation, and that this effect is conferred through a functional PPARα-response element (PPRE) located in the distal 5′-flanking region of the SULT2A1 gene [53]. The treatment of primary cultured human hepatocytes with ciprofibrate induced SULT2A1 mRNA, protein and enzyme activity levels by ~2-fold [53]. This finding was in marked contrast to the rat counterpart of SULT2A1, which was not inducible in primary cultured rat hepatocytes following treatment with a PPARα agonist [53]. Analysis of a series of SULT2A1 5′-flanking region-luciferase reporter constructs in HepG2 cells revealed the presence of a functional direct repeat with one intervening nucleotide (DR-1) located at nucleotides −5949 to −5929 relative to the transcription start site [53] (Figure 2). Further site-directed mutagenesis, EMSA and chromatin immunoprecipitation analyses confirmed the functionality of this PPRE in the human SULT2A1 gene [53]. These investigations reveal that SULT2A1 represents a target for lipid signaling in human hepatocytes and suggest that rodent models do not capture the significance of PPARα as a modulator of SULT2A expression.
3.2.3. Regulation of SULT2B1b in Human Keratinocytes by PPAR Agonists
Maintenance of the skin requires a well orchestrated program of keratinocyte proliferation and differentiation during which the cells pass through several phenotypic phases identifiable as distinct layers (Figure 3). As keratinocytes progress through this differentiation program, a major change that occurs is the production of progressively larger amounts of lipid. Cholesterol 3-sulfate has been detected at a level of 2.4% (weight percent of total lipids) in a combined preparation of basal/spinous cells, and 5.5% in the granular layer [54]. The amount of cholesterol 3-sulfate is then reduced in the stratum corneum, due to the conversion of cholesterol 3-sulfate to free cholesterol through the action of steroid sulfatase [54]. Cholesterol 3-sulfate production in the lower levels of the epidermis and its hydrolysis in the stratum corneum has been termed the epidermal cholesterol cycle [55].
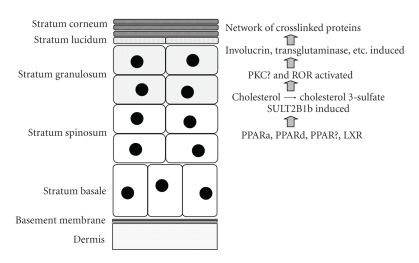
Figure 3.
Representation of keratinocyte differentiation and role of SULT2B1b-mediated cholesterol sulfonation. During formation of the epidermis, replicating basal keratinocytes give rise to progeny that move progressively upwards and pass through several histologically distinct strata. During this process, SULT2B1b expression becomes activated at approximately the level of the stratum granulosum. SULT2B1b-catalyzed cholesterol sulfonation produces cholesterol 3-sulfate that, in addition to its role as a lipid component of the outer barrier, functions as a signaling molecule that activates PKCη and possibly RORα. These signaling events result in the induction of proteins that are involved in formation of the barrier. PPARα, PPARδ, PPARγ, and LXR have all been reported to positively regulate SULT2B1b expression in cultured keratinocytes.
In 1984, an enzyme capable of sulfonating cholesterol was reported to be present in mouse epidermis [56], and several subsequent pieces of evidence have supported the concept that “cholesterol sulfotransferase” expression and cholesterol 3-sulfate production are events that are closely linked to keratinocyte differentiation. For example, the specific activity of cholesterol sulfotransferase, but not of steroid sulfatase, paralleled both the accumulation of cholesterol 3-sulfate and the formation of the multilayered structure of the epidermis during mouse development [57, 58]. Cholesterol sulfotransferase activity was also induced during the culture of epidermal tissues isolated from 13.5-days, postcoitus mouse embryos with a time course that paralleled in vitro stratification [58]. In monolayer cultures of normal human keratinocytes, confluence-mediated differentiation was accompanied by increased cholesterol sulfotransferase activity and accumulation of cholesterol 3-sulfate, with parallel increases in transglutaminase-1 [59]. Calcium-mediated differentiation of normal human keratinocytes was accompanied by the induction of two distinct sulfotransferase activities, cholesterol sulfotransferase, and minoxidil sulfotransferase [60]. Exposure of fetal rat skin explants to air caused the accelerated expression of cholesterol sulfotransferase along with several other markers of keratinocyte differentiation, including filaggrin, loricrin and involucrin [61]. A single topical administration of a tumor promoter (e.g., phorbol ester) to mouse skin caused the induction of cholesterol sulfotransferase, the accumulation of cholesterol 3-sulfate, and the induction of transglutaminase-1 [62, 63]. Cholesterol sulfotransferase activity was also elevated during epidermal wound healing [62].
SULT2B1b preferentially sulfonates cholesterol and is the chief SULT2B1 enzyme that is expressed in human keratinocytes [64]. Higashi et al. [35] reported that SULT2B1b, but not SULT2B1a or SULT2A1, was present in normal human epidermal tissue, as well as in cultures of normal human keratinocytes following calcium-induced differentiation. By immunohistochemistry, SULT2B1b was largely localized to the granular layer of normal skin where cholesterol sulfate content is highest [35]. Altogether, the combination of substrate specificity and expression properties establish SULT2B1b as the “cholesterol sulfotransferase” of differentiating keratinocytes [35].
In addition to its role as structural component, cholesterol 3-sulfate is an important signaling molecule that plays an essential role in keratinocyte differentiation (Figure 3). For example, cholesterol 3-sulfate has been proposed to be a physiological activator of the novel protein kinase C (PKC) isoform, PKCη, which is localized to the granular layer where cholesterol sulfate concentrations are the highest [65–67]. Activation of PKCη signals the continuance of the keratinocyte differentiation program, resulting in the expression of late differentiation markers, such as transglutaminase1 [65, 66, 68, 69]. In addition, it has been demonstrated that cholesterol sulfate is a preferred ligand for the retinoid-related orphan nuclear receptor-α (ROR-α). This nuclear receptor, particularly the RORα4 splice variant, is robustly expressed in the epidermis, where it is present in suprabasal differentiating keratinocytes but not in proliferating basal keratinocytes [70]. Until recently, RORα was considered to be a true orphan receptor, since no firm ligand activator for RORα had been identified. However, recent X-ray crystallography data revealed the presence of either cholesterol or cholesterol sulfate within the RORα ligand binding pocket domain [70]. Furthermore, pharmacological manipulation of cellular cholesterol levels altered RORα transcriptional activity in human osteosarcoma cells [70]. These studies led to the recognition that RORα may represent a novel regulatory pathway in the control of cholesterol homeostasis [71]. Compared to cholesterol, cholesterol sulfate demonstrated a greater affinity for the RORα ligand binding domain and more profound effects on RORα-mediated transcriptional activation, suggesting a major role for cholesterol sulfate as a physiologic activator of RORα in tissues such as the epidermis where both cholesterol sulfate and RORα are abundantly coexpressed [71, 72].
In light of the functional role of SULT2B1b in keratinocyte differentiation, corneoctye desquamation and dermal lipid homeostasis, it is reasonable to expect that SULT2B1b should be a target for regulation by lipid-sensing nuclear receptors in the skin. The effects, on SULT2B1b expression, of treating cultured human keratinocytes with chemical activators of PPARα, PPARδ, PPARγ, and of the liver X receptor (LXRα and LXRβ) have been examined [73]. Measurable levels of PPAR and LXR transcripts were detected in cultured human keratinocytes, and the levels of PPARγ, LXRα, and LXRβ increased following calcium-induced differentiation [73]. By contrast, PPARα and PPARδ mRNA levels did not change demonstrably with keratinocyte differentiation [73]. Treatment with PPARα, PPARδ, PPARγ, and LXR agonists significantly induced SULT2B1b mRNA and enzyme activity in cultured human keratinocytes [73]. The PPARα agonist clofibric acid increased SULT2B1b mRNA content by ~39.9% [73]. Activators of PPARδ (GW501516) and PPARγ (ciglitazone) produced even greater increases (~9.8-fold and ~25.1-fold, resp.) in SULT2B1b mRNA content, and these increases were further augmented by calcium-induced keratinocyte differentiation [73]. As modulators of SULT2B1b expression in differentiating keratinocytes, these results underscore the central role of PPAR transcription factors as integrators of skin physiology and barrier function (Figure 3).
