Abstract
Distributions of proton MR-detected metabolites have been mapped throughout the brain in a group of normal subjects using a volumetric MR spectroscopic imaging (MRSI) acquisition with an interleaved water reference. Data were processed with intensity and spatial normalization to enable voxel-based analysis methods to be applied across a group of subjects. Results demonstrate significant regional, tissue, and gender-dependent variations of brain metabolite concentrations, and variations of these distributions with normal aging. The greatest alteration of metabolites with age was observed for white-matter choline and creatine. An example of the utility of the normative metabolic information is then demonstrated for analysis of data acquired from a subject who suffered a traumatic brain injury. This study demonstrates the ability to obtain proton spectra from a wide region of the brain and to apply fully automated processing methods. The resultant data provide a normative reference for subsequent utilization for studies of brain injury and disease.
Keywords: brain, metabolite distributions, age, proton MR spectroscopic imaging, data processing
Proton MR spectroscopy (MRS) enables the detection of a number of tissue metabolites that provide sensitive markers of disease or injury, making these techniques of considerable interest for clinical diagnostic purposes and particularly for studies in the brain. The acquisition and analysis of MRS data have several technical challenges that compromise the spatial resolution and accuracy of the resultant metabolite values. Furthermore, metabolic changes with disease and injury can frequently be subtle and diffuse, with the result that metabolite images may not be visually interpretable in the sense of a structural MRI. Therefore, the analysis of MRS data greatly benefits from comparison against a known reference signal.
Since in vivo MRS measurements are dependent on the acquisition method used, the reference data must be acquired in an identical manner to that of the data under analysis. It is also necessary to take into account normal variations in metabolite concentrations; for example, variations in metabolite concentrations between tissue type, across different brain regions, and changes with age are well documented (1–14). Other reports have indicated differences in metabolite concentrations with gender, lateralization (15), intelligence quotient (IQ) (16), and associations with smoking and alcohol consumption (17). To account for these factors, results are commonly compared against data obtained from the same location in a group of control subjects matched to the study group under investigation. However, although many studies have reported metabolite values from normal control subjects, these values can rarely be used as the reference information for other investigations. For example, published values using single-voxel spectroscopy (SVS) measurements are limited to only a few brain regions, and acquisition parameters, analysis methods, and subject selection criteria, such as age, vary considerably, making it unlikely that existing data can be used as reference information for a new investigation. Additionally, a sufficient number of measurements must be acquired to account for the acquisition-dependent variability of the MRS measurement, which primarily arises from low signal-to-noise ratio (SNR), line-shape distortions, spectral contamination from unsuppressed water and lipid signals, and variable partial volume contributions from surrounding regions. This requirement to obtain the matched reference metabolite information with a sufficient number of subjects to address the experimental variability of the measurement represents a considerable effort and cost for each new study.
MRS analysis methods have commonly made use of metabolite ratios, which conveniently removes concerns for signal calibration and correction for detection sensitivity variability. The use of ratios, however, also results in loss of information, and an analysis based on individual metabolite concentrations may be more desirable, for which a calibration procedure is required. Although absolute quantitation methods for MR spectroscopic imaging (MRSI) have been implemented (2,18) with the aim of reporting results in standardized metabolite concentration units, their implementation requires that metabolite and reference relaxation be accounted for, which involves a significant modification to the acquisition protocol that becomes impractical for routine clinical studies. Therefore, methods for signal normalization to “institutional units” (19) are more conveniently implemented in the clinical setting.
In this report, results obtained from mapping proton MRS-observed metabolite distributions throughout a large portion of the brain are presented. In addition to providing information on spatial variations and subject-dependent differences of metabolite distributions, these data constitute a brain metabolite image database for healthy adult subjects that can provide the normative information required for comparisons against other study data, for the same acquisition protocol. The approach demonstrated in this report is to use a standardized MRI and MRSI acquisition protocol that incorporates signal normalization and spatial transformation of volumetric metabolite images into a common spatial reference frame. The method takes into account variable tissue contribution to each MRSI voxel location, to enable analysis of regional and subject-dependent differences in metabolite distributions. To illustrate the potential advantage gained by this approach, one example based on MRSI following brain injury is presented.
MATERIALS AND METHODS
Subject Selection
A total of 126 healthy subjects were recruited for this study. Subjects were screened to exclude any history of brain disease or injury, substance abuse, or psychiatric history. The study was approved for human subjects research, and written informed consent was obtained before participation. Of the acquired data, 40 studies were excluded, of which approximately half were of inadequate data quality, primarily due to motion artifacts, and half due to normal variants in brain structure. The resultant studies used for the analysis included 41 male and 47 female subjects ranging in age from 18 to 59 years (mean age = 33 years). Of these, a subset of 22 males and 22 females was used for additional analyses on an age-restricted group from 18 to 30 years old (mean age = 24 years).
To provide a demonstration of how the normal brain metabolite data can benefit the analysis of MRSI data acquired in a single subject, a single study was selected from a subject who had a traumatic brain injury (TBI). This subject was a 28-year-old male who was studied 10 days following admission to the trauma center following an assault. The injury was assessed as a moderate closed-head brain injury with a Glasgow coma score of 11.
Data Acquisition
All subjects were scanned at 3T (Siemens Trio) with an eight-channel head array coil (In Vivo Corporation). The MRI protocol consisted of a T1-weighted magnetization-prepared rapid gradient echo (MPRAGE) sequence, TR/TE/TI = 2150/4.38/1100 ms, 256 × 256 × 144 points, 1 × 1 × 1 mm resolution; and either a double-spin echo proton density (PD) and T2-weighted sequence or a PD-only sequence. The double spin-echo sequence used TR/TE1/TE2 = 4200/20/80 ms, 256 × 192 × 44 acquisition, 3-mm slice thickness, and the PD-only sequence used spin-echo acquisition with 4200/8 ms, 128 × 128 × 44 points, and 3-mm slice thickness.
The MRSI acquisition used a volumetric echo-planar sequence, with 50 × 50 × 18 k-space points, and 500 spectral sample points after removal of the ×4 oversampling prior to Fourier reconstruction. The resultant MRSI data covered a spatial region of 280 × 280 × 180 mm3 and had a spectral sweep width of 1250 Hz. The sequence used chemical shift-selective (CHESS) water suppression, lipid inversion nulling with TI = 198 ms, an excitation angle of 73°, TR = 1710 ms, spin-echo acquisition with TE = 70 ms, and included an interleaved water reference acquisition obtained using a gradient-echo acquisition with 20° excitation angle and TE = 6.3 ms. The reconstructed MRIs and acquired multichannel MRSI data were copied to local computers for further processing.
Over the course of this study the MR instrument received two system upgrades and the data obtained with each configuration were maintained under separate projects. Prior to the onset of the final analysis, a comparison was made between each of the study groups that indicated no detectable systematic differences. All data were therefore combined for the following analyses.
Data Processing
All data were processed using the fully automated MRSI processing pipeline provided by the MIDAS software package, which was previously described (20). This included lipid k-space extrapolation, spectral lineshape and B0 correction, and parametric spectral analysis using Gaussian lineshape for fitting signals from N-acetylaspartate (NAA), total creatine (Cre), and total choline (Cho). Gaussian line-broadening of 2 Hz was applied prior to spectral fitting, and voxels were excluded from the spectral fitting if the linewidth of the water spectroscopic image (SI) signal at the corresponding voxel exceeded 15 Hz. The metabolite images were reconstructed to 64 × 64 × 32 points with the nominal voxel volume of 0.31 ml that was increased to approximately 1 ml following spatial smoothing. Modifications from the earlier description include combination of multichannel data using phase and amplitude maps generated from the water-reference SI; derivation of the mask for lipid k-space extrapolation from the coregistered MRI data; exclusion of voxels for spectral fitting based on the water-reference linewidth; and signal normalization of the reconstructed metabolite maps was based on the tissue water signal derived from the interleaved water-reference MRSI. The signal normalization procedure (21) used tissue water as an internal reference, which has been widely used for single-voxel MRS measurements and also applied to MRSI (22,23). Knowledge of the tissue water distribution was obtained by convolution of the MRI-derived tissue segmentations to the SI spatial response function and using calculation of the water content for gray matter (GM) and white matter (WM), which was derived using the PD MRI. This procedure then derived a 100% water-equivalent reference image that corrected for the variable receiver sensitivity function and normalized the metabolite images. The resultant individual metabolite images therefore represent the metabolite signal obtained following spectral fitting relative to a reference signal equivalent to that of 100% water at the same voxel location. The signal normalization procedure included an estimate on the water T1 based on previous reports (24,25), but did not account for metabolite relaxation rates. MRI tissue segmentation used the FSL/FAST program (26,27) with the T1 image only.
