Abstract
The lipid A of Gram-negative bacteria plays a major role in the pathogenesis of bacterial infections. Lipid A diversity is observed both in the number and length of fatty-acid side chains and in the presence of terminal phosphate residues and associated modifications. In this report, we describe a new sample preparation method based on microwave-assisted enzymatic digestion and detergent-free mild hydrolysis, in conjunction with a MALDI-time-of-flight (TOF)/TOF analysis, to determine the structures of lipid A from Helicobacter pylori. The total time for sample preparation and mass spectrometric analysis is within 2 h and applicable to profiling the lipid A structures from dried bacterial cells on as little as 1 μg. The reliability of the technique was further demonstrated through the analysis of the lipid A from bacterial cells of different H. pylori strains. The phosphorylation and acylation patterns of lipid A could be elucidated using material from a single colony. Furthermore, we found unusual heptaacyl lipid A species present in H. pylori mutant that have not been previously reported, although the abundance was relatively low. The present study provides the first characterization of the lipid A component from a single bacterial colony sample by mass spectrometry.
Keywords: Helicobacter pylori, acylation, microwave-assisted digestion, mass spectrometry, MALDI-TOF/TOF
Helicobacter pylori, the only Gram-negative bacterium capable of colonizing the human stomach, is the primary cause of active chronic gastritis in humans. Humans were already infected by H. pylori before their migrations from Africa and H. pylori has remained intimately associated with their human host populations ever since (1, 2). Similar to the cell surface structures of other Gram-negative bacteria, lipopolysaccharide (LPS) is a major component of H. pylori outer membrane. The H. pylori LPS consists of a lipid A region, a core region, and an O-chain polysaccharide (also known as the O-antigen) (3). Lipid A, the hydrophobic moiety of LPS and a glucosamine-based saccharolipid, is the principal structural component responsible for the range of biological activities of LPS (4, 5).
Generally, lipid A is a glucosamine disaccharide that carries phosphates at positions 1 and 4′ and usually has four primary (glucosamine-linked) hydroxyacyl chains and one or more secondary acyl chains (3, 6, 7). However, H. pylori lipid A is different from that of other bacterial species, including both phosphorylation and acylation patterns (8). The lipid A of H. pylori contains a phosphoethanolamine (PEtn) group directly linked to the 1-position of the disaccharide backbone. This is in contrast to the PEtn groups found in other pathogenic Gram-negative bacteria, which are attached to the lipid A phosphate group to form a pyrophosphate linkage (9, 10). In addition, the predominant absence of ester-bound 4’-phosphate and the presence of tetraacyl lipid A with fatty acids of 16 to 18 carbons in length differentiate H. pylori lipid A from that of other Gram-negative bacteria. H. pylori synthesizes two types of lipid A molecules: hexaacyl- and tetraacyl-lipid A. Hexaacyl-lipid A has two phosphates or phosphoethanolamines on the lipid A disaccharide backbone, whereas tetraacyl-lipid A contains only one phosphate (8). It has also been reported that H. pylori does not survive long before it is deacylated at the 3′ position from hexaacyl structure to form the tetraacyl major lipid A species (11). The toxicity of H. pylori tetraacyl-lipid A on human monocytes is ∼4-fold lower than that of the hexaacyl form (12). It has been suggested that the phosphorylation and acylation patterns in lipid A of H. pylori LPS are responsible for its low biological activity (13, 14).
Lipid A diversity is observed both in the number and length of fatty-acid side chains and in the presence of terminal phosphate residues and associated modifications (6). Pathogenic bacteria modify the lipid A portion of their LPS to help evade the host innate immune response. The variability of lipid A could have profound implications for disease, particularly in humans, owing to altered recognition by the Toll-like receptor-4 complex. Through binding to the specific receptors of the mammalian innate immune system, lipid A is recognized by immune cells as pathogen-associated molecular pattern and stimulates secretion of proinflammatory cytokines (6, 7, 14). Immune detection of lipid A is so sensitive and robust that a bloodstream infection can cause a severe complication called endotoxic shock (6). Therefore, an exquisitely sensitive analytical method is required to determine not only the major lipid A components, but also minor components directly from bacterial cells. Mass spectrometry has been widely used to gain knowledge about lipid A heterogeneity, i.e., the number of different species of the lipid A families and distribution of the fatty acids on each glucosamine group (11, 15–19). We have previously characterized lipid A species through analysis of the intact LPS (20). Lipid A is generally prepared from the isolated LPS by harsh hydrolysis conditions, such as 0.1–2 M HCl or 1–2% acetic acid at 100°C for 1–2 h, or milder hydrolysis conditions, such as 0.01 M sodium acetate (pH 4.5) with the addition of 1% SDS at 100°C for 1 h (21). Because at least 7–10 days are required for a large-scale extraction and purification of LPS, classical isolation of lipid A is considerably laborious and time-consuming. Recently El Hamadi et al. (17) developed a method based on a hot ammonium-isobutyrate procedure to isolate lipid A directly from whole bacterial cells. This method allows obtaining the mass spectrometric data in a working day with a detection limit of 50–100 μg of lyophilized cells. The relatively moderate detection limit offered by the method, however, precludes its application to profiling lipid A structures from the material sources as small as a single colony. In an effort to study the relationship between the lipid A structure and the role of H. pylori LPS in pathogenesis, we developed a fast and sensitive analytical method to analyze lipid A based on microwave-assisted enzymatic digestion and mild acid hydrolysis. We used five strains: H. pylori strain 26695, 26695/hp1191::kan mutant, 26695/hp0479::kan mutant, 26695/hp0826::kan mutant, and Sydney (SS1) strain, to demonstrate the application of the proposed technique. These five strains are of significant importance in H. pylori research: the complete genome sequence of strain 26695 has been published, the Sydney (SS1) strain is widely employed in a mouse model of H. pylori infection, and 26695/hp1191::kan isogenic mutant strain expresses a deep-rough inner core LPS. Their LPS structures have been revealed (22).
