Abstract
Multiple, complex molecular events characterize cancer development and progression1,2. Deciphering the molecular networks that distinguish organ-confined disease from metastatic disease may lead to the identification of critical biomarkers for cancer invasion and disease aggressiveness. Although gene and protein expression have been extensively profiled in human tumors, little is known about the global metabolomic alterations that characterize neoplastic progression. Using a combination of high throughput liquid and gas chromatography-based mass spectrometry, we profiled more than 1126 metabolites across 262 clinical samples related to prostate cancer (42 tissues and 110 each of urine and plasma). These unbiased metabolomic profiles were able to distinguish benign prostate, clinically localized prostate cancer, and metastatic disease. Sarcosine, an N-methyl derivative of the amino acid glycine, was identified as a differential metabolite that was highly elevated during prostate cancer progression to metastasis and can be detected non-invasively in urine. Sarcosine levels were also elevated in invasive prostate cancer cell lines relative to benign prostate epithelial cells. Knockdown of glycine-N-methyl transferase (GNMT), the enzyme that generates sarcosine from glycine, attenuated prostate cancer invasion. Addition of exogenous sarcosine or knockdown of the enzyme that leads to sarcosine degradation, sarcosine dehydrogenase (SARDH), induced an invasive phenotype in benign prostate epithelial cells. Androgen receptor and the ERG gene fusion product coordinately regulate components of the sarcosine pathway. Taken together, we profiled the metabolomic alterations of prostate cancer progression revealing sarcosine as a potentially important metabolic intermediary of cancer cell invasion and aggressivity.
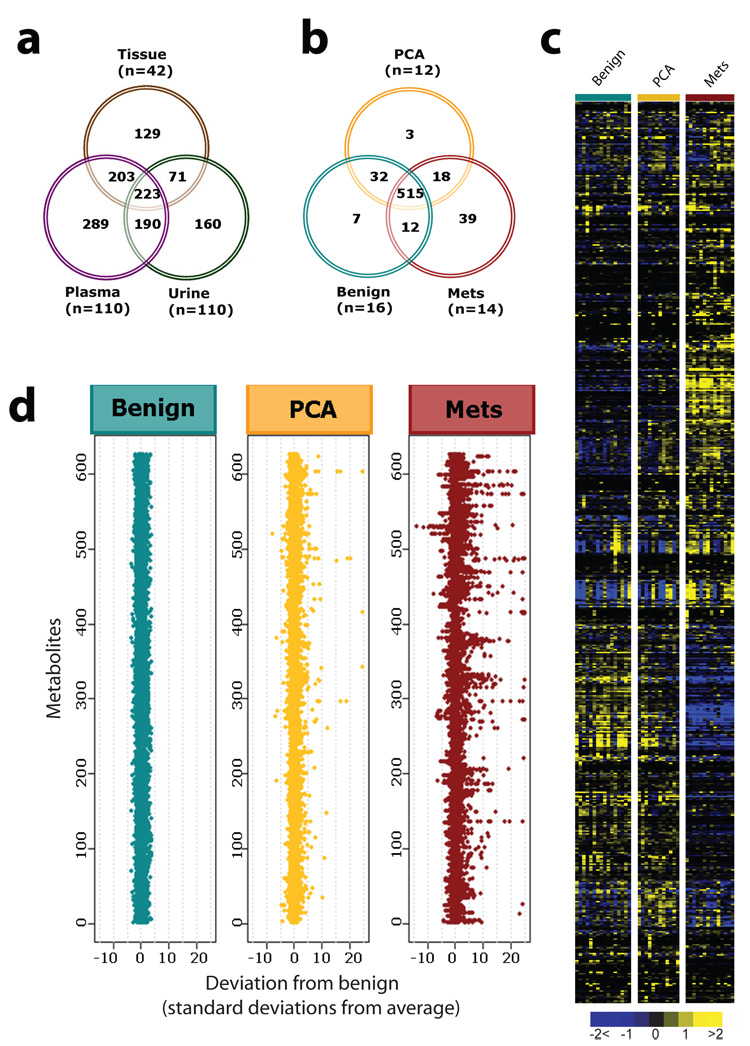
To profile the “metabolome” during prostate cancer progression, we used both liquid and gas chromatography coupled with mass spectrometry3 to interrogate the relative levels of metabolites across 262 prostate-related biospecimens (outlined in Supplementary Fig. 1). Specifically 42 tissue samples and 110 matched specimens of plasma and post-digital rectal exam (DRE) urine from biopsy positive cancer patients (n=59) and biopsy negative control individuals (n=51) were assayed (Fig. 1a). A total of 1126 metabolites were quantified and as expected only a small percentage of these metabolites (10.6%) were shared across the disparate biospecimen types (Fig. 1a).
Figure 1. Metabolomic profiling of prostate cancer.
a, Venn diagram of the total metabolites detected across 42 prostate-related tissues and 110 matched plasma and urine samples. b, Venn diagram of 626 metabolites in tissues measured across 16 benign adjacent prostate, 12 clinically localized prostate cancers (PCA), and 14 metastatic prostate cancers (Mets). c, Heat map representation of unsupervised hierarchical clustering of b (rows) grouped by sample type (columns). Shades of yellow and blue represent elevation and decrease of a metabolite respectively relative to the median metabolite levels (see color scale). d, Z-score plots for b normalized to the mean of the benign prostate samples (truncated at 25 SD for clarity, see Supplementary Methods for procedural details).
