The connection between function and folding among proteins has inspired a growing number of efforts to identify unnatural oligomers that adopt discrete tertiary and/or quaternary structures.1 Recently we have shown that modification of a self-assembling α-amino acid sequence by systematic replacement of some α-residues with analogous β3-amino acid residues (identical side chains) can generate α/β-peptide “foldamers” that display protein-like helix-bundle quaternary structure. The designs reported thus far have placed the β3-residues mostly or entirely on the quaternary structure periphery.2 Here we describe a new α/β-peptide that forms a helix bundle with a hydrophobic core comprised exclusively of β3-amino acid residues; this unique quaternary structure displays unprecedented features.
Much of our previous work on α/β-peptide helix bundles has focused on the dimerization domain of yeast transcription factor GCN4 and an engineered variant designated GCN4-pLI.3 The wild type sequence encodes a parallel coiled-coil dimer, while GCN4-pLI forms a parallel helix-bundle tetramer. The sequence of GCN4-pLI features a typical abcdefg heptad repeat pattern, with hydrophobic residues at the a and d positions (Leu and Ile respectively; Figure 1a,c). The a and d side chains align upon folding, resulting in an amphipathic α-helix.4 Burial of hydrophobic side chains provides the driving force for assembly. In the new α/β-peptide βad, residues at all a and d positions of GCN4-pLI have been replaced with the homologous β3 residues (e.g., Leu→β3-hLeu, Ile→β3-hIle; Figure 1).
Figure 1.
(a) Sequences of GCN4-derived α/β–peptides. Heptad positions are shown in italics. (b) Helical wheel diagrams of α/β–peptides βad, βae, and βdg. Letters refer to substituted heptad positions. (c) Helical wheel diagram of the GCN4-pLI sequence. (d) Structures of an α–amino acid and a β3-amino acid.
Circular dichroism (CD) data for 5, 10, 25, and 100 μM βad in aqueous buffer show a strong minimum at 207 nm, which is consistent with extensive α/β-peptide helicity.5 Little change in CD intensity occurs upon heating to 98°C or dilution from 100 μM to 5 μM, suggesting a very stable assembly. Analytical ultracentrifugation (AU) data for 200 μM βad at 25°C are consistent with a tetrameric species. We crystallized βad and solved the structure to gain insight on the tetramer assembly.
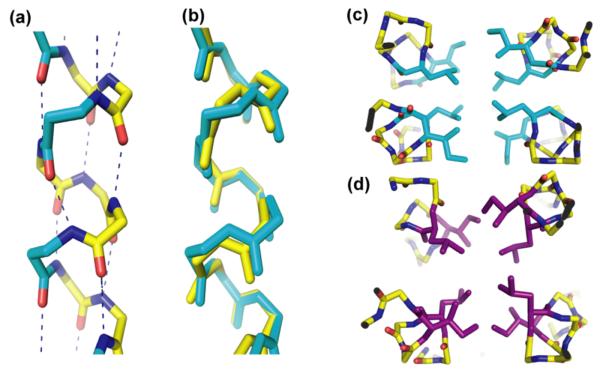
βad forms a four-helix bundle in the crystalline state; the hydrophobic core is comprised entirely of β3-residues (Figure 2). The conformation of each βad molecule closely mimics an α-helix, as illustrated by the overlay with GCN4-pLI (Figure 3a,b); βad retains the i→i+4 C=O--H-N hydrogen bonding pattern that is characteristic of the α-helix. Despite the similarities between βad and α-peptide GCN4-pLI in terms of stoichiometry and helical secondary structure, the quaternary structures are quite dissimilar. Neighboring helices are antiparallel in the βad tetramer, while all α-helices are parallel in the GCN4-pLI tetramer. Previously, only parallel orientations have been observed in α/β-peptide helix bundles.2a,c,3 Furthermore, the hydrophobic packing arrangement within the core of the βad tetramer has no precedent among known proteins.
Figure 2.
(a) 2.0 Å x-ray crystal structure of α/β-peptide βad shown as cartoon helices displaying amino acid side chains (PDB code: 3F86). β3-amino acids are shown in cyan, α-amino acids are shown in yellow. (b) Single layer of βad hydrophobic core residues fit into 2FO-FC electron density at a map level of 1.2 σ.
Figure 3.
(a) i to i+4 hydrogen bonding in βad. β3-amino acids are shown in cyan. (b) Overlay of βad (cyan) with GCN4-pLI (yellow). CαRMSD for residues 4-30 = 0.55 Å. (c) and (d) Hydrophobic cores of βad and an antiparallel GCN4-pLI derivative (PDB: 2CCF),5 respectively. β3–amino acids are shown in cyan, α–residues are shown in purple.