4. UDP-Glucuronosyltransferases
4.1. The Role of UGTs in Metabolism
The UGTs catalyze the transfer of glucuronic acid from a high-energy cofactor, UDP-glucuronic acid, to a xenobiotic or endogenous substrate containing an available nucleophilic center such as a hydroxyl, carboxyl, amino, or thiol group [74–77] (Figure 1). The UGTs are membrane-bound enzymes localized on the luminal surface of the endoplasmic reticulum [74]. Relative to the parent substrate, the end-products of glucuronidation are typically more polar and better suited for excretion and elimination through the urine or bile [74]. Endogenous UGT substrates include bilirubin, neutral steroids, bile acids, fatty acids, and retinoids [74, 78, 79]. Xenobiotic UGT substrates range from environmental toxicants such as benzo[a]pyrene to common pharmaceuticals such as acetaminophen, nonsteroidal anti-inflammatory agents, fibrates, thiazolidinedione-class insulin sensitizers, and opioids [75, 78–84]. Individual UGT enzymes display distinctive patterns of substrate specificity and inducible regulation, but as with the SULTs, some UGTs display overlapping substrate specificities [74, 85]. Different UGTs are expressed in a species- and tissue-specific manner [86, 87]. Overall, the broad metabolic range of the UGTs distinguishes this class of conjugating enzyme as a major detoxicating system in rodents and humans.
Though over 50 individual UGT enzymes have been described [86, 88], comparisons of cDNA and amino acid sequences have revealed two major UGT gene families, UGT1 and UGT2 [75, 86, 88, 89]. The UGT1A multigenic locus is unusual in that it is comprised of a tandem series of thirteen promoter regions on human chromosome 2 [75, 86, 90]. Each of nine functional UGT1A proteins is produced as a result of transcription initiation at a particular promoter, which results in the transcription and splicing of a unique exon 1 sequence to a cassette of common exons (exons 2–5) that share the same 3′ end [75, 86, 88, 90, 91]. As an additional layer of complexity, recent evidence indicates that alternative splicing events generate UGT1A isoforms with different exon 5 sequences. For each of these UGT1A proteins, isoform 1 (containing exon 5a) is the classical catalytically active enzyme, while isoform 2 (containing exon 5b) is a smaller protein that lacks UGT activity but can inhibit the activity of the corresponding isoform 1 [92, 93]. By contrast to the UGT1A locus, UGT2 enzymes are products of individual genes [75].
Like sulfonation, glucuronidation plays a physiological role in the modulation of biologically active endogenous hormones and metabolic intermediates. For example, the activity of UGT1A1 conjugation in the tight control of bilirubin metabolism has established the importance of UGT1A1 genetic polymorphisms in the pathogenesis of toxic hyperbilirubinemias such as the Crigler-Najjar and Gilbert's syndromes in humans [75, 89, 94]. Thyroxine is metabolized by O-glucuronidation in addition to deiodination and sulfonation [95]. Studies using recombinant UGTs and human liver microsomes revealed that human hepatic UGT1A1 and UGT1A3 are the UGT principals most catalytically active toward thyroxine [95]. Bile acids represent the end-products of hepatic cholesterol metabolism and in the absence of sufficient detoxication metabolism, particularly in the face of cholestasis, the detergent properties of bile acids produce significant hepatotoxicity [96]. Human UGT2B4 [97], UGT2B7 [96, 98], UGT1A4 [96, 98], and UGT1A3 [98, 99] are all bile acid-metabolizing enzymes. The formation of chenodeoxycholic acid (CDCA) glucuronide by UGT1A3 has been shown to decrease farnesoid X receptor (FXR) activation by CDCA, the prototypical FXR ligand [99], suggesting that UGT1A3 induction in human liver would be expected to have down-stream consequences for an array of gene expression networks that are transcriptionally regulated by FXR.
4.2. Regulation of UGTs by PPAR
4.2.1. Studies of the Effects of Clofibrate and Other Perixosome Proliferation-Inducing Agents on UGT Activities
It has been more than 40 years since the discovery that clofibrate treatment causes peroxisome proliferation in rat liver [100, 101]. After clofibrate was subsequently shown to cause hepatic induction of a unique cytochrome P450 with a reduced carbon monoxide-bound absorbance peak at 452 nm and catalytic activity toward lauric acid 12-hydroxylation, clofibrate became accepted as the prototype of a novel class of microsomal enzyme inducers [102]. Clofibrate, therefore, was used in a number of studies designed to evaluate the effects of microsomal enzyme inducer treatments on glucuronidation activities. The most commonly reported finding in these studies was that treatment with clofibrate or a structurally related compound increased hepatic glucuronidation activity toward bilirubin in rats [103–110], mice [111], and primary cultured rat hepatocytes [112], with the clofibrate-mediated increases generally in the range of two-fold over control. In some of these studies, use of multiple compounds provided evidence for correspondence between induction of rat hepatic bilirubin conjugation activity and induction of lauric acid 12-hydroxylation [104, 105], implying probable identity of the induction mechanism. In addition, human subjects with Gilbert's syndrome treated with clofibrate had decreased serum total bilirubin concentrations [113], and hepatic microsomes prepared from humans who had received clofibrate contained elevated bilirubin UGT activity [114].
Clofibrate treatment of rats has also been reported to increase hepatic UGT activity toward 4′-hydroxy-N,N-dimethyl-4-aminoazobenzene [106], thyroxine and reverse triiodothyronine [109, 115], retinoic acid [116], and the antithrombotic drug LF 4.0212 [117]. In agreement with these findings, clofibric acid treatment also increased thyroxine UGT activity in primary cultured rat hepatocytes [112]. However, a species difference in UGT induction was suggested by the finding that clofibrate treatment of male OF-1 mice displayed no increase in hepatic microsomal thyroxine UGT activity [111]. Likewise, clofibric acid treatment of primary cultured mouse hepatocytes (OF-1 strain) did not increase bilirubin or thyroxine UGT activity [112]. In a porcine model, in vivo treatment with clofibrate induced hepatic glucuronidation of thyroid hormones sufficiently to reduce circulating plasma 3,3′,5-triiodothyronine and thyroxine concentrations [118].
Clofibrate treatment has been consistently reported not to increase UGT activity toward p-nitrophenol in rat liver [106, 109, 110], mouse liver [111], primary cultured rat hepatocytes [112, 119], or primary cultured mouse hepatocytes [112]. Clofibrate treatment of rats or mice (OF-1 strain) has also been shown to have little effect on hepatic microsomal UGT activities toward triiodothyronine and androsterone [109, 111].
In another study, treatment of rats with a single dose of the nonfibrate peroxisome proliferator, the fully-fluorinated ten-carbon fatty acid perfluorodecanoic acid, induced hepatic bilirubin UGT activity two-fold, and this induced a state persisted for 3 weeks [120]. This single-dose treatment also decreased hepatic UGT activities toward 1-naphthol, morphine, and testosterone, with maximal reductions occurring 12 days after treatment and recovery to control activities occurring at 3 weeks [120].
These early findings, in which peroxisome proliferator treatments produced differential effects on various UGT activities, provided a clear indication of the multiplicity of the UGT superfamily. As seen for the cytochromes P450, only certain UGT activities displayed peroxisome proliferator inducibility, predicting the later demonstration that particular UGT genes would be targets of PPARα-mediated transactivation.
4.2.2. UGT1A Regulation by PPARs
UGT1A1 is the major catalyst of bilirubin glucuronidation [121] (Figure 1). Therefore, based on the above-described observation that clofibrate treatment consistently caused induction of hepatic bilrubin UGT activity, UGT1A1 is expected to be a PPARα target gene, possibly along with other UGTs of the “bilirubin-like” portion of the UGT1A subfamily (i.e., UGTs 1A2-1A5).
In a study using antipeptide antibodies to examine the effects of drug treatments on rat hepatic microsomal UGT levels, clofibrate treatment was found to increase the immunoreactive protein levels of UGT1A1 and UGT1A5 (termed UGT1B1 and UGT1B5 in that study) along with bilirubin UGT activity [110]. In a more recent study, four-day treatments of male rats with PPARα activators produced modest increases in hepatic UGT1A1 and UGT1A3 mRNA levels [122]. In addition, clofibric acid treatment of primary cultured rat hepatocytes has been reported to increase the amount of UGT1A1 protein by western blot analysis [119] and the amount of UGT1A1 mRNA by microarray analysis [123]. Therefore, as predicted by the bilirubin glucuronidation activity data, UGT1A1 is a target of PPARα activation. Clofibrate treatment has also been reported to decrease the amount of rat hepatic microsomal UGT1A6 protein (termed UGT1A1 in that study) [110]. This finding is also in agreement with the above-mentioned earlier finding that clofibrate treatment did not induce p-nitrophenol glucuronidation activity in rodent liver or cultured hepatocytes, since UGT1A6 is a major catalyst of that activity [124] (Figure 1).