A nonlinear spatial transform (28,29) was applied to all signal-normalized metabolite images, metabolite ratio images, and additional images reflecting quality criteria of the spectral analysis to enable voxel-based image analysis. The BrainWeb simulated MRI from the Montreal Neurological Institute (30) was used as the spatial reference, which was associated with a brain atlas that identified nine anatomical regions defining the left and right cerebral lobes and the cerebellum. Spatial transformation included interpolation to 2-mm isotropic voxels.
Data Analysis
All data were analyzed using the project review module (PRANA) in the MIDAS package with additional statistical analysis following export into Excel (Microsoft) and SAS (SAS Institute) and image display using MRIcro (www.mricro.com). Processing and voxel selection options included exclusion of voxels with a linewidth result from the spectral fitting outside of a specified range, which was set to 3 Hz and 13 Hz (0.1 ppm). Voxels with outlying data values were also excluded using a threshold based on the mean value and standard deviation (SD) of all voxels within the selection, with values ≥3 times the SD away from the mean being omitted from the analysis. For voxel-based analyses performed across all subjects, valid data from a minimum of 10 subjects were required at a voxel location to generate a result.
A correction for signal loss due to cerebral spinal fluid (CSF) partial volume was applied to all individual metabolite images. The amount of correction was limited to CSF fractions within the SI voxel of less than 0.2, and no correction was applied for CSF fractions above this value. This threshold level was found to apply full correction for voxels within the bulk of the cerebrum, whereas increasing the threshold level resulted in an apparent overcorrection and an edge-enhancement effect at the edges of the ventricles. Average metabolite values corresponding to 100% GM and 100% WM within each brain region were obtained by regression of each parameter against the tissue content determined from the SI-resolution tissue segmentation images, for all voxels over each of the atlas-defined regions. For regressions using the individual metabolite images, a comparison was first made between using a one-parameter regression of WM content against the CSF-corrected metabolite image, and using a two-parameter regression for WM and CSF against the uncorrected metabolite image. This indicated no significant differences between the two types of analyses, and the one-parameter regression was subsequently used because the processing time was slightly faster.
Image-based analyses were carried out to determine mean value and SD over the subject group at each voxel location for each metabolite. Since changes of metabolite concentrations are known to occur with age, this analysis was performed on a subset of the subject group that was limited to an age range of 18–30 years.
For evaluation of changes in metabolite distributions with age, two approaches were used. The first approach was to perform tissue regressions for each brain region defined in the atlas to obtain the average metabolite values corresponding to 100% WM and GM within each brain region, and then to examine the variation of these parameters as a function of age and gender. The second was an image-based analysis that used a regression of the metabolite value at each voxel across all subjects following spatial smoothing of each subject image with 10-mm full-width at half-maximum (FWHM) Gaussian. Additional tests were carried out to examine the significance of differences between the left and right hemispheres for each brain region.
RESULTS AND DISCUSSION
Spectroscopic Image Observations
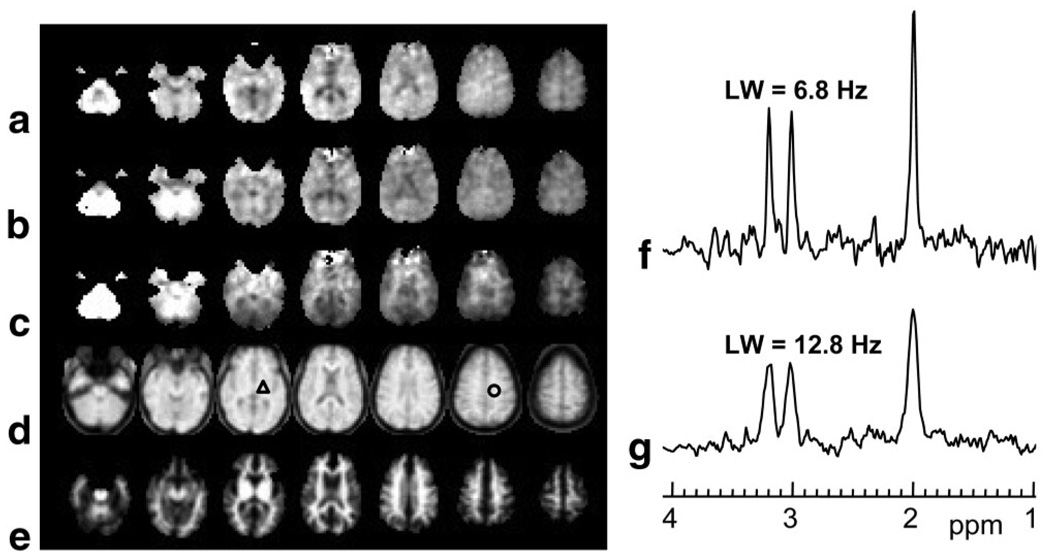
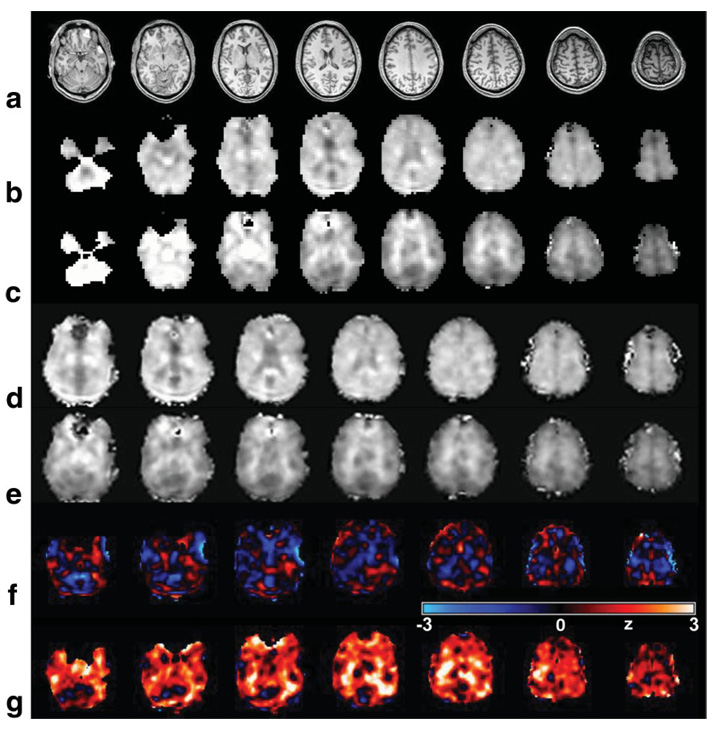
In Fig. 1 are shown examples of the image quality obtained from a single-subject study, together with two sample spectra showing the quality for a linewidth typical of the narrowest obtained to the broadest accepted for inclusion in the subsequent analyses. These spectra reflect the applied line-broadening of 2 Hz, and are shown following intensity normalization and with fixed scaling, which also illustrates the varying noise level due to the variation of the phased-array coil sensitivity. As shown in Fig. 1a the metabolite images (top three rows) commonly exhibited moderate noise patterns and a small number of missing or outlying voxel amplitudes. Larger regions with no metabolite value, e.g., in the fronto-orbital regions, indicate those regions where the water resonance linewidth exceed the 15-Hz threshold, and were excluded from the spectral analysis. A small number of other clearly incorrect results from the spectral fitting occur in regions that passed the water linewidth threshold, but may have reduced quality due to B0 inhomogeneity or unsuppressed water. While the quality of the spectral analysis at individual voxels can be improved by repeating the fitting with a larger number of iterations, this required manual operation and was not performed in this study. Further analysis indicated that voxels with outlying data values occurred in regions of increased inhomogeneity or large CSF volume contribution, and that these were effectively excluded from the subsequent data analyses by the spectral linewidth exclusion test and outlier removal. Also shown in Fig. 1 are the images obtained from the interleaved SI acquisition and one of the SI-resolution tissue maps, which is included to illustrate the availability of this information. This latter image was derived from the WM result of the MRI tissue segmentation and convolved to match the SI spatial response function. The water-reference SI data exhibit T1 weighting with sufficient structural information to enable affine registration with the high-resolution T1-weighted MRI. Note that these last two image sets show values for all voxels within the brain, whereas the metabolite images do not.