MATERIALS AND METHODS
Chemicals
Proteinase K was purchased from Promega (Madison, WI). Isobutyric acid was from Eastman Chemical Co. (Kingsport, TN). SDS, sodium acetate, and 5-chloro-2-mercaptobenzothiazole (CMBT) were purchased from Sigma-Aldrich (St. Louis, MO). Chloroform, methanol, and acetic acid were from EMD Science (Gibbstown, NJ).
Bacterial strains and growth conditions
H. pylori strain 26695 was obtained from Dr. R. Alm at AstraZeneca (Boston, MA). H. pylori strain SS1 was from Dr. A. Lee (University of New South Wales, Sydney, Australia). Cells were grown at 37°C on antibiotic-supplemented Columbia Blood agar (Difco) plates containing 7% horse blood in microaerophilic environment for 72 h as previously described (23). Kanamycin (10 mg/l) was used for mutant strains in addition to regular antibiotics (23). The bacterial lawns were harvested into sterile PBS, pH 7.4, and brought to an OD600 of 0.5. Dilutions of 1:100 (v/v) in PBS were made, and volumes of 5, 10, and 25 µl were spread onto the plates for colonies and incubated at 37°C for 2–3 days in a microaerophilic environment. Two- or 3-day-old colonies (one randomly picked) were suspended in 1% phenol (w/v) in PBS, washed with deionized water, and subjected to analysis.
To prepare small scale samples, dried cells (∼2 mg) were suspended in water at a concentration 1 mg/ml. Serial dilutions were then prepared from the stock solution, vigorously vortexed before each pipetting, and lyophilized prior to being subjected to various hydrolysis procedures.
Lipid A isolation from cells using isobutyric acid-ammonium hydroxide procedure
Lyophilized cells were suspended in 200 µl of isobutyric acid-1 M ammonium hydroxide (5:3, v/v) and kept for 2 h at 100°C in a 1.5-ml eppendorf tube. The mixture was cooled in ice water and centrifuged (2000 g for 15 min at 4°C). The supernatant was dried by speed vac. The sample was then washed twice with 400 µl of methanol and centrifuged at 10000 g for 5 min. Finally, the insoluble lipid A was extracted once with 200 µl of a mixture of chloroform and methanol (1:1, v/v). After centrifugation at 8000 g for 5 min, the supernatant was transferred to another 0.5-ml eppendorf tube and dried under a stream of nitrogen.
Lipid A isolation from cells using SDS-promoted hydrolysis procedure
The cells were suspended in 200 µl of 1% SDS in 10 mM sodium acetate (pH 4.5) and incubated at 100°C for 1 h. The reaction mixture was dried by speed vac. SDS was removed by washing the mixture with 50 µl of distilled water and 250 µl of acidified ethanol (prepared by adding 100 µl 4 M HCl to 20 ml 95% ethanol) followed by centrifugation (10000 g for 5 min). The samples were then washed twice with 400 µl of 95% ethanol and centrifuged (10000 g for 5 min). The lipid A was extracted from the pellets with 200 µl of a mixture of chloroform and methanol (1:1), followed by centrifugation at 8000 g for 5 min. The supernatant was transferred to another 0.5-ml eppendorf tube and dried under a stream of nitrogen.
Lipid A isolation from cells using microwave-assisted enzymatic digestion and sodium acetate hydrolysis
The lyophilized cells were placed in a 1.5-ml eppendorf tube and suspended in 200 µl of 50 mM sodium acetate buffer (pH 4.5) containing proteinase K (60 µg/ml). Proteinase K digestion was used to help break down cells. Under microwave irritation at 50 W (Discovery System, CEM Corp., Mathews, NC), the enzymatic digestion was carried out for 5 min at 58°C. The suspension was then kept for 1 h at 100°C. The reaction mixture was centrifuged at 10000 g for 5 min. The supernatant was discarded. The pellets were then washed twice with 400 µl methanol and centrifuged at 10000 g for 5 min. Finally, the lipid A was dissolved and extracted once with 200 µl of a mixture of chloroform and methanol (1:1, v/v), followed by centrifugation at 8000 g for 5 min. The supernatant was transferred to another 0.5-ml eppendorf tube and dried under a stream of nitrogen.
Lipid A isolation from a single colony
Bacteria of each single colony were carefully scraped off the culture plate and collected in a 1.5-ml eppendorf tube containing 1% phenol in PBS. Following washings with deionized water by centrifugation, the killed cells were suspended in 100 µl of 50 mM sodium acetate buffer (pH 4.5) containing proteinase K (60 µg/ml). After microwave irritation, the suspension was heated at 100° for 1 h. The mixture was centrifuged at 10000 g for 5 min and the pellets were washed twice with methanol. Lipid A was extracted from the pellets with 100 µl mixture of chloroform and methanol (1:1, v/v). Following centrifugation at 8000 g for 5 min, the supernatant was transferred to another 0.5-ml eppendorf tube and dried under a stream of nitrogen.