In a recent study by Senekeo-Effenberger et al. [125], Wy-14,643 treatment of primary cultured human hepatocytes increased the levels of UGT1A1, UGT1A3, UGT1A4, and UGT1A6, but not UGT1A9, mRNAs. In the same study, experiments conducted with transgenic mice engineered to express the complete human UGT1 gene locus demonstrated that oral Wy-14,643 treatment resulted in prominent induction of human UGT1A1 and UGT1A6, and observable induction of UGT1A4, immunoreactive protein content in liver microsomes. At the mRNA level in liver, very strong (>100-fold) induction of UGT1A1 and UGT1A3, significant (~3- to 4-fold) induction of UGT1A4 and UGT1A6, and no induction of UGT1A9 were seen, in general agreement with the above-described effects in the human hepatocyte cultures [125]. In small intestine, Wy-14,643 treatment produced induction of UGT1A1 and UGT1A4, but not UGT1A6, while in kidney only UGT1A6 was induced [125]. Additional experiments confirmed the presence of functional PPREs in the 5′-flanking regions of UGT1A1, UGT1A3 and UGT1A6 [125] (Figure 2).
In another study, treatment of primary cultured human hepatocytes with activators of PPARα increased the expression of UGT1A3 and UGT1A3-catalyzed glucuronidation of chenodeoxycholic acid (CDCA) and demonstrably tempered the transactivation of FXR by CDCA [99]. Promoter analysis of the human UGT1A3 gene revealed coregulation by two lipid-sensing transcription factors, LXR [126] and PPARα [125].
UGT1A4 was also identified as a significantly upregulated gene in clofibric acid-treated primary cultured human hepatocytes by Affymetrix microarray analysis [123]. In this same study, clofibric acid treatment of primary cultured mouse hepatocytes failed to cause significant alteration of any UGT transcripts [123], consistent with the above-described apparent lack of sensitivity of mouse to PPARα-mediated regulation of UGT activities.
By comparison to the results described by Senekeo-Effenberger et al. [125], Barbier et al. [78] previously reported that mouse and human UGT1A9 are transcriptionally activated by PPARα and PPARγ, and that human UGT1A9 contains a functional PPRE located at nucleotides −719 to −706 (Figure 2). UGT1A9 mRNA induction was observed in primary cultured human hepatocytes treated with fenofibric acid, HepG2 cells treated with Wy-14,643, 3T3-L1 adipocytes treated with rosiglitazone, primary human macrophages treated with Wy-14,643, or differentiated THP-1 macrophages treated with Wy-14,643 or rosiglitazone [78]. As expected, the inducing effect of fenofibric acid on mouse hepatic UGT1A9 was ablated in PPARα-null mice [78].
4.2.3. UGT2B Regulation by PPARα
In human hepatocytes, treatment with a PPARα agonist, fenofibric acid or Wy-14643, increased the expression of UGT2B4 mRNA and stimulated the glucuronidation of hyodeoxycholic acid, a model substrate for UGT2B4 [97] (Figure 1). Transient transfection and EMSA studies revealed a functional DR-1 PPRE at nucleotides −1199 to −1175 nt relative to the UGT2B4 transcription start site and solidified UGT2B4 as a transcriptional target of PPARα [97] (Figure 2).
5. Regulation of SULTs and UGTs by Other Nuclear Receptors
In addition to regulation by PPARs, the SULTs and UGTs receive transcriptional input from multiple other nuclear receptors. Here, we do not attempt to be comprehensive but present several findings related to the regulation of hepatic SULT2A expression as an example. Our previous investigations suggested roles for both the glucocorticoid receptor (GR) and the pregnane X receptor (PXR) in the mediation of glucocorticoid-inducible rat hepatic SULT2A expression [127]. GR-activating concentrations of glucocorticoid transactivated SULT2A transcription indirectly, through intermediary steps involving GR-inducible liver-enriched CCAAT enhancer binding protein [128], while pharmacological concentrations of dexamethasone induced rat hepatic SULT2A expression via a PXR-mediated mechanism [127]. Rodent and human SULT2A are differentially regulated by the xenobiotic-sensing receptor, PXR. In mice and rats, hepatic SULT2A transcription is activated by PXR through the direct binding of PXR to the 5′-flanking regions of SULT2A genes [127, 129]. However, unlike rodent hepatic SULT2As, treatment of human liver cells with rifampicin, the prototypical ligand activator of human PXR [130], produces a biphasic effect on SULT2A1 expression [131]. Treatment with PXR-activating concentrations of rifampicin causes PXR-dependent suppression of SULT2A1 expression, whereas treatment with higher rifampicin concentrations induces SULT2A1 expression through a PXR-independent mechanism [131]. In addition, the nuclear receptor hepatocyte nuclear factor 4α (HNF4α) is a positive regulator of SULT2A1 expression, and both the suppressive and activating effects of rifampicin appear to be transduced through interactions with HNF4α [131].
The constitutive androstane receptor (CAR) also partners with RXR and transactivates murine hepatic SULT2A, and possibly human SULT2A1 [132]. The vitamin D receptor (VDR, NR1I1) is activated not only by 1α,25-dihydroxyvitamin D3 but also by bile acids [133], and emerging evidence suggests that the VDR regulates murine hepatic SULT2A transcription, and can also drive the transcription of SULT2A1 in vitro [134]. The role of SULT2A as an integrator of endogenous lipid metabolism is just emerging. Oxysterol intermediates of cholesterol metabolism are physiological ligands for LXR, an RXR heterodimer transcription factor that regulates a number of genes involved in the maintenance of lipid homeostasis [135]. LXR-inducible murine hepatic SULT2A gene transcription has been described and has been shown to confer a protective effect against bile acid toxicity during cholestasis [136]. Certain sulfonated auto-oxidized sterols, such as 5α,6α epoxycholesterol-3-sulfate and 7-ketocholesterol-3-sulfate, have been shown to interact with the LXRα ligand-binding domain and inhibit LXRα-mediated transactivation in vitro [137]. A suppressive role for FXR in the regulation of hepatic SULT2A expression has also been identified, as SULT2A expression was increased 8 fold in FXR-null mice as compared to wild-type mice, and CDCA feeding decreased SULT2A expression in wild-type mice [138]. Also, SULT2A1 expression was reduced following treatment of HepG2 cells with FXR agonists [138].
6. Conclusions
Emerging evidence supports an under-appreciated physiological role for members of the SULT and UGT gene families to serve as modulators of biologically active lipids and to undergo transactivation by lipid-sensing transcription factors such as the PPARs. Particularly in keratinocytes which rely on lipid signaling for the progression of programmed cellular differentiation, the inducible expression of cholesterol sulfotransferase (SULT2B1b) by PPAR activators has been demonstrated. Studies in primary cultured human but not rat hepatocytes clearly demonstrate that PPARα-inducible human hepatic SULT2A1 expression occurs through a distal PPRE in the 5′-flanking region of the SULT2A1 gene. Several human hepatic UGTs also demonstrate inducible transcription in response to PPARα activation, and transgenic mice expressing the human UGT1 gene locus display transcriptional regulation of human UGT1A transgenes in liver and intestine by PPARα. In view of the coordinated integration between phase II metabolism and PPAR lipid signaling networks, future investigations will likely focus on the disturbances in hepatic and gastrointestinal lipid homeostasis that significantly alter SULT and UGT expression sufficiently to disrupt the down-stream metabolism of environmental xenobiotics, pharmaceuticals, or biologically active intermediates of metabolism.