FIG. 1.
Representative images and spectra for a single-subject study. The figure shows every second slice from the volumetric SI-resolution data for (a) NAA, (b) Cre, (c) Cho, (d) the water-reference, and (e) the WM segmentation at the MRSI spatial resolution. f,g: Example spectra for a typical narrow linewidth and at the broadest linewidth accepted for further analysis, obtained from the regions indicated by the triangle and circle, respectively.
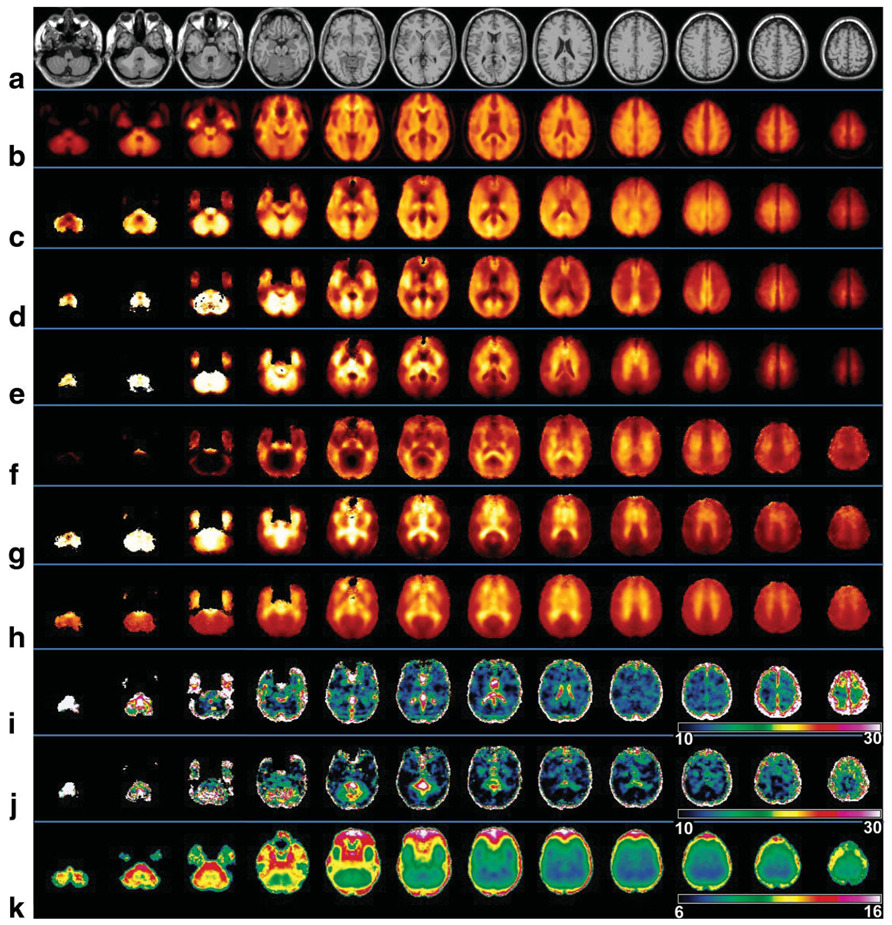
In Fig. 2 are shown images obtained following analysis of multiple subject data, together with the spatial reference image (Fig. 2a), which shows the slice corresponding to the center of the SI resolution images. For the sole purpose of creating the mean-value images for display in Fig. 2, image data from the whole subject group were used since this made available more voxels at the edges of the brain, thereby slightly improving the appearance of the mean-value image. All of the mean-value images for the individual metabolites (Fig. 2c–e) and the metabolite ratio images (Fig. 2f–h) show a wide range of image intensity that benefits from display using a color scale. Also shown in Fig. 2i and j are the results of the variance calculations over the 18–30 age group for NAA and NAA/Cre, respectively. These show a variance over most of the central brain in the range of 10–14%, with higher values in the neighborhood of the ventricles and edges of the brain. Taking an average value over a central WM region, the intrasubject coefficients of variance (COVs) for NAA, Cre, and Cho were found to be 9%, 10%, and 12%, respectively. For the ratio images average COVs for NAA/Cre, Cho/Cre, and Cho/NAA were 8%, 11%, and 13%, respectively. The average linewidth image obtained over all subjects, shown in Fig. 2k, indicates that a large volume of the brain is consistently obtained with a linewidth of under 9 Hz.
FIG. 2.
Selected axial slices at 10-mm spacing following the voxel-based group analysis. Shown are (a) the spatial-reference MRI, (b) mean water-reference SI, (c) mean NAA, (d) mean Cre, (e) mean Cho, (f) NAA/Cre, (g) Cho/NAA, (h) Cho/Cre, (i) COV for NAA (%), (j) COV for NAA/Cre (%), and (k) mean linewidth (Hz). The images shown for i and j were obtained for the 18–30-year age group.
Examination of the mean metabolite images indicates considerable spatial variation of all individual metabolites. Summary values for each of the atlas-defined brain regions and tissue type are given in Table 1 and Table 2. Table 1 shows the average values over the whole subject group for each region from the right side of the brain, and the ratio between the right and left sides (R/L). For this analysis, the signal-normalized and CSF-corrected metabolite values were first obtained by linear regression over all voxels within each brain regions against the WM content. The significance of the difference between right and left cerebral hemispheres was tested using a two-tailed paired t-test. Significant right-left differences are evident for most values and regions, ranging to as much as 10% for temporal GM NAA. This analysis was repeated to test for the effects of gender and age, but this was not found to alter the number of significant lateralization differences.
Table 1.
Average Metabolite Values (in Institutional Units) and Metabolite Ratios Over All Subjects, for Gray Matter and White Matter
| Frontala | Temporala | Parietala | Occipitala | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Mean ± SE | R/L | Mean ± SE | R/L | Mean ± SE | R/L | Mean ± SE | R/L | Cerebellum (mean)b |
|
| Gray matter | All voxels | ||||||||
| NAA | 4523 ± 59 | 0.99* | 4942 ± 83 | 0.91* | 4356 ± 65 | 0.98* | 4948 ± 75 | 0.98 | 5057 |
| Cre | 3145 ± 42 | 0.97* | 3429 ± 59 | 0.93* | 3071 ± 43 | 0.95* | 3461 ± 53 | 1.01 | 4496 |
| Cho | 679 ± 12 | 0.99 | 700 ± 14 | 0.96* | 492 ± 8 | 0.96* | 544 ± 13 | 1.10* | 1054 |
| NAA/Cre | 1.44 ± 0.01 | 1.02* | 1.40 ± 0.012 | 0.98* | 1.42 ± 0.01 | 0.97* | 1.40 ± 0.013 | 0.96* | 1.03 |
| Cho/Cre | 0.22 ± 0.002 | 1.02* | 0.20 ± 0.002 | 1.05* | 0.16 ± 0.002 | 1.02** | 0.15 ± 0.002 | 1.09* | 0.21 |
| Cho/NAA | 0.15 ± 0.002 | 1.01 | 0.14 ± 0.002 | 1.08* | 0.12 ± 0.002 | 1.05* | 0.11 ± 0.002 | 1.13* | 0.20 |
| White matter | |||||||||
| NAA | 4857 ± 67 | 1.00 | 4941 ± 71 | 1.01 | 5053 ± 69 | 0.98* | 5165 ± 81 | 0.92* | — |
| Cre | 2936 ± 42 | 1.00 | 2903 ± 44 | 1.01 | 2985 ± 42 | 0.98* | 3048 ± 48 | 0.92* | — |
| Cho | 858 ± 15 | 0.97* | 838 ± 15 | 0.92* | 824 ± 14 | 0.96* | 671 ± 12 | 0.91* | — |
| NAA/Cre | 1.67 ± 0.014 | 1.00 | 1.75 ± 0.017 | 1.01 | 1.71 ± 0.015 | 1.00 | 1.73 ± 0.016 | 1.00 | — |
| Cho/Cre | 0.29 ± 0.003 | 0.98* | 0.29 ± 0.003 | 0.93* | 0.28 ± 0.003 | 0.98* | 0.22 ± 0.003 | 0.99 | — |
| Cho/NAA | 0.18 ± 0.002 | 0.97* | 0.17 ± 0.002 | 0.91* | 0.16 ± 0.002 | 0.98* | 0.13 ± 0.002 | 0.98 | — |
Mean values and standard errors (SE) for the right side of the atlas-defined brain region are given in the first column, followed by the ratio of the right brain to the left brain (R/L).