MALDI-TOF/TOF analysis
Lipid A was analyzed using a 4800 MALDI-TOF/TOF in the negative ion mode (Applied Biosystems). Samples were dissolved in a mixture of chloroform and methanol (1:1, v/v) and then mixed with equal volume of CMBT (20 mg/ml) in chloroform-methanol-water 4:4:1 (v/v/v), from which 0.2 µl samples were loaded on MALDI target. Both MS and MS/MS data were acquired in reflection mode. Four hundred shots were accumulated for each MS spectrum and 1,250 shots were accumulated for each MS/MS spectrum. The precursor isolation window was set to ±2 Da. MS/MS spectra were acquired with collision energies of 1 keV, and air was used as the collision gas. Data were acquired and processed with Data Explorer (Applied Biosystems).
RESULTS AND DISCUSSION
Analysis of lipid A prepared using previously reported methods
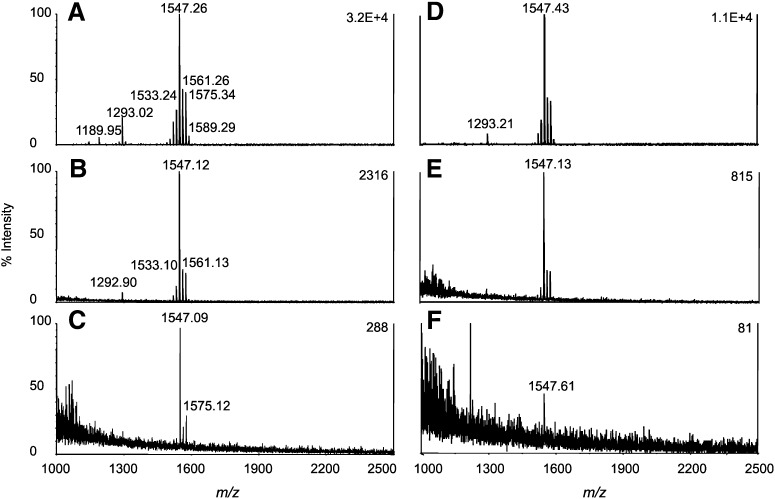
The isolation of lipid A directly from bacterial cells has been developed by using an isobutyric acid-ammonium hydroxide hydrolysis procedure (17, 19). We first prepared the H. pylori lipid A using this method and the MALDI-TOF mass spectra obtained from 100 μg and 10 μg of dried cells are presented in Fig. 1A and C. As shown in Fig. 1C, this method is suitable for analysis of lipid A from H. pylori cells on quantities as small as 10 μg. On the other hand, an SDS-promoted hydrolysis method has been widely used for lipid A preparation from isolated LPS/lipooligosaccharide (19, 24). Thus, we also used the SDS-promoted hydrolysis method and found that it offers a higher yield of lipid A than that of isobutyric acid-ammonium hydroxide hydrolysis. The MALDI-TOF mass spectra obtained from 100 μg and 10 μg of H. pylori cells are presented in Fig. 1B and D. It is noteworthy that we observed the exact opposite phenomena when comparing these two methods for the preparation of Campylobacter jejuni lipid A (data not shown). Whereas lipid A preparation using SDS-promoted procedure can be completed in one working day, the clean up of SDS can be very time-consuming, in which the step of drying the SDS-containing lipid A suspension alone takes 1–2 h. In other words, we could significantly reduce the time for sample preparation if the use of SDS could be avoided.
Fig. 1.
MALDI-TOF mass spectra of lipid A from H. pylori Sydney (SS1) strain. Samples prepared from 100 μg of dried cells using method (A) isobutyric acid-ammonium hydroxide hydrolysis and (B) 1% SDS in 10 mM sodium acetate, pH 4.5. Samples prepared from 10 μg of dried cells using method (C) isobutyric acid-ammonium hydroxide hydrolysis and (D) 1% SDS in 10 mM sodium acetate, pH 4.5.
Analysis of lipid A using microwave-assisted enzymatic digestion and sodium acetate hydrolysis
The combination of microwave-assisted digestion and MS analysis has allowed us to rapidly characterize C. jejuni intact lipooligosaccharide in order to accurately determine the glycan composition (25). This method is not only rapid but also very sensitive and requires minimal material, as demonstrated by successful analysis using single colony samples. We then attempted to develop a rapid sample method to extract lipid A directly from bacterial cells based on microwave-assisted enzymatic digestion and detergent-free hydrolysis. In this case, proteinase K, instead of SDS, was used to disrupt the cells. The microwave irradiation can accelerate the enzymatic digestion of proteins, such that the reactions requiring several hours under conventional conditions can be reduced to only a few minutes with high yields and reaction selectivity. The sample was then treated using a detergent-free hydrolysis procedure, in which 200 μl of 50 mM sodium acetate, pH 4.5, was added and the solution was heated at 100°C for 1 h. The entire process for lipid A preparation from cells takes about 2 h. The MALDI-TOF mass spectra for the lipid A from 100, 10, and 1 μg of dried cells are presented in Fig. 2. The analysis results revealed that the sensitivity of the method is much higher than that of the previously reported methods (see Fig. 1). The mass spectrum ( also suggested that the proposed method could be used to profile the lipid A structures from less than 1 μg of dried cells. For comparison, 100, 10, and 1 μg of dried cells were also subjected to proteinase K digestion for 5 min, without application of microwave irradiation, followed by acidic hydrolysis (Fig. 2C–E). As indicated in Fig. 2, the microwave-assisted digestion can significantly enhance the yield of lipid A and thus improve the sensitivity.