Evaluation of the unbiased metabolomic profiles of plasma or urine did not identify robust differences between biopsy positive and biopsy negative individuals. For plasma, 19 of 478 (4%) metabolites were differential (Wilcoxon P < 0.05) with a false discovery rate (FDR) of 99%. Likewise, for urine 34 of 583 (6%) metabolites were differential (Wilcoxon P < 0.05) with an FDR of 64%. Thus, our initial focus was directed towards understanding the tissue metabolomic profiles as they exhibited more robust alterations.
Tissue samples were derived from benign adjacent prostate (n=16), clinically localized prostate cancer (n=12, PCA) and metastatic prostate cancer patients (n=14, Mets). Selection of metastatic tissue samples from different sites (see Supplementary Table 2) minimized characterization of analytes specific to cells of non-prostatic origin. In total, high throughput profiling of the tissues quantitatively detected 626 metabolites (175 named, 19 isobars, and 432 metabolites without identification), of which 82.3% (515/626) were shared by the three diagnostic classes (Fig. 1b). Notably, there were 60 metabolites found in PCA and/or metastatic tumors but not in benign prostate. These profiles were displayed as a heat map (Fig.1c) and z-score plot (Fig. 1d). In the latter benign-based z-scores were plotted for each of the 626 metabolites. The plots revealed robust metabolic alterations in metastatic tumors (z-score range: −13.6 to 81.9) compared to fewer changes in clinically localized prostate cancer samples (z-score range: −7.7 to 45.8).
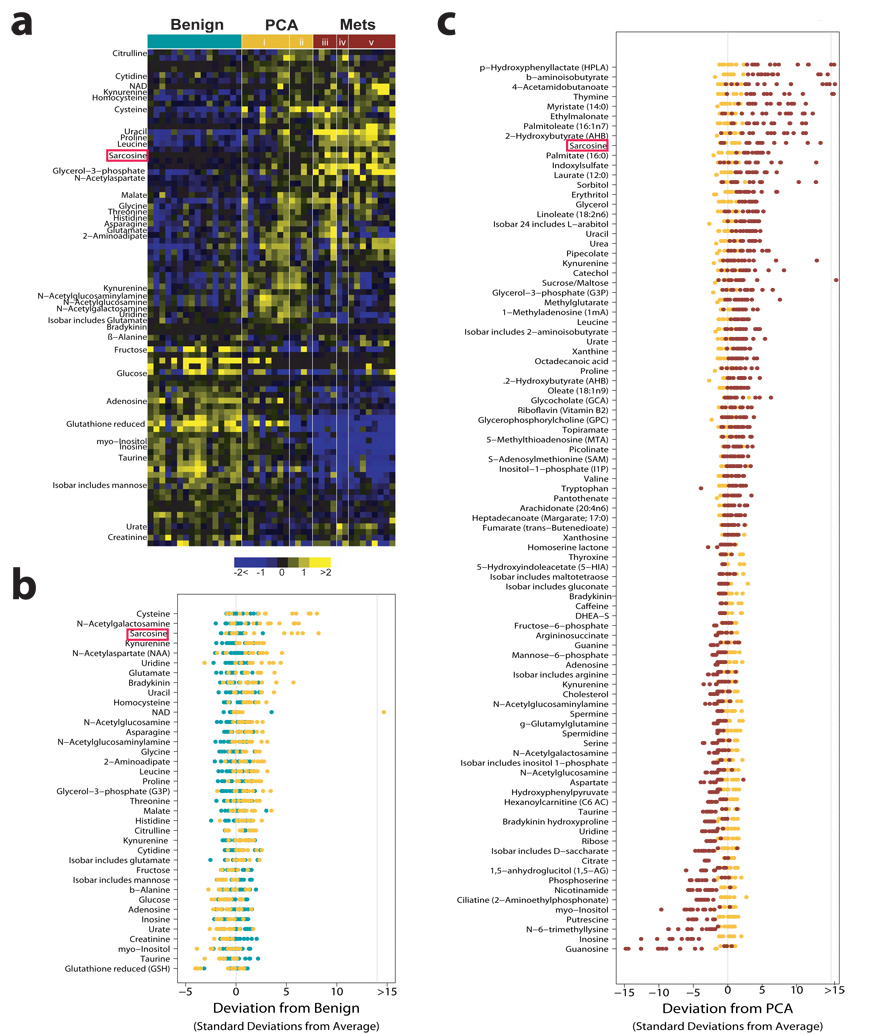
We identified the differential metabolites between the PCA and benign samples using a two-sided Wilcoxon rank-sum test coupled with a permutation test (n = 1,000). A total of 87/518 metabolites were differential across these two classes (P<0.05, corresponding to a 23% FDR). For visualizing the relationship between the 87 altered metabolites hierarchical clustering was used to arrange the metabolites based on their relative levels across samples (Fig. 2a). Among the perturbed metabolites, 50 were elevated in PCA while 37 were down-regulated. Fig. 2b displays the relative levels of the 37 named metabolites that were differential between benign prostate and PCA. Similarly, 124/518 metabolites were found to be elevated in the metastatic samples compared to the localized tumors with 102 compounds down-regulated (P<0.05, corresponding to a 4% FDR). Fig. 2c displays the levels of the 91 named metabolites altered in metastatic samples. A subset of six metabolites including sarcosine, uracil, kynurenine, glycerol-3-phosphate, leucine and proline were significantly elevated upon disease progression from benign to PCA to Mets. These metabolites could potentially serve as biomarkers for progressive disease, one of the factors that motivated us to examine sarcosine in greater detail.