Mean values for the cerebellum, without separation into gray matter and white matter.
Significant difference between the left and the right sides for P < 0.01
Significant difference between the left and the right sides for P < 0.05.
Table 2.
Gray Matter to White Matter Ratio, by Brain Region, of Metabolite Values and Ratios of Metabolites for the 18–30-Year Age Group*
| Metabolite | Frontal GM/WM | Temporal GM/WM | Parietal GM/WM | Occipital GM/WM |
|---|---|---|---|---|
| NAA | 1.00 ± 0.08 | 1.11 ± 0.10 | 0.98 ± 0.08 | 1.00 ± 0.11 |
| Creatine | 1.17 ± 0.08 | 1.34 ± 0.13 | 1.17 ± 0.09 | 1.20 ± 0.15 |
| Choline | 0.87 ± 0.09 | 0.90 ± 0.14 | 0.67 ± 0.08 | 0.83 ± 0.17 |
| NAA/Cre | 0.85 ± 0.05 | 0.79 ± 0.07 | 0.81 ± 0.05 | 0.79 ± 0.07 |
| Cho/Cre | 0.73 ± 0.07 | 0.64 ± 0.11 | 0.56 ± 0.07 | 0.67 ± 0.12 |
| Cho/NAA | 0.88 ± 0.08 | 0.82 ± 0.13 | 0.71 ± 0.08 | 0.89 ± 0.20 |
Values are given as mean ± SD of the ratio for all voxels in each brain region.
Of the single metabolite images, NAA (Fig. 2c) exhibits the smallest average GM/WM contrast ratio, but with distinct local changes as well as a trend to increasing values from the anterior to posterior cerebrum. Also visible is increased intensity along the cortical-spinal tracts, corona radiata, and internal capsule (WM structures). In the corpus callosum, there is lower NAA in the genu (anterior corpus callosum) in comparison to the splenium (posterior corpus callosum), consistent with the anterior–posterior trend of increasing values, as previously reported (10). NAA appears to be greater in the insular cortex, which is a structure that overlaps with each of the temporal, frontal, and parietal lobe regions defined in the atlas and was not separately defined in the region-defined analysis.
The average values of the GM/WM signal ratios shown in Table 2 indicate small differences between the average CSF-corrected NAA values in GM and WM. Visual observation of Fig. 2c does give some indication of locally reduced GM NAA (for example, in the upper three slices); however, this may also result from incomplete compensation for CSF partial volume contribution in the image data. Averaged over the whole cerebrum, NAA is 3.7% lower in GM relative to WM. Previous studies have indicated both increased (2,11) and decreased (13) GM/WM NAA concentrations, and it is apparent that the location from which the data were obtained may account for these different findings, together with differences in the correction for CSF volume, acquisition methods, and tissue-dependent spin relaxation times.
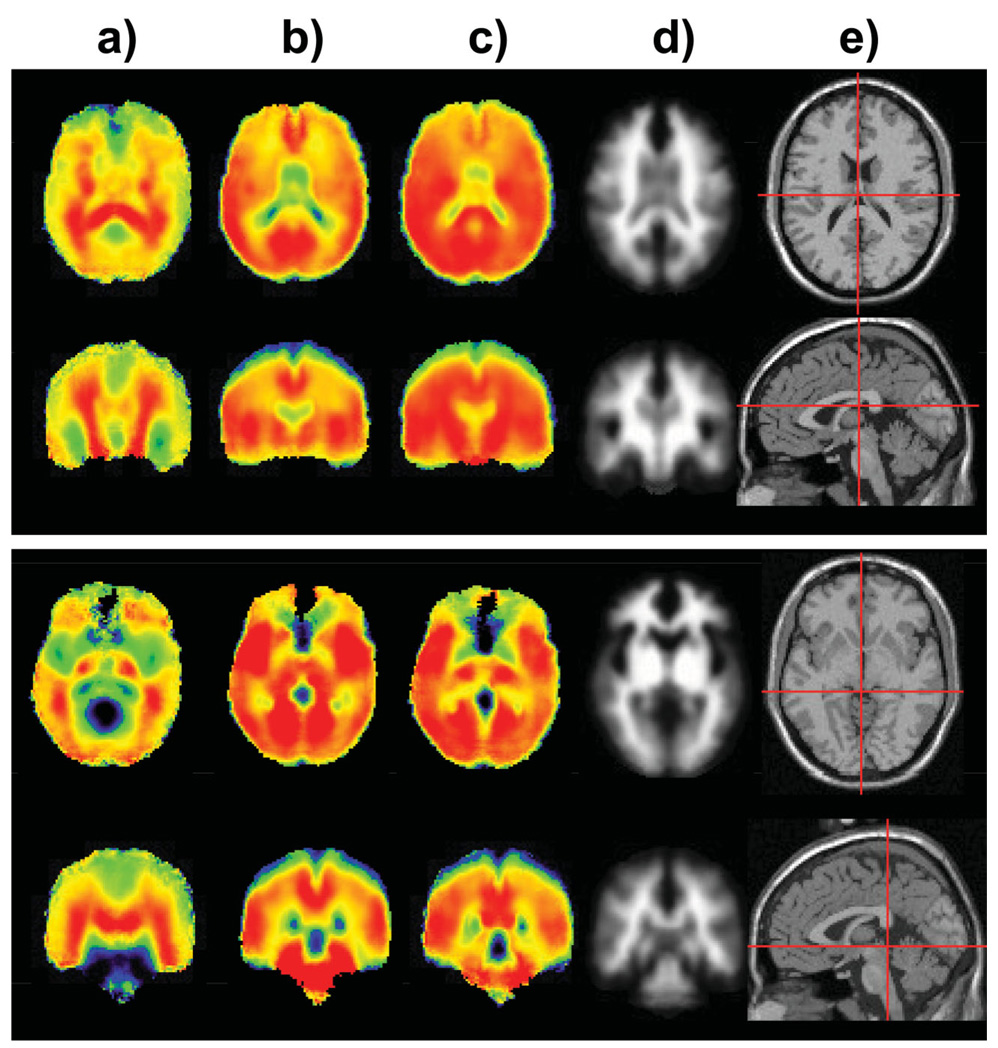
Both the Cre and Cho images show greater GM/WM differentiation, which is also indicated in the results of the tissue-regression analysis shown in Table 2, which was obtained using data from the 18–30 age group. These results indicate that GM/WM ratios differ across regions, and that there are locally increased values, such as in the region of the insula, the central gyrus, and in the cerebellum, in agreement with a previous report (31). Cre was significantly decreased in the temporal lobe WM as compared to the WM in other regions. Averaged over the whole cerebrum, GM Cre is 13% higher and Cho is 21% lower relative to WM. The GM/WM differences are enhanced in the ratio images, with differences reaching as much as 44% for parietal GM in comparison to the WM in that region. As additional examples of the features identified from the metabolite distributions, images in axial and coronal orientation are shown in Fig. 3. Here, the NAA/Cre image (Fig. 3a) highlights the WM distributions and the NAA image (Fig. 3c) also illustrates the anterior–posterior increase, as previously described.
FIG. 3.
Selected axial and coronal sections of the mean images for (a) NAA/Cre, (b) Cre, (c) NAA, (d) WM, and (e) the reference MRI in axial and sagittal sections showing the slice selection.
The magnitude of local changes evident in these images will be underestimated in the mean value results in Table 1 since the atlas-defined brain regions are relatively large. Similarly, the relative GM and WM ratios shown in Table 2 also represent an average over the selected group, since these vary as a function of age, as shown in a following section.
Age- and Gender-Dependent Changes
Multiple linear regression was used to determine the relation of age and gender to each metabolite and metabolite ratio, for WM and GM in each brain region, and the results are shown in Table 3. For this calculation the average individual metabolite values were first obtained by performing the tissue regression for each brain region in each subject and then examining the linear regression of the values obtained for individual GM and WM tissue against the subject age and gender. The model assumed the same age dependence for male and female. These results indicate a small decrease of NAA with increasing age, although they do not reach significance in any one brain region. Averaged over the whole brain, NAA decreased by 2.3%/decade for GM and 0.5%/decade for WM. The magnitude of the age-dependent changes of Cre and Cho were much larger and greatest in WM, with whole-brain average increases of 3.5%/decade for Cre and 5.8%/decade for Cho. While previous reports of age-related changes have been mixed, these results are consistent with some previous studies (1,3,12) and comparable to a quantitative measurement for frontal lobe GM by Brooks et al. (7) that accounted for metabolite T2 and brain water content. That study found a decrease of 3%/decade for NAA, compared to our result of 2.2%/decade, and increased Cho at 5%/decade, compared to 2%/decade in this study.