Fig. 2.
MALDI-TOF mass spectra of lipid A from H. pylori Sydney (SS1) strain isolated from different amounts of dried cells: (A) 100 μg; (B) 10 μg; (C) 1 μg. Samples were prepared using the combination of microwave-assisted enzymatic digestion and sodium acetate hydrolysis. The mass spectra for the samples prepared without microwave irradiation: (D) 100 μg; (E) 10 μg; (F) 1 μg.
To further validate the new method, we applied it to isolating the lipid A from four strains of H. pylori strain 26695 and three corresponding mutants: 26695/hp0826::kan, 26695/hp0479::kan, and 26695/hp1191::kan (Fig. 3). The mass spectrum from H. pylori 26695/hp0826::kan was similar to that from H. pylori 26695/hp0479::kan. The most abundant ion detected at m/z 1547 corresponded to a singly deprotonated lipid A with four 3-hydroxyhexadecanoic [16:0 (3-OH)], octadecanoic (18:0), and 3-hydroxyoctadecanoic [18:0 (3-OH)] fatty acid residues and one PEtn group attached to the glucosamine backbone. The mass spectra clearly indicated that the lipid A structures from H. pylori 26695/hp1191::kan mutant (Fig. 3B), in which the ion corresponding to a pentaacyl lipid A was also detected (i.e., m/z 1801), were significantly different from those of H. pylori 26695/hp0826::kan and H. pylori 26695/hp0479::kan. In addition, the observation of ion at m/z 1561 in the mass spectrum of lipid A from H. pylori Sydney strain (Fig. 2) made it distinguishable from H. pylori 26695 and its mutants. Mutations to HP0479 ORF truncate the LPS at the branching DD-heptose residue, leaving the core backbone oligosaccharide intact and resulting in an LPS structure that lacks the O-chain polysaccharide and the outer core region (25). We have previously confirmed that H. pylori 26695/hp0479::kan expressing a truncated LPS was deficient in its ability to successfully colonize the murine stomach (23). We have also constructed H. pylori 26695/hp0826::kan mutant and demonstrated that this mutation resulted in formation of truncated LPS lacking the O-chain polysaccharide but maintaining the outer core region (26). Finally, we have previously described 26695/hp1191::kan mutant expressing a deep-rough inner core LPS structure lacking both the O-chain and the outer core region and containing a single Kdo residue attached to LD-heptose residue substituted by a PEtn group. The total fatty acid analysis of LPS from H. pylori strains 26695 and the corresponding 26695/hp1191::kan mutant indicated the presence of 3-hydroxyoctadecanoic acid, n-octadecanoic acid, and 3-hydroxyhexadecanoic acid as main components. Subsequent analysis of the O-linked fatty acids in 26695/hp1191::kan LPS confirmed that both 3-hydroxyhexadecanoic acid and n-octadecanoic acid were O-linked; traces of O-linked 3-hydroxyoctadecanoic acid were also present (data not shown).
Fig. 3.
MALDI-TOF mass spectra of lipid A prepared from 100 μg of bacterial cells. A: H. pylori 26695; B: H. pylori 26695/hp1191::kan; C: H. pylori 26695/hp0479::kan; D: H. pylori 26695/hp0826::kan.
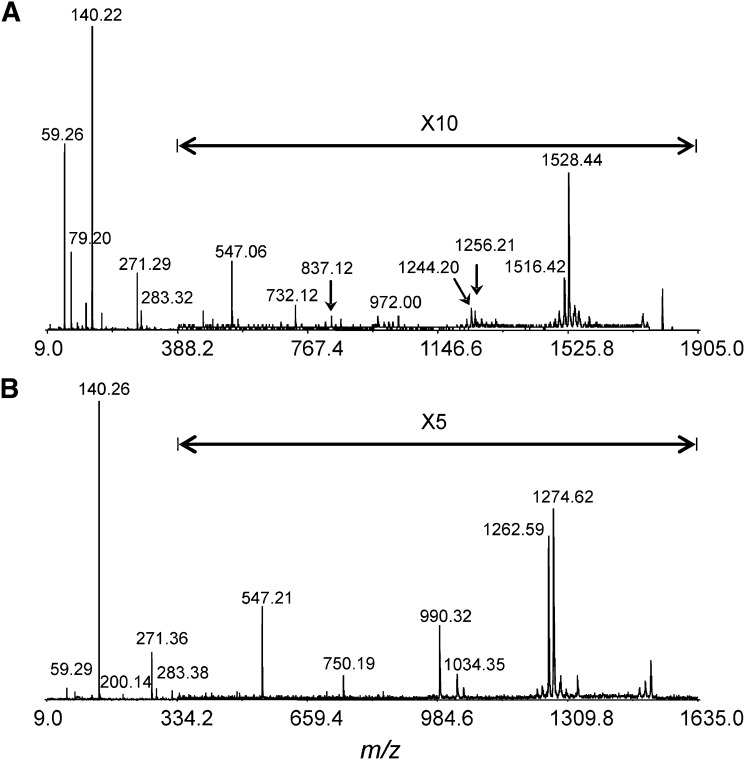
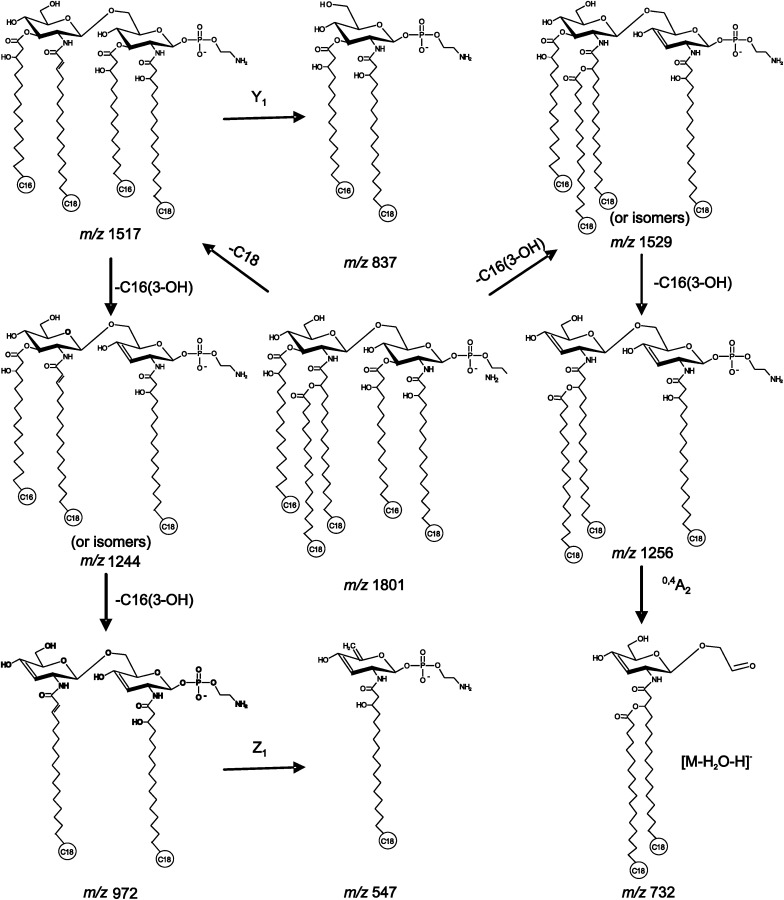