Figure 2. Metabolomic alterations of prostate cancer progression.
a, Heat map showing 87 differential metabolites in PCA relative to benign samples (Wilcoxon P ≤ 0.05). Localized PCA samples are grouped as i., low grade (Gleason<6) and ii., high grade (Gleason>=7). Metastatic samples are grouped by the site of tissue procurement namely iii., soft tissue, iv., rib/diaphragm or v., liver. b, Benign-based z-score plot of named metabolites from a. Each point represents one metabolite in one sample, colored by tissue type (jade=benign, yellow=PCA). c, As in b except for the comparison between Mets (red) and PCA (yellow), with data represented relative to the mean of the PCA samples. For clarity, the plots in b and c have been truncated at 15 standard deviations above the mean of the benign and PCA samples, respectively.
Mapping the differential metabolomic profiles to their respective biochemical pathways as outlined in the Kyoto Encyclopedia of Genes and Genomes (KEGG, release 41.1, Supplementary Fig. 8) revealed an increase in amino acid metabolism and nitrogen breakdown pathways during cancer progression to metastatic disease. A similar enrichment network of amino acid metabolism was also identified by the bioinformatics tool Oncomine Concept Map4,5(OCM, www.oncomine.org, P = 6 × 10−13, Supplementary Fig. 9, 10a), supporting our earlier gene-expression-based prediction of androgen-induced protein synthesis as an early event during prostate cancer development5. Additionally, OCM found strong enrichment for elevated “methyltransferase activity” (Supplementary Fig. 10b, P = 7.7 × 10−8) among metabolites up-regulated in metastatic samples. This corroborates previous studies from our group and others showing elevation of the histone methyltransferase EZH2 in metastatic tumors 6–11.
As amino acid metabolism and methylation were enriched during prostate cancer progression, we focused on differential metabolites that characterize these processes and additionally show a progressive elevation from benign to PCA to metastatic disease. The amino acid metabolite sarcosine, an N-methyl derivative of glycine, fit these criteria. Notably, metastatic samples showed markedly elevated levels of sarcosine in 79% of the specimens analyzed (Chi-square test, P = 0.0538), whereas 42% of the PCA samples showed an increase in the levels of this metabolite (Fig. 2a–c). Importantly, none of the benign samples had detectable levels of sarcosine. Taken together this suggested the possible utility of sarcosine in monitoring disease progression and aggressiveness.
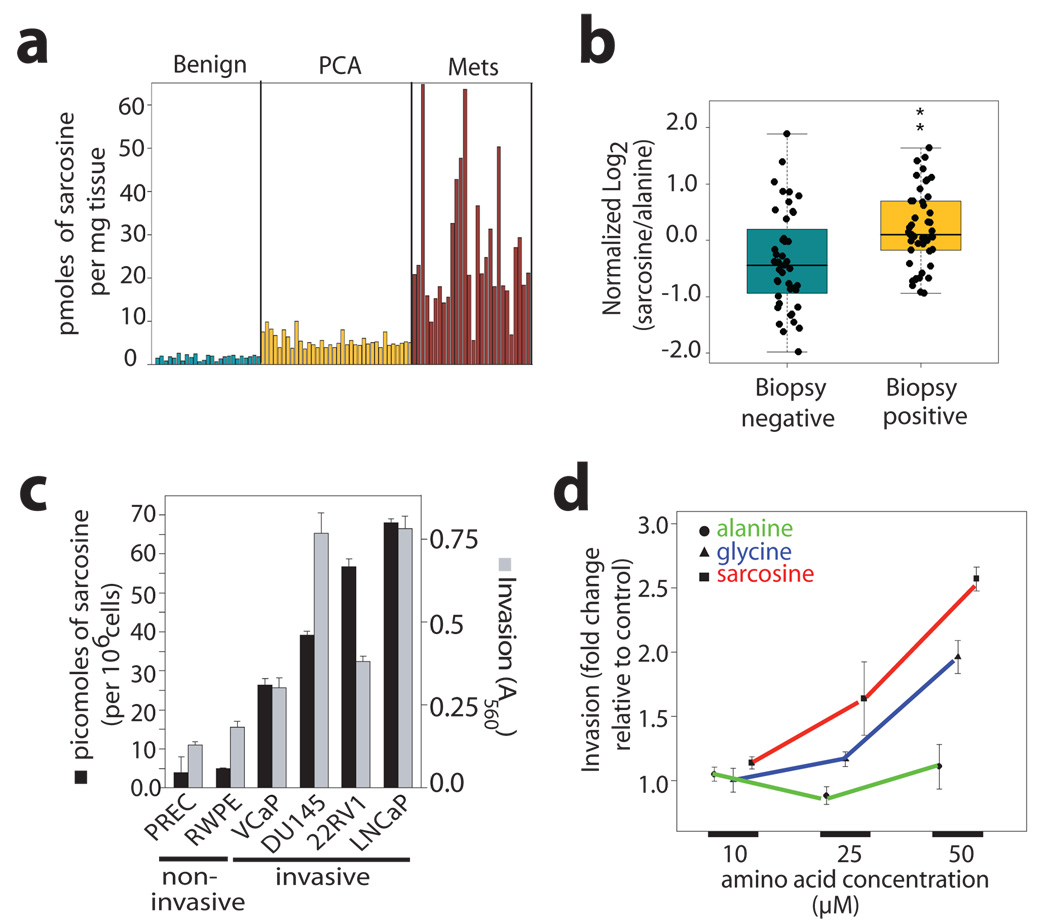