Table 3.
Results of Multiple Linear Regression Analysis Showing Metabolite Values and Ratio of Metabolites and Rate of Change in These Values by Age and Difference by Gender
| Metabolite | Type of brain matter |
Brain region | Value ± SEa | % Change per decadeb |
% Difference by between male and femaleb |
R2c |
|---|---|---|---|---|---|---|
| NAA | GM | Frontal | 4642 ± 196 | −2.17 | 2.08 | 0.05 |
| Temporal | 5244 ± 269 | −2.30 | 3.85 | 0.05 | ||
| Parietal | 4564 ± 219 | −2.41 | 4.88 | 0.08 | ||
| Occipital | 4981 ± 244 | −1.91 | 5.71* | 0.07 | ||
| Cerebellum | 4968 ± 268 | −2.67 | 3.27 | 0.05 | ||
| WM | Frontal | 4823 ± 220 | −0.67 | 3.50 | 0.02 | |
| Temporal | 4868 ± 237 | −1.13 | 4.70 | 0.04 | ||
| Parietal | 5090 ± 229 | −0.69 | 2.47 | 0.01 | ||
| Occipital | 5393 ± 264 | −2.00 | 4.57 | 0.06 | ||
| Cerebellum | 5425 ± 398 | 1.74 | 5.15 | 0.02 | ||
| Cre | GM | Frontal | 3111 ± 143 | 1.95 | −0.20 | 0.03 |
| Temporal | 3445 ± 193 | 1.45 | 3.00 | 0.02 | ||
| Parietal | 3053 ± 147 | 1.45 | 2.18 | 0.02 | ||
| Occipital | 3309 ± 174 | 1.10 | 4.97 | 0.04 | ||
| Cerebellum | 4440 ± 341 | −0.39 | 4.95 | 0.01 | ||
| WM | Frontal | 2774 ± 129 | 4.24** | 1.93 | 0.11 | |
| Temporal | 2718 ± 145 | 3.69* | 4.13 | 0.08 | ||
| Parietal | 2851 ± 131 | 4.68** | 0.30 | 0.12 | ||
| Occipital | 3076 ± 161 | 1.93 | 2.27 | 0.03 | ||
| Cerebellum | 4596 ± 502 | 3.21 | −8.64 | 0.03 | ||
| Cho | GM | Frontal | 678 ± 36 | 1.86 | −3.88 | 0.03 |
| Temporal | 711 ± 47 | 0.57 | −0.69 | 0.00 | ||
| Parietal | 504 ± 26 | 1.42 | −4.40 | 0.03 | ||
| Occipital | 506 ± 41 | −0.75 | 6.47 | 0.02 | ||
| Cerebellum | 1009 ± 98 | −1.57 | −2.73 | 0.01 | ||
| WM | Frontal | 796 ± 45 | 7.27** | 2.03 | 0.17 | |
| Temporal | 807 ± 48 | 6.17** | 1.85 | 0.12 | ||
| Parietal | 778 ± 43 | 6.55** | 1.10 | 0.15 | ||
| Occipital | 675 ± 38 | 3.72* | −1.07 | 0.06 | ||
| Cerebellum | 1318 ± 134 | 5.41 | −10.02 | 0.06 | ||
| Cho/Cre | GM | Frontal | 0.217 ± 0.008 | 0.19 | −3.64 | 0.03 |
| Temporal | 0.198 ± 0.007 | −1.18 | −4.04 | 0.06 | ||
| Parietal | 0.164 ± 0.006 | 0.58 | −5.81* | 0.07 | ||
| Occipital | 0.147 ± 0.007 | −1.48 | 0.09 | 0.02 | ||
| Cerebellum | 0.186 ± 0.011 | −1.22 | −0.83 | 0.01 | ||
| WM | Frontal | 0.290 ± 0.009 | 2.20* | −0.40 | 0.07 | |
| Temporal | 0.300 ± 0.010 | 2.14* | −2.19 | 0.07 | ||
| Parietal | 0.278 ± 0.010 | 1.37 | 0.17 | 0.02 | ||
| Occipital | 0.222 ± 0.009 | 1.60 | −2.93 | 0.04 | ||
| Cerebellum | 0.285 ± 0.023 | 1.22 | −6.73 | 0.02 | ||
| Cho/NAA | GM | Frontal | 0.148 ± 0.005 | 5.13** | −7.04** | 0.31 |
| Temporal | 0.137 ± 0.005 | 3.35** | −5.31* | 0.14 | ||
| Parietal | 0.114 ± 0.004 | 4.83** | −10.11** | 0.30 | ||
| Occipital | 0.104 ± 0.007 | 2.08 | −0.08 | 0.02 | ||
| Cerebellum | 0.187 ± 0.014 | 2.08 | −6.27 | 0.03 | ||
| WM | Frontal | 0.167 ± 0.006 | 8.03** | −2.69 | 0.36 | |
| Temporal | 0.168 ± 0.006 | 7.68** | −4.60 | 0.34 | ||
| Parietal | 0.154 ± 0.006 | 7.42** | −2.36 | 0.30 | ||
| Occipital | 0.125 ± 0.006 | 6.34** | −6.76* | 0.23 | ||
| Cerebellum | 0.231 ± 0.023 | 6.92* | −14.02* | 0.10 | ||
| NAA/Cr | GM | Frontal | 1.494 ± 0.029 | −3.82** | 2.55* | 0.43 |
| Temporal | 1.474 ± 0.034 | −3.46** | 1.69 | 0.31 | ||
| Parietal | 1.489 ± 0.028 | −3.31** | 3.28** | 0.40 | ||
| Occipital | 1.471 ± 0.041 | −2.61** | 1.27 | 0.14 | ||
| Cerebellum | 0.921 ± 0.074 | −1.09 | 5.09 | 0.02 | ||
| WM | Frontal | 1.750 ± 0.038 | −3.68** | 1.50 | 0.36 | |
| Temporal | 1.826 ± 0.043 | −3.78** | 1.19 | 0.33 | ||
| Parietal | 1.801 ± 0.039 | −4.11** | 1.84 | 0.41 | ||
| Occipital | 1.786 ± 0.043 | −3.53** | 2.82* | 0.31 | ||
| Cerebellum | 1.260 ± 0.116 | −3.15 | 8.32 | 0.05 |
The metabolite values are in “institutional units” and the metabolite ratios are calculated for age 20 years.
Values are shown as percent change per decade and percent difference between male and female subjects.
R2 values represent the coefficient of difference.
Significant at P < 0.05.
Significant at P < 0.01.
The age-dependent changes for individual metabolite values reach statistical significant only for Cre and Cho in three regions, and the gender difference reaches significance only for NAA in occipital GM. For these parameters the coefficient of determination, R2, values are small, indicating that there remains a significant fraction of the intersubject variability that cannot be attributed the factors considered for this model. However, due to the trend of an age-dependent decrease in NAA, and a similar trend of opposite differences between NAA and Cre and Cho between male and female subjects, the age and gender differences in the metabolite ratio values reach significance in more brain regions, although again most R2 values are still relatively small. The gender differences indicate that averaged over all regions there is increased NAA in females relative to males, with an average of 4% for both GM and WM, and a trend to lower Cho values. However, the gender differences were highly variable, for example, ranging from −10% in the cerebellum to 6.5% in occipital GM, and many regions with no significant differences. Similarly, previous reports of gender differences have been mixed. Our results show the same trend to increased frontal NAA as reported by Braun et al. (15), and although our result did not reach significance, the value for frontal GM NAA/Cre did. Similarly, our observation of decreased Cho/NAA in occipital lobe in females is consistent with the report of Wilkinson et al. (14). Other reports have found no differences with gender (8,9,16).