References
- 1.Robinson E, Grieve DJ. Significance of peroxisome proliferator-activated receptors in the cardiovascular system in health and disease. Pharmacology & Therapeutics. 2009;122(3):246–263. doi: 10.1016/j.pharmthera.2009.03.003. [DOI] [PubMed] [Google Scholar]
- 2.Töröcsik D, Szanto A, Nagy L. Oxysterol signaling links cholesterol metabolism and inflammation via the liver X receptor in macrophages. Molecular Aspects of Medicine. 2009;30(3):134–152. doi: 10.1016/j.mam.2009.02.002. [DOI] [PubMed] [Google Scholar]
- 3.Chapman E, Best MD, Hanson SR, Wong C-H. Sulfotransferases: structure, mechanism, biological activity, inhibition, and synthetic utility. Angewandte Chemie International Edition. 2004;43(27):3526–3548. doi: 10.1002/anie.200300631. [DOI] [PubMed] [Google Scholar]
- 4.Strott CA. Sulfonation and molecular action. Endocrine Reviews. 2002;23(5):703–732. doi: 10.1210/er.2001-0040. [DOI] [PubMed] [Google Scholar]
- 5.Dooley TP, Haldeman-Cahill R, Joiner J, Wilborn TW. Expression profiling of human sulfotransferase and sulfatase gene superfamilies in epithelial tissues and cultured cells. Biochemical and Biophysical Research Communications. 2000;277(1):236–245. doi: 10.1006/bbrc.2000.3643. [DOI] [PubMed] [Google Scholar]
- 6.Falany CN, Krasnykh V, Falany JL. Bacterial expression and characterization of a cDNA for human liver estrogen sulfotransferase. Journal of Steroid Biochemistry and Molecular Biology. 1995;52(6):529–539. doi: 10.1016/0960-0760(95)00015-r. [DOI] [PubMed] [Google Scholar]
- 7.Kester MHA, van Dijk CH, Tibboel D, et al. Sulfation of thyroid hormone by estrogen sulfotransferase. Journal of Clinical Endocrinology and Metabolism. 1999;84(7):2577–2580. doi: 10.1210/jcem.84.7.5975. [DOI] [PubMed] [Google Scholar]
- 8.Kitada H, Miyata M, Nakamura T, et al. Protective role of hydroxysteroid sulfotransferase in lithocholic acid-induced liver toxicity. The Journal of Biological Chemistry. 2003;278(20):17838–17844. doi: 10.1074/jbc.M210634200. [DOI] [PubMed] [Google Scholar]
- 9.Engel SR, Purdy RH, Grant KA. Characterization of discriminative stimulus effects of the neuroactive steroid pregnanolone. Journal of Pharmacology and Experimental Therapeutics. 2001;297(2):489–495. [PubMed] [Google Scholar]
- 10.Akwa Y, Ladurelle N, Covey DF, Baulieu E-E. The synthetic enantiomer of pregnenolone sulfate is very active on memory in rats and mice, even more so than its physiological neurosteroid counterpart: distinct mechanisms? Proceedings of the National Academy of Sciences of the United States of America. 2001;98(24):14033–14037. doi: 10.1073/pnas.241503698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shimizu C, Fuda H, Yanai H, Strott CA. Conservation of the hydroxysteroid sulfotransferase SULT2B1 gene structure in the mouse: pre- and postnatal expression, kinetic analysis of isoforms, and comparison with prototypical SULT2A1. Endocrinology. 2003;144(4):1186–1193. doi: 10.1210/en.2002-221011. [DOI] [PubMed] [Google Scholar]
- 12.Shimada M, Yoshinari K, Tanabe E, et al. Identification of ST2A1 as a rat brain neurosteroid sulfotransferase mRNA. Brain Research. 2001;920(1-2):222–225. doi: 10.1016/s0006-8993(01)03061-x. [DOI] [PubMed] [Google Scholar]
- 13.Lewis AJ, Walle UK, King RS, Kadlubar FF, Falany CN, Walle T. Bioactivation of the cooked food mutagen N-hydroxy-2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine by estrogen sulfotransferase in cultured human mammary epithelial cells. Carcinogenesis. 1998;19(11):2049–2053. doi: 10.1093/carcin/19.11.2049. [DOI] [PubMed] [Google Scholar]
- 14.Banoglu E. Current status of the cytosolic sulfotransferases in the metabolic activation of promutagens and procarcinogens. Current Drug Metabolism. 2000;1(1):1–30. doi: 10.2174/1389200003339234. [DOI] [PubMed] [Google Scholar]
- 15.Glatt H, Boeing H, Engelke CEH, et al. Human cytosolic sulphotransferases: genetics, characteristics, toxicological aspects. Mutation Research. 2001;482(1-2):27–40. doi: 10.1016/s0027-5107(01)00207-x. [DOI] [PubMed] [Google Scholar]
- 16.Petrotchenko EV, Pedersen LC, Borchers CH, Tomer KB, Negishi M. The dimerization motif of cytosolic sulfotransferases. FEBS Letters. 2001;490(1-2):39–43. doi: 10.1016/s0014-5793(01)02129-9. [DOI] [PubMed] [Google Scholar]
- 17.Nagata K, Yamazoe Y. Pharmacogenetics of sulfotransferase. Annual Review of Pharmacology and Toxicology. 2000;40:159–176. doi: 10.1146/annurev.pharmtox.40.1.159. [DOI] [PubMed] [Google Scholar]
- 18.Rutgers M, Heusdens FA, Visser TJ. Deiodination of iodothyronine sulfamates by type I iodothyronine deiodinase of rat liver. Endocrinology. 1991;129(3):1375–1381. doi: 10.1210/endo-129-3-1375. [DOI] [PubMed] [Google Scholar]
- 19.Kester MHA, Kaptein E, Roest TJ, et al. Characterization of human iodothyronine sulfotransferases. Journal of Clinical Endocrinology and Metabolism. 1999;84(4):1357–1364. doi: 10.1210/jcem.84.4.5590. [DOI] [PubMed] [Google Scholar]
- 20.Li X, Clemens DL, Cole JR, Anderson RJ. Characterization of human liver thermostable phenol sulfotransferase (SULT1A1) allozymes with 3,3′,5-triiodothyronine as the substrate. Journal of Endocrinology. 2001;171(3):525–532. doi: 10.1677/joe.0.1710525. [DOI] [PubMed] [Google Scholar]
- 21.Sakakibara Y, Yanagisawa K, Takami Y, Nakayama T, Suiko M, Liu M-C. Molecular cloning, expression, and functional characterization of novel mouse sulfotransferases. Biochemical and Biophysical Research Communications. 1998;247(3):681–686. doi: 10.1006/bbrc.1998.8872. [DOI] [PubMed] [Google Scholar]
- 22.Tsoi C, Falany CN, Morgenstern R, Swedmark S. Identification of a new subfamily of sulphotransferases: cloning and characterization of canine SULT1D1. Biochemical Journal. 2001;356(3):891–897. doi: 10.1042/0264-6021:3560891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Falany JL, Falany CN. Expression of cytosolic sulfotransferases in normal mammary epithelial cells and breast cancer cell lines. Cancer Research. 1996;56(7):1551–1555. [PubMed] [Google Scholar]
- 24.Falany JL, Azziz R, Falany CN. Identification and characterization of cytosolic sulfotransferases in normal human endometrium. Chemico-Biological Interactions. 1998;109(1–3):329–339. doi: 10.1016/s0009-2797(97)00143-9. [DOI] [PubMed] [Google Scholar]
- 25.Tong MH, Jiang H, Liu P, Lawson JA, Brass LF, Song W-C. Spontaneous fetal loss caused by placental thrombosis in estrogen sulfotransferase-deficient mice. Nature Medicine. 2005;11(2):153–159. doi: 10.1038/nm1184. [DOI] [PubMed] [Google Scholar]
- 26.Javitt NB, Lee YC, Shimizu C, Fuda H, Strott CA. Cholesterol and hydroxycholesterol sulfotransferases: identification, distinction from dehydroepiandrosterone sulfotransferase, and differential tissue expression. Endocrinology. 2001;142(7):2978–2984. doi: 10.1210/endo.142.7.8244. [DOI] [PubMed] [Google Scholar]
- 27.Falany CN, Vazquez ME, Kalb JM. Purification and characterization of human liver dehydroepiandrosterone sulphotransferase. Biochemical Journal. 1989;260(3):641–646. doi: 10.1042/bj2600641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Her C, Szumlanski C, Aksoy IA, Weinshilboum RM. Human jejunal estrogen sulfotransferase and dehydroepiandrosterone sulfotransferase Immunochemical characterization of individual variation. Drug Metabolism and Disposition. 1996;24(12):1328–1335. [PubMed] [Google Scholar]
- 29.Parker CR, Jr., Falany CN, Stockard CR, Stankovic AK, Grizzle WE. Immunohistochemical localization of dehydroepiandrosterone sulfotransferase in human fetal tissues. Journal of Clinical Endocrinology and Metabolism. 1994;78(1):234–236. doi: 10.1210/jcem.78.1.8288708. [DOI] [PubMed] [Google Scholar]