To confirm this pattern of sarcosine elevation in cancer progression we developed a highly sensitive and specific isotope dilution GC/MS method for accurately quantifying the metabolite from biospecimens (limit of detection = 10 femtomoles, Supplementary Fig. 11). In an independent set of 89 tissue samples (Supplementary Table 6), sarcosine levels were significantly elevated in PCA specimens (n=36) compared to benign adjacent prostate (n=25, Wilcoxon P = 4.34 ×10−11, Fig 3a). Additionally, there was an even greater elevation of sarcosine in metastatic samples (n=28) compared to organ-confined disease (Wilcoxon P = 6.02 × 10−11, Fig 3a). By contrast, sarcosine was undetectable in adjacent non-neoplastic tissues from patients with metastatic disease (Supplementary Fig. 12a–c).
Figure 3. Sarcosine levels in prostate cancer and its association with cell invasion.
a, Sarcosine levels in prostate cancer related tissue specimens (n=89). b, Sarcosine levels in post-DRE urine sediments from men with biopsy-proven prostate cancer (n=49) and prostate biopsy negative controls (n=44). c, Elevated levels of sarcosine (black bars) were found in invasive prostate cancer cells compared to non-invasive benign prostate epithelial cell lines. Mean +/− s.e.m. of sarcosine levels (n=3, except for PrEC cells where n=2). Cell invasion (grey bars) was also measured (mean +/− s.e.m.). e, Assessment of cell invasiveness of prostate epithelial cells upon exogenous administration of alanine, glycine, or sarcosine (mean +/− s.e.m., n=3).
The above findings led us to explore the potential of sarcosine as a candidate for future development in biomarker panels for early disease detection and aggressivity prediction. Towards this end, we monitored its levels in urine specimens from biopsy positive and negative individuals, a majority of whom have elevated PSA levels (>4.0 ng/ml), and in which prostate needle biopsy was used for diagnosis. This is a particularly challenging cohort as not only are these men at high risk for prostate cancer, even a negative needle biopsy does not rule out the presence of cancer due to sampling issues. Sarcosine was found to be significantly higher in urine sediments (n=49, Wilcoxon P = 0.0004, Fig 3b) and supernatants (n=59, Wilcoxon P = 0.0025, Supplementary Fig. 14a) derived from biopsy positive prostate cancer patients as compared to biopsy negative controls (n = 44 and n= 51, respectively, Supplementary Table 7 and Table 8). The overall receiver operator characteristic (ROC) curves for sarcosine suggest its predictive value is modest with AUCs of 0.71 and 0.67 for urine sediments and supernatants, respectively (Supplementary Fig. 14b, c). Notably an AUC of 1.0 indicates perfect prediction and an AUC of 0.5 indicates prediction equivalent to random selection. Interestingly, when restricted to samples having PSA in the clinical grey zone of 2–10 ng/ml (n=53), sarcosine performed better than PSA in delineating the two diagnostic classes with an AUC of 0.69 (95% CI: 0.55, 0.84) compared to an AUC of 0.53 (95% CI: 0.37, 0.69) for PSA (Supplementary Fig. 15). Thus, sarcosine may have potential to identify patients with modestly elevated PSA that are likely to have a positive prostate biopsy.