MALDI-TOF/TOF tandem mass spectrometry was employed for the elucidation of the lipid A structures. The tandem mass spectra of the two abundant lipid A ions, m/z 1547 and 1801, from H. pylori 26695/hp1191::kan are presented in Fig. 4. The tandem mass spectrum for the precursor ion at m/z 1547 is presented in Fig. 4B, in which the most abundant product ion, m/z 140, was assigned to the PEtn group. The ion at m/z 750, although of low abundance, corresponded to glycosidic bond cleavage. The fragment ion at m/z 547 was identified as a C1/Z1 ion. Fragment ions at m/z 1274 and 1264 were attributed to the ion at m/z 1547 minus one 3-hydroxyhexadecanoic acid residue or one octadecanoic acid residue, respectively. Cleavage of both residues produced the fragment ion at m/z 990. The major fragment ions from the precursor ion at m/z 1801 are illustrated in Scheme 1. The results suggested that tandem mass spectrometry is a valuable tool for elucidation of lipid A structures. However, this technique might encounter challenges for de novo sequencing of unknown species, especially when dealing with structural isomers. In such cases, separation of isomers prior to analysis or in combination with selective hydrolysis might be required (19).
Fig. 4.
MALDI-TOF/TOF tandem mass spectra of lipid A from H. pylori 26695/hp1191::kan. All experimental conditions were the same as those in Fig 2. A: Product ion spectrum of ion at m/z 1801; B: product ion spectrum of ion at m/z 1547.
Scheme 1.
Proposed fragmentation pathways of the precursor ion at m/z 1801.
Application to single colony sample analysis
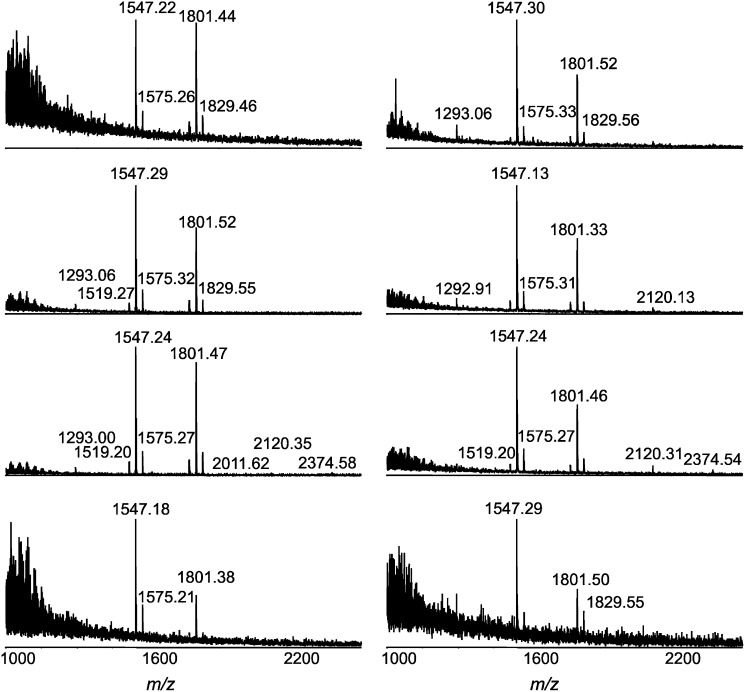
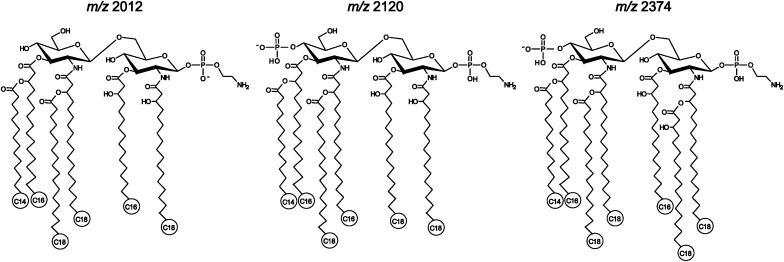
The significant difference observed in the lipid A pattern of H. pylori 26695/hp1191::kan mutant has prompted us to analyze single colony samples of this strain. As expected, all the mass spectra showed two dominant ions at m/z 1547 and 1801 (Fig. 5). Although some heterogeneity among individual colony samples is expected, the difference in intensities of the two ions arises primarily from a different number of bacterial cells per colony. Actually, the colonies of H. pylori are naturally very small, making it hard to collect individual colonies containing equal amounts of bacterial cells. Nevertheless, we were able to detect the major lipid A species in all samples. The most striking discovery from the analysis of those single colony samples is detection of hexa- and even heptaacyl lipid A species, m/z 2012, m/z 2120, and m/z 2374, in some samples. It is noteworthy that these minor components have much higher relative abundance in the mass spectra for lipid A isolated from single colony samples than those from large-scale cell samples. We believe that fluctuations in expression levels of certain enzymes due to phase variation were probably responsible for the observation. In other words, not all the colonies/cells expressed these lipid A species. As a result, the relative abundance of the species in a large-scale sample was lower than that in the individual colony. Furthermore, the ion suppression in MALDI ionization could also contribute to the observation. Therefore, the tandem mass spectrometry experiments were carried out for the precursor ions at m/z 2012, 2120, and 2374 using the pooled material from samples of the individual colonies (see supplementary Fig. I). The lipid A species of m/z 2012 was interpreted to be a hexaacyl form, previously detected in H. pylori 26695/hp0694::cam mutant which lacked the enzyme to remove an acyl chain (11). Its structure was evident by the observation of several characteristic fragment ions in the tandem MS spectrum, e.g., m/z 547, 732, 1529, which were also detected in the MS/MS of m/z 1547 (Fig. 4A). Similarly, the lipid A of m/z 2120 was assigned to be a hexaacyl form with an additional phosphate group at position 4′ of the backbone disaccharide (Scheme 2). It has been reported that, in addition to the predominant monophosphorylated tetraacyl lipid A, H. pylori smooth-form LPS also contained a minor constituent, a bisphosphorylated hexaacyl lipid that is distinguishable