- 30.Duanmu Z, Weckle A, Koukouritaki SB, et al. Developmental expression of aryl, estrogen, and hydroxysteroid sulfotransferases in pre- and postnatal human liver. Journal of Pharmacology and Experimental Therapeutics. 2006;316(3):1310–1317. doi: 10.1124/jpet.105.093633. [DOI] [PubMed] [Google Scholar]
- 31.Labrie F, Bélanger A, Luu-The V, et al. DHEA and the intracrine formation of androgens and estrogens in peripheral target tissues: its role during aging. Steroids. 1998;63(5-6):322–328. doi: 10.1016/s0039-128x(98)00007-5. [DOI] [PubMed] [Google Scholar]
- 32.Her C, Wood TC, Eichler EE, et al. Human hydroxysteroid sulfotransferase SULT2B1: two enzymes encoded by a single chromosome 19 gene. Genomics. 1998;53(3):284–295. doi: 10.1006/geno.1998.5518. [DOI] [PubMed] [Google Scholar]
- 33.Kohjitani A, Fuda H, Hanyu O, Strott CA. Cloning, characterization and tissue expression of rat SULT2B1a and SULT2B1b steroid/sterol sulfotransferase isoforms: divergence of the rat SULT2B1 gene structure from orthologous human and mouse genes. Gene. 2006;367(1-2):66–73. doi: 10.1016/j.gene.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 34.Dongning HE, Meloche CA, Dumas NA, Frost AR, Falany CN. Different subcellular localization of sulphotransferase 2B1b in human placenta and prostate. Biochemical Journal. 2004;379(3):533–540. doi: 10.1042/BJ20031524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Higashi Y, Fuda H, Yanai H, et al. Expression of cholesterol sulfotransferase (SULT2B1b) in human skin and primary cultures of human epidermal keratinocytes. Journal of Investigative Dermatology. 2004;122(5):1207–1213. doi: 10.1111/j.0022-202X.2004.22416.x. [DOI] [PubMed] [Google Scholar]
- 36.Geese WJ, Raftogianis RB. Biochemical characterization and tissue distribution of human SULT2B1. Biochemical and Biophysical Research Communications. 2001;288(1):280–289. doi: 10.1006/bbrc.2001.5746. [DOI] [PubMed] [Google Scholar]
- 37.He D, Frost AR, Falany CN. Identification and immunohistochemical localization of Sulfotransferase 2B1b (SULT2B1b) in human lung. Biochimica et Biophysica Acta. 2005;1724(1-2):119–126. doi: 10.1016/j.bbagen.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 38.Koizumi M, Momoeda M, Hiroi H, et al. Expression and regulation of cholesterol sulfotransferase (SULT2B1b) in human endometrium. doi: 10.1016/j.fertnstert.2009.01.075. Fertility and Sterility. In press. [DOI] [PubMed] [Google Scholar]
- 39.Dumas NA, He D, Frost AR, Falany CN. Sulfotransferase 2B1b in human breast: differences in subcellular localization in African American and Caucasian women. Journal of Steroid Biochemistry and Molecular Biology. 2008;111(3–5):171–177. doi: 10.1016/j.jsbmb.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yanai H, Javitt NB, Higashi Y, Fuda H, Strott CA. Expression of cholesterol sulfotransferase (SULT2B1b) in human platelets. Circulation. 2004;109(1):92–96. doi: 10.1161/01.CIR.0000108925.95658.8D. [DOI] [PubMed] [Google Scholar]
- 41.Falany CN, He D, Dumas N, Frost AR, Falany JL. Human cytosolic sulfotransferase 2B1: isoform expression, tissue specificity and subcellular localization. Journal of Steroid Biochemistry and Molecular Biology. 2006;102(1–5):214–221. doi: 10.1016/j.jsbmb.2006.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fuda H, Lee YC, Shimizu C, Javitt NB, Strott CA. Mutational analysis of human hydroxysteroid sulfotransferase SULT2B1 isoforms reveals that exon 1B of the SULT2B1 gene produces cholesterol sulfotransferase, whereas exon 1A yields pregnenolone sulfotransferase. The Journal of Biological Chemistry. 2002;277(39):36161–36166. doi: 10.1074/jbc.M207165200. [DOI] [PubMed] [Google Scholar]
- 43.Baulieu EE. Neurosteroids: a novel function of the brain. Psychoneuroendocrinology. 1998;23(8):963–987. doi: 10.1016/s0306-4530(98)00071-7. [DOI] [PubMed] [Google Scholar]
- 44.Kohjitani A, Fuda H, Hanyu O, Strott CA. Regulation of SULT2B1a (pregnenolone sulfotransferase) expression in rat C6 glioma cells: relevance of AMPA receptor-mediated NO signaling. Neuroscience Letters. 2008;430(1):75–80. doi: 10.1016/j.neulet.2007.10.023. [DOI] [PubMed] [Google Scholar]
- 45.Takahashi S, Sakakibara Y, Mishiro E, et al. Molecular cloning, expression, and characterization of mouse amine N-sulfotransferases. Biochemical and Biophysical Research Communications. 2008;375(4):531–535. doi: 10.1016/j.bbrc.2008.08.051. [DOI] [PubMed] [Google Scholar]
- 46.Falany CN, Xie X, Wang J, Ferrer J, Falany JL. Molecular cloning and expression of novel sulphotransferase-like cDNAs from human and rat brain. Biochemical Journal. 2000;346, part 3:857–864. [PMC free article] [PubMed] [Google Scholar]
- 47.Liyou NE, Buller KM, Tresillian MJ, et al. Localization of a brain sulfotransferase, SULT4A1, in the human and rat brain: an immunohistochemical study. Journal of Histochemistry and Cytochemistry. 2003;51(12):1655–1664. doi: 10.1177/002215540305101209. [DOI] [PubMed] [Google Scholar]
- 48.Freimuth RR, Wiepert M, Chute CG, Wieben ED, Weinshilboum RM. Human cytosolic sulfotransferase database mining: identification of seven novel genes and pseudogenes. Pharmacogenomics Journal. 2004;4(1):54–65. doi: 10.1038/sj.tpj.6500223. [DOI] [PubMed] [Google Scholar]
- 49.Gamage N, Barnett A, Hempel N, et al. Human sulfotransferases and their role in chemical metabolism. Toxicological Sciences. 2006;90(1):5–22. doi: 10.1093/toxsci/kfj061. [DOI] [PubMed] [Google Scholar]
- 50.Lindsay J, Wang L-L, Li Y, Zhou S-F. Structure, function and polymorphism of human cytosolic sulfotransferases. Current Drug Metabolism. 2008;9(2):99–105. doi: 10.2174/138920008783571819. [DOI] [PubMed] [Google Scholar]
- 51.Alnouti Y, Klaassen CD. Regulation of sulfotransferase enzymes by prototypical microsomal enzyme inducers in mice. Journal of Pharmacology and Experimental Therapeutics. 2008;324(2):612–621. doi: 10.1124/jpet.107.129650. [DOI] [PubMed] [Google Scholar]
- 52.Fan L-Q, You L, Brown-Borg H, Brown S, Edwards RJ, Corton JC. Regulation of phase I and phase II steroid metabolism enzymes by PPARα activators. Toxicology. 2004;204(2-3):109–121. doi: 10.1016/j.tox.2004.06.018. [DOI] [PubMed] [Google Scholar]
- 53.Fang H-L, Strom SC, Cai H, Falany CN, Kocarek TA, Runge-Morris M. Regulation of human hepatic hydroxysteroid sulfotransferase gene expression by the peroxisome proliferator-activated receptor α transcription factor. Molecular Pharmacology. 2005;67(4):1257–1267. doi: 10.1124/mol.104.005389. [DOI] [PubMed] [Google Scholar]
- 54.Lampe MA, Williams ML, Elias PM. Human epidermal lipids: characterization and modulations during differentiation. Journal of Lipid Research. 1983;24(2):131–140. [PubMed] [Google Scholar]
- 55.Epstein EH, Jr., Williams ML, Elias PM. The epidermal cholesterol sulfate cycle. Journal of the American Academy of Dermatology. 1984;10(5):866–868. doi: 10.1016/s0190-9622(84)80144-9. [DOI] [PubMed] [Google Scholar]
- 56.Epstein EH, Jr., Bonifas JM, Barber TC, Haynes M. Cholesterol sulfotransferase of newborn mouse epidermis. Journal of Investigative Dermatology. 1984;83(5):332–335. doi: 10.1111/1523-1747.ep12264131. [DOI] [PubMed] [Google Scholar]
- 57.Kagehara M, Tachi M, Harii K, Iwamori M. Programmed expression of cholesterol sulfotransferase and transglutaminase during epidermal differentiation of murine skin development. Biochimica et Biophysica Acta. 1994;1215(1-2):183–189. doi: 10.1016/0005-2760(94)90109-0. [DOI] [PubMed] [Google Scholar]