To determine whether sarcosine elevation in prostate cancer has biological relevance, we measured its levels in prostate cancer cell lines VCaP, DU145, 22RV1 and LNCaP (n=3 each) and their benign epithelial counterparts, primary benign prostate epithelial cells (PrEC, n=2) and immortalized benign RWPE prostate cells (n=3). Significantly elevated levels of the metabolite were found in prostate cancer cells compared to the benign cells (ANOVA P=0.0218, Fig. 3c). Additionally, sarcosine levels correlated well with cell invasiveness (Spearman’s correlation coefficient: 0.943, P=0.0048). Based on our earlier findings that EZH2 over-expression in benign cells could mediate cell invasion and neoplastic progression6,10,11, sarcosine levels were assessed upon modulation of EZH2 expression. Interestingly, over-expression of EZH2 in benign prostate epithelial cells increased sarcosine levels (4.5 fold, Supplementary Fig. 16a) while its knock-down in DU145 prostate cancer cells diminished the levels of the metabolite (Supplementary Fig. 16 a–c). To determine if sarcosine may play a more direct role in this process, we added the metabolite to non-invasive benign prostate epithelial cells. Alanine, an isomer of sarcosine, was used as a control for these experiments. Remarkably, the mere addition of exogenous sarcosine imparted an invasive phenotype to benign prostate epithelial cells (Fig. 3d, Supplementary Fig. 17). Furthermore, the number of motile prostate epithelial cells were significantly higher upon sarcosine treatment (t-test P=6.997e-06, n=10) compared to alanine-treated controls (Supplementary Fig. 18). Sarcosine treatment however did not affect the ability of these cells to progress through the different stages of cell cycle (Supplementary Fig. 19a–d) or impair cell proliferation (Supplementary Fig. 19e). Notably, glycine, a precursor of sarcosine, induced invasion in these cells, although to a lesser degree than sarcosine (Fig. 3e). This invasion could result from the conversion of glycine to sarcosine by the enzyme glycine-N-methyltransferase (GNMT) (Fig. 4a).
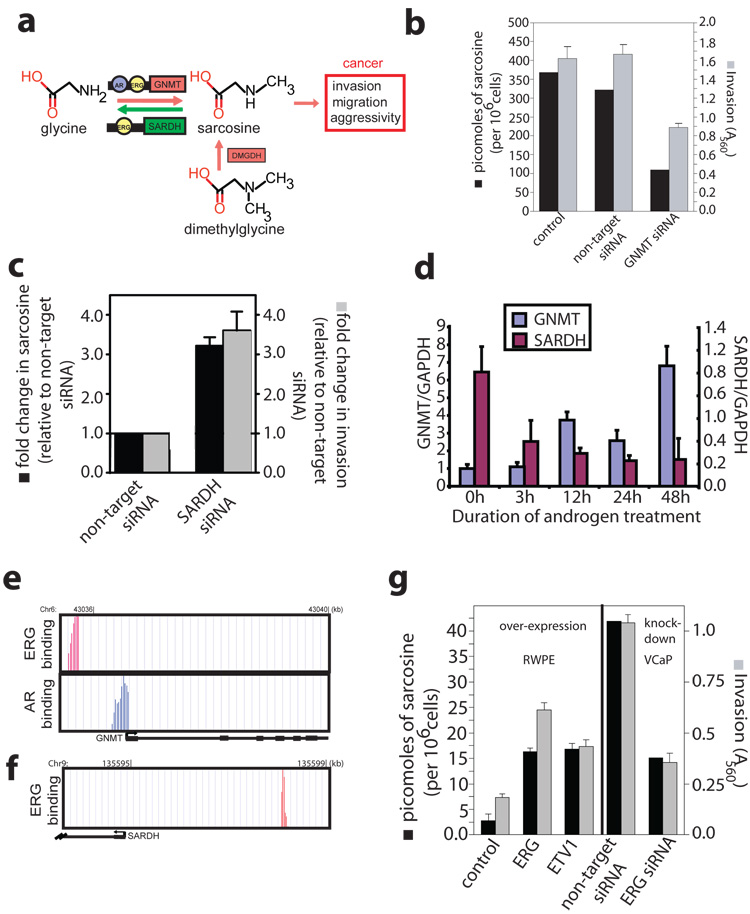
Figure 4. A role for sarcosine in androgen signaling and prostate cancer cell invasion.
a, Schematic of the sarcosine pathway and its potential link to prostate cancer. b, Assessment of sarcosine levels and cell invasiveness after knockdown of GNMT in DU145 cells by RNA interference. c, As in b except knockdown of SARDH in RWPE cells (n=6). d, QRT-PCR analysis of GNMT and SARDH mRNA expression in androgen stimulated VCaP cells. e, AR and ERG binding sites on the promoter of GNMT as determined by ChIP-seq. The Y axes display the number of reads in a 25 bp sliding window. f, As in e, except ERG binding sites in the promoter of SARDH. g, Left panel, overexpression of ERG or ETV1 in RWPE cells and measurement of sarcosine levels and cell invasiveness. Right panel, as in left, except knockdown of TMPRSS2-ERG in VCaP cells by RNA interference. All error bars represent mean +/− s.e.m., n=3 unless indicated otherwise.
In addition to GNMT, sarcosine levels are regulated by sarcosine dehydrogenase (SARDH), the enzyme that converts sarcosine back to glycine and dimethylglycine dehydrogenase (DMGDH) which generates sarcosine from dimethylglycine (Fig. 4a). By virtue of their ability to control sarcosine levels in cells, these enzymes may assume a critical role in modulating prostate cancer invasion. To test this hypothesis, a series of RNA interference-mediated knock-down experiments were carried out. Attenuation of GNMT (Fig. 4b, Supplementary Fig. 20) in DU145 prostate cancer cells resulted in a significant reduction in cell invasion (t-test P= 0.0073, n=3) with a concomitant 3-fold decrease in the intracellular sarcosine levels compared to control non-target siRNA-transfected cells. Similar knockdown experiments performed in benign RWPE cells significantly hampered the ability of exogenous glycine (t-test P=0.0082, n=3), but not sarcosine, to induce invasion (Supplementary Fig. 21a,b). Comparable loss of cell invasion and reduction in sarcosine levels were also apparent in DU145 cancer cells upon knock-down of DMGDH (Supplementary Fig. 22a,b). In contrast, knock-down of SARDH in benign prostate epithelial cells resulted in approximately a 3-fold increase in endogenous sarcosine levels with a concomitant > 3.5 fold increase in invasion (Fig. 4c, Supplementary Fig. 23).