Two of the antiparallel helix pairings within the βad tetramer involve very close backbone contacts (Figure 3c), which results in an unusual rectangular arrangement of the four subunits about the helix bundle axis. In contrast, a more symmetrical (square) arrangement about the helix bundle axis is typical of both parallel and antiparallel coiled-coils (Figure 3c,d).6 The closely interacting βad helices have an 8.1 Å separation (center-to-center); the other interhelical separation is 13.2 Å. In contrast, a typical α-helix tetramer displays uniform 10-11 Å interhelical separation.5,6 The short interhelical distance in βad is a result of a “stripe” of backbone methylene groups that is created by alignment of β3-hLeu residues (a positions) along the helical axis. The backbone-backbone interactions between close-packed helix pairs cause their a and d side chains to generate a relatively flat hydrophobic surface. Packing of two of these flat surfaces against one another leads to tetramer formation. Thus, the core side chain arrangement in the βad tetramer is quite different from the “knobs-into-holes” packing that is characteristic α-helical coiled-coil quaternary structures (Figure 3c,d)7a,b and previously reported α/β-peptide helix bundles (which have mostly or entirely α-residues in their cores).
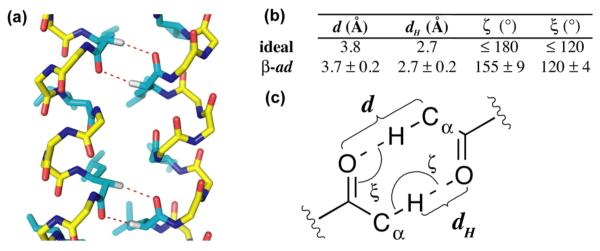
The backbone methylene stripes displayed by helical βad molecules represent patches of non-polar surface, and it is possible that burial of these patches stabilizes the βad tetramer via a hydrophobic effect. Another structural role is possible as well: formation of multiple Cα-H--O=C hydrogen bonds between the close-packed helices. Cα-H--O=C hydrogen bonds have been proposed to play a role in the folding and association of integral membrane proteins, specifically, the dimerization of helical domains that contain a GxxxG motif.8 Whether or not these interhelical Cα-H--O=C interactions contribute to dimer stability, however, remains a subject of debate.9 For each close antiparallel pairing within the βad four-helix bundle there are 10 Cα-H--O=C interactions (i.e., 20 such interactions per tetramer). The interatomic distances and angles for these interactions are within the parameters proposed based upon membrane protein structural data.8b An extended Cα-H--O=C interaction array of the type seen in the βad crystal structure would appear to be impossible for α-peptides because of the large crossing angle dictated by GxxxG-mediated α-helix association.5
In order to determine whether the unique quaternary structure observed for βad requires β3-residues at both the a and d positions of the heptad repeat we examined two isomers, α/β-peptides βae and βdg (Figure 1a,b). These isomers have the same ααβαααβ backbone pattern and side chain sequence of βad, but the locations of the β3-residues differ. For βae, CD data indicate extensive helix formation at room temperature, but helicity is disrupted at higher temperatures and at concentrations below 25 mM.5 AU data suggest that βae forms a trimer. Isomer βdg is relatively unstructured according to CD data, and AU data suggest indiscrete aggregation.5 For a previously reported GCN4-based, α/β-peptide crystal structure2b, we observed that β3-subsitution significantly altered the orientation of a or d side chain projection from the helix. Extension of this analysis to e and g position β3-residues reveals comparable effects and may explain why global a-e β3-substitution of GCN4-pLI allows assembly, while global d-g β3-substitution nearly abolishes assembly.5 We conclude that the placement of β3-residues at all the hydrophobic core positions of the GCN4-pLI sequence is necessary for formation of the unique quaternary structure observed for βad.
The asymmetry of interaction within the βad helix bundle and the face-to-face side-chain packing motif in the βad tetramer core are, to our knowledge, unprecedented among naturally occurring or designed α-helical assemblies or among β- or α/β-peptide helix bundles.4a,5,10a,b Although we do not have high-resolution structural information for βad in solution, AU and CD data are consistent with the hypothesis that the tetrameric assembly observed in the crystalline state forms in aqueous solution as well, and that this assembly is quite stable. The occurrence of antiparallel helix orientations within the βad tetramer raises the exciting prospect that foldamer tertiary structures could be generated by linking helix-forming α/β-peptide segments.11
Supplementary Material
Figure 4.
(a) Side view of close-packed helices with potential Cα-H-O hydrogen bonds shown as red dashes. (b) Comparison of idealized Cα-H-O hydrogen bond parameters with average values calculated from the crystal structure of βad. (c) Diagram defining the geometric parameters of the Cα-H-O hydrogen bond.9
Acknowledgement
This research was supported by NIH grant GM61238. M. G. was supported in part by the UW-Madison NSEC (NSF DMR-0425880), and W. S. H. was supported in part by an NIH fellowship (CA119875). We thank Professors J. Keck and K. Forest for the use of x-ray crystallography facilities, Dr. D. McCaslin for assistance with AU experiments, and Peptech for providing discounted Fmoc-β3-amino acids.