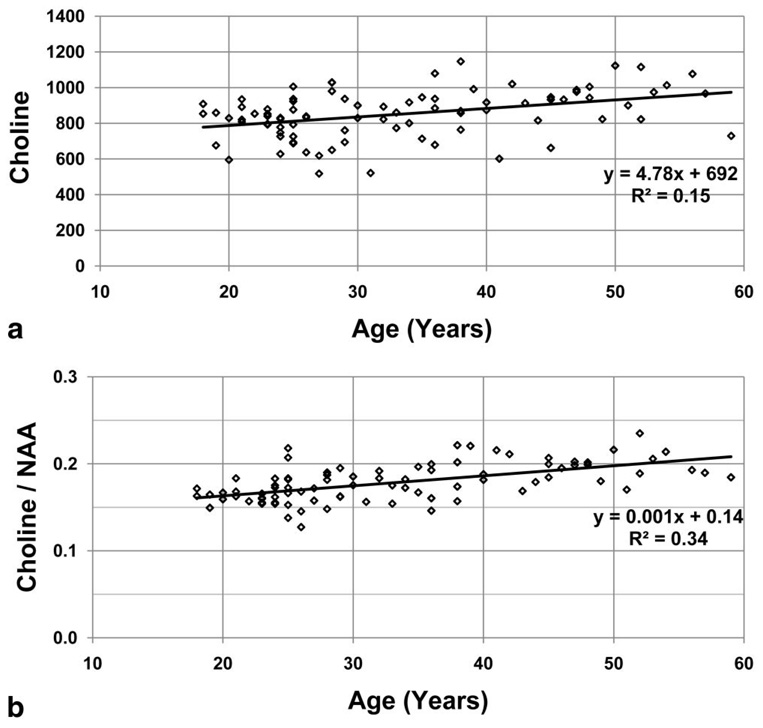
Example regressions for frontal WM Cho and Cho/NAA are shown in Fig. 4. While the increased correlation coefficient for the ratio of two metabolites with opposite slope over the single metabolite measures is expected, there is also some indication of more outlying values in the normalized metabolite values, suggesting that additional variability is caused by the signal normalization to tissue water.
FIG. 4.
Example results for the regression of the average frontal WM Cho in institutional units (a) and Cho/NAA ratio (b) as a function of the subject age. The linear regression equations are shown in the inset with their coefficient of determination, R2.
In Fig. 5 are shown representative slices for the voxel-based regression of the individual metabolites as a function of age. These images are presented with a dual-contrast image scale, with black representing no change, and support the results in Table 3, with the small decrease of NAA, and increases of Cho and Cre that are greatest in WM. The local variability in these images is considered to be within the error of the measurement and there is no evidence of localized changes corresponding to specific brain structures, as was previously indicated in studies using short-TE measurements (4).
FIG. 5.
Selected slices showing the results of the image voxel-based age regression analysis. Shown are (a) the reference MRI, (b) NAA, (c) Cre, and (d) Cho. Images are shown in a dual color scale running from −6%/decade (blue) to +6%/decade (white).
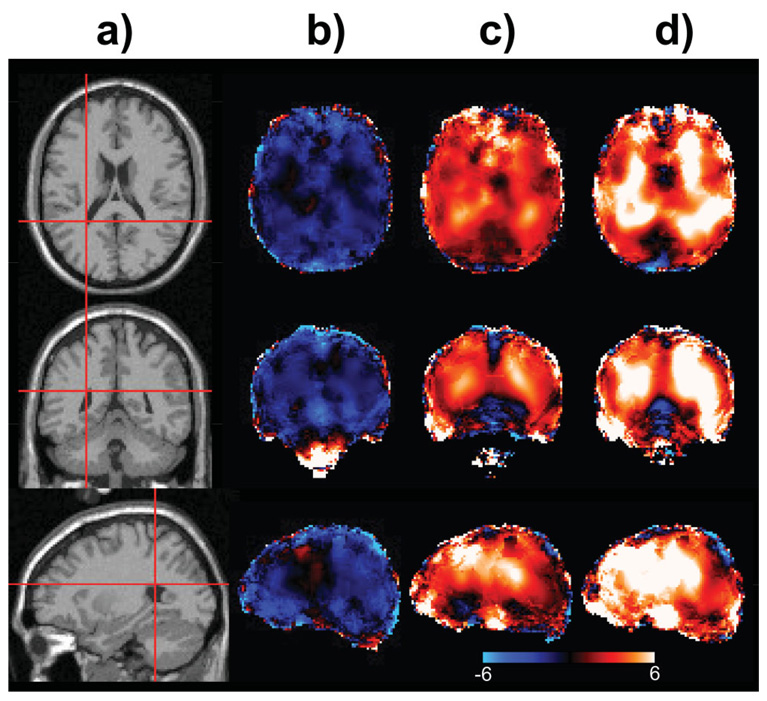
In Fig. 6 are shown metabolite images and image analysis results for the TBI subject study. The T1-weighted MRI (Fig. 6a) indicates the presence of small contusions in left frontal and temporal regions that were associated with a loss of all metabolite signals in those specific regions due to the presence of blood products. A visual analysis of the NAA and Cho images (shown in Fig. 6b and c) throughout the rest of the parenchyma does not indicate any metabolic abnormality, with no indication of any structure within the NAA image and the presence of slightly increased Cho in WM being consistent with normal values (e.g., compare with Fig. 2e). Following spatial transformation (Fig. 6d and e), these metabolite images were then compared against the normal subject values using a z-score analysis that accounted for the GM and WM content in each voxel (32). Voxels with a linewidth greater than 12 Hz were excluded from the analysis. These results show the difference of the metabolite image data from the single TBI subject relative to the mean values for an age-matched group of the control subject data, which consisted of 19 male and 23 female subjects selected for the age range of 20–30 years. The result is then scaled by the SD, σ, of the control group data and presented with a color scale ranging from −3σ to +3σ, with black indicating no change. This analysis indicates some variability of the NAA z-score map (Fig. 6f) that is within an expected range of ±2σ and therefore not considered significant. The Cho z-score result (Fig. 6g) shows a consistent trend of increased values over much of the brain that reaches significantly increased levels of +3σ in some regions. In this example, both the similarity of the NAA image and the difference of the Cho image relative to normal values, over a wide region of the brain, have been indicated using an automated analysis approach. This specific example also demonstrates the utility of the reference metabolite image information for evaluation of diffuse metabolic changes.
FIG. 6.
Example metabolite image analysis based on comparison against normative values for a subject with a TBI. Shown are axial slices at 11-mm intervals from the T1 MRI (a) and metabolite images for NAA (b) and Cho (c) from the acquired study data. These same metabolite images are then shown following spatial transformation to the standard reference frame in d and e at a 10-mm slice interval. The corresponding z-score maps indicating the relative difference from the normal subject reference data are then shown for (f) NAA and (g) Cho. These last two images are shown in a color scale running from −3σ (blue) to +3σ (white).
CONCLUSIONS
A primary finding of this report is the considerable heterogeneity of metabolite concentrations across all regions of the brain, which are additionally modified by tissue type and subject variables of age and gender. This variability means that any MRS analyses that use comparisons between subjects or groups must have the comparative information available for the same region, age, and gender. While the results of this study confirm several previous reports of region-, gender-, and age-dependent changes of brain metabolites, these findings have been extended to a much larger brain region and simultaneously detecting spatial variations of differences attributed to age and gender. The complexity of the metabolite distributions suggested by the images in Fig. 2, together with the regional dependencies of lateralization and changes with age and gender, strongly indicate that the most sensitive method for evaluation of metabolic alterations in single subjects will be a voxel-based comparative analysis between the subject data and a comparison subject group, which must also take into account the tissue content at each voxel.
It is evident that any between-subject comparisons must account for tissue content; however, the relative importance of laterality, age, and gender is less clear. For the experimental methods used in this study, the difference between right and left sides for the same region and gender remained below the across-subject variance for the single metabolite measures, but became comparable to the COV for the metabolite ratios in some regions. Similarly, the changes with age were larger for the metabolite ratios, and for measurements that include Cho, a general recommendation is to limit the comparison of a single subject data set to comparison data obtained from a subject group matched to within ±10 years.
While this study provides evidence for differences of single metabolite values between tissue types and regions, there are several limitations to these findings. Observations of changes for left and right of the same region can be affected by alterations in detection sensitivity that were not completely removed by the signal normalization procedure. Additionally, information on handedness was not available to include in the analysis. The observation of tissue differences and changes with age is also clearly dependent upon the accuracy of the MRI tissue segmentation method, and an additional area for further optimization is undoubtedly in the development of tissue segmentation methods that can accurately quantify CSF volume in the narrow sulcal spaces.