With the understanding that androgen signaling and ETS gene (ERG, ETV1) fusions are key factors for prostate cancer progression12, we investigated their role in regulating GNMT and SARDH. Treatment with androgen for 48h in VCaP (ERG positive) and LNCaP (ETV1 positive) prostate cancer cells resulted in a step-wise increase in GNMT expression and a concomitant decrease in SARDH levels as assessed by digital gene expression and QPCR (Fig 4d, Supplementary Fig. 25e). This finding was supported by chromatin immunoprecipitation sequencing (ChiP-Seq) which revealed direct binding of AR and ERG to the promoter of GNMT in VCaP cells (Fig. 4e), while only ERG binding was seen on the SARDH promoter (Fig. 4f). In ETV1 positive LNCaP cells, AR, but not ERG as expected, was bound to both GNMT and SARDH promoters (Supplementary Figs. 25a,b). The binding data was validated by ChIP-PCR (Supplementary Fig 24 a,b,d and 25 c,d) that additionally revealed weak binding of AR to the SARDH promoter in VCaP cells (Supplementary Fig. 24 c). These findings together directly link activation of the sarcosine pathway to AR and ETS gene fusion regulation, two key mediators of prostate cancer progression. Remarkably, both ERG-and ETV1-induced invasion were associated with a 3-fold sarcosine elevation in benign RWPE cells (Fig. 4h). Similarly, knock-down of the TMPRSS2-ERG gene fusion in VCaP cells (Supplementary Fig. 26) resulted in > 3-fold decrease in sarcosine with a similar decrease in the invasive phenotype (Fig 4h).
Taken together, we explored the “metabolome” of prostate cancer progression. This led to the characterization of metabolomic signatures, which in the context of other molecular alterations, may lead to a more complete understanding of disease progression. Specifically, we identified sarcosine as a key metabolite elevated most robustly in metastatic prostate cancer and detectable in the urine of men with organ-confined disease. Interestingly, sarcosine, and its proximal regulatory enzymes, appear to play an intermediary role in neoplastic progression modulating cell invasion and migration. The master transcriptional regulators of prostate cancer progression, AR and the ETS gene fusions, appear to directly regulate sarcosine levels via transcriptional control of its regulatory enzymes. Thus, components of the sarcosine pathway may have potential as biomarkers of prostate cancer progression as well as serve as novel avenues for therapeutic intervention.
METHODS SUMMARY
Biospecimens and associated clinical data related to the study were collected with written consent from the University of Michigan and approved for use by the Internal Review Board. Unbiased metabolomic profiling using liquid/gas-chromatography coupled to mass spectrometry (LC/GC MS) was performed as described 3 using a Thermofisher Linear Ion Trap mass spectrometer with Fourier Transform and Mat-95 XP mass spectrometers respectively (Supplementary Fig.1). Target metabolites were assessed in tissue and urine samples using isotope dilution GC-MS. Metabolomic data analysis is detailed in Supplementary Fig. 4. All Wilcoxon rank-sum tests and t-tests are two-sided using a threshold of P<0.05 for significance. Repeated measures ANOVA is used for the cell line data with p-values from the model F-test. Class-specific metabolomic patterns were visualized using Z-score plots and heat maps. Unsupervised clustering of samples using metabolomic signatures was performed using cluster13 and tree view14 and visualized using heat maps. Network relationship among various molecular concepts and metabolomic data was performed using Oncomine Concept Map4,5(www.oncomine.org) as outlined in Supplementary Fig. 9. Invasion was measured using a modified Boyden Chamber assay as described10. Cell motility assay was performed as previously reported using blue flourescent microsphere beads15. Targeted knock-down of candidate genes 16 using gene–specific siRNA sequences are listed in Supplementary Table 9. QPCR for enzymes regulating sarcosine levels, EZH2 and ETS were performed as described12 using indicated oligonucleotide primers (Supplementary Table 10). Chromatin immunoprecipitation to interrogate regulatory role of androgen and ETS was performed using published protocols17. ChIP-Seq and digital gene expression were measured using the Genomic DNA sample prep kit and the NIaIII kit on a Genome Analyzer (Illumina) as per manufacturer’s instructions.
Full methods and any associated references are available in the online version of the paper at www.nature.com/nature.
METHODS
Biospecimens and cell lines
Prostate tissues, urine and plasma were obtained from the the University of Michigan SPORE and EDRN Tissue Core. All samples were collected with informed consent as per the approval of the Institutional Review Board.