from tetraacyl lipid A by carrying 3-(12:0-O)-16:0 or 3-(14:0-O)-16:0 at position-3′ and an extra phosphate group at position-4’ (8). Although of a relatively low intensity, the lipid A species of m/z 2374 were repeatedly detected in several individual colony samples. We believe that this is a heptaacyl lipid A component (Scheme 2). Although the tandem mass spectrum was not enough to decipher the structure of the ion at m/z 2374 due to its low abundance, it could be evident by the detection of fragment ions at m/z 79 and 140, corresponding to phosphate and phosphoethanolamine groups, and ions at m/z 227, 271, and 283, corresponding to C14:0, 3-OH-C16:0, and C18:0, respectively (see supplementary ). Whereas the heptaacyl form of lipid A has been detected in other Gram-negative bacteria (27), this study has demonstrated, for the first time, that H. pylori lipid A can synthesize heptaacyl species.
Fig. 5.
MALDI-TOF mass spectra of lipid A from single colony samples of H. pylori 26695/hp1191::kan.
Scheme 2.
Proposed structures for hexa- and heptaacyl lipid A species.
CONCLUSIONS
The acylation and phosphorylation of lipid A has long been recognized as a key determinant of the toxicity of LPS or LOS (7, 28). Lipid A is generally the most potent immunostimulatory component responsible for inflammatory immune response (7). The level of acylation is critical and pentaacylated species are known to be less effective in eliciting proinflammatory cytokine responses (29). The unusual structure of H. pylori lipid A is believed to be responsible for its low endotoxicity as compared with lipid A of Salmonella and Escherichia coli (12). Generally, it takes a nine-step enzymatic pathway to synthesize a “typical” hexaacyl lipid A (11). However, the lipid A synthesis in H. pylori has only eight homologs to the nine lipid A biosynthesis enzymes, which offers an excellent model for studying H. pylori lipid A modifications (7). The presence of palmitate, heptaacyl form, in a minor fraction of H. pylori lipid A has been known since the chemical structure of lipid A was first elucidated, but the functional importance in bacterial pathogenesis of regulated lipid A palmitoylation has become clear only recently (30). For example, it has been shown that, in Bordetella bronchiseptica, heptaacylated lipid A species acted as an antagonist toward the activity of hexaacylated lipid A species with regard to the induction of tumor necrosis factor-α and nuclear factor kappa B activation of human-derived THP-1 cells (31). Similarly, in Pseudomonas aeruginosa, palmitoylated LPS induced significantly higher IL-8 response (29). In E. coli, the modification enzyme, the palmitoyl transferase (PagP), catalyzes the addition of a palmitate to one of the primary linked acyl chains of lipid, resulting in a heptaacyl lipid A structure (27). The heptaacyl form observed in H. pylori 26695/hp1191::kan mutant suggest the possibility that a PagP-like enzyme may exist in H. pylori. Furthermore, this is the first characterization of the lipid A structure from a single bacterial colony sample by mass spectrometry. We have previously shown that the level of invasion of gastric cancer cells was unaffected for an H. pylori 26695/hp1191::kan deep-rough mutant as compared with the wild-type 26695 with an extended LPS structure (32). The role of heptaacyl species of H. pylori lipid A in vivo is unknown and warrants further investigation. It is tempting to speculate that synthesis of the heptaacyl lipid A in vivo could be linked to observed invasiveness of H. pylori and the persistent nature of H. pylori infection (33).