This study also demonstrates the utility of fully automated processing procedures for MRSI studies (20), in contrast to the more typical implementation of MRS processing methods that rely on operator-supervised methods and frequently ad hoc methods of data analysis. By using a rapid volumetric MRSI acquisition at 3T, spectral information was obtained over a large fraction of the brain with a relative intersubject variance that is comparable to similar studies from more limited regions of the brain (33,34) and only slightly larger than single-voxel measurements (35). Furthermore, the availability of the volumetric MRSI data along with the corresponding water reference SI enabled spatial normalization methods to be applied. This, together with the availability of the corresponding reference information, has enabled voxel-based statistical analysis methods to be applied across multiple subjects. Although the final statistical analyses presented in this report still relied on operator-directed software programs, the use of spatial normalization methods means that analyses such as the detection of significant differences between subjects or between different brain regions could also be implemented in an automated manner.
There remain several limitations of the processing methods used. First, the processing procedures still required visual inspection to exclude results degraded by subject motion or indications of normal brain anatomical variations that did not match the reference brain used in this study. Second, the tissue segmentation and spatial normalization methods are optimized for normal-appearing brain, and may fail in subjects with normal variants of brain anatomy. These methods may also fail in the presence of significant pathology, although this was not a concern for this study. Another potential limitation of the implemented processing methods is increased variability introduced by the signal and spatial normalization procedures. The intrasubject variances obtained in this study, for both individual metabolite values and metabolite ratios, are comparable to those reported in single-voxel (36–38) and MRSI studies (23). However, it is of interest to note the similar intersubject variance for the intensity-normalized NAA image in comparison to the NAA/Cre image, indicating that the normalization to tissue water is itself subject to some variability across the subject group, even though the water reference signal clearly has a greater SNR than a metabolite image. These relative COVs also match those of Ratai et al. (39) using water-referenced MRSI at 3T. Using a calibrated spectral analysis approach for a short-TE single-voxel measurement at 1.5T, Schirmer and Auer (36) obtained slightly lower COVs for individual metabolite values than for ratios (e.g., 7.6% for NAA and 8.5% for NAA/Cr), indicating that the use of tissue water as a signal reference may be adding some variability. Given the smaller voxel volume of the MRSI measurement in this study, the variability in the water-referenced metabolite values may simply reflect SNR and normal variance of metabolite concentrations across the subject group, but there are potentially additional sources of variability in the signal normalization method itself. These have also been discussed by Gasparovic et al. (23) and include regional differences in water content, spin-lattice relaxation, and errors in the tissue segmentation procedure. The larger COV values found in this study for Cho, and ratios including Cho, likely reflect increased variability in the spectral analysis due to baseline variations associated with variable water suppression, which also reflects a limitation of the volumetric MRSI approach used.
A primary outcome of this study is the development of a reference database for voxel-based comparative analysis of individual subject MRSI results, which is currently being evaluated for studies of brain injury and disease (40,41). This database is made available on request. As illustrated in Fig. 6, one clear advantage that can be obtained from a comparison of single-subject data against the information obtained from the reference database is for visualization of diffuse metabolic alterations, as opposed to focal changes that may be visualized directly in a metabolite image, which for example may occur with well-localized tumors or following stroke. In all cases, however, the generation of metabolic images that indicate abnormal values facilitates review by personnel who may not have specific expertise with MRS data analysis, and it removes the necessity of viewing individual voxel spectra, which can become time-consuming for review of these large data sets. An additional benefit is the availability of quantitative information on the relative difference from normal values, which may potentially be of benefit for classification of the degree of metabolic abnormality.
ACKNOWLEDGMENTS
Components of the MIDAS software package were developed by Dr. L. Hall, University of South Florida, and Dr. N. Schuff, University of California, San Francisco.
Grant sponsor: National Institutes of Health; Grant numbers: EB000822; NS041946; NS055107.
REFERENCES
- 1.Chang L, Ernst T, Poland RE, Jenden DJ. In vivo proton magnetic resonance spectroscopy of the normal aging human brain. Life Sci. 1996;58:2049–2056. doi: 10.1016/0024-3205(96)00197-x. [DOI] [PubMed] [Google Scholar]
- 2.McLean MA, Woermann FG, Barker GJ, Duncan JS. Quantitative analysis of short echo time 1H-MRSI of cerebral gray and white matter. Magn Reson Med. 2000;44:401–411. doi: 10.1002/1522-2594(200009)44:3<401::aid-mrm10>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- 3.Angelie E, Bonmartin A, Boudraa A, Gonnaud PM, Mallet JJ, Sappey-Marinier D. Regional differences and metabolic changes in normal aging of the human brain: proton MR spectroscopic imaging study. AJNR Am J Neuroradiol. 2001;22:119–127. [PMC free article] [PubMed] [Google Scholar]
- 4.Harada M, Miyoshi H, Otsuka H, Nishitani H, Uno M. Multivariate analysis of regional metabolic differences in normal ageing on localised quantitative proton MR spectroscopy. Neuroradiology. 2001;43:448–452. doi: 10.1007/s002340000513. [DOI] [PubMed] [Google Scholar]
- 5.Schuff N, Ezekiel F, Gamst A, Amend D, Capizzano AA, Maudsley AA, Weiner MW. Region and tissue differences of metabolites in normally aged brain using 1H magnetic resonance spectroscopic imaging. Magn Reson Med. 2001;45:899–907. doi: 10.1002/mrm.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kadota T, Horinouchi T, Kuroda C. Development and aging of the cerebrum: assessment with proton MR spectroscopy. AJNR Am J Neuroradiol. 2001;22:128–135. [PMC free article] [PubMed] [Google Scholar]
- 7.Brooks JC, Roberts N, Kemp GJ, Gosney MA, Lye M, Whitehouse GH. A proton magnetic resonance spectroscopy study of age-related changes in frontal lobe metabolite concentrations. Cereb Cortex. 2001;11:598–605. doi: 10.1093/cercor/11.7.598. [DOI] [PubMed] [Google Scholar]
- 8.Komoroski RA, Heimberg C, Cardwell D, Karson CN. Effects of gender and region on proton MRS of normal human brain. Magn Reson Imaging. 1999;17:427–433. doi: 10.1016/s0730-725x(98)00186-6. [DOI] [PubMed] [Google Scholar]
- 9.Nagae-Poetscher LM, Bonekamp D, Barker PB, Brant LJ, Kaufmann WE, Horska A. Asymmetry and gender effect in functionally lateralized cortical regions: a proton MRS imaging study. J Magn Reson Imaging. 2004;19:27–33. doi: 10.1002/jmri.10429. [DOI] [PubMed] [Google Scholar]
- 10.Degaonkar MN, Pomper MG, Barker PB. Quantitative proton magnetic resonance spectroscopic imaging: regional variations in the corpus callosum and cortical gray matter. J Magn Reson Imaging. 2005;22:175–179. doi: 10.1002/jmri.20353. [DOI] [PubMed] [Google Scholar]