RWPE, DU145, LnCAP, and PC3 cells were obtained from ATCC, PrEC cells from Cambrex BioScience, 22-RV1 was provided by J. Macoska and VCaP by K. Pienta. VCaP and LnCAP were grown in charcoal stripped serum containing media for 24h, before treatment for further 24 h with vehicle or 1nM methyltrienolone (R1881, NEN) dissolved in ethanol.
Metabolomic Profiling was performed using the platform described in Lawton et al. (2008)3 and outlined in Supplementary Fig 1. The LC/MS portion of the platform is based on a Surveyor HPLC and a Thermo-Finnigan LTQ-FT mass spectrometer (Thermo Fisher Corporation, Waltham, MA) with the instrument set for continuous monitoring of both positive and negative ions. Samples that were analyzed by GC MS were derivatized under dried nitrogen using bistrimethyl-silyl-triflouroacetamide (BSTFA) and analyzed on a Thermo-Finnigan Mat-95 XP using electron impact ionization (EI) and high resolution. For both LC and GC mass spectrometry, spectral files were searched using metabolomic libraries created by Metabolon that contain about 800 commercially available compounds.
Quantitation of target metabolites was performed using isotope dilution GC MS using selected ion monitoring (SIM). The samples were modified to their t-butyl dimethylsilyl derivatives and analyzed with an Agilent 5975 MSD mass detector using EI. For SIM analysis the m/z for native and labeled molecular peaks for various target metabolites quantified were: 158 and 161 (sarcosine), 406 and 407 (cysteine), 432 and 437 (glutamic acid), 297 and 301 (thymine), and 218 and 219 (glycine), respectively. Assessment of citric acid was performed on the GC-MS in the full scan mode.
Statistical Analysis
Missing data were imputed with zero for metabolites with mean measure >100,000 across samples (i.e., missingness likely due to absence), otherwise one half of the sample minimum was imputed (i.e., missingness likely due to censoring). Imputed data were median centered and inter-quartile range (IQR) scaled per sample. Plotted z-scores were calculated based on the mean and standard deviation of a reference set (benign samples, unless otherwise stated). Hierarchical clustering14 based on Pearson’s correlation was performed on the log transformed normalized data after median centering per metabolite. A small value (unity) was added to each normalized value to allow log transformation. Per-metabolite chi-square tests were used to assess class specific metabolite patterns of present and absent (undetected) measurements. Per-metabolite two-tailed Wilcoxon rank sum tests were used for two-sample tests of association between classes. Kruskal-Wallis tests were used for three-way comparisons between all diagnosis groups. Non-parametric tests were chosen to reduce the influence of the imputed values. Tests were run on those metabolites with detectable expression in at least 20% of the samples. Significance was determined using 1000 sample permutations False discovery rates were calculated using the q-value conversion algorithm of Storey18. Pairwise differences in expression in the cell line data and small scale tissue data were tested using two-tailed t-tests with Satterthwaite variance estimation. Comparisons involving multiple cell lines used repeated measures analysis of variance (ANOVA) to adjust for the multiple measures per cell line. Fold change was estimated using ANOVA on a log scale, following the model log(Y) = A + B*Treatment + E. In this way exp(B) is an estimate of (Y | Treatment = 1)/(Y | Treatment = 0) and the standard error of exp(B) can be estimated from SE(B) using the delta method. The threshold for significance was P<0.05 for all tests.
ChIP-PCR
ChIP was carried out as previously described19 using antibodies against AR (Millipore), ERG (Santa Cruz) and rabbit IgG (Santa Cruz). AR ChIP was performed in paired ethanol-treated and R1881-treated samples. ChIP-enriched chromatin as well as the whole-cell extract was amplified by ligation-mediated PCR. When examining AR binding on target genomic regions, equal amount of ethanol-treated and R1881-treated ChIP amplicons were subjected to QPCR and the fold enrichment (R1881/ethanol) was determined based on the cycle differences after normalization to input DNA. For ERG ChIP assays, VCaP cells grown in regular medium were used for ChIP using antibodies against ERG and rabbit IgG control. ChIP products were directly analyzed by QPCR assay and ERG binding was evaluated based on the cycle difference between ChIP-enriched chromatin by ERG and corresponding IgG. The primers used are listed in Supplemental Table 9.
ChIP-Seq
ChIP samples were prepared for sequencing using the Genomic DNA sample prep kit following manufacturers protocols. ChIP-Sequencing was performed using Illumina Genome Analyzer according to standard manufacturer’s procedures. The raw sequencing image data were analyzed by our analysis pipeline, aligned to the unmasked human reference genome (NCBI v36, hg18) using ELAND software to generate sequence reads of 25–32 bps.
Digital gene expression analysis
Trizol extracted RNA from samples with 0h and 48h androgen treatment were prepared for sequencing using the Digital Gene Expression-Tag Profiling with NIaIII kit (Illumina) and sequenced by the Genome Analyzer. Sequencing reads were mapped back to the human reference genome using the ELAND software. The number of sequencing reads for genes of interest was counted. The expression level of each gene was measured as the number of transcripts per million of total sequencing reads.