- 58.Kashiwagi M, Kuroki T, Huh N-H. Specific inhibition of hair follicle formation by epidermal growth factor in an organ culture of developing mouse skin. Developmental Biology. 1997;189(1):22–32. doi: 10.1006/dbio.1997.8650. [DOI] [PubMed] [Google Scholar]
- 59.Jetten AM, George MA, Nervi C, Boone LR, Rearick JI. Increased cholesterol sulfate and cholesterol sulfotransferase activity in relation to the multi-step process of differentiation in human epidermal keratinocytes. Journal of Investigative Dermatology. 1989;92(2):203–209. doi: 10.1111/1523-1747.ep12276731. [DOI] [PubMed] [Google Scholar]
- 60.Johnson GA, Baker CA, Knight KA. Minoxidil sulfotransferase, a marker of human keratinocyte differentiation. Journal of Investigative Dermatology. 1992;98(5):730–733. doi: 10.1111/1523-1747.ep12499930. [DOI] [PubMed] [Google Scholar]
- 61.Kömüves LG, Hanley K, Jiang Y, et al. Induction of selected lipid metabolic enzymes and differentiation-linked structural proteins by air exposure in fetal rat skin explants. Journal of Investigative Dermatology. 1999;112(3):303–309. doi: 10.1046/j.1523-1747.1999.00511.x. [DOI] [PubMed] [Google Scholar]
- 62.Kiguchi K, Kagehara M, Higo R, Iwamori M, DiGiovanni J. Alterations in cholesterol sulfate and its biosynthetic enzyme during multistage carcinogenesis in mouse skin. Journal of Investigative Dermatology. 1998;111(6):973–981. doi: 10.1046/j.1523-1747.1998.00404.x. [DOI] [PubMed] [Google Scholar]
- 63.Yamamoto S, Jiang H, Kato R. Involvement of prostaglandin E2 in the tumor promoter phorbol ester-caused increase in epidermal cholesterol sulfotransferase activity. Carcinogenesis. 1991;12(6):1145–1147. doi: 10.1093/carcin/12.6.1145. [DOI] [PubMed] [Google Scholar]
- 64.Lee KA, Fuda H, Lee YC, Negishi M, Strott CA, Pedersen LC. Crystal structure of human cholesterol sulfotransferase (SULT2B1b) in the presence of pregenenolone and 3'-phosphoadenosine 5′-phosphate. The Journal of Biological Chemistry. 2003;278:44953–44599. doi: 10.1074/jbc.M308312200. [DOI] [PubMed] [Google Scholar]
- 65.Denning MF, Kazanietz MG, Blumberg PM, Yuspa SH. Cholesterol sulfate activates multiple protein kinase C isoenzymes and induces granular cell differentiation in cultured murine keratinocytes. Cell Growth and Differentiation. 1995;6(12):1619–1626. [PubMed] [Google Scholar]
- 66.Kuroki T, Ikuta T, Kashiwagi M, et al. Cholesterol sulfate, an activator of protein kinase C mediating squamous cell differentiation: a review. Mutation Research. 2000;462(2-3):189–195. doi: 10.1016/s1383-5742(00)00036-3. [DOI] [PubMed] [Google Scholar]
- 67.Denning MF. Epidermal keratinocytes: regulation of multiple cell phenotypes by multiple protein kinase C isoforms. International Journal of Biochemistry and Cell Biology. 2004;36(7):1141–1146. doi: 10.1016/j.biocel.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 68.Kawabe S, Ikuta T, Ohba M, et al. Cholesterol sulfate activates transcription of transglutaminase 1 gene in normal human keratinocytes. Journal of Investigative Dermatology. 1998;111(6):1098–1102. doi: 10.1046/j.1523-1747.1998.00441.x. [DOI] [PubMed] [Google Scholar]
- 69.Ohba M, Ishino K, Kashiwagi M, et al. Induction of differentiation in normal human keratinocytes by adenovirus-mediated introduction of the η and δ isoforms of protein kinase C. Molecular and Cellular Biology. 1998;18(9):5199–5207. doi: 10.1128/mcb.18.9.5199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kallen JA, Schlaeppi J-M, Bitsch F, et al. X-ray structure of the hRORα LBD at 1.63 Å: structural and functional data that cholesterol or a cholesterol derivative is the natural ligand of RORα . Structure. 2002;10(12):1697–1707. doi: 10.1016/s0969-2126(02)00912-7. [DOI] [PubMed] [Google Scholar]
- 71.Kallen J, Schlaeppi J-M, Bitsch F, Delhon I, Fournier B. Crystal structure of the human RORα Ligand binding domain in complex with cholesterol sulfate at 2.2 Å. The Journal of Biological Chemistry. 2004;279(14):14033–14038. doi: 10.1074/jbc.M400302200. [DOI] [PubMed] [Google Scholar]
- 72.Bitsch F, Aichholz R, Kallen J, Geisse S, Fournier B, Schlaeppi J-M. Identification of natural ligands of retinoic acid receptor-related orphan receptor α ligand-binding domain expressed in Sf9 cells—a mass spectrometry approach. Analytical Biochemistry. 2003;323(1):139–149. doi: 10.1016/j.ab.2003.08.029. [DOI] [PubMed] [Google Scholar]
- 73.Jiang YJ, Kim P, Elias PM, Feingold KR. LXR and PPAR activators stimulate cholesterol sulfotransferase type 2 isoform 1b in human keratinocytes. Journal of Lipid Research. 2005;46(12):2657–2666. doi: 10.1194/jlr.M500235-JLR200. [DOI] [PubMed] [Google Scholar]
- 74.Little JM, Drake RR, Vonk R, Kuipers F, Lester R, Radominska A. Characterization of human liver microsomal UDP-glycosyltransferases using photoaffinity analogs. Journal of Pharmacology and Experimental Therapeutics. 1995;273(3):1551–1559. [PubMed] [Google Scholar]
- 75.Owens IS, Basu NK, Banerjee R. UDP-glucuronosyltransferases: gene structures of UGT1 and UGT2 families. Methods in Enzymology. 2005;400:1–22. doi: 10.1016/S0076-6879(05)00001-7. [DOI] [PubMed] [Google Scholar]
- 76.Caldwell J. Glucuronic acid conjugation in the context of the metabolic conjugation of xenobiotics. In: Matern S, Bock SW, Gerock W, editors. Advances in Glucuronide Conjugation. Boston, Mass, USA: MTP Press; 1985. pp. 7–20. [Google Scholar]
- 77.Dutton GJ. Glucuronidation of Drugs and Other Compounds. Boca Raton, Fla, USA: CRC Press; 1980. [Google Scholar]
- 78.Barbier O, Villeneuve L, Bocher V, et al. The UDP-glucuronosyltransferase 1A9 enzyme is a peroxisome proliferator-activated receptor α and γ target gene. The Journal of Biological Chemistry. 2003;278(16):13975–13983. doi: 10.1074/jbc.M300749200. [DOI] [PubMed] [Google Scholar]
- 79.King CD, Rios GR, Green MD, Tephly TR. UDP-glucuronosyltransferases. Current Drug Metabolism. 2000;1(2):143–161. doi: 10.2174/1389200003339171. [DOI] [PubMed] [Google Scholar]
- 80.Loi C-M, Young M, Randinitis E, Vassos A, Koup JR. Clinical pharmacokinetics of troglitazone. Clinical Pharmacokinetics. 1999;37(2):91–104. doi: 10.2165/00003088-199937020-00001. [DOI] [PubMed] [Google Scholar]
- 81.Watanabe Y, Nakajima M, Yokoi T. Troglitazone glucuronidation in human liver and intestine microsomes: high catalytic activity of UGT1A8 and UGT1A10. Drug Metabolism and Disposition. 2002;30(12):1462–1469. doi: 10.1124/dmd.30.12.1462. [DOI] [PubMed] [Google Scholar]
- 82.Yoshigae Y, Konno K, Takasaki W, Ikeda T. Characterization of UDP-glucuronosyltransferases (UGTS) involved in the metabolism of troglitazone in rats and humans. Journal of Toxicological Sciences. 2000;25(5):433–441. doi: 10.2131/jts.25.5_433. [DOI] [PubMed] [Google Scholar]
- 83.Prueksaritanont T, Zhao JJ, Ma B, et al. Mechanistic studies on metabolic interactions between gemfibrozil and statins. Journal of Pharmacology and Experimental Therapeutics. 2002;301(3):1042–1051. doi: 10.1124/jpet.301.3.1042. [DOI] [PubMed] [Google Scholar]
- 84.Prueksaritanont T, Tang C, Qiu Y, Mu L, Subramanian R, Lin JH. Effects of fibrates on metabolism of statins in human hepatocytes. Drug Metabolism and Disposition. 2002;30(11):1280–1287. doi: 10.1124/dmd.30.11.1280. [DOI] [PubMed] [Google Scholar]
- 85.Burchell B, Coughtrie MWH. UDP-glucuronosyltransferases. Pharmacology and Therapeutics. 1989;43(2):261–289. doi: 10.1016/0163-7258(89)90122-8. [DOI] [PubMed] [Google Scholar]
- 86.Zhou J, Zhang J, Xie W. Xenobiotic nuclear receptor-mediated regulation of UDP-glucuronosyltransferases. Current Drug Metabolism. 2005;6(4):289–298. doi: 10.2174/1389200054633853. [DOI] [PubMed] [Google Scholar]