- 11.Helms G. A precise and user-independent quantification technique for regional comparison of single volume proton MR spectroscopy of the human brain. NMR Biomed. 2000;13:398–406. doi: 10.1002/1099-1492(200011)13:7<398::aid-nbm660>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- 12.Pfefferbaum A, Adalsteinsson E, Spielman D, Sullivan EV, Lim KO. In vivo spectroscopic quantification of the N-acetyl moiety, creatine, and choline from large volumes of brain gray and white matter: effects of normal aging. Magn Reson Med. 1999;41:276–284. doi: 10.1002/(sici)1522-2594(199902)41:2<276::aid-mrm10>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 13.Wiedermann D, Schuff N, Matson GB, Soher BJ, Du AT, Maudsley AA, Weiner MW. Short echo time multislice proton magnetic resonance spectroscopic imaging in human brain: metabolite distributions and reliability. Magn Reson Imaging. 2001;19:1073–1080. doi: 10.1016/s0730-725x(01)00441-6. [DOI] [PubMed] [Google Scholar]
- 14.Wilkinson ID, Paley MN, Miszkiel KA, Hall-Craggs MA, Kendall BE, Chinn RJ, Harrison MJ. Cerebral volumes and spectroscopic proton metabolites on MR: is sex important? Magn Reson Imaging. 1997;15:243–248. doi: 10.1016/s0730-725x(96)00334-7. [DOI] [PubMed] [Google Scholar]
- 15.Braun CM, Boulanger Y, Labelle M, Khiat A, Dumont M, Mailloux C. Brain metabolic differences as a function of hemisphere, writing hand preference, and gender. Laterality. 2002;7:97–113. doi: 10.1080/13576500143000212. [DOI] [PubMed] [Google Scholar]
- 16.Jung RE, Haier RJ, Yeo RA, Rowland LM, Petropoulos H, Levine AS, Sibbitt WL, Brooks WM. Sex differences in N-acetylaspartate correlate of general intelligence: an 1H-MRS study of normal human brain. Neuroimage. 2005;26:965–972. doi: 10.1016/j.neuroimage.2005.02.039. [DOI] [PubMed] [Google Scholar]
- 17.Durazzo TC, Meyerhoff DJ. Neurobiological and neurocognitive effects of chronic cigarette smoking and alcoholism. Front Biosci. 2007;12:4079–4100. doi: 10.2741/2373. [DOI] [PubMed] [Google Scholar]
- 18.Soher BJ, van Zijl PC, Duyn JH, Barker PB. Quantitative proton MR spectroscopic imaging of the human brain. Magn Reson Med. 1996;35:356–363. doi: 10.1002/mrm.1910350313. [DOI] [PubMed] [Google Scholar]
- 19.Kreis R, Ernst T, Ross BD. Absolute quantitation of water and metabo-lites in the human brain. II. Metabolite concentrations. J Magn Reson B. 1993;102:9–19. [Google Scholar]
- 20.Maudsley AA, Darkazanli A, Alger JR, Hall LO, Schuff N, Studholme C, Yu Y, Ebel A, Frew A, Goldgof D, Gu Y, Pagare R, Rousseau F, Siva-sankaran K, Soher BJ, Weber P, Young K, Zhu X. Comprehensive processing, display and analysis for in vivo MR spectroscopic imaging. NMR Biomed. 2006;19:492–503. doi: 10.1002/nbm.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maudsley AA, Domenig C. Signal normalization for MR spectroscopic imaging using an interleaved water-reference; Proceedings of the 16th Annual Meeting of ISMRM; Toronto, Ontario, Canada. 2008. (Abstract 1613) [Google Scholar]
- 22.Alger JR, Symko SC, Bizzi A, Posse S, DesPres DJ, Armstrong MR. Absolute quantitation of short TE brain 1H-MR spectra and spectroscopic imaging data. J Comput Assist Tomogr. 1993;17:191–199. doi: 10.1097/00004728-199303000-00006. [DOI] [PubMed] [Google Scholar]
- 23.Gasparovic C, Song T, Devier D, Bockholt HJ, Caprihan A, Mullins PG, Posse S, JungC RE, Morrison LA. Use of tissue water as a concentration reference for proton spectroscopic imaging. Magn Reson Med. 2006;55:1219–1226. doi: 10.1002/mrm.20901. [DOI] [PubMed] [Google Scholar]
- 24.Wansapura JP, Holland SK, Dunn RS, Ball WSJ. NMR Relaxation times in the human brain at 3.0 Tesla. J Magn Reson Imaging. 1999;9:531–538. doi: 10.1002/(sici)1522-2586(199904)9:4<531::aid-jmri4>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 25.Rooney WD, Johnson G, Li X, Cohen ER, Kim SG, Ugurbil K, Springer CS., Jr Magnetic field and tissue dependencies of human brain longitudinal H2O relaxation in vivo. Magn Reson Med. 2007;57:308–318. doi: 10.1002/mrm.21122. [DOI] [PubMed] [Google Scholar]
- 26.Smith SM, Jenkinson M, Woolrich MW, Beckmann CF, Behrens TE, Johansen-Berg H, Bannister PR, De Luca M, Drobnjak I, Flitney DE, Niazy RK, Saunders J, Vickers J, Zhang Y, De Stefano N, Brady JM, Matthews PM. Advances in functional and structural MR image analysis and implementation as FSL. Neuroimage. 2004;23 (Suppl 1):S208–S219. doi: 10.1016/j.neuroimage.2004.07.051. [DOI] [PubMed] [Google Scholar]
- 27.Zhang Y, Brady M, Smith S. Segmentation of brain MR images through a hidden Markov random field model and the expectation-maximization algorithm. IEEE Trans Med Imaging. 2001;20:45–57. doi: 10.1109/42.906424. [DOI] [PubMed] [Google Scholar]
- 28.Studholme C, Hill DL, Hawkes DJ. Automated three-dimensional registration of magnetic resonance and positron emission tomography brain images by multiresolution optimization of voxel similarity measures. Med Phys. 1997;24:25–35. doi: 10.1118/1.598130. [DOI] [PubMed] [Google Scholar]
- 29.Rousseau F, Maudsley AA, Ebel A, Darkazanli A, Weber P, Sivasankaran K, Yu Y, Studholme C. Evaluation of sub-voxel registration accuracy between MRI and 3D MR spectroscopy of the brain. Proc Soc Photo Opt Instrum Eng. 2005;5747:1213–1221. [Google Scholar]
- 30.Collins DL, Zijdenbos AP, Kollokian V, Sled JG, Kabani NJ, Holmes CJ, Evans AC. Design and construction of a realistic digital brain phantom. IEEE Trans Med Imaging. 1998;17:463–468. doi: 10.1109/42.712135. [DOI] [PubMed] [Google Scholar]
- 31.Jacobs MA, Horska A, van Zijl PC, Barker PB. Quantitative proton MR spectroscopic imaging of normal human cerebellum and brain stem. Magn Reson Med. 2001;46:699–705. doi: 10.1002/mrm.1248. [DOI] [PubMed] [Google Scholar]
- 32.Maudsley AA, Studholme C, Govindaraju V. Tissue-dependent analysis of metabolic alterations in the brain by MR spectroscopic imaging; Proceedings of the 16th Annual Meeting of ISMRM; Toronto, Ontario, Canada. 2008. (Abstract 1604) [Google Scholar]
- 33.DeVito TJ, Nicolson Y, Bureau Y, Williamson PC, Drost DJ. Reproducibility of multi-slice 2D proton MRSI at 3.0T; Proceedings of the 12th Annual Meeting of ISMRM; Kyoto, Japan. 2004. (Abstract 2459) [Google Scholar]
- 34.Inglese M, Spindler M, Babb JS, Sunenshine P, Law M, Gonen O. Field, coil, and echo-time influence on sensitivity and reproducibility of brain proton MR spectroscopy. AJNR Am J Neuroradiol. 2006;27:684–688. [PMC free article] [PubMed] [Google Scholar]
- 35.Komoroski RA, Kotrla KJ, Lemen L, Lindquist D, Diaz P, Foundas A. Brain metabolite concentration ratios in vivo: multisite reproducibility by single-voxel 1H MR spectroscopy. Magn Reson Imaging. 2004;22:721–725. doi: 10.1016/j.mri.2003.12.001. [DOI] [PubMed] [Google Scholar]
- 36.Schirmer T, Auer DP. On the reliability of quantitative clinical magnetic resonance spectroscopy of the human brain. NMR Biomed. 2000;13:28–36. doi: 10.1002/(sici)1099-1492(200002)13:1<28::aid-nbm606>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 37.Knight-Scott J, Haley AP, Rossmiller SR, Farace E, Mai VM, Christopher JM, Manning CA, Simnad VI, Siragy HM. Molality as a unit of measure for expressing 1H MRS brain metabolite concentrations in vivo. Magn Reson Imaging. 2003;21:787–797. doi: 10.1016/s0730-725x(03)00179-6. [DOI] [PubMed] [Google Scholar]
- 38.Safriel Y, Pol-Rodriguez M, Novotny EJ, Rothman DL, Fulbright RK. Reference values for long echo time MR spectroscopy in healthy adults. AJNR Am J Neuroradiol. 2005;26:1439–1445. [PMC free article] [PubMed] [Google Scholar]
- 39.Ratai EM, Hancu I, Blezek DJ, Turk KW, Halpern E, Gonzalez RG. Automatic repositioning of MRSI voxels in longitudinal studies: impact on reproducibility of metabolite concentration measurements. J Magn Reson Imaging. 2008;27:1188–1193. doi: 10.1002/jmri.21365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maudsley AA, Govindaraju V, Sharma K. Image-based analysis of metabolic alterations with ALS; Proceedings of the 16th Annual Meeting of ISMRM; Toronto, Ontario, Canada. 2008. (Abstract 2212) [Google Scholar]
- 41.Govindaraju V, Jagid J, Falcone S, Maudsley AA. Volumetric MRSI of metabolic alterations with traumatic brain injury; Proceedings of the 16th Annual Meeting of ISMRM; Toronto, Ontario, Canada. 2008. (Abstract 2274) [Google Scholar]