Supplementary Material
Footnotes
Abbreviations:
- CMBT
- 5-chloro-2-mercaptobenzothiazole
- LPS
- lipopolysaccharide
- PEtn
- phosphoethanolamine
- TOF
- time-of-flight
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of a figure.
REFERENCES
- 1.Moodley Y., Linz B., Yamaoka Y., Windsor H. M., Breurec S., Wu J. Y., Maady A., Bernhoft S., Thiberge J. M., Phuanukoonnon S., et al. 2009. The peopling of the Pacific from a bacterial perspective. Science. 323: 527–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Linz B., Balloux F., Moodley Y., Manica A., Liu H., Roumagnac P., Falush D., Stamer C., Prugnolle F., van der Merwe S. W., et al. 2007. An African origin for the intimate association between humans and Helicobacter pylori. Nature. 445: 915–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moran A. P. 2007. Lipopolysaccharide in bacterial chronic infection: insights from Helicobacter pylori lipopolysaccharide and lipid A. Int. J. Med. Microbiol. 297: 307–319. [DOI] [PubMed] [Google Scholar]
- 4.Rutten L., Mannie J. P., Stead C. M., Raetz C. R., Reynolds C. M., Bonvin A. M., Tommassen J. P., Egmond M. R., Trent M. S., Gros P. 2009. Active-site architecture and catalytic mechanism of the lipid A deacylase LpxR of Salmonella typhimurium. Proc. Natl. Acad. Sci. USA. 106: 1960–1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Raetz C. R., Reynolds C. M., Trent M. S., Bishop R. E. 2007. Lipid A modification systems in gram-negative bacteria. Annu. Rev. Biochem. 76: 295–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miller S. I., Ernst R. K., Bader M. W. 2005. LPS, TLR4 and infectious disease diversity. Nat. Rev. Microbiol. 3: 36–46. [DOI] [PubMed] [Google Scholar]
- 7.Trent M. S., Stead C. M., Tran A. X., Hankins J. V. 2006. Diversity of endotoxin and its impact on pathogenesis. J. Endotoxin Res. 12: 205–223. [DOI] [PubMed] [Google Scholar]
- 8.Moran A. P., Lindner B., Walsh E. J. 1997. Structural characterization of the lipid A component of Helicobacter pylori rough- and smooth-form lipopolysaccharides. J. Bacteriol. 179: 6453–6463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tran A. X., Karbarz M. J., Wang X., Raetz C. R., McGrath S. C., Cotter R. J., Trent M. S. 2004. Periplasmic cleavage and modification of the 1-phosphate group of Helicobacter pylori lipid A. J. Biol. Chem. 279: 55780–55791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tran A. X., Whittimore J. D., Wyrick P. B., McGrath S. C., Cotter R. J., Trent M. S. 2006. The lipid A 1-phosphatase of Helicobacter pylori is required for resistance to the antimicrobial peptide polymyxin. J. Bacteriol. 188: 4531–4541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stead C. M., Beasley A., Cotter R. J., Trent M. S. 2008. Deciphering the unusual acylation pattern of Helicobacter pylori lipid A. J. Bacteriol. 190: 7012–7021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ogawa T., Suda Y., Kashihara W., Hayashi T., Shimoyama T., Kusumoto S., Tamura T. 1997. Immunobiological activities of chemically defined lipid A from Helicobacter pylori LPS in comparison with Porphyromonas gingivalis lipid A and Escherichia coli-type synthetic lipid A (compound 506). Vaccine. 15: 1598–1605. [DOI] [PubMed] [Google Scholar]
- 13.Muotiala A., Helander I. M., Pyhala L., Kosunen T. U., Moran A. P. 1992. Low biological activity of Helicobacter pylori lipopolysaccharide. Infect. Immun. 60: 1714–1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mattsby-Baltzer I., Mielniczuk Z., Larsson L., Lindgren K., Goodwin S. 1992. Lipid A in Helicobacter pylori. Infect. Immun. 60: 4383–4387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aussel L., Therisod H., Karibian D., Perry M. B., Bruneteau M., Caroff M. 2000. Novel variation of lipid A structures in strains of different Yersinia species. FEBS Lett. 465: 87–92. [DOI] [PubMed] [Google Scholar]
- 16.Casabuono A. C., D'Antuono A., Sato Y., Nonami H., Ugalde R., Lepek V., Erra-Balsells R., Couto A. S. 2006. A matrix-assisted laser desorption/ionization mass spectrometry approach to the lipid A from Mesorhizobium loti. Rapid Commun. Mass Spectrom. 20: 2175–2182. [DOI] [PubMed] [Google Scholar]
- 17.El Hamadi A., Tirsoaga A., Novikov A., Hussein A., Caroff M. 2005. Microextraction of bacterial lipid A: easy and rapid method for mass spectrometric characterization. J. Lipid Res. 46: 1773–1778. [DOI] [PubMed] [Google Scholar]