- 87.Shelby MK, Cherrington NJ, Vansell NR, Klaassen CD. Tissue mRNA expression of the rat UDP-glucuronosyltransferase gene family. Drug Metabolism and Disposition. 2003;31(3):326–333. doi: 10.1124/dmd.31.3.326. [DOI] [PubMed] [Google Scholar]
- 88.Tukey RH, Strassburg CP. Human UDP-glucuronosyltransferases: metabolism, expression, and disease. Annual Review of Pharmacology and Toxicology. 2000;40:581–616. doi: 10.1146/annurev.pharmtox.40.1.581. [DOI] [PubMed] [Google Scholar]
- 89.Mackenzie PI, Owens IS, Burchell B, et al. The UDP glycosyltransferase gene superfamily: recommended nomenclature update based on evolutionary divergence. Pharmacogenetics. 1997;7(4):255–269. doi: 10.1097/00008571-199708000-00001. [DOI] [PubMed] [Google Scholar]
- 90.Ritter JK, Chen F, Sheen YY, et al. A novel complex locus UGT1 encodes human bilirubin, phenol, and other UDP-glucuronosyltransferase isozymes with identical carboxyl termini. The Journal of Biological Chemistry. 1992;267(5):3257–3261. [PubMed] [Google Scholar]
- 91.Gong Q-H, Cho JW, Huang T, et al. Thirteen UDPglucuronosyltransferase genes are encoded at the human UGT1 gene complex locus. Pharmacogenetics. 2001;11(4):357–368. doi: 10.1097/00008571-200106000-00011. [DOI] [PubMed] [Google Scholar]
- 92.Lévesque E, Girard H, Journault K, Lépine J, Guillemette C. Regulation of the UGT1A1 bilirubin-conjugating pathway: role of a new splicing event at the UGT1A locus. Hepatology. 2007;45(1):128–138. doi: 10.1002/hep.21464. [DOI] [PubMed] [Google Scholar]
- 93.Girard H, Lévesque E, Bellemare J, Journault K, Caillier B, Guillemette C. Genetic diversity at the UGT1 locus is amplified by a novel 3′ alternative splicing mechanism leading to nine additional UGT1A proteins that act as regulators of glucuronidation activity. Pharmacogenetics and Genomics. 2007;17(12):1077–1089. doi: 10.1097/FPC.0b013e3282f1f118. [DOI] [PubMed] [Google Scholar]
- 94.Owens IS, Ritter JK. The novel bilirubin/phenol UDP-glucuronosyltransferase UGT1 gene locus: implications for multiple nonhemolytic familial hyperbilirubinemia phenotypes. Pharmacogenetics. 1992;2(3):93–108. doi: 10.1097/00008571-199206000-00001. [DOI] [PubMed] [Google Scholar]
- 95.Kato Y, Ikushiro S-I, Emi Y, et al. Hepatic UDP-glucuronosyltransferases responsible for glucuronidation of thyroxine in humans. Drug Metabolism and Disposition. 2008;36(1):51–55. doi: 10.1124/dmd.107.018184. [DOI] [PubMed] [Google Scholar]
- 96.Barbier O, Trottier J, Kaeding J, Caron P, Verreault M. Lipid-activated transcription factors control bile acid glucuronidation. Molecular and Cellular Biochemistry. 2009;326(1-2):3–8. doi: 10.1007/s11010-008-0001-5. [DOI] [PubMed] [Google Scholar]
- 97.Barbier O, Duran-Sandoval D, Pineda-Torra I, Kosykh V, Fruchart J-C, Staels B. Peroxisome proliferator-activated receptor α induces hepatic expression of the human bile acid glucuronidating UDP-glucuronosyltransferase 2B4 enzyme. The Journal of Biological Chemistry. 2003;278(35):32852–32860. doi: 10.1074/jbc.M305361200. [DOI] [PubMed] [Google Scholar]
- 98.Gall WE, Zawada G, Mojarrabi B, et al. Differential glucuronidation of bile acids, androgens and estrogens by human UGT1A3 and 2B7. Journal of Steroid Biochemistry and Molecular Biology. 1999;70(1–3):101–108. doi: 10.1016/s0960-0760(99)00088-6. [DOI] [PubMed] [Google Scholar]
- 99.Trottier J, Verreault M, Grepper S, et al. Human UDP-glucuronosyltransferase (UGT)1A3 enzyme conjugates chenodeoxycholic acid in the liver. Hepatology. 2006;44(5):1158–1170. doi: 10.1002/hep.21362. [DOI] [PubMed] [Google Scholar]
- 100.Hess R, Staubli W, Riess W. Nature of the hepatomegalic effect produced by ethyl-chlorophenoxy-isobutyrate in the rat. Nature. 1965;208(5013):856–858. doi: 10.1038/208856a0. [DOI] [PubMed] [Google Scholar]
- 101.Reddy JK. Possible properties of microbodies (peroxisomes). Microbody proliferation and hypolipidemic drugs. Journal of Histochemistry and Cytochemistry. 1973;21(11):967–971. doi: 10.1177/21.11.967. [DOI] [PubMed] [Google Scholar]
- 102.Gibson GG, Orton TC, Tamburini PP. Cytochrome P-450 induction by clofibrate. Purification and properties of a hepatic cytochrome P-450 relatively specific for the 12- and 11-hydroxylation of dodecanoic acid (lauric acid) Biochemical Journal. 1982;203(1):161–168. doi: 10.1042/bj2030161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Foliot A, Drocourt JL, Etienne JP, Housset E, Fiessinger JN, Christoforov B. Increase in the hepatic glucuronidation and clearance of bilirubin in clofibrate treated rats. Biochemical Pharmacology. 1977;26(6):547–549. doi: 10.1016/0006-2952(77)90333-1. [DOI] [PubMed] [Google Scholar]
- 104.Fournel S, Magdalou J, Thomassin J, et al. Structure-dependent induction of bilirubin glucuronidation and lauric acid 12-hydroxylation by arylcarboxylic acids chemically related to clofibrate. Biochimica et Biophysica Acta. 1985;842(2-3):202–213. doi: 10.1016/0304-4165(85)90204-1. [DOI] [PubMed] [Google Scholar]
- 105.Fournel S, Magdalou J, Pinon P, Siest G. Differential induction profile of drug-metabolizing enzymes after treatment with hypolipidaemic agents. Xenobiotica. 1987;17(4):445–457. doi: 10.3109/00498258709043951. [DOI] [PubMed] [Google Scholar]
- 106.Raza H, Levine WG, Chowdhury NR, Chowdhury JR. Microsomal azoreduction and glucuronidation in the metabolism of dimethylaminoazobenzene by the rat liver. Xenobiotica. 1987;17(6):669–677. doi: 10.3109/00498258709043974. [DOI] [PubMed] [Google Scholar]
- 107.Mounie J, Champion L, Goudonnet H, Escousse A, Truchot RC. Inductive effects of fenofibrate and metabolism of phenobarbital. Fundamental and Clinical Pharmacology. 1988;2(4):259–265. doi: 10.1111/j.1472-8206.1988.tb00637.x. [DOI] [PubMed] [Google Scholar]
- 108.Boiteux-Antoine A-F, Magdalou J, Fournel-Gigleux S, Siest G. Comparative induction of drug-metbolizing enzymes by hypolipidaemic compounds. General Pharmacology. 1989;20(4):407–412. doi: 10.1016/0306-3623(89)90187-0. [DOI] [PubMed] [Google Scholar]
- 109.Visser TJ, Kaptein E, van Toor H, et al. Glucuronidation of thyroid hormone in rat liver: effects of in vivo treatment with microsomal enzyme inducers and in vitro assay conditions. Endocrinology. 1993;133(5):2177–2186. doi: 10.1210/endo.133.5.8404669. [DOI] [PubMed] [Google Scholar]
- 110.Ikushiro S-I, Emi Y, Iyanagi T. Identification and analysis of drug-responsive expression of UDP- glucuronosyltransferase family 1 (UGT1) isozyme in rat hepatic microsomes using anti-peptide antibodies. Archives of Biochemistry and Biophysics. 1995;324(2):267–272. doi: 10.1006/abbi.1995.0039. [DOI] [PubMed] [Google Scholar]
- 111.Viollon-Abadie C, Lassere D, Debruyne E, Nicod L, Carmichael N, Richert L. Phenobarbital, β-naphthoflavone, clofibrate, and pregnenolone-16α-carbonitrile do not affect hepatic thyroid hormone UDP-glucuronosyl transferase activity, and thyroid gland function in mice. Toxicology and Applied Pharmacology. 1999;155(1):1–12. doi: 10.1006/taap.1998.8558. [DOI] [PubMed] [Google Scholar]
- 112.Viollon-Abadie C, Bigot-Lasserre D, Nicod L, Carmichael N, Richert L. Effects of model inducers on thyroxine UDP-glucuronosyl-transferase activity in vitro in rat and mouse hepatocyte cultures. Toxicology in Vitro. 2000;14(6):505–512. doi: 10.1016/s0887-2333(00)00044-8. [DOI] [PubMed] [Google Scholar]
- 113.Kutz K, Kandler H, Gugler R, Fevery J. Effect of clofibrate on the metabolism of bilirubin, bromosulphophthalein and indocyanine green and on the biliary lipid composition in Gilbert's syndrome. Clinical Science. 1984;66(4):389–397. doi: 10.1042/cs0660389. [DOI] [PubMed] [Google Scholar]