Footnotes
Supporting Information Available: Coordinates and structure factors for βad were deposited in the PDB with ID codes 3F86 and 3F87. Experimental protocols, biophysical data, and crystallographic statistics are available free of charge online at http://pubs.acs.org
References
- 1.(a) Raguse TL, Lai JR, LePlae PR, Gellman SH. Org. Lett. 2001;3:3963–3966. doi: 10.1021/ol016868r. [DOI] [PubMed] [Google Scholar]; (b) Cheng RP, DeGrado WF. J. Am. Chem. Soc. 2002;124:11564–11565. doi: 10.1021/ja020728a. [DOI] [PubMed] [Google Scholar]; (c) Daniels DS, Petersson EJ, Qiu JX, Schepartz A. J. Am. Chem. Soc. 2007;129:1532–1533. doi: 10.1021/ja068678n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.(a) Horne WS, Price JL, Keck JL, Gellman SH. J. Am. Chem. Soc. 2007;129:4178–4180. doi: 10.1021/ja070396f. [DOI] [PubMed] [Google Scholar]; (b) Price JL, Horne WS, Gellman SH. J. Am. Chem. Soc. 2007;129:6376–6377. doi: 10.1021/ja071203r. [DOI] [PubMed] [Google Scholar]; (c) Horne WS, Price JL, Gellman SH. Proc. Natl. Acad. Sci. U.S.A. 2008;105:9151–9156. doi: 10.1073/pnas.0801135105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harbury PB, Zhang T, Kim PS, Alber T. Science. 1993;262:1401–1407. doi: 10.1126/science.8248779. [DOI] [PubMed] [Google Scholar]
- 4.(a) Oakley MG, Hollenbeck JJ. Curr. Opin. Struct. Biol. 2001;11:450–457. doi: 10.1016/s0959-440x(00)00232-3. [DOI] [PubMed] [Google Scholar]; (b) Lupas AN, Gruber M. Adv. Prot. Chem. 2005;70:37–78. doi: 10.1016/S0065-3233(05)70003-6. [DOI] [PubMed] [Google Scholar]; (c) Woolfson DN. Adv. Prot. Chem. 2005;70:79–112. doi: 10.1016/S0065-3233(05)70004-8. [DOI] [PubMed] [Google Scholar]; (d) Grigoryan G, Keating AE. Curr. Opin. Struct. Biol. 2008;18:477–483. doi: 10.1016/j.sbi.2008.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.See Supporting Information
- 6.Yadav MK, Leman LJ, Price DJ, Brooks CL, Stout CD, Ghadiri MR. Biochemistry. 2006;45:4463–73. doi: 10.1021/bi060092q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.(a) Crick FHC. Nature. 1952;170:882–883. doi: 10.1038/170882b0. [DOI] [PubMed] [Google Scholar]; (b) Crick FHC. Acta Cryst. 1953;6:689–697. [Google Scholar]
- 8.(a) Derewenda ZS, Lee L, Derewenda U. J. Mol. Biol. 1995;252:248–62. doi: 10.1006/jmbi.1995.0492. [DOI] [PubMed] [Google Scholar]; (b) Senes A, Ubarretxene-Belandia I, Engelman DM. Proc. Natl. Acad. Sci. U.S.A. 2001;98:9056–9061. doi: 10.1073/pnas.161280798. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Mackenzie KR. Chem. Rev. 2006;106:1931–1977. doi: 10.1021/cr0404388. [DOI] [PubMed] [Google Scholar]
- 9.(a) Arbely E, Arkin. IT. J. Am. Chem. Soc. 2004;126:5362–5363. doi: 10.1021/ja049826h. [DOI] [PubMed] [Google Scholar]; (b) Dunitz JD, Gavezzotti A. Angew. Chem. Int. Ed. 2005;44:1766–1787. doi: 10.1002/anie.200460157. [DOI] [PubMed] [Google Scholar]
- 10.(a) Schnarr NA, Kennan AJ. J. Am. Chem. Soc. 2002;124:9779–83. doi: 10.1021/ja0174940. [DOI] [PubMed] [Google Scholar]; (b) Yoder NC, Kumar K. J. Am. Chem. Soc. 2006;128:188–91. doi: 10.1021/ja055494k. [DOI] [PubMed] [Google Scholar]
- 11.Petersson EJ, Schepartz A. J. Am. Chem. Soc. 2008;130:821–823. doi: 10.1021/ja077245x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.