- 18.Jones J. W., Shaffer S. A., Ernst R. K., Goodlett D. R., Turecek F. 2008. Determination of pyrophosphorylated forms of lipid A in Gram-negative bacteria using a multivaried mass spectrometric approach. Proc. Natl. Acad. Sci. USA. 105: 12742–12747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tirsoaga A., El H. A., Perry M. B., Caroff M., Novikov A. 2007. A rapid, small-scale procedure for the structural characterization of lipid A applied to Citrobacter and Bordetella strains: discovery of a new structural element. J. Lipid Res. 48: 2419–2427. [DOI] [PubMed] [Google Scholar]
- 20.Dzieciatkowska M., Schweda E. K., Moxon E. R., Richards J. C., Li J. 2008. Characterization of intact lipopolysaccharides from the Haemophilus influenzae strain RM 118 using electrophoresis-assisted open-tubular liquid chromatography-mass spectrometry. Electrophoresis. 29: 2171–2181. [DOI] [PubMed] [Google Scholar]
- 21.Caroff M., Tacken A., Szabo L. 1988. Detergent-accelerated hydrolysis of bacterial endotoxins and determination of the anomeric configuration of the glycosyl phosphate present in the “isolated lipid A” fragment of the Bordetella pertussis endotoxin. Carbohydr. Res. 175: 273–282. [DOI] [PubMed] [Google Scholar]
- 22.Monteiro M. A., Appelmelk B. J., Rasko D. A., Moran A. P., Hynes S. O., MacLean L. L., Chan K. H., Michael F. S., Logan S. M., O'Rourke J., et al. 2000. Lipopolysaccharide structures of Helicobacter pylori genomic strains 26695 and J99, mouse model H. pylori Sydney strain, H. pylori P466 carrying sialyl Lewis X, and H. pylori UA915 expressing Lewis B classification of H. pylori lipopolysaccharides into glycotype families. Eur. J. Biochem. 267: 305–320. [DOI] [PubMed] [Google Scholar]
- 23.Hiratsuka K., Logan S. M., Conlan J. W., Chandan V., Aubry A., Smirnova N., Ulrichsen H., Chan K. H., Griffith D. W., Harrison B. A., et al. 2005. Identification of a D-glycero-D-manno-heptosyltransferase gene from Helicobacter pylori. J. Bacteriol. 187: 5156–5165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tirsoaga A., Novikov A., Adib-Conquy M., Werts C., Fitting C., Cavaillon J. M., Caroff M. 2007. Simple method for repurification of endotoxins for biological use. Appl. Environ. Microbiol. 73: 1803–1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dzieciatkowska M., Liu X., Heikema A. P., Houliston R. S., van Belkum A., Schweda E. K., Gilbert M., Richards J. C., Li J. 2008. Rapid method for sensitive screening of oligosaccharide epitopes in the lipooligosaccharide from Campylobacter jejuni strains isolated from Guillain-Barre syndrome and Miller Fisher syndrome patients. J. Clin. Microbiol. 46: 3429–3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Logan S. M., Conlan J. W., Monteiro M. A., Wakarchuk W. W., Altman E. 2000. Functional genomics of Helicobacter pylori: identification of a beta-1,4 galactosyltransferase and generation of mutants with altered lipopolysaccharide. Mol. Microbiol. 35: 1156–1167. [DOI] [PubMed] [Google Scholar]
- 27.Smith A. E., Kim S. H., Liu F., Jia W., Vinogradov E., Gyles C. L., Bishop R. E. 2008. PagP activation in the outer membrane triggers R3 core oligosaccharide truncation in the cytoplasm of Escherichia coli O157:H7. J. Biol. Chem. 283: 4332–4343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Swords W. E., Chance D. L., Cohn L. A., Shao J., Apicella M. A., Smith A. L. 2002. Acylation of the lipooligosaccharide of Haemophilus influenzae and colonization: an htrB mutation diminishes the colonization of human airway epithelial cells. Infect. Immun. 70: 4661–4668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Backhed F., Normark S., Schweda E. K., Oscarson S., Richter-Dahlfors A. 2003. Structural requirements for TLR4-mediated LPS signalling: a biological role for LPS modifications. Microbes Infect. 5: 1057–1063. [DOI] [PubMed] [Google Scholar]
- 30.Bishop R. E. 2005. The lipid A palmitoyltransferase PagP: molecular mechanisms and role in bacterial pathogenesis. Mol. Microbiol. 57: 900–912. [DOI] [PubMed] [Google Scholar]
- 31.Kwok T., Backert S., Schwarz H., Berger J., Meyer T. F. 2002. Specific entry of Helicobacter pylori into cultured gastric epithelial cells via a zipper-like mechanism. Infect. Immun. 70: 2108–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chandan V., Logan S. M., Harrison B. A., Vinogradov E., Aubry A., Stupak J., Li J., Altman E. 2007. Characterization of a waaF mutant of Helicobacter pylori strain 26695 provides evidence that an extended lipopolysaccharide structure has a limited role in the invasion of gastric cancer cells. Biochem. Cell Biol. 85: 582–590. [DOI] [PubMed] [Google Scholar]
- 33.Preston A., Maxim E., Toland E., Pishko E. J., Harvill E. T., Caroff M., Maskell D. J. 2003. Bordetella bronchiseptica PagP is a Bvg-regulated lipid A palmitoyl transferase that is required for persistent colonization of the mouse respiratory tract. Mol. Microbiol. 48: 725–736. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.