- 114.Dragacci S, Hamar-Hansen C, Fournel-Gigleux S, Lafaurie C, Magdalou J, Siest G. Comparative study of clofibric acid and bilirubin glucuronidation in human liver microsomes. Biochemical Pharmacology. 1987;36(22):3923–3927. doi: 10.1016/0006-2952(87)90459-x. [DOI] [PubMed] [Google Scholar]
- 115.Barter RA, Klaassen CD. Rat liver microsomal UDP-glucuronosyltransferase activity toward thyroxine: characterization, induction, and form specificity. Toxicology and Applied Pharmacology. 1992;115(2):261–267. doi: 10.1016/0041-008x(92)90331-l. [DOI] [PubMed] [Google Scholar]
- 116.Genchi G, Wang W, Barua A, Bidlack WR, Olson JA. Formation of β-glucuronides and of β-galacturonides of various retinoids catalyzed by induced and noninduced microsomal UDP-glucuronosyltransferases of rat liver. Biochimica et Biophysica Acta. 1996;1289(2):284–290. doi: 10.1016/0304-4165(95)00168-9. [DOI] [PubMed] [Google Scholar]
- 117.Pless D, Gouze JN, Senay C, et al. Characterization of the UDP-glucuronosyltransferases involved in the glucuronidation of an antithrombotic thioxyloside in rat and humans. Drug Metabolism and Disposition. 1999;27(5):588–595. [PubMed] [Google Scholar]
- 118.Luci S, Kluge H, Hirche F, Eder K. Clofibrate increases hepatic triiodothyronine (T3)- and thyroxine (T4)-glucuronosyltransferase activities and lowers plasma T3 and T4 concentrations in pigs. Drug Metabolism and Disposition. 2006;34(11):1887–1892. doi: 10.1124/dmd.106.011379. [DOI] [PubMed] [Google Scholar]
- 119.Jemnitz K, Veres Z, Monostory K, Vereczkey L. Glucuronidation of thyroxine in primary monolayer cultures of rat hepatocytes: in vitro induction of UDP-glucuronosyltranferases by methylcholanthrene, clofibrate, and dexamethasone alone and in combination. Drug Metabolism and Disposition. 2000;28(1):34–37. [PubMed] [Google Scholar]
- 120.Arand M, Coughtrie MWH, Burchell B, Oesch F, Robertson LW. Selective induction of bilirubin UDP-glucuronosyl-transferase by perfluorodecanoic acid. Chemico-Biological Interactions. 1991;77(1):97–105. doi: 10.1016/0009-2797(91)90008-u. [DOI] [PubMed] [Google Scholar]
- 121.Clementi M, Di Gianantonio E, Fabris L, et al. Inheritance of hyperbilirubinemia: evidence for a major autosomal recessive gene. Digestive and Liver Disease. 2007;39(4):351–355. doi: 10.1016/j.dld.2006.12.019. [DOI] [PubMed] [Google Scholar]
- 122.Shelby MK, Klaassen CD. Induction of rat UDP-glucuronosyltransferases in liver and duodenum by microsomal enzyme inducers that activate various transcriptional pathways. Drug Metabolism and Disposition. 2006;34(10):1772–1778. doi: 10.1124/dmd.106.010397. [DOI] [PubMed] [Google Scholar]
- 123.Richert L, Lamboley C, Viollon-Abadie C, et al. Effects of clofibric acid on mRNA expression profiles in primary cultures of rat, mouse and human hepatocytes. Toxicology and Applied Pharmacology. 2003;191(2):130–146. doi: 10.1016/s0041-008x(03)00231-x. [DOI] [PubMed] [Google Scholar]
- 124.Luukkanen L, Taskinen J, Kurkela M, Kostiainen R, Hirvonen J, Finel M. Kinetic characterization of the 1A subfamily of recombinant human UDP-glucuronosyltransferases. Drug Metabolism and Disposition. 2005;33(7):1017–1026. doi: 10.1124/dmd.105.004093. [DOI] [PubMed] [Google Scholar]
- 125.Senekeo-Effenberger K, Chen S, Brace-Sinnokrak E, et al. Expression of the human UGT1 locus in transgenic mice by 4-chloro-6-(2,3-xylidino)-2-pyrimidinylthioacetic acid (WY-14643) and implications on drug metabolism through peroxisome proliferator-activated receptor α activation. Drug Metabolism and Disposition. 2007;35(3):419–427. doi: 10.1124/dmd.106.013243. [DOI] [PubMed] [Google Scholar]
- 126.Verreault M, Senekeo-Effenberger K, Trottier J, et al. The liver X-receptor alpha controls hepatic expression of the human bile acid-glucuronidating UGT1A3 enzyme in human cells and transgenic mice. Hepatology. 2006;44(2):368–378. doi: 10.1002/hep.21259. [DOI] [PubMed] [Google Scholar]
- 127.Runge-Morris M, Wu W, Kocarek TA. Regulation of rat hepatic hydroxysteroid sulfotransferase (SULT2-40/41) gene expression by glucocorticoids: evidence for a dual mechanism of transcriptional control. Molecular Pharmacology. 1999;56(6):1198–1206. doi: 10.1124/mol.56.6.1198. [DOI] [PubMed] [Google Scholar]
- 128.Fang H-L, Abdolalipour M, Duanmu Z, et al. Regulation of glucocorticoid-inducible hydroxysteroid sulfotransferase (SULT2A-40/41) gene transcription in primary cultured rat hepatocytes: role of CCAAT/enhancer-binding protein liver-enriched transcription factors. Drug Metabolism and Disposition. 2005;33(1):147–156. doi: 10.1124/dmd.104.000281. [DOI] [PubMed] [Google Scholar]
- 129.Sonoda J, Xie W, Rosenfeld JM, Barwick JL, Guzelian PS, Evans RM. Regulation of a xenobiotic sulfonation cascade by nuclear pregnane X receptor (PXR) Proceedings of the National Academy of Sciences of the United States of America. 2002;99(21):13801–13806. doi: 10.1073/pnas.212494599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lehmann JM, McKee DD, Watson MA, Willson TM, Moore JT, Kliewer SA. The human orphan nuclear receptor PXR is activated by compounds that regulate CYP3A4 gene expression and cause drug interactions. Journal of Clinical Investigation. 1998;102(5):1016–1023. doi: 10.1172/JCI3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Fang H-L, Strom SC, Ellis E, et al. Positive and negative regulation of human hepatic hydroxysteroid sulfotransferase (SULT2A1) gene transcription by rifampicin: roles of hepatocyte nuclear factor 4α and pregnane X receptor. Journal of Pharmacology and Experimental Therapeutics. 2007;323(2):586–598. doi: 10.1124/jpet.107.124610. [DOI] [PubMed] [Google Scholar]
- 132.Assem M, Schuetz EG, Leggas M, et al. Interactions between hepatic Mrp4 and Sult2a as revealed by the constitutive androstane receptor and Mrp4 knockout mice. The Journal of Biological Chemistry. 2004;279(21):22250–22257. doi: 10.1074/jbc.M314111200. [DOI] [PubMed] [Google Scholar]
- 133.Drocourt L, Ourlin J-C, Pascussi J-M, Maurel P, Vilarem M-J. Expression of CYP3A4, CYP2B6, and CYP2C9 is regulated by the vitamin D receptor pathway in primary human hepatocytes. The Journal of Biological Chemistry. 2002;277(28):25125–25132. doi: 10.1074/jbc.M201323200. [DOI] [PubMed] [Google Scholar]
- 134.Song CS, Echchgadda I, Seo Y-K, et al. An essential role of the CAAT/enhancer binding protein-α in the vitamin D-induced expression of the human steroid/bile acid-sulfotransferase (SULT2A1) Molecular Endocrinology. 2006;20(4):795–808. doi: 10.1210/me.2005-0428. [DOI] [PubMed] [Google Scholar]
- 135.Tontonoz P, Mangelsdorf DJ. Liver X receptor signaling pathways in cardiovascular disease. Molecular Endocrinology. 2003;17(6):985–993. doi: 10.1210/me.2003-0061. [DOI] [PubMed] [Google Scholar]
- 136.Uppal H, Saini SPS, Moschetta A, et al. Activation of LXRs prevents bile acid toxicity and cholestasis in female mice. Hepatology. 2007;45(2):422–432. doi: 10.1002/hep.21494. [DOI] [PubMed] [Google Scholar]
- 137.Song C, Hiipakka RA, Liao S. Auto-oxidized cholesterol sulfates are antagonistic ligands of liver X receptors: implications for the development and treatment of atherosclerosis. Steroids. 2001;66(6):473–479. doi: 10.1016/s0039-128x(00)00239-7. [DOI] [PubMed] [Google Scholar]
- 138.Miyata M, Matsuda Y, Tsuchiya H, et al. Chenodeoxycholic acid-mediated activation of the farnesoid X receptor negatively regulates hydroxysteroid sulfotransferase. Drug Metabolism and Pharmacokinetics. 2006;21(4):315–323. doi: 10.2133/dmpk.21.315. [DOI] [PubMed] [Google Scholar]