Quantitative RT-PCR
Q-PCR was performed using SYBR Green Mastermix on an Applied Biosystems 7300 PCR machine as previously described 19. All primers were designed using Primer 3 and synthesized by Integrated DNA Technologies and are listed in Supplementary Table 9.
RNA interference
DU145 or RWPE cells were treated with non-targeting siRNA (Dharmacon) and gene-specific siRNA sequences as listed in Supplementary Table 10.
Cell invasion assay
Cell invasion assays were carried out using a modified basement membrane chamber assay as previously described19.
Cell motility assay
For cell motility assays, we used the Cellomics Cell Motility kit as per manufacturer’s instructions.
Supplementary Material
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Acknowledgments
We thank Jill Granger for help in manuscript preparation, Javed Siddiqui and Radhika Varambally for help with the clinical database and Adaikkalam Vellaichamy and Sailaja Pullela for technical assistance. We thank Dr. Kenneth Pienta for access to metastatic prostate cancer samples from the University of Michigan Prostate SPORE rapid autopsy program. This work is supported in part by the Early Detection Research Network (to A.M.C.), National Institutes of Health (to A.S., S.P., J.B., T.M.R , D.G., G.S.O. and A.M.C.) and an MTTC grant (G.O. and A.S.). A.M.C. is supported by a Clinical Translational Science Award from the Burroughs Welcome Foundation. A.S. is supported by a grant from the Fund for Discovery of the University of Michigan Comprehensive Cancer Center. L.M.P. is supported by the University of Michigan Cancer Biostatistics Training Grant. A.M.C and S.P. are supported by the Doris Duke Charitable Foundation.
Footnotes
Author Information Reprints and permissions information is available at www.nature.com/reprints. C.B. was previously an employee of Metabolon. C.B., D.C.A., J.R.S. and A.B. own equity in Metabolon. A.M.C. joined the Scientific Advisory Board of Metabolon in July 2008. The University of Michigan has licensed the diagnostic field of use of the metabolomic biomarkers discussed in this manuscript to Metabolon (A.M.C. and A.S. are named as inventors).
References
- 1.Abate-Shen C, Shen MM. Molecular genetics of prostate cancer. Genes Dev. 2000;14:2410–2434. doi: 10.1101/gad.819500. [DOI] [PubMed] [Google Scholar]
- 2.Ruijter E, et al. Molecular genetics and epidemiology of prostate carcinoma. Endocr Rev. 1999;20:22–45. doi: 10.1210/edrv.20.1.0356. [DOI] [PubMed] [Google Scholar]
- 3.Lawton KA, et al. Analysis of the adult human plasma metabolome. Pharmacogenomics. 2008;9:383–397. doi: 10.2217/14622416.9.4.383. [DOI] [PubMed] [Google Scholar]
- 4.Rhodes DR, et al. Molecular concepts analysis links tumors, pathways, mechanisms, and drugs. Neoplasia. 2007;9:443–454. doi: 10.1593/neo.07292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tomlins SA, et al. Integrative molecular concept modeling of prostate cancer progression. Nat Genet. 2007;39:41–51. doi: 10.1038/ng1935. [DOI] [PubMed] [Google Scholar]
- 6.Varambally S, et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419:624–629. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- 7.van der Vlag J, Otte AP. Transcriptional repression mediated by the human polycomb-group protein EED involves histone deacetylation. Nat Genet. 1999;23:474–478. doi: 10.1038/70602. [DOI] [PubMed] [Google Scholar]
- 8.Laible G, et al. Mammalian homologues of the Polycomb-group gene Enhancer of zeste mediate gene silencing in Drosophila heterochromatin and at S. cerevisiae telomeres. Embo J. 1997;16:3219–3232. doi: 10.1093/emboj/16.11.3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cao R, et al. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science (New York, N.Y. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 10.Kleer CG, et al. EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:11606–11611. doi: 10.1073/pnas.1933744100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Varambally S, et al. Genomic loss of microRNA-101 leads to overexpression of histone methyltransferase EZH2 in cancer. Science (New York, N.Y. 2008;322:1695–1699. doi: 10.1126/science.1165395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tomlins SA, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science (New York, N.Y. 2005;310:644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 13.Eisen MB, Brown PO. DNA arrays for analysis of gene expression. Methods Enzymol. 1999;303:179–205. doi: 10.1016/s0076-6879(99)03014-1. [DOI] [PubMed] [Google Scholar]
- 14.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klemke RL, et al. Regulation of cell motility by mitogen-activated protein kinase. The Journal of cell biology. 1997;137:481–492. doi: 10.1083/jcb.137.2.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Varambally S, et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419:624–629. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- 17.Yu J, et al. A polycomb repression signature in metastatic prostate cancer predicts cancer outcome. Cancer research. 2007;67:10657–10663. doi: 10.1158/0008-5472.CAN-07-2498. [DOI] [PubMed] [Google Scholar]
- 18.Storey J. J Royal Stat Soc. 2002;64:479. [Google Scholar]
- 19.Yu J, et al. Integrative genomics analysis reveals silencing of beta-adrenergic signaling by polycomb in prostate cancer. Cancer Cell. 2007;12:419–431. doi: 10.1016/j.ccr.2007.